Abstract
Apicoplast, an essential organelle of human malaria parasite Plasmodium falciparum contains a ∼35 kb circular genome and is a possible target for therapy. Proteins required for the replication and maintenance of the apicoplast DNA are not clearly known. Here we report the presence of single–stranded DNA binding protein (SSB) in P falciparum. PfSSB is targeted to the apicoplast and it binds to apicoplast DNA. A strong ssDNA binding activity specific to SSB was also detected in P. falciparum lysate. Both the recombinant and endogenous proteins form tetramers and the homology modelling shows the presence of an oligosaccharide/oligonucleotide-binding fold responsible for ssDNA binding. Additionally, we used SSB as a tool to track the mechanism of delayed death phenomena shown by apicoplast targeted drugs ciprofloxacin and tetracycline. We find that the transport of PfSSB is severely affected during the second life cycle following drug treatment. Moreover, the translation of PfSSB protein and not the transcription of PfSSB seem to be down-regulated specifically during second life cycle although there is no considerable change in protein expression profile between drug-treated and untreated parasites. These results suggest dual control of translocation and translation of apicoplast targeted proteins behind the delayed death phenomena.
INTRODUCTION
Plasmodium falciparum, the causative agent of most severe form of human malaria belongs to the phylum apicomplexa (1). The apicomplexan parasites possess a non-photosynthetic plastid like organelle called apicoplast. The apicoplast is the site for several biosynthetic pathways (fatty acid, isoprenoid, heme biosynthesis) that are central to the core cellular functioning of the parasite (2). The biosynthetic pathways located within the apicoplast provide novel sites of drug intervention against the malaria disease (3). Moreover, the house keeping metabolic processes like DNA replication, transcription and translation operative within the apicoplast are prokaryotic in nature and therefore they are good targets of existing antibacterials or novel drugs (4).
The ∼35kb, extremely A+T rich (∼86%) apicoplast DNA is circular and replicates via a D-loop/bi-directional theta mode in late trophozoite/early schizont stages of intraerythrocytic life cycle (5). The origins of apicoplast DNA replication have been mapped in the inverted repeat (IR) regions (6). The apicoplast genome houses only 68 open reading frames (ORFs) for rRNAs, tRNAs, ribosomes, RNA polymerase, translational elongation factor (EF-Tu), ClpC chaperone and Fe–S cluster protein (SufB) (1,7). None of the ORFs on the apicoplast DNA codes for replication protein. Most of the proteins involved in the apicoplast metabolic pathways are encoded in the nucleus, synthesized in the cytoplasm and subsequently imported into the apicoplast (2). The nuclear encoded proteins are targeted to apicoplast by the apicoplast localization signal that is cleaved off from the protein once inside the apicoplast (8). Recently, a ∼220 kDa nuclear encoded multifunctional enzyme PfPRex (P. falciparum plastidic DNA replication/repair enzyme complex) has been shown to localize in the apicoplast. This enzyme has three important activities (helicase, primase and polymerase) associated with DNA replication (9). A histone like protein (HU) exhibiting the DNA condensation property is imported into apicoplast suggesting its role in organization of apicoplast genome (10). We and others have shown that the gyrase subunits present in the parasite are targeted to apicoplast where they might be involved in negative supercoiling of the DNA circle, an essential step for the replication process (11,12). Consistent with the above findings, quinolone (ciprofloxacin) or coumarin (coumermycin, novobiocin) antibiotics target the parasitic gyrase and inhibit the apicoplast DNA replication leading to the parasite death (12,13).
Since apicoplast is of prokaryotic origin, several antibiotics against bacterial replication, transcription and translation processes have been used successfully to block parasitic growth. However, majority of these antibiotics show typical delayed growth phenotype, characterized by defect in parasite growth and decrease in parasitemia only during second life cycle following the addition of these drugs (4). It has been suggested that these drugs affect apicoplast morphology, segregation and most importantly the transport of essential proteins in the apicoplast. Using fusion protein containing apicoplast signal sequence of acyl carrier protein (ACP) and GFP, the effect of these drugs on protein translocation have been studied (14,15). However, no endogenous apicoplast targeted protein involved in house keeping function has been followed after drug treatment in the above studies. Neither the transcription and translation status of apicoplast targeted proteins was investigated simultaneously in the presence of these drugs.
To gain further insight into the enzymology of P. falciparum apicoplast DNA replication, we wanted to study another essential key protein, the homologue of bacterial single-stranded DNA binding protein (SSB) (16). The prokaryotic type circular P. falciparum apicoplast DNA suggests a possible requirement of bacterial type SSB for various DNA metabolic processes in that organelle. The analysis of the parasite genome indeed reveals the presence of a bacterial type ssb on the chromosome V of the nuclear DNA (PFE0435c). The N-terminal extension in the primary sequence of the protein is predicted to be a potential apicoplast targeting sequence.
SSBs are known to play essential roles in many aspects of nucleic acid metabolism including DNA replication, recombination and repair. SSBs protect and stabilize the single-stranded DNA (ssDNA) intermediates as well as remove secondary structures in the DNA. Any misstep in DNA replication or a failure to properly recombine or repair DNA can lead to gross aberrations in the genome, signifying the role of SSBs in these processes (17).
In eukaryotes, a heterotrimeric complex called replication protein A (RPA) (18) carries out the ssDNA binding activity in the nucleus. Mitochondrial DNA replication involves the ssDNA binding protein (mtSSB) that is quite distinct from nuclear RPA. mtSSB shows a great degree of sequence homology with bacterial SSB (19). In P. falciparum, however, ssDNA binding activity in the nucleus has been reported earlier corresponding to the large subunit (∼55 kDa) of RPA (20).
SSB proteins from different organisms share sequence homology as well as distinct biochemical and structural characteristics. The SSBs have a common oligosaccharide/oligonucleotide-binding fold (OB fold) which binds to ssDNA. The SSBs from all prokaryotic organisms have an acidic C-terminal tail that is essential for DNA replication by mediating protein–protein interactions at the replication fork (17). In solution, SSBs are found in different oligomeric states. They are found as homodimers (Bacteriophages, Thermus thermophilus, T. aquatics and Deinococcus radiodurans), heterotrimer (eukaryotic RPAs) and homotetramers (mitochondrial and most prokaryotic SSBs) (21).
We report here the presence of a prokaryotic SSB homologue in the protozoan parasite P falciparum (PfSSB) that is encoded in the nucleus and targeted to the apicoplast. PfSSB is expressed in all the three stages of intraerythrocytic life cycle of parasite and binds specifically to apicoplast DNA. Both recombinant as well as endogenous PfSSB proteins form tetramers in solution like Escherichia coli SSB protein although it fails to complement E. coli ssb mutant cells suggesting subtle differences between these proteins possibly due to the differences in the C-terminal region. Consistent with the expression of PfSSB in the parasites, a strong ssDNA binding activity specific to PfSSB was found in the parasite lysate. Further, we show that inhibitors against apicoplast replication and translation machinery modulate PfSSB expression and function significantly, possibly by affecting the translocation of the protein to the apicoplast as well as intervening with the translation of the protein during the second life cycle following drug treatment. These results suggest a mechanism for delayed death phenomena shown by parasites treated with drugs that affect apicoplast house keeping functions.
MATERIALS AND METHODS
Construction of recombinant expression vector
The expression vector pET28a (Novagen) with N-terminal His6-tag was used to clone ssb. Pfssb was amplified (omitting apicoplast targeting signal) from the genomic DNA by means of polymerase chain reaction using triple master mix PCR enzyme (Eppendorf) and specific primers (P1–P2). Please see Table 1 for all primer sequences. The amplified DNA product was double digested with BamHI and XhoI restriction enzymes (New England Biolabs), ligated into pET28a expression vector and used to transform DH10β bacterial cells. The recombinant clone was subsequently sequenced and the sequence perfectly matched with the sequence reported in PLASMODB.ORG (PFE0435c).
Table 1.
List of primers
| Primer number | Name of the primer | Primer sequence from 5′ to 3′ |
|---|---|---|
| P1 | Pf SSB fw BamH1 | CGGGATCCATGAATGAGAAATCATTAAAT |
| P2 | Pf SSB rv Xho1 | CC GCTCGAGTCATTCAAATTCTTGG |
| P3 | Pf SSBΔ28 rvXho1 | CCGCTCGAGTTCATTATTAYCATCTAACCTAT |
| P4 | Pf SSB fw RT | CAGTCAACCGAAACAAATAAC |
| P5 | Pf SSB rv RT | CATTATTATCATCTAACCTAC |
| P6 | Pf GAPDH fw RT | ATGCCAAGTAGATGTTGTATGTGAA |
| P7 | Pf GAPDH rv RT | TCGTACCATGAAACTAATTTGAAGA |
| P8 | IRA Fw | CAATATTTTAATACTGTC |
| P9 | IRA Rv | TAGCTCAGAATTAACGCTA |
| P10 | TufA Fw | GCAACACCTAATAAATTAAAG |
| P11 | TufA Rv | TAATTTTTTATTTCTGTTATAATAC |
| P12 | Ori Fw | CTTAATGATCCGATAATTATTTAG |
| P13 | Ori Rv | TTATACTTAACTACTCAACTTTAC |
| P14 | Sir2 Fw | CGTCTACTGTATCAACAGCT |
| P15 | Sir2 Rv | CACTTGACCCTTTTAATATATTT |
| P16 | Orc5 Fw | AGTTTAATATGTCAACAAATTAATAC |
| P17 | Orc5 Rv | TTATATTATCAACTCATCTAGAGG |
Protein purification
His6-PfSSB was purified using Ni–NTA resin (QIAGEN) as per the protocol suggested by the vendor. The details of the protocol are described in the supplementary section (‘Materials and Methods’ section). GST-PfSSB and GST proteins were purified using glutathione sepharose 4B beads as described in the Supplementary Data (‘Materials and Methods’ section).
Immunodepletion assay
GST-PfSSB (50–60 µg) or GST bound glutathione sepharose 4B beads were incubated overnight with 30 µl crude anti-SSB sera at 4°C. These beads were separated by low-speed centrifugation (3000 r.p.m.) from immunodepleted sera. Each immunodepleted sera (3 µl) were taken out and the rest were further incubated with respective fresh protein bound beads for another 8 h at 4°C. After the second round of immunodepletion process, the sera were separated from the beads as described earlier. Western blot experiments were performed using four (1–4) different PVDF strips having equal amount of parasite lysate. The first strip was treated with untreated anti-SSB sera (1:5000 dilution) as control, second strip was treated with second round GST-depleted anti-sera whereas third and fourth strips were treated with first and second round GST-PfSSB depleted anti-sera with same dilution as control. All the blots were retreated with anti-Pf Actin sera to confirm equal loading.
Electrophoretic mobility shift assay
To investigate the ssDNA binding activity of PfSSB, EMSA was performed either using a mixture of M13mp18 circular ssDNA and pUC18 circular dsDNA or radiolabelled 70 base synthetic oligonucleotide as described elsewhere (22,26). The details of the EMSA protocol are described in the Supplementary Data (‘Materials and Methods’ section).
Generation of antibodies
Polyclonal antibodies against purified His6-PfSSB were raised in mice using essentially the protocol described previously by Harlow and Lane (23).
Western blotting and immunoprecipitation
Either mixed stage or synchronized parasites were released from the host RBCs by saponin lysis and the parasites were resuspended in 50 mM Tris–Cl (pH 7.5) containing protease inhibitor cocktail (Sigma) followed by addition of SDS–PAGE loading buffer and boiling for 5 min at 95°C. The cell lysates were resolved in 12% SDS–PAGE and subsequently used for western blot analysis. The immunoprecipitation was carried out using the protocol described elsewhere (10).
Parasite culture and drug treatment
Plasmodium falciparum (3D7) strain was cultured as described earlier (11). Synchronization of parasite culture was obtained by treating the RBC-infected parasites in 5% Sorbitol (W/V) solution in every 48 h at ring stage. Study of effect of different drugs on parasites and the modulation of PfSSB and other proteins was conducted by treatment with ciprofloxacin (5, 10 and 15 µM) or tetracyclin (10 µM) or artemisinin (10, 20 and 30 nM) in parasite culture containing 0.5% parasitemia in 6-well culture plate. Fresh medium with different drugs were replaced in every 24 h. Finally, parasites were harvested in different stages (first or second life cycle). MG132 (100 nM) was added in the ciprofloxacin treated culture during the second life cycle.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed essentially following the protocol described by Ram et al. (10) with appropriate modifications. Please see the Supplementary Data (‘Materials and Methods’ section) for details.
RT-PCR analysis
Total RNA from the parasite was isolated by Trizol method (11) RNA was treated with DNase I (Fermentas) to avoid DNA contamination. Synthesis of cDNA was performed by using Superscript II invitrogen Kit as per the manufacture’s instruction. RNA without reverse transcriptase was considered as negative control during cDNA synthesis. PCR was carried out with 4 µl cDNA sample or negative control by using primers for PfGAPDH (P6–P7) or PfSSB (P4–P5). PCR products were further analysed by agarose gel elctrophoresis.
Sucrose gradient ultracentrifugation
Sucrose gradient ultracentrifugation assay was done following the protocol described by Choudhury et al. (24) Recombinant PfSSB (150 µg) or 150 µl P. falciparum whole cell extract (in lysis buffer) obtained from 50 µl mixed stage parasite pellet or different marker proteins (∼200 µg) were layered on top of 4.9 ml 10–40% (w/v) sucrose step gradient in a buffer containing 25 mM Tris–HCl (pH 7.5), 250 mM NaCl, 2 mM sodium bisulphate and 0.05% Triton X-100. Samples were centrifuged in a Beckman MLS-50 rotor at 35 000 r.p.m. for 18 h at 4°C. Several fractions (∼170 µl) were collected for each protein. Fractions of recombinant protein and marker proteins were subjected to 10% SDS–PAGE analysis where as fractions of parasite extract were TCA precipitated and subjected to western blot analysis by anti-PfSSB antibodies. Standard curve was generated using Microsoft excel application program by plotting molecular mass as the function of sedimentation distance for various marker proteins, e.g. ovalbumin (43 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa) and thyroglobulin (669 kDa) (Amarsham Biosciences, USA). Regression analysis using the Microsoft excel application programme yielded the equation y = 2.0155x + 0.108. ‘y’ represents the log of the molecular mass in kDa and ‘x’ represents log of fraction number. The peak fraction numbers of recombinant PfSSB (fraction no. 9) and endogenous PfSSB (fraction no. 8) were fitted to the standard curve and relative molecular masses were estimated.
Dynamic light-scattering study of Pf SSB
The particle size measurement was performed by a dynamic light scattering (DLS) technique (mostly at scattering angel = 90° and laser wave length = 660.00 nm) on a digital correlator (Photocor instrument, USA). Scattered light obtained from the sample kept in a quartz cuvette was detected by a photo multiplier tube (Hamamatsu). The signal was converted to an intensity autocorrelation function by a digital correlator. The data analysis was done using CONTIN software provided by Brookhaven instrument. Further details about DLS and data analysis can be found elsewhere (25).
Fluorescence and confocal microscopy
Smears of parasitized RBC of different developmental stages were made and fixed in chilled methanol. Blocking was done by incubating the slide with a solution containing 3% BSA and 0.05% saponin in a humidified chamber for 2 h at 37°C. The slides were washed with 1× PBS and incubated with anti-PfSSB/anti-PfACP antibodies at 1 : 2000 dilutions for overnight at 4°C. After washing the slides thoroughly with 1× PBS, they were again incubated with 1× PBS containing anti-mouse/anti rabbit secondary antibodies and DAPI (Molecular probe, Invitrogen) for 1 h at room temperature. Finally, the slides were washed and mounted with anti-fed reagent and viewed under 100 × oil immersion objective. Confocal images were collected using Laser scanning Olympus microscope or Carl Ziess Apotome fluorescence microscope and the figures were prepared using Adobe photoshop.
Complementation assay—plasmid bumping assay
For complementation assay, E. coli RDP317 strain (a kind gift from Dr Umesh Varshney, Indian Institute of Science, Bangalore, India) was used. In this strain, the chromosomal ssb gene is replaced by a kanamycin resistance (ssb::Kan) marker and it harbours a support plasmid pRPZ150 (ColEl ori, TcR) coding wild-type SSB protein. The complementation assay was performed essentially following the protocol described elsewhere (26) with some modifications as described in the Supplementary Data (‘Materials and Methods’ section).
Metabolic labelling of parasite proteins
Synchronized parasites (∼12–14 h post-invasion) were treated with ciprofloxacin (10 µM). Untreated and ciprofloxacin treated trophozoite stage parasites during second life cycle (∼68 h post drug treatment) were incubated with 35S-methionine (specific activity = 15 mCi/ml, added to a final concentration of 150 µCi/ml) for 3 h. The parasites were then released by saponin lysis, washed with 1 × PBS and subsequently lysed in lysis buffer (30 mM Tris–HCl, pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.7% Triton X, Protease inhibitor cocktail). The lysate was used for immunoprecipitation assay with anti-PfSSB and anti-PfActin antibodies or respective pre-immune sera as described earlier (10). Immunoprecipitated samples were further subjected to analysis by SDS–PAGE and autoradiography.
RESULTS
PfSSB is constitutively expressed in intra-erythrocytic life cycle of P. falciparum and it is targeted to the apicoplast
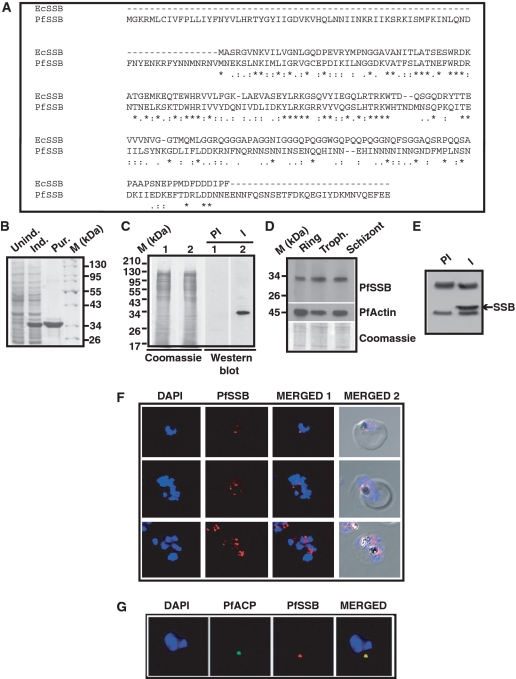
PLASMODB contains an ORF (PFE0435c) that shows 39% identity and 66% homology with E. coli SSB (Figure 1A). It has a long stretch of 76 amino acid residues at the N-terminus that shows putative apicoplast targeting sequence. PfSSB also contains an extension of 28 amino acid residues at the C-terminus that does not show homology with EcSSB. Overall, the N-terminal ssDNA binding region (excluding the apicoplast targeting sequence) shows more homology than the C-terminal protein–protein interacting domain (excluding the extreme C-terminal 28 residues) that is rich in asparagine residues in contrast to the similar domain of EcSSB that is rich in glycine residues (Figure 1A). Analysis of primary amino acid sequence of PfSSB reveals evolutionary conserved oligosaccharide/oligonucleotide binding domain (from amino acid 80–184) (OBD) implicated in ssDNA binding. The three tryptophan residues (Trp 40, Trp 54, Trp 58) which are necessary for ssDNA binding in EcSSB (27) are conserved in OBD of PfSSB (Trp-117, Trp-131, Trp-167) suggesting their possible involvement in ssDNA binding. Likewise, His-132, an amino acid residue involved in oligomerization (28) is conserved in both the species. The crucial extreme C-terminal conserved hydrophobic residues proline and isoleucine found in EcSSB (involved in protein–protein interaction) (17) are replaced by glutamic acid residues in PfSSB suggesting difference in interacting partners between these two proteins.
Figure 1.
Amino acid sequence alignment, protein expression profile and immunolocalization of PfSSB. (A) Clustal W amino acid sequence alignment of PfSSB with E. coli SSB (EcSSB). Identical residues are marked with ‘asterisk’ whereas ‘colon’ and ‘end note’ signs are used to indicate strongly and weekly similar residues, respectively. (B) Coomassie-stained gel picture of induction and purification of His6-tagged Pf SSB. Protein markers (M) are indicated on the right. (C) The right panel shows the western blot analysis using pre-immune (PI) or immune (I) sera against PfSSB (lanes 1 and 2) against 3D7 parasite lysate. The left panel shows the coomassie stained gel following transfer of the proteins on the membrane (loading control) (D) Expression profile of PfSSB at the different erythrocytic stages as indicated on top is shown by western blot analysis using polyclonal antibodies against PfSSB. The same blot was reprobed with antibodies against PfActin (middle panel). The bottom panel shows the coomassie stained gel following transfer. (E) Immunoprecipitation of PfSSB protein from parasite lysate using immune (I) or pre-immune sera (PI) followed by western blot analysis using polyclonal antibodies against SSB. Arrowhead indicates the position of PfSSB protein. (F) Localization of PfSSB by immunofluorosecnce analysis using glass slide containing fixed parasites obtained from trophozoite and schizont stages in the presence of anti-PfSSB antibodies. DAPI indicates nuclei whereas merged panel shows the presence of PfSSB with respect to the nuclei. The last panel shows the phase picture of parasite infected RBC. (G) Immuno-colocalization of PfSSB and apicoplast marker acyl carrier protein (ACP). Merged panel shows the co-localization of both the proteins.
The ssb ORF excluding the signal peptide was amplified by using specific primers (as shown in Table 1) and genomic DNA from 3D7 strain of P. falciparum. The amplified PCR product was subsequently cloned in the expression vector pET28a and the recombinant construct was further sequenced completely. The sequence of the cloned ssb fragment was found to be identical with the sequence reported in PLASMODB.ORG. BL21 codon plus cells were transformed with the recombinant clone and His6-PfSSB protein was purified using Ni–NTA affinity purification as described in the ‘Materials and Methods’ section (Figure 1B). Polyclonal antibodies were generated in mice using the purified protein as antigen.
In order to investigate whether PfSSB is expressed in the parasites, western blot analysis was performed using parasite lysate obtained from mixed stage parasites in the presence of pre-immune and immune sera against PfSSB. A specific band (∼30 kDa) was obtained only in the immune sera treated lane and not in the pre-immune sera treated lane (right panel, Figure 1C). The left panel shows commassie stained gel following transfer of the proteins onto the membrane as loading control. To crosscheck the specificity of the anti-PfSSB serum, immunodepletion assay was performed. For this purpose, anti-PfSSB sera were incubated overnight with glutathione sepharose 4B beads containing GST proteins or GST-PfSSB proteins. GST or GST-PfSSB treated sera were further immunodepleted using fresh GST or GST-PfSSB proteins containing beads for another 8 h. These different immunodepleted sera and untreated sera were used for western blot analysis against parasite lysate obtained from mixed stage parasites. We find that both untreated and GST protein depleted sera show a strong band corresponding to PfSSB. However, the intensity of the PfSSB band was reduced drastically when GST-PfSSB imuunodepleted sera were used for western blot analysis (Supplementary Figure S1A). The same membranes were treated with anti-PfActin sera resulting in a strong band corresponding to PfActin in all the lanes confirming the loading of equivalent amount of proteins in all the lanes (Supplementary Figure S1A). The immunodepletion results clearly indicate the specificity of the anti-PfSSB sera. Further, the expression pattern of the endogenous protein at the different developmental stages were investigated by western blot analysis using anti-PfSSB antibodies against parasite lysate obtained from synchronized ring, trophozoite and schizont stage parasites. A specific band (∼30 kDa) was obtained in all the stages suggesting that PfSSB is expressed during all the stages of the asexual developmental cycle (Figure 1D). The deduced molecular mass of PfSSB is ∼24.6 kDa. We find that PfSSB runs anomalously in SDS–PAGE consistent with many other Plasmodium proteins (10). As a loading control, the same blot was used to track the expression of PfActin at different stages (Figure 1D). To further investigate the authenticity of the band obtained in western blot analysis, immunoprecipitation experiment was performed using parasite lysate and pre-immune and immune sera against PfSSB. Western blot analysis following immunoprecipitation indicates the presence of a specific band in the immune sera precipitated lysate but not in pre-immune sera treated samples (Figure 1E). These results truly reflect the endogenous expression of PfSSB and the specificity of the antibodies.
We further investigated the subcellular localization of PfSSB during asexual developmental cycle by immunofluroscence assay using antibodies against PfSSB. Distinct subnuclear spot corresponding to apicoplast was observed in late ring/trophozoite stage parasites with undivided nucleus (Figure 1F, first row, top panel,) whereas multiple subnuclear spots were observed in multinucleated schizont stage parasites (Figure 1F, bottom two rows, top panel). These results are consistent with the multiplication of apicoplast organelle with parasite growth. To further confirm whether PfSSB is located in the apicoplast organelle of the parasite, immuno-colocalization experiment was performed using antibodies against P. falciparum ACP (a marker for apicoplast) and PfSSB (11). We find that PfACP is localized in the sub-nuclear apicoplast compartment. Interestingly, PfSSB signal was completely merged with the PfACP signal suggesting that PfSSB is truly localized in the apicoplast (Figure 1F, the bottom panel).
Oligomeric status PfSSB in vitro and in vivo and homology modelling
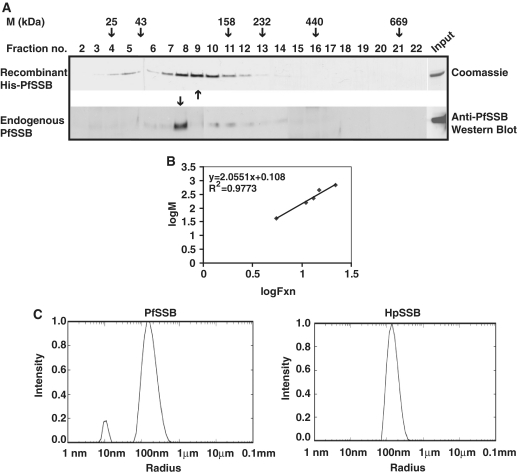
EcSSB forms homo-tetramer in solution (28). It will be interesting to see the oligomeric status of recombinant PfSSB and endogenous PfSSB from parasite lysate in solution. For this purpose, both recombinant PfSSB and Plasmodium lysate from mixed stage parasites were subjected to sucrose density gradient centrifugation as described in the ‘Materials and Methods’ section. Different molecular mass markers were also used for centrifugation under the same experimental conditions. Several fractions were collected for each molecular mass marker and peak fraction was identified (shown with arrow heads on top, Figure 2A). Subsequently, the peak fraction of recombinant PfSSB was identified by SDS–PAGE and coomassie staining of the gel containing different fractions following centrifugation (Figure 2A, top panel). The peak fraction of endogenous PfSSB was identified by western blot analysis of each fraction using anti-PfSSB antibodies (Figure 2A, bottom panel). A standard curve was plotted using the log fraction number (peak fraction) against molecular mass of the standard proteins (Figure 2B). This helped us to estimate the molecular mass of recombinant PfSSB (∼104 kDa) and native PfSSB (∼92 kDa). The apparent difference in the calculated molecular masses between the recombinant and endogenous proteins could be due to the presence of six histidine residues and some extra amino acid residues in recombinant PfSSB contributed by the expression vector.
Figure 2.
Oligomeric properties of PfSSB. (A) Sucrose density gradient analysis of recombinant Pf SSB and endogenous Pf SSB. Recombinant PfSSB was subjected to sucrose density gradient ultracentrifugation and different fractions were collected. Upper panel shows the coomassie stained SDS–PAGE gel of recombinant protein of different fractions following sucrose density gradient. The lower panel shows the western blot analysis of different fractions following sucrose density gradient ultracentrifugation of Plasmodium extract using anti-PfSSB antibodies. Arrowheads indicate peak fractions of elution profile of recombinant and endogenous PfSSB. Peak fractions of molecular mass markers following ultracentrifugation are shown on the top. (B) The graph represents the standard curve of log of peak fraction number against log of molecular mass of known marker proteins. (C) Dynamic light-scattering analysis of HpSSB and PfSSB. The right panel shows particle size distribution of HpSSB that indicates the accumulation of a single species population of average hydrodynamic diameter of 139.1 ± 4.125 nm. The major population of Pf SSB is found to be accumulated in the average hydrodynamic diameter of 152.133 ± 2.65 nm (left panel).
Comparison of the calculated molecular mass of monomeric form of recombinant PfSSB (25.2 kDa) and endogenous PfSSB (24.6 kDa) with the estimated molecular mass obtained from sucrose gradient experiments revealed that both the recombinant PfSSB and endogenous PfSSB would be in tetrameric form.
Further, DLS experiment was performed to evaluate whether tetrameric form of PfSSB were present predominantly in the solution. For this purpose, SSB protein from gastric pathogen Helicobacter pylori was taken as positive control. The molecular mass of HpSSB is ∼25 kDa (close to ∼His6-PfSSB, ∼25.2 kDa) and it has been shown recently that HpSSB exists as tetramer in solution (26). DLS allows us to study the average particle size (hydrodynamic diameter) and its distribution in solution. The average hydrodynamic diameter of Pf SSB was found to be 152.1 ± 2.65 nm which is quite close to the hydrodynamic diameter 139.1 ± 4.12 nm of HpSSB (Figure 2C). Thus, it can be inferred that majority of PfSSB remains as tetramer as HpSSB in solution. These results also reconfirm sucrose density gradient ultracentrifugation results.
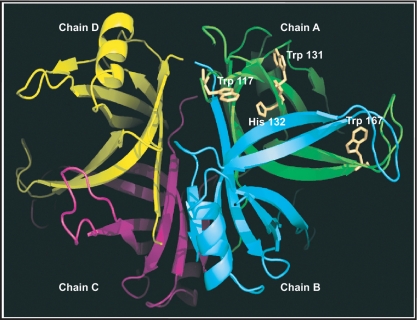
Consistent with the tetrameric from of PfSSB as obtained using various methods described earlier, we performed the homology modelling of the protein using EcSSB structure as template (29). Monomer model of putative Oligo nucleotide Binding domain of PfSSB (80–193 aa) is modelled using Modeller 9v7 software with the Oligo nucleotide binding domain of EcSSB (ssDNA bound subunit A1001–1112 of PDBID:1EYG) as a template since it has significant sequence homology (39% identity). Modelled monomer of PfSSB was superimposed using Dali pairwise structure alignment program (http://ekhidna.biocenter.helsinki.fi/dali/start) onto individual monomer of tetramer (A–D subunits in PDBID:1EYG) of EcSSB to generate the tetramer of PfSSB with root mean square deviation (RMSD) of 1.8, 1.6, 2.0 and 1.8 Å, respectively. The model revealed three conserved tryptophan residues (Trp-117, Trp-131 and Trp-166) which are essential for ssDNA interaction. His 55 residue in EcSSB structure where it has been implicated for oligomerization of SSB tetramer is also conserved in PfSSB (His132) (29). The homology model of PfSSB homotetrameric oligo binding domain is shown in Figure 3 where different monomers (chains/subunits) (A–D) are shown in different colours using Pymol software. A homology model was also generated using tetrameric form of PfSSB along with two stretches of ssDNA (chains A and B) that shows the interaction of ssDNA with the oligonucleotide binding domain of PfSSB (Supplementary Figure S1B).
Figure 3.
Homology modelling of PfSSB. Pf SSB structure was modelled using the crystal structure of oligonucleotide binding domain of E. coli SSB as template. The positions of the conserved amino acid residues (Trp-117, Trp-131, Trp-167 and His-132) are shown in the figure. Four monomers of SSB that form the tetramer are shown in different colours (chains A–D). The model also shows two ssDNA molecules (A and B) interacting with the tetramer model.
Complementation of E. coli ssb mutant cells with wild-type and deletion mutant of PfSSB
In order to investigate whether PfSSB is a true homologue of SSB in vivo, we performed genetic complementation experiments using an E. coli ssb mutant strain with the help of plasmid bumping experiment using EcSSB and H. pylori SSB as positive control (26). The details of this method are described in the ‘Materials and Methods’ section and the results are shown in Table 2. We found that both EcSSB and HpSSB could complement this mutant strain whereas PfSSB could not complement under the same experimental conditions. Further, a construct deleting the unique extreme C-terminal 28 residues present in PfSSB also cannot complement the E. coli ssb mutant strain suggesting that PfSSB is inherently different from EcSSB. It has been reported earlier that the extreme 10 amino acid residues of EcSSB are essential for its in vivo function (30). Poor homology at the extreme C-terminal regions of these two proteins may contribute to the inability of PfSSB to complement E. coli ssb mutant strain.
Table 2.
Complementation experiments results
| Test ssb | Total no of patches | AmpR | AmpR+TcR | Percentage of plasmid bumping efficiency (%) |
|---|---|---|---|---|
| pTRC + Ecossb(+ve) | 40 | 40 | 4 | 90 |
| pTRC(−ve) | 40 | 40 | 38 | 5 |
| pTRC + Hpssb | 40 | 40 | 6 | 85 |
| pTRC + Pfssbwt | 40 | 40 | 37 | 7.5 |
| pTRC + PfssbΔC28 | 40 | 40 | 38 | 5 |
In vitro and in vivo ssDNA binding activity of PfSSB
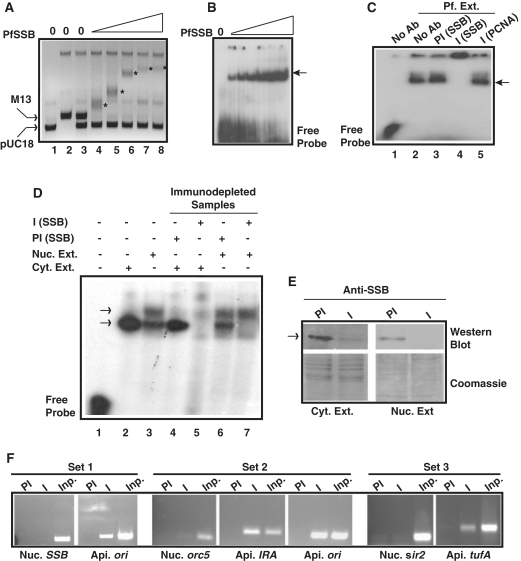
During DNA replication, recombination and repair, SSB protein stabilizes the ssDNA intermediates. ssDNA binding activity of purified recombinant PfSSB was monitored using agarose gel mobility shift assay in the presence of a mixture of M13 ssDNA and pUC18 dsDNA. The results indicated that the mobility of single-stranded M13 DNA was retarded significantly with increasing amount of PfSSB protein compared to pUC18 DNA whose mobility did not change at all (Figure 4A). Further, gel mobility shift assay was performed using radioactive ssDNA probe and purified PfSSB. We find similar results as above where increasing amount of PfSSB results major shift of the ssDNA probe (Figure 4B). We also investigated whether P. falciparum extract would contain ssDNA binding activity contributed by PfSSB. For this purpose, radiolabelled ssDNA probe (70 mer) was incubated with P. falciparum parasite extract followed by PAGE analysis of the reaction mixture. We find that P. falciparum extract cause a considerable shift of the ssDNA probe (lane 2, Figure 4C). To identify whether this binding is specific for PfSSB, the reaction mixture was incubated further in the presence of undiluted anti-PfSSB or pre-immune sera as described in the ‘Materials and Methods’ section. We find, while pre-immune sera do not affect the ssDNA binding activity, immune sera causes a supershift suggesting that the binding is specific for PfSSB (lanes 3 and 4, Figure 4C). Non-immune PfPCNA antibodies also do not affect the ssDNA binding activity further confirming the presence of specific PfSSB like ssDNA binding activity in the P. falciparum extract.
Figure 4.
In vitro and in vivo DNA binding property of Pf SSB. (A) PfSSB binds specifically to ssDNA. Different amount of Pf SSB was incubated with the mixture of M13 mp18 ssDNA and pUC18 dsDNA followed by separation of the protein-DNA complex by agarose gel electrophoresis. ssDNA was progressively retarded (asterisk) with increasing concentration of PfSSB whereas mobility of dsDNA was not affected. (B) Electrophoretic mobility shift assay using short radiolablled single-stranded oligonucleotide probe with increasing amount of Pf SSB. The arrowhead indicates the protein bound DNA. (C) Gel retardation analysis of single stranded DNA binding activity from P. falciparum cell extract. Labelled ssDNA probe was incubated with P. falciparum cell extract in the absence or presence of anti-PfSSB or anti-PCNA antibodies or pre-immune sera against SSB followed by PAGE analysis of the protein–DNA complexes (shown by arrowhead). Supershift of the DNA–protein complex was observed only in the presence of immune sera against PfSSB (lane 4) but not in the presence of pre-immune sera (lane 3) or non-immune sera (anti-PCNA) (lane 5). (D) Single-stranded DNA binding activity of cytoplasmic or nuclear extract or immunodepleted samples from each fraction in the presence of preimmune and immune sera against PfSSB. A single shifted band of labelled probe was observed in cytoplasmic fraction (lane 2) whereas two such bands were observed in nuclear extract treated samples (lane 3). No band was observed in the presence of immunodepleted cytoplasmic extract (lane 5) whereas the bottom band was not visible in the presence of immunodepleted nuclear extract treated sample (lane 7). Pre-immune depleted samples did not affect the pattern of bands (lanes 4 and 6) found in extract only lanes (lanes 2 and 3). (E) The upper part of the left panel shows the western blot analysis of immune and pre-immune depleted Plasmodium cytoplasmic extract whereas the lower part represents the coomassie stained gel as loading control following transfer of the proteins on the membrane. Similarly, the upper part of the right panel shows the western blot analysis of the immunodepleted nuclear extract (immune and pre-immune treated) and the lower part of the same panel represents the coomassie stained gel as loading control. The arrowhead indicates the position of the PfSSB protein. (F) Specific binding of PfSSB to apicoplast DNA in ChIP assay. PCR amplification of ori region of apicoplast (api.) DNA was found only in the immunoprecipitated samples using immune sera (I) against PfSSB but not with the pre-immune (PI) sera whereas nuclear (nuc.) ssb gene could not be amplified using ssb gene specific primers (set 1). Further, PCR amplification of immunoprecipitated DNA along with input (inp.) genomic DNA were performed using specific primer sets from nuclear orc5, apicoplast ori and IRA region (set 2) or nuclear sir2 and apicoplast tufA regions (set 3). Specific products were found only in the immunoprecipitated samples using immune sera and primer sets from apicoplast DNA. No significant amplification was found using primer sets from nuclear DNA. PCR products were obtained in all the control input lanes.
Specific interaction of the high molecular mass containing tetrameric form of PfSSB with high titer antibodies against PfSSB may form huge complex causing supershift of the DNA–protein complex that may not enter the polyacrylamide gel properly as shown above. The supershift experiments were repeated with some modifications like using different concentrations of diluted PfSSB antibodies (1–5 μl of 1:1000 diluted anti-PfSSB sera in 1× PBS) and resolving the DNA–protein complex for a longer time during PAGE analysis. We find that pre-immune sera against PfSSB do not show any supershifted band even at the highest concentration whereas immune sera show specific supershifted bands at all concentrations. However, with increasing concentration of immune sera, the intensity of the original DNA–protein complex is reduced significantly with the appearance of a higher molecular mass containing population that does not enter the gel properly apart from the supershifted band that entered the gel (Supplementary Figure S2). The complex that does not enter the gel may be the result of the accumulation of huge complex containing ssDNA–Protein-Ab at the higher concentration of antibodies.
To confirm whether the ssDNA binding activity present in P. falciparum parasite extract is due to the presence of PfSSB, sub-cellular fractionation of the parasite extract was performed to obtain cytoplasmic extract and nuclear extract as described earlier (31). Each fraction was immunodepleted using immune and pre-immune sera against PfSSB. Immunodepleted cytoplasmic and nuclear extract were used for gel retardation assay using labelled ssDNA probe. We find that cytoplasmic extract yields one shifted band whereas nuclear extract results in two shifted bands. An extra band was obtained on top of the band corresponding to the band obtained from the cytoplasmic extract (lanes 2 and 3, Figure 4D). Immunodepleted cytoplasmic extract using PfSSB immune sera resulted in complete loss of DNA binding activity (lane 5). Interestingly, immunodepleted nuclear extract resulted in the loss of band corresponding to the band obtained in cytoplasmic extract. However, the upper band obtained in nuclear extract was not affected following immunodepletion suggesting the shift represented by upper band would not be due to the presence of PfSSB (lanes 6 and 7, Figure 4D). This ssDNA binding activity could be due to the presence of specific nuclear protein present in the nuclear extract. This could be due to the presence of RPA in the nuclear fraction as it has been reported earlier (20). Immunodepletion of cytoplasmic extract was further confirmed by western blot analysis of pre-immune and immune sera depleted cytoplasmic extract using anti-PfSSB antibodies (Figure 4E, left panel). The results indicate the presence of a strong band corresponding to PfSSB in the pre-immune sera depleted sample compared to the very weak band present in the immune-sera depleted sample. The similar loading in both the lanes is indicated by the coomassie stained gel following transfer of the proteins onto the membrane. Immunodepletion of nuclear extract using pre-immune sera and PfSSB antibodies followed by western blot analysis indicate the presence of PfSSB in the nuclear extract possibly as a contamination of nuclear fraction with cytoplasmic proteins (Figure 4E, right panel) that may have resulted the appearance of the lower band in the gel shift assay using nuclear fraction as shown earlier. These results indicate that the specific immunodepletion of PfSSB from the cytoplasmic extract may have caused the inability of the immunodepleted extract to bind to ssDNA.
We further investigated whether PfSSB binds specifically to apicoplast DNA in vivo. For this purpose, ChIP experiments were performed using pre-immune or immune sera against PfSSB. Recently a similar approach has worked nicely to show that PfGyrB binds specifically to its target apicoplast DNA with no affinity towards nuclear DNA (22). ChIP results indicate that PfSSB binds strongly to the apicoplast ori region compared to the nuclear ssb gene as control (Figure 4F, set 1). ChIP experiments were repeated several times followed by PCR using primer sets from different nuclear loci and apicoplast loci. We find that the intensity of the PCR product using primer set from apicoplast DNA is always higher many fold than that obtained from nuclear DNA in different sets of experiments (Figure 4F, sets 2 and 3). These results indeed suggest that PfSSB binds specifically to apicoplast DNA as compared to nuclear DNA.
Modulation of PfSSB expression and activity in the presence of inhibitors against apicoplast machinery
Plasmodium contains both the subunits of prokaryote type gyrase (a and b) that are nuclear encoded but targeted to the apicoplast (11). Gyrase is required for the maintenance of the apicoplast DNA as positive supercoils are generated ahead of the replication fork that should be relieved intermittently for the faithful replication of apicoplast DNA. It has been shown earlier that ciprofloxacin, a gyrase A specific inhibitor affects the apicoplast DNA replication specifically without affecting nuclear DNA replication (32). Interestingly, ciprofloxacin shows typical delayed death phenotype of the parasites where parsitemia drops significantly during the second life cycle of the parasites following the addition of the drug without affecting the parasites during the first life cycle (13). We were interested to study whether ciprofloxacin would modulate PfSSB function since it is also a key molecule for apicoplast DNA replication.
We first investigated the effect of ciprofloxacin on parasite growth during first and second life cycle of parasite development. Although ciprofloxacin caused ∼40% decline in parasitemia at 15 µM concentration, it did not cause any significant effect at lower drug concentration (5/10 µM) at the end of the first life cycle (∼48 h). However, at the end of the second life cycle (∼92 h), there was a dramatic drop in parasitemia at 5/10 µM drug concentration that resembled the delayed death effect of ciprofloxacin as described earlier (Figure 5A) (13).
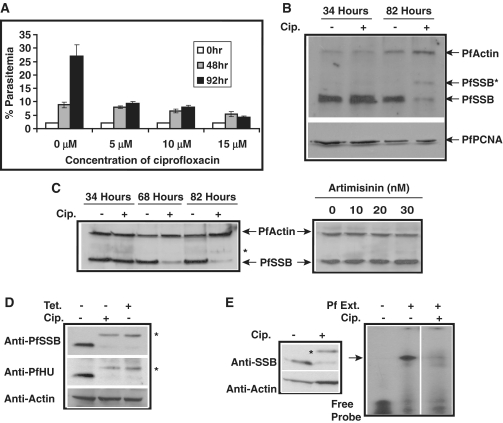
Figure 5.
Modulation of PfSSB expression and activity in the presence of apicoplast inhibitors. (A) The graph shows the parasitemia (%) at different time points in the absence and presence of different concentration of gyrase inhibitor ciprofloxacin. (B) Western blot analysis using anti-PfSSB antibodies to show the effect of ciprofloxacin on the expression of PfSSB at protein level during first life cycle (34 h) and second life cycle (82 h). There was overall decrease in PfSSB protein level as well as appearance of an extra band (asterisk) during second life cycle in ciprofloxacin treated samples. The expression profile of PfActin and nuclear PfPCNA are also shown as controls. (C) The left panel shows the expression profile PfSSB at different time points along with control PfActin in the absence and presence of ciprofloxacin. ‘Asterisk’ indicates the unprocessed form of SSB. The right panel shows the effect of artimisinin at different concentration on PfSSB and PfActin expression. Artimisinin does not seem to have any effect on PfSSB expression. (D) Western blot analysis using parasite lysate obtained from ciprofloxacin (cip) or tetracycline (tet) treated or untreated samples in the presence of anti-PfSSB or anti-PfHU (histone like protein) or PfActin antibodies. ‘Asterisk’ indicates the unprocessed form of HU and PfSSB. (E) Effect of ciprofloxacin on ssDNA binding activity from Plasmodium extract. The left panel shows the western blot analysis using anti-PfSSB antibodies with or without drug treatment along with actin as control. The right panel shows the gel retardation assay using parasite lysate from cip. treated and untreated samples.
It has been reported earlier that drugs that affect apicoplast housekeeping functions (e.g. tetracycline), inhibit the translocation of the proteins to the apicoplast (during second life cycle) using signal sequence of ACP fused to GFP as marker (14,15). No endogenous apicoplast targeted protein has been tracked in the above studies. We were interested to investigate whether treatment of the parasites with ciprofloxacin would modulate PfSSB translocation and function. Subsequently, we performed western blot analysis using ciprofloxacin (10 μM) treated parasite lysate obtained from first life cycle (∼34 h post-drug treatment) and second life cycle (∼82 h post-drug treatment) by using antibodies raised against PfSSB. Antibodies against PfActin and PfPCNA were used as control for western blot experiments. We find that during the first life cycle, addition of ciprofloxacin does not change the expression of PfSSB protein compared to the untreated parasites (Figure 5B). However, during second life cycle, the expression of PfSSB is found to be reduced in ciprofloxacin treated parasites compared to the untreated parasites. In contrary, the expression level of control protein PfActin and nuclear replication protein PfPCNA did not change significantly. Moreover, a higher molecular weight band is observed in drug treated samples that is absent in the untreated samples (Figure 5B). This may be the unprocessed form of PfSSB before the cleavage of signal peptide and transit peptide required for the faithful translocation of apicoplast proteins. Apicoplast specific drugs often result in accumulation of the immature form of the apicoplast targeted proteins during second life cycle (15). However, the decrease in overall PfSSB protein level during second life cycle following ciprofloxacin treatment is very striking.
The previous experiment did not allow us to investigate the timing of the effect of ciprofloxacin on PfSSB protein level during second life cycle. For this purpose, ciprofloxacin treated or untreated parasite pellets were obtained from the trophozoite (∼68 h post-drug treatment) and schizont stages (∼82 h post-drug treatment) during the second life cycle followed by analysis of the PfSSB protein level by western blot analysis using anti-PfSSB antibodies (Figure 5C, left panel). Parasite lysate from the first life cycle was also taken as control. We find that the expression level of PfSSB protein is reduced even at the trophozoite stage that continues until schizont stage during second life cycle whereas the PfActin level does not change at all. A faint band corresponding to unprocessed form of PfSSB was also visible at the latter stage.
We further show that the decrease in overall PfSSB protein level and the appearance of the mature form of PfSSB following ciprofloxacin treatment is specific for apicoplast targeted drugs. Use of increasing amount of artemisinin that cause considerable parasitic death resulting in huge decrease in parasitemia does not affect the level of PfSSB at all. Neither the mature form of PfSSB could be detected even at the highest concentration of artimisinin used (30 nM) (Figure 5C, right panel).
We are interested to find out whether modulation of PfSSB expression and maturation is ciprofloxacin-specific or it is a manifestation of apicoplast targeted drugs to modulate apicoplast targeted proteins in general. For this purpose, we performed western blot analysis using ciprofloxacin and tetracycline (protein translation inhibitor that also affects apicoplast) treated parasite extract in the presence of PfSSB and PfHU (histone like protein) antibodies. Western blot analysis indicates that the treatment of ciprofloxacin and tetracycline affects the expression and maturation of both PfSSB and PfHU protein to the similar extent (Figure 5D). However, these drugs did not have any effect on control protein PfActin.
Further, to investigate whether ciprofloxacin truly affects the translocation of apicoplast targeted proteins like PfSSB during second life cycle, we have performed immunofluorescence assay using untreated and drug treated parasites in the presence of PfSSB antibodies. We find specific apicoplast localized PfSSB signal in the untreated parasites whereas the apicoplast localization of PfSSB is severely affected in the drug-treated parasites under the same experimental conditions (Supplementary Figure S3). These results strongly suggest that cirpofloxacin inhibits the translocation of SSB during second life cycle.
The above results prompted us to investigate whether parasite extract obtained from ciprofloxacin treated parasites would yield ssDNA binding activity. This would also help us to confirm whether the ssDNA binding activity from the parasite extract is truly dependent on PfSSB. Gel retardation assay was performed using parasite extract obtained from ciprofloxacin treated and untreated control parasites. We find a strong shift of the radiolabelled probe in the presence of untreated parasite extract compared to the free probe. Interestingly, ciprofloxacin treated parasite extract fails to show any prominent shifted band suggesting that the inability of ssDNA binding may be attributed to the poor presence of the mature form of PfSSB in ciprofloxacin treated samples (Figure 5E, right panel). Western blot analysis using extract obtained from ciprofloxacin treated and untreated parasites in the presence of PfSSB antibodies further confirm the above results (Figure 5E, left panel).
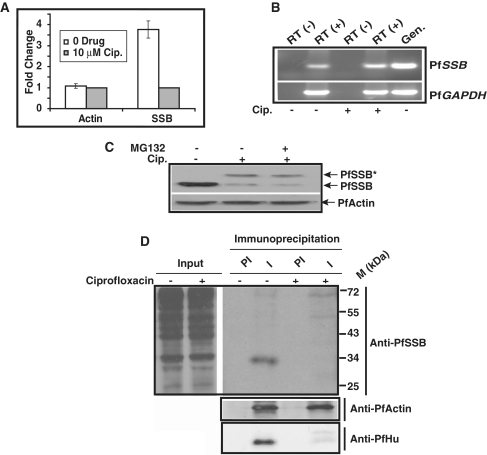
Synthesis of SSB protein is affected during second life cycle following ciprofloxacin treatment
We find that apicoplast targeted drug, ciprofloxacin affects the translocation of the proteins to the apicoplast as evidenced by the accumulation of the unprocessed form of the protein in the presence of the drug. It is also interesting that there is an overall decrease (∼4-fold) in PfSSB protein level compared to control protein PfActin following drug treatment (Figure 6A). Transcription of ssb is up-regulated in E. coli cells in the presence of ciprofloxacin (33). It is possible that the decrease in SSB protein level following ciprofloxacin treatment during second life cycle is caused by the reduced transcription of Pfssb in the nucleus. To address this issue, we performed semi-quantitative RT-PCR analysis using cDNA prepared from ciprofloxacin treated and untreated samples. We find that there is no significant difference in the transcript level of ssb in the presence or absence of ciprofloxacin (Figure 6B). PfGAPDH was used as control that also did not change its expression as ssb. These results indicate that down-regulation of SSB protein in the ciprofloxacin treated parasites may not be due to the transcriptional defect. This is consistent with the earlier published results where the microarray analysis of tetracyclin treated parasites do not show change in transcription level of nuclear genes (including genes required for nuclear encoded apicoplast-targeted proteins) compared to the untreated parasites (14). The other possibility is that SSB protein was degraded with time leading to the decrease in protein level. We used MG132, a proteosomal degradation pathway inhibitor during the second life cycle following ciprofloxacin treatment. Recently, we have shown that MG132 can stabilize PfORC1 by inhibiting degradation of PfORC1 at the late schizont stage (34). Western blot analysis using parasite lysate obtained from 100 nM MG132 treated and untreated samples (during second cycle following ciprofloxacin treatment) reveals that MG132 does not change the SSB protein level (Figure 6C) suggesting that the decrease in SSB protein level would not be due to the degradation of the protein. MG132 was active in the parasites as addition of similar concentration of drug could stabilize PfORC1 from degradation at the late schizont stage as shown by immunofluorescence and western blot analysis (Supplementary Figure S4A and B).
Figure 6.
(A) Comparison of expression level of PfSSB and PfActin in the absence and presence of ciprofloxacin for three different set of experiments. Western blot experiments were performed using parasite lysate obtained from ciprofloxacin (10 μM) treated or untreated parasites in the presence of antibodies against PfSSB and PfActin. The relative intensity of the bands was measured using densitometry scanning followed by graphical representation of them to show the fold change in the expression level. (B) RT-PCR analysis of PfSSB and PfGAPDH transcript level from the parasites treated with cip. or untreated parasites. RT (−) indicates the cDNA prepared from total RNA samples treated without reverse transcriptase. (C) Western blot analysis to evaluate the effect of proteosome inhibitor MG132 on PfSSB expression. Parasites treated with ciprofloxacin were further treated with MG132 during second life cycle before harvesting for western blot analysis. The results indicate that treatment with MG132 does not change the PfSSB protein level. The same blot was reprobed with anti-PfActin antibodies as control. (D) Status of newly synthesized PfSSB during second life cycle following ciprofloxacin treatment. Ciprofloxacin treated or control parasites were incubated in the presence of 35S-methionine for 3 h during the trophozoite stage of the second life cycle (∼68 h post-drug treatment) followed by immunoprecipitation analysis of PfSSB or PfActin or PfHU proteins using respective pre-immune (PI) or immune (I) sera from control or drug treated parasite lysate. The total parasite lysate obtained from untreated and drug treated 35S-methionine labelled parasites were loaded as input (inp.). The results indicate that the expression of newly synthesized PfSSB and PfHU is affected significantly without affecting the expression of PfActin under the same experimental conditions.
These results suggest that the decrease in the SSB protein level during the second life cycle following ciprofloxacin treatment might be attributed to the translational defect. To address this issue further, ciprofloxacin-treated or untreated parasites were incubated in the presence of 35S-methionine during second life cycle (trophozoite stage, ∼68 h post-drug treatment) for 3 h. The parasite lysate were obtained from both control and drug-treated parasites followed by SDS–PAGE analysis and autoradiography to visualize the labelled proteins. The results indicate that there is no gross change in the metabolic labelling of the proteins during second life cycle in the presence or absence of the drug (Figure 6D, left panel). The above lysate were used to immunoprecipitate newly synthesized SSB protein during second life cycle using pre-immune and immune sera against SSB. We find that SSB can be immunoprecipitated specifically using immune sera but not pre-immune sera during second life cycle from the untreated sample only (Figure 6D, right panel). Interestingly, PfActin can be immunoprecipitated from both the untreated and drug-treated sample to the similar extent using specific antibodies against PfActin under the similar experimental conditions (Figure 6D, middle panel). We also performed immunoprecipitation experiments using pre-immune and immune sera against PfHU under the same experimental conditions. We find that the translation of newly synthesized PfHU protein during second life cycle following drug treatment is also significantly reduced compared to the untreated samples (Figure 6D, bottom panel). These results suggest although there is no significant change in protein expression profile during trophozoite stage of the second life cycle between drug-treated and untreated samples, there is a specific down-regulation of translation of nuclear encoded apicoplst targeted proteins like SSB and HU during second life cycle following ciprofloxacin treatment. It is important to note that all apicoplast targeted proteins may not be affected to the similar extent at a particular time since there may be additional mechanisms to control their expression profile that need to be explored further.
DISCUSSION
The replication of apicoplast DNA circle begins in trophozoite stage and continues into schizogony (5). Very little is known about the process and enzymology involved in the replication of apicoplast genome. So far, few enzymes including gyrase and PfPRex have been reported that could be the members of active replication/recombination machinery within the apicoplast. This study is an attempt to identify and functionally characterize another key DNA replication protein, the ssDNA binding protein in P. falciparum. To our knowledge, the identification of SSB in P. falciparum is the first report of the presence of bacterial type SSB in any eukaryotic parasite. In E. coli and other prokaryotes, in addition to DNA replication the role of SSB has been extended to DNA recombination and repair events. PfSSB may be involved in these processes to maintain the apicoplast genome.
It is known that SSB protein binds to circular supercoiled DNA containing ssDNA stretches that have been induced by topological stress generated by different cellular processes like DNA replication, transcription and recombination (35). Our data suggest that PfSSB is exclusively targeted to the apicoplast and it binds to the apicoplast DNA and not to the nuclear DNA. Together, these data confirm apicoplast specific role of PfSSB. The apicoplast DNA replication occurs in the late trophozoite-early schizont stages. However, PfSSB is constitutively expressed and localized into the apicoplast during all the three stages of intraerythrocytic developmental cycle of P. falciparum. This may explain the possible involvement of PfSSB in different DNA transactions other than DNA replication during the non-replicative phases of intra-erythrocytic parasitic life cycle (ring, early trophozoite).
Earlier, ssDNA binding activity from P. falciparum had been assigned to large subunit of RPA. The ssDNA binding activity exhibited by RPA has been shown to be present in both nuclear and cytoplasmic fractions. However, specificity of the ssDNA binding activity could not be tested due to the non-availability of specific antibodies against RPA. In contrast, we show that the ssDNA binding activity from cytoplasmic fraction is largely contributed by PfSSB. The loss of ssDNA binding activity in immunodepleted cytosolic fraction using specific anti-PfSSB antibodies strongly substantiates our claim.
There are two ssDNA binding activities in nuclear fraction and only one of them is contributed by PfSSB. We show that upon immunodepletion of PfSSB from the nuclear extract, the ssDNA binding activity contributed by PfSSB is completely lost whereas the other activity remains unaffected. The unaffected ssDNA activity in the nuclear extract may probably be due to RPA as reported earlier (20). However, we cannot rule out the ssDNA binding activity by PfSSB in the nuclear extract by cross-contamination of cytoplasmic PfSSB during the cell fractionation because our ChIP experiment using specific anti-PfSSB antibodies yield robust amplification of apicoplast DNA only and no significant amplification of nuclear DNA.
Generally, SSB modulates the functions of other replication/recombination/repair proteins by interacting with them via acidic C-terminus region. It has been reported that the last 10 amino acid residues of SSB are highly conserved among different bacteria and they are essential for in vivo protein–protein interaction (30). The C-terminus of PfSSB shows little sequence conservation and there is an extension of 28 amino acid residues. Poor sequence conservation at the C-terminus of PfSSB may explain its failure to complement E. coli ssb function in vivo. The identification of the proteins interacting with PfSSB C-terminus would be an essential step towards understanding the role of PfSSB during different developmental stages of the parasite erythrocytic life cycle, particularly during the ring stage where DNA replication does not take place.
PfSSB forms tetramer in solution similar to EcSSB. His-55 and the region between amino acid residues 89–105 have been suggested to have a role in tetramerization of EcSSB. In PfSSB, His-132 is equivalent to His-55 of EcSSB and the region between 165 and 194 shows a significant homology with the region 89–105 of EcSSB. These similarities may explain the tetrameric nature of the protein both in vitro and in vivo as shown here. Our ongoing crystallization studies of PfSSB may provide a clear insight into oligomerization interfaces of the protein.
Ciprofloxacin is a quinolone antibiotic that principally targets DNA gyrase and results in the breakage of the P. falciparum apicoplast DNA without affecting the nuclear DNA during second life cycle at an IC50 of ∼7 μM. Surprisingly, our results indicate that blocking apicoplast replication with ciprofloxacin results in the drastic reduction in the levels of PfSSB and its translocation to the apicoplast as evidenced by the accumulation of PfSSB with the signal peptide sequence. However, ciprofloxacin treatment does not change the levels of cytoplasmic actin and nuclear replication protein PCNA1. The level of PfSSB is not affected when the parasites are treated with commonly used non-apicoplast targeted anti-malarial drug artemisinin, suggesting that the drug stress does not lead to the decrease in PfSSB levels. However, treatment of the parasites with tetracycline that affects apicoplast translation also modulates PfSSB expression and maturation like ciprofloxacin suggesting that these drugs may follow general mechanism that interfere with the translocation of the proteins to the apicoplast and the protein level. The similar fate of HU, another protein targeted to the apicoplast, following ciprofloxacin and tetracyclin treatment further confirms the generalized mechanism of apicoplast targeted drugs. Simultaneous treatment of parasites with ciprofloxacin and proteosome blocker MG132 clearly indicates that protein is not degraded in the presence of ciprofloxacin during the second cycle. Moreover, the inhibition of apicoplast DNA replication does not affect the level of the ssb transcripts produced in the nucleus housing ssb gene on ChrV as shown by the semi-quantitative RT-PCR analysis. Incubation of the parasites with 35S-methionine to track the newly synthesized proteins during second life cycle suggests that translation of SSB and HU transcripts are specifically affected during second life cycle following drug treatment although both untreated and drug-treated parasites show comparable protein expression profile and there is no change in expression level of control PfActin protein. Like PfSSB, the expression level of newly synthesized PfHU protein during the second life cycle following drug treatment was severely affected. It seems that apicoplast targeted drug ciprofloxacin may selectively inhibit the translation of apicoplast targeted proteins during second life cycle without affecting the translation of other proteins. It remains to be seen further, how this differential regulation of translation is maintained in the parasites.
This work shed some light to understand the mechanism behind the delayed death phenomena showed by some drugs like ciprofloxacin and tetracycline that affect apicoplast function. The parasites undergo normal division and segregation of organelles during the first life cycle. However, severe defects in apicoplast growth, morphology and cytokinesis take place in the second life cycle. Consistent with the above results, we find that the translocation of apicoplast targeted proteins like SSB is not affected during the first life cycle. It is possible that the accumulation of the drugs in the four membrane bound apicoplast is a slow process and the parasites can bypass the first life cycle since the drug may be limiting and it may not reach the critical concentration that may affect the apicoplast function immediately. There is also a constant translocation of fresh proteins from the cytoplasm to the apicoplast during this stage. However, during the second life cycle, the parasites may acquire the critical concentration of the drugs that may affect the apicoplast function, growth and morphology, which in turn affect the protein translocation in the apicoplast, leading to the accumulation of the unprocessed form of the proteins in the cytoplasm. This, in turn may be sensed by the cytoplasmic translational machinery involved in synthesis of apicoplast targeted proteins, to selectively reduce their synthesis. Latter, the complete inhibition of apicoplast function and growth may affect overall growth of the parasite leading to the parasitic death since apicoplast is the indispensable organelle of the parasite involved in various important pathways like fatty acid synthesis and heam biosynthesis. Therefore, dual mechanism consisting of translocation and translation defect of apicoplast targeted proteins during second life cycle may contribute to the delayed death phenomena shown by some apicoplast targeted drugs like ciprofloxacin and tetracyclin (Figure 7). Although ciprofloxacin is a specific inhibitor of gyrase, it remains to be seen further whether ciprofloxacin has a direct effect on the translational machinery during second life cycle following drug treatment.
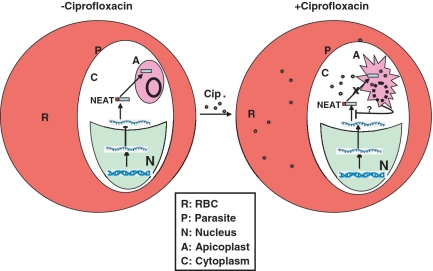
Figure 7.
Model showing the effect of ciprofloxacin on parasites. In the absence of apicoplast targeted drug like ciprofloxacin, nuclear encoded apicoplast targeted (NEAT) genes are transcribed in the nucleus (N) and transported into the cytoplasm (C) where they are translated as unprocessed protein that are translocated to the apicoplast (A) and gets processed. During second life cycle following drug treatment, the apicoplast achieve critical concentration of the drug that affects apicoplast function and morphology that inhibits translocation of NEAT proteins into the apicoplast which in turn may down-regulate the translation of these proteins. Therefore, a dual mechanism of inhibition of translocation of NEAT proteins and modulation of translation level of these proteins may contribute to the delayed death effect exerted by such drugs like ciprofloxacin. The solid and broken circles in apicoplast indicate the apicoplast genome in the absence and presence of drug.
To understand the fundamental process of apicoplast DNA replication completely, it is essential to identify all the key enzymes and study their role for the replication of 35 kb DNA circle. Our results provide conclusive evidence that apicoplast imports a bacterial homologue of nuclear encoded ssDNA binding protein that could be required in ssDNA intermediate stabilization during the process of DNA replication. The apicoplast localized PfSSB can be used as a tool to identify other components of the replication machinery as its homologue in bacteria has been shown to interact and modulate the activities of different members of replication proteins during the fork progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust, London (070046); Swarnajayanti Fellowship awarded by Department of Science and Technology, Government of India and Signalling in Malaria Parasite (MALSIG) project funded by European Union. Funding for Open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr Saman Habib for providing polyclonal antibodies against PfHU protein. Prof. H. Bohidar and Mr Pradip Gangwar are greatly acknowledged for their help with dynamic light scattering experiments. D.P., A.S. and S.D. acknowledge Council of Scientific and Industrial Research, India and A.D. acknowledges University Grant Commission, India for fellowships.
REFERENCES
- 1.Foth BJ, McFadden GI. The apicoplast: a plastid in Plasmodium falciparum and other apicomplexan parasites. Int. Rev. Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. [DOI] [PubMed] [Google Scholar]
- 2.Ralph SA, Van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Metabolic maps and function of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 3.McFadden GI, Roos DS. Apicomplexan plastids as drug targets. Trends Microbiol. 1999;7:328–333. doi: 10.1016/s0966-842x(99)01547-4. [DOI] [PubMed] [Google Scholar]
- 4.Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24:279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Williamson DH, Preiser PR, Moore PW, McCredy S, Strath M, Wilson RJM. The plastid DNA of the malaria parasite Plasmodium falciparum is replicated by two mechanisms. Mol Mirobiol. 2002;45:533–542. doi: 10.1046/j.1365-2958.2002.03033.x. [DOI] [PubMed] [Google Scholar]
- 6.Singh D, Chaubey S, Habib S. Replication of the Plasmodium falciparum apicoplst DNA initiates within the inverted repeat region. Mol. Biochem. Parasitol. 2003;126:9–14. doi: 10.1016/s0166-6851(02)00251-7. [DOI] [PubMed] [Google Scholar]
- 7.Wilson RJM, Williamson DH. Extrachromosomal DNA in apicomplexa. Micro. Mol. Boil. Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Ross DS, Cowman AF, McFadden GI. Dissecting apicoplst targeting in the malaria parasite Plasmodium faliciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 9.Seow F, Sato S, Janssen CS, Riehle MO, Mukhopadhyay A, Phillip RS, Wilson RJM(Iain), Barrett MP. The plastidic DNA replication complex of Plasmodium falciparum. Mol Biochem Parasitol. 2005;141:145–153. doi: 10.1016/j.molbiopara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Ram EVSR, Naik R, Ganguli M, Habib S. DNA organization by the apicoplast-targeted bacterial histone-like protein of Plasmodium falciparum. Nucleic Acids Res. 2008;36:5061–5073. doi: 10.1093/nar/gkn483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast targeted Plasmodium falciparum DNA gyrase genes: Unique intrinsic ATPase activity and ATP dependent dimerization of Pf GyrB subunit. Eukaryot. Cell. 2007;6:398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ram EVSR, Kumar A, Biswas S, Kumar A, Chaubey S, Siddiqi MI, Habib S. Nuclear gyrB encodes a functional subunits of the Plasmodium falciparum gyrase that is involved in apicoplast DNA replication. Mol. Biochem. Parasitol. 2007;154:30–39. doi: 10.1016/j.molbiopara.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Fichera ME, Roos DS. A Plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 14.Dahl EI, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Tetracyclines specifically target the apicoplast of the malaria Parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 2007;152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA- binding protein required for eukaryotic DNA metabolism. Ann. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 19.Dyck EV, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S.cerevisiae is homologous to E.coli SSB. EMBO J. 1992;1:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voss TS, Mini T, Jenoe P, Beck HP. Plasmodium falciparum possesses a cell cycle regulated short type replication protein A large subunit encoded by unusual transcript. J. Biol. Chem. 2002;227:17493–17501. doi: 10.1074/jbc.M200100200. [DOI] [PubMed] [Google Scholar]
- 21.Kur J, Olszewski M, Długołęcka A, Filipkowski P. Single-stranded DNA-binding proteins (SSBs) sources and applications in molecular biology. Acta Biochemi. Pol. 2005;52:569–574. [PubMed] [Google Scholar]
- 22.Dar A, Prusty D, Mondal N, Dhar SK. A unique 45-amino-acid region in the toprim domain of Plasmodium flaciparum gyraseB is essential for its activity. Eukt. Cell. 2009;8:1759–1769. doi: 10.1128/EC.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 24.Choudhury NR, Malik PS, Singh DK, Islam MN, Kaliappan K, Mukherjee SK. The oligomeric Rep protein Mungbean yellow mosaic virus (MYMIV) is a likely replicative helicase. Nucleic Acid Res. 2006;34:6362–6377. doi: 10.1093/nar/gkl903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohanty B, Verma AK, Claesson P, Bohidar HB. Physical and anti-microbial characteristics of carbon nano-particles prepared from lamp soot. Nanotechnology. 2007;18:445102–445110. [Google Scholar]
- 26.Sharma A, Nitharwal RG, Singh B, Dar A, Dasgupta S, Dhar SK. Helicobacter pylori single-stranded DNA binding protein-functional characterization and modulation of H. pylori DnaB helicase activity. FEBS J. 2009;276:519–531. doi: 10.1111/j.1742-4658.2008.06799.x. [DOI] [PubMed] [Google Scholar]
- 27.Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single stranded DNA binding protein to single stranded polynucleotide. A study by optical detection of magnetic resonance and site- selected mutagenesis. J. Biol. Chem. 1987;262:10938–10945. [PubMed] [Google Scholar]
- 28.Curth U, Bayer I, Greipel J, Mayer F, Urbanke C, Maass G. Amino acid 55 plays a central role in tetramerization and function of Escherichia coli single stranded DNA binding protein. Eur J. Biochem. 1991;196:87–93. doi: 10.1111/j.1432-1033.1991.tb15789.x. [DOI] [PubMed] [Google Scholar]
- 29.Ragunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E.coli SSB bound to ssDNA. Nature struct. Biol. 2000;8:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 30.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of C-terminus of Escherichia coli single stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roch KGL, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across Plasmodium falciparum Life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissig V, Vetro-Widenhouse TS, Rowe TC. Topoisomerase II inhibitors induce cleavage of nuclear and 35-kb plastid DNAs in the malarial parasite Plasmodium falciparum. DNA Cell Biol. 1997;16:1483–1492. doi: 10.1089/dna.1997.16.1483. [DOI] [PubMed] [Google Scholar]
- 33.Gmuender H, Kuratil K, Padova KD, Gray CP, Keck W, Evers S. Gene Expression Changes Triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta A, Mehra P, Dhar SK. Plasmodium flaciparum origin recognition complex subunit 5: functional characterization and role in DNA replication foci formation. Mol. Microbiol. 2008;69:646–665. doi: 10.1111/j.1365-2958.2008.06316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langowski J, Benight AS, Fujimoto BS, Schurr JM, Schomburg U. Change of conformation and internal dynamics of supercoiled DNA binding of Escherichia coli single stranded binding protein. Biochemistry. 1985;15:4022–4028. doi: 10.1021/bi00336a033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.