Abstract
The GGCC-specific restriction endonuclease BspRI is one of the few Type IIP restriction endonucleases, which were suggested to be a monomer. Amino acid sequence information obtained by Edman sequencing and mass spectrometry analysis was used to clone the gene encoding BspRI. The bspRIR gene is located adjacently to the gene of the cognate modification methyltransferase and encodes a 304 aa protein. Expression of the bspRIR gene in Escherichia coli was dependent on the replacement of the native TTG initiation codon with an ATG codon, explaining previous failures in cloning the gene using functional selection. A plasmid containing a single BspRI recognition site was used to analyze kinetically nicking and second-strand cleavage under steady-state conditions. Cleavage of the supercoiled plasmid went through a relaxed intermediate indicating sequential hydrolysis of the two strands. Results of the kinetic analysis of the first- and second-strand cleavage are consistent with cutting the double-stranded substrate site in two independent binding events. A database search identified eight putative restriction-modification systems in which the predicted endonucleases as well as the methyltransferases share high sequence similarity with the corresponding protein of the BspRI system. BspRI and the related putative restriction endonucleases belong to the PD-(D/E)XK nuclease superfamily.
INTRODUCTION
Type IIP restriction endonucleases (REase) are characterized by recognition sequences displaying dyad axes of symmetry (palindromes), and constitute the most abundant class of characterized restriction enzymes (1). The first Type IIP REases, which were biochemically characterized, were shown to consist of two identical subunits: EcoRI (2), BclI (3), BstI (4) BamHI (5). Recognition of a symmetric recognition sequence by a homodimeric protein and cutting the two strands simultaneously using two active sites was an attractive model also because of the economy of the required protein synthesis, as first pointed out by Kelly and Smith (6). For a long time, the results of crystallographic studies supported the generalization that Type IIP REases are homodimers, e.g. (7–11) or tetramers (12).
To our knowledge, the first Type IIP REase, which was suggested to exist as a monomer was BspRI (13). BspRI of Bacillus sphaericus recognizes the sequence GGCC and cuts after the second G to produce blunt ends (14). The conclusion that the enzyme consists of a single subunit was derived from a comparison of molecular masses determined under native (gel filtration) and denaturing (SDS–polyacryamide gel electrophoresis) conditions. Later, based mostly on similar biochemical evidence as for BspRI, a few other Type IIP REases were also reported to consist of a single polypeptide chain, such as BsuRI (GG/CC) (15), BcnI (CC/SGG) (16) DpnI (GmA/TC) (17), Sau96I G/GNCC (18), BshFI (GG/CC) (19). However, because of a lack of supporting structural data, the notion of monomeric Type IIP REases received little attention.
This changed when the X-ray structure of an MspI–DNA specific recognition complex was reported in 2004. MspI was shown to interact with its symmetric recognition sequence (C/CGG) as a monomer (20,21). Soon other articles describing structures of similar asymmetric complexes of three other Type IIP enzymes followed: HinPI (G/CGC) (22,23), MvaI (CC/WGG) (24) and BcnI (CC/SGG) (25) establishing a new paradigm to think about this class of REases.
The gene of the BspRI methyltransferase (bspRIM), the cognate methyltransferase (MTase) of BspRI endonuclease, was cloned and expressed in Escherichia coli (26), but attempts to clone the BspRI REase gene (bspRIR) were not successful (A. Kiss, unpublished). The acceptance of the idea of monomeric Type IIP REases prompted us to revisit the project and to try to clone the bspRIR gene by an approach, which was not dependent on the expression of R.BspRI in E. coli.
Here we report the results of experiments, in which we used amino acid sequence information to identify the silent BspRI endonuclease gene on the previously cloned fragment, in the vicinity of the MTase gene. Replacing the native TTG start codon with ATG, and cloning the gene in an expression vector providing a strong promoter and Shine–Dalgarno sequence resulted in high-level expression of BspRI REase in E.coli. A database search identified eight putative restriction-modification systems, in which the predicted REases as well as the predicted MTases share high sequence similarity with the corresponding protein of the BspRI system. Secondary-structure prediction was used to determine whether R.BspRI can be assigned to any family of characterized metal-dependent REases. The DNA cleavage mechanism of BspRI was studied using a plasmid containing a single BspRI recognition site. This substrate allowed us to analyze kinetically both nicking and double-strand cleavage at the unique target site.
MATERIALS AND METHODS
Strains and growth conditions
Bacillus sphaericus R, originally isolated as a culture contaminant, is the native host of the BspRI R-M system (14). Bacillus sphaericus has recently been reclassified as Lysinibacillus sphaericus (27). Escherichia coli ER1821 F− glnV44, e14− (McrA−) endA1 thi-1 Δ(mcrC-mrr)114::IS10 obtained from New England Biolabs was used as cloning host. ER1821(DE3) was made by lysogenizing ER1821 with λDE3 using the λDE3 lysogenization kit of Novagen. ER1821(DE3) expresses T7 RNA polymerase upon induction with isopropyl-β-d-thiogalactopyranoside (IPTG). Bacteria were grown in LB medium (28) at 30°C (B. sphaericus) or at 37°C (E. coli). Ampicillin (Ap) and chloramphenicol (Cm) were used at 100 and 25 µg/ml, respectively. For BspRI overproduction ER1821(DE3 + pLysS + pET3H-BspRI) was grown to OD550 ∼ 0.5, then BspRI production was induced by adding 0.4 mM IPTG to the culture and growth was continued for 4–5 h at 30°C.
DNA preparations
Baccillus sphaericus R genomic DNA was prepared from a 50 ml dense culture. Cells were sedimented by centrifugation, washed with 10 ml 20 mM Tris–HCl pH 8.0, then resuspended and lysed in a solution containing 50 mM Tris–HCl pH 7.5, 50 mM EDTA, 0.2% SDS and 200 µg/ml proteinase-K. After incubation at 37°C overnight, the DNA solution was extracted three times with phenol/chloroform and precipitated with ethanol. The precipitated DNA was collected by a glass rod, dried and dissolved in TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA).
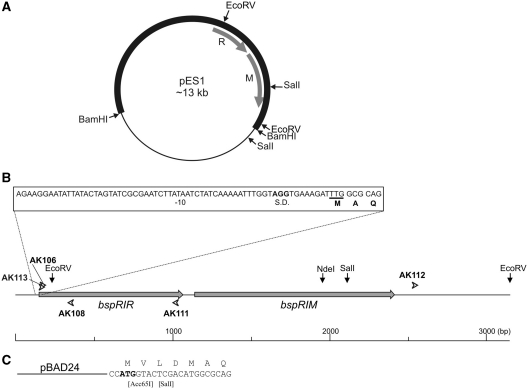
Plasmid pES1 contains the gene of the BspRI MTase on a ∼9kb BamHI fragment of B.sphaericus DNA cloned in pBR322 (26) (Figure 1A). pTZ-Bsp1 carries the segment of the bspRIR gene corresponding to the Q11–K97 peptide. It was constructed by PCR amplification using B. sphaericus genomic DNA as template and AK106/AK108 as primers (Figure 1B), and subsequent cloning of the PCR product in the commercial plasmid vector pTZ57R/T (Fermentas). pTZ-Bsp3 encodes the N-terminal M1–K97 peptide of R.BspRI. It was constructed by PCR-synthesis using pES1 as template and AK113/AK108 as primers (Figure 1B), and cloning the PCR product in pTZ57R/T. Plasmid pTZ-Bsp5, which contains the complete BspRI system, was made by inserting the 2920 bp EcoRV fragment of pES1 carrying part of the bspRIR gene and the intact bspRIM gene (Figure 1A) into the unique EcoRV site of pTZ-Bsp3. The orientation of the BspRI genes in pTZ-Bsp5 is opposite to the lac transcription on the plasmid.
Figure 1.
Schematic map of the B. sphaericus DNA region carrying the genes of the BspRI R-M system. Restriction sites used in plasmid constructions are shown. (A), pES1 Plasmid. Thin line, pBR322 vector; heavy line, B. sphaericus DNA; R, bspRIR gene; M, bspRIM gene. (B) Open arrowheads show the positions of the oligonucleotide primers used in PCR reactions. (C) Beginning of the ORF encoding R.BspRI in pBAD-Bsp3. The ATG start codon of the pBAD24 vector is shown in bold. Square brackets indicate remnants of restriction sites used in construction of pBAD-Bsp3.
To construct a plasmid overexpressing R.BspRI, first the EcoRV fragment of pES1 (Figure 1) was cloned into the SmaI site of pBAD24 (29) to yield pBAD-Bsp2. pBAD-Bsp2 lacks the beginning of the bspRIR gene. To reconstruct the complete BspRI system, the SalI-NdeI fragment of pTZ-Bsp5 was inserted between the Acc65I and NdeI sites of pBAD-Bsp2. The SalI site in pTZ-Bsp5, added by the AK113 PCR primer, immediately precedes the ATG start codon of R.BspRI, whereas the Acc65I site is in the pBAD24 polylinker upstream of the inserted fragment. Before ligation, the Acc65I and SalI ends were filled-in by Klenow polymerase (Figure 1C). The resulting plasmid (pBAD-Bsp3) encodes a BspRI variant, which carries a four amino acid extension at the N-terminus (MVLDMAQRKY…). To facilitate purification of R.BspRI, the SalI-NcoI fragment of pTZ-Bsp5 carrying the BspRI restriction and modification genes was cloned between the XhoI and NcoI sites of the T7 expression vector pET3-His (30) to yield pET3H-BspRI. The R.BspRI variant encoded by pET3H-BspRI consists of 313 amino acids and has the following N-terminal extension (underlined): MHHHHHHLDMAQ… Plasmid pLysS (31) served to stabilize pET3H-BspRI.
Plasmid pC194 (32) containing single sites for BspRI/BsuRI and HinP1I (33) was used as substrate to analyze single- and double-strand cleavage by these enzymes.
Plasmid DNA was prepared from E. coli cells by standard methods (28) or using commercial kits. For preparation of pC194, B. subtilis BD364(pC194) cells were treated with 5 mg/ml lysozyme before starting purification with the GenElute HP Plasmid Midiprep kit (Sigma).
Oligonucleotides
Oligonucleotides were purchased from Integrated DNA Technologies (IDT) or were synthesized in the BRC, Szeged. For approximate positions of the PCR primers see Figure 1B. AK106 and AK108 are pools of degenerate oligonucleotides, where R = A, G; Y = C, T; I = inosine.
AK106 (5′-CAR AAR GTI GCI AAY ATI TTY ATI AAY) corresponds to the Q11KVANIFIN19 peptide of BspRI endonuclease (sense strand).
AK108 (5′-YTT YTG CCA RTT) corresponds to the N94WQK97 peptide of BspRI endonuclease (anti-sense strand).
AK111 (5′-GAT GGG TCT AAG ATA CTA TT) corresponds to the N291SILDPS297 peptide of BspRI endonuclease (anti-sense strand).
AK112 (5′-GAA ATG ATT TAT ATG ATG TG) hybridizes down-stream of the bsprIM gene stop codon (sense strand).
AK113 (5′-GTC GAC ATG GCG CAG AGA AAA TAT GGT GCA) corresponds to the A2QRKYGA8 peptide of BspRI endonuclease, and carries an ATG start codon and a SalI site (underlined) as 5′-extension (sense strand).
Other DNA techniques
Restriction digestion, polymerase chain reaction, agarose gel electrophoresis and cloning in E. coli plasmid vectors were carried out using standard procedures (28). Restriction endonucleases, DNA polymerase large (Klenow) fragment, Taq DNA polymerase and T4 DNA ligase were purchased from Fermentas or New England Biolabs. DNA sequence was determined by an automated sequencer (ABI).
Purification of BspRI endonuclease
The following method was used to purify BspRI from B. sphaericus R as well as from arabinose-induced E. coli ER1821(pBAD-Bsp3) cells. Cells (30 g) were suspended in 50 ml buffer A (10 mM potassium phosphate pH 7.4, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, 10% glycerol and 100 mM sodium chloride) and disrupted by sonication. After removing cell debris by centrifugation (18 000 r.p.m., 20 min), the cell extract was loaded onto a 150 ml phosphocellulose (Whatman P11) column equilibrated with the same buffer. Proteins were eluted by a 0.1–1.0 M NaCl gradient. Peak fractions were pooled, diluted with 10 mM Tris–HCl pH 7.4, and loaded onto a 30 ml heparin–agarose column equilibrated with buffer A. After elution with a 0.1-1.0 M NaCl gradient peak fractions were pooled and loaded directly onto a 30 ml hydroxyapatite column equilibrated with buffer A. BspRI was eluted with a 10–300 mM potassium-phosphate gradient. Peak fractions were pooled and dialysed against a buffer containing 10 mM Tris–HCl pH 7.5, 75 mM NaCl, 5% glycerol and loaded onto a 6 ml Resource-S column (Pharmacia). BspRI was eluted with a gradient 0–1 M NaCl in 20 mM potassium-phosphate (pH 7.5) buffer.
For purification of N-terminally His-tagged BspRI, ER1821(DE3 + pLysS + pET3H-BspRI) cells obtained from 1 l IPTG-induced culture were resuspended in 50 ml buffer E (50 mM potassium phosphate, pH 7.4, 0.15 M NaCl, 5% glycerol, 10 mM 2-mercaptoethanol) containing 10 mM imidazole and disrupted by sonication. Cell debris was removed by centrifugation, and the supernatant was applied onto a 5 ml Ni–agarose column (His-Select Nickel Affinity Gel, Sigma) previously equilibrated with buffer E/10 mM imidazole. Proteins were eluted with a step gradient of imidazole (50, 125, 200 and 250 mM) in buffer E. BspRI endonuclease eluted in the 200 mM imidazole step. The enzyme preparation was diluted 5-fold with a buffer containing 10 mM potassium phosphate pH 6.9, 100 mM KCl, 10 mM 2-mercaptoethanol and 5% glycerol and loaded onto a 19 ml ceramic hydroxyapatite CHT (BioRad) column equilibrated with the same buffer. Proteins were eluted with a gradient containing 10–300 mM potassium phosphate, 100 mM KCl, 10 mM 2-mercaptoethanol and 5% glycerol.
Both methods yielded enzyme preparations that looked at least 99% pure by SDS–polyacrylamide gel electrophoresis (34) after Coomassie staining. Protein concentration was determined by the Bradford method (35) using bovine serum albumin standard.
Edman sequencing
Purified BspRI was dialyzed against 10 mM sodium–phosphate pH 7.5, 0.05% SDS, then concentrated by evaporation in a SpeedVac instrument but avoiding drying of the sample. Protein samples (10–30 µl) were applied to polybrene-coated glass fiber filters and dried under argon. Filters with dried protein sample were acidified with neat trifluoroacetic acid vapor and extracted with n-heptane to remove excess SDS. The filters were subjected to Edman degradation on an Applied Biosystems 470A protein sequencer, and the resulting phenylthiohydantoin (PTH) amino acid derivatives were analyzed by reverse phase HPLC (Applied Biosystems) according to the manufacturer’s specifications. PTH-amino acids were quantitated by comparison to standards using UV absorbance (36).
Mass spectrometry analysis
In-gel digestion
Gel pieces containing the R.BspRI band were cut out from SDS–polyacrylamide gels and soaked in 50% acetonitrile containing 25 mM NH4HCO3 to remove the Commassie stain and salts. Disulfide bridges were reduced with dithiothreitol and the free sulfhydryl groups were alkylated with iodoacetamide. After additional washing steps, the protein was digested in-gel with side-chain protected porcine trypsin for 4.5 h at 37°C. The resulting peptides were extracted with 2% formic acid in 50% acetonitrile.
Peptide derivatization
A portion of the digest was derivatized as described earlier (37). Briefly, to 5 μl of the digest 30 μl SPITC reagent (4-sulfophenyl isothiocyanate, 20 μg/ μl in 25 mM NH4HCO3) was added and the pH of the reaction mixture was adjusted to pH 9.0 by the addition of NH4OH. After 30 min at 55°C, the reaction was terminated with formic acid, then the peptides were purified on a C18 ZipTip according to the manufacturer’s instructions.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) MS
The tryptic digest was analyzed unfractionated prior to and after the derivatization using 2,5-dihydroxybenzoic acid as the matrix. Both mixtures were also fractionated by reversed phase HPLC (C18, 180 μm × 150 mm column, flow rate 1 μl/min, gradient: 5–40% B in 35 min, then up to 80% B in 10 min. Solvent A: 0.1 % TFA/5 % acetonitrile in water; solvent B: 0.085% TFA in 95 % acetonitrile). Fractions were collected directly on the MALDI target. Post source decay (PSD) data were acquired in 10–12 segments, lowering the reflectron voltage by 25% in each step, then stitching the data together.
LC-MS/MS
The underivatized digest was also subjected to on-line LC-MS/MS analysis on an ABI QSTAR ESI-QqTOF mass spectrometer in information dependent acquisition (IDA) mode: 1 s MS acquisitions were followed by 5 s collision-induced dissociation (CID) analyses on computer-selected multiply charged ions. Nano-HPLC: C18, 75 μm × 150 mm column, flow rate 300 nl/min, gradient: 5–50% B in 30 min, solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile.
Database searches were performed against the NCBI non-redundant protein database using the Protein Prospector software package (http://prospector.ucsf.edu). De novo sequencing was performed manually.
DNA cleavage analysis
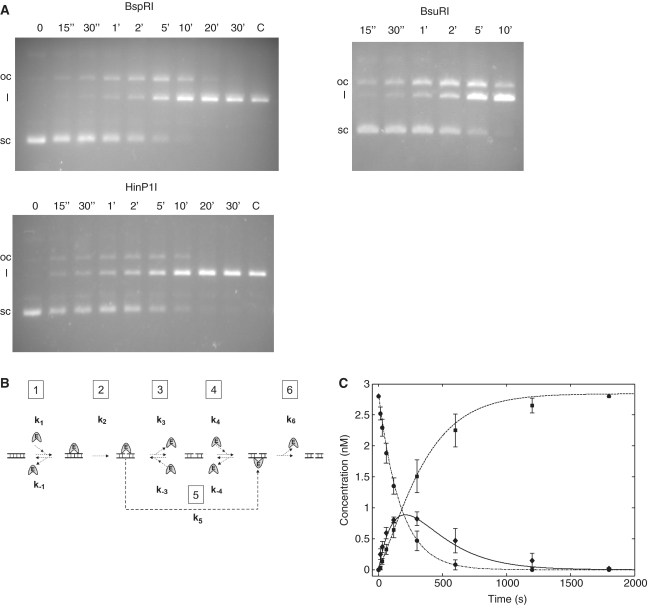
For kinetic analysis, supercoiled pC194 plasmid DNA (2.84 nM) was incubated in 33 mM Tris–acetate pH 7.9, 10 mM Mg–acetate, 66 mM K–acetate, 0.1 mg/ml BSA (Fermentas Tango buffer) with His-tagged BspRI endonuclease (0.0054 pM) at 37°C. Aliquots were withdrawn at timed intervals and added to excess EDTA to stop digestion. Digestion of pC194 with HinP1I (New England Biolabs) and BsuRI (Fermentas) was tested using buffers recommended by the manufacturers. HinP1I and BsuRI were used at 0.016 and 0.05 U/µl concentrations, respectively. BsuRI digestions were performed at room temperature. Plasmid isoforms were separated by electrophoresis in 1% agarose gel at low voltage (1.25 V/cm) and stained with ethidium bromide after the run. Amounts of DNA in the individual bands were determined by densitometry of the gel photograph using the GeneTools software (version 4.01, Synoptics). The kinetics of the cleavage reactions were analyzed in the framework of the reaction scheme shown in Figure 4B using the MATLAB Program Package (MathWorks Inc.,). The differential equation system was solved numerically, using the Newton method (iteration step 0.001 s).
Figure 4.
Digestion of the single-site plasmid pC194 with BspRI, HinP1I and BsuRI endonucleases. (A) Agarose gel electrophoresis of the digestion products. Reaction times are shown above the lanes. Digestions were performed as described in ‘Materials and Methods’ section. Sc, l and oc denote supercoiled, linear and open circular forms, respectively. (B) Proposed kinetic scheme for DNA cleavage by BspRI endonuclease. Step 1, equilibrium binding to uncut DNA; step 2, nicking of the first strand; step 3, equilibrium binding to nicked DNA, BspRI facing the nicked strand; step 4, equilibrium binding to nicked DNA, BspRI facing the intact strand; step 5, transposition of BspRI from the nicked strand to the uncut strand; step 6, cleaving the second strand. (C) Time course of digestion of supercoiled plasmid DNA with BspRI. pC194 plasmid DNA was digested for the indicated times and the plasmid forms were quantitated as described in ‘Materials and Methods’ section. Experimental data are denoted by symbols (circle, supercoiled; diamond, nicked; square, linear). Lines represent results of the simulation (dashed–dotted, supercoiled; solid, nicked; dashed, linear). Error bars indicate the standard error of the data calculated from four parallel experiments.
Bioinformatic tools
DNA and protein sequence similarity searches were performed using the BLAST (38,39) or the ClustalW2 (40) programs. Secondary structure predictions were performed with the Jpred 3 program (41). In all cases default settings were used.
RESULTS
Partial amino acid sequence of the BspRI REase
The gene of the BspRI MTase was originally cloned on a ∼9 kb BamHI fragment. E. coli cells carrying pES1 (Figure 1A) expressed BspRI MTase, but did not show phage restriction and no BspRI endonuclease activity was detectable in the cell extracts (26). Cloning of longer overlapping fragments or screening a plasmid gene library for clones restricting non-modified phage failed to yield a clone expressing BspRI endonuclease.
To use an approach that is not dependent on the expression of R.BspRI in E. coli, the enzyme was purified from B. sphaericus, and a short N-terminal amino acid sequence (AQRKYGALEQKVANIFINEQVFTFKG) was determined by Edman sequencing.
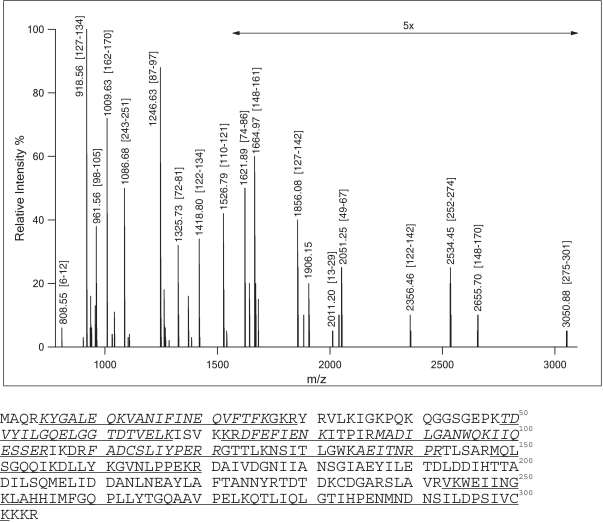
To obtain additional sequence information, purified BspRI was digested with trypsin and the peptides were subjected to mass spectrometry (MS) analysis as described in ‘Materials and Methods’ section. The tryptic digest was extensively analyzed by MALDI and electrospray mass spectrometry following off- or on-line HPLC fractionation. No proteins could be identified from the PSD and CID spectra by database search. To aid de novo sequencing, a portion of the digest was sulfonated on the N-termini of the peptides. Such derivatization usually leads to almost exclusive y-ion formation in PSD analysis. Peptide sequences determined manually from the MS/MS (PSD and/or CID) data are summarized in Table 1.
Table 1.
Peptide sequences determined from MS/MS data
| MH+ | Sequence |
|---|---|
| 961.4 | [IL]a[IL]QESSER |
| 808.4 | YGA[IL]EQK |
| 956.6 | AE[IL]TNRPR |
| 936.52 | KYGA[IL]EQK |
| 1041.48 | {DF}bEF[IL]ENK |
| 1370.63 | FADC*S[IL][IL]YPER |
| 1262.6 | M(O)cAD[IL][IL]GANWQK |
| 1246.58 | MAD[IL][IL]GANWQK |
| 1278.6 | MAD[IL][IL]GANW(O2)cQK |
| 2051.08 | {DT/ESd}VY[IL][IL]G[QK]eE[IL]GGTDTVE[IL]K |
| 1526.75 | RFADC*S[IL][IL]YPER |
| 1385.76 | [IL]F[IL]NE[QK]VFTFK |
| 1669.9 | VAN[IL]F[IL]NE[QK] VFTFK |
C* stands for carbamidomethyl Cys.
aLow-energy CID analysis cannot differentiate between isomeric Ile and Leu residues.
bThe order of the amino acids could not be determined.
cOxidation of methionine and tryptophan are common side-reactions.
dAmino acid combinations corresponding to the mass difference observed.
eGln and Lys are isobaric amino acids with identical nominal masses, but different elemental composition. Since the QSTAR mass spectrometer affords a 50 ppm mass accuracy, in most cases these residues could be identified, however there are exceptions.
The bspRIR gene
The peptide sequences obtained by Edman degradation and MS analysis were used to design primers for PCR-amplification of a section of the bspRIR gene. Two primers (AK106 and AK108) were synthesized. Primer AK106 corresponded to amino acids Q10KVANIFIN18, whereas AK108 corresponded to the tetrapeptide NWQK (Table 1, Figure 1B). To reduce complexity of the AK106 pool, the neutral base inosine, which can form stable base pairs with all four bases (42), was used at positions with greater ambiguity. PCR amplification, using B. sphaericus DNA as template, produced an ∼250 bp fragment, which was cloned in pTZ57R/T to yield pTZ-Bsp1. Sequencing of the insert revealed that the cloned fragment encodes several peptides previously detected by MS indicating that pTZ-Bsp1 carries a portion of the bspRIR gene. Unexpectedly, PCR synthesis using the same primers but pES1 plasmid DNA as template produced a similar fragment, suggesting that at least a part of the bspRIR gene was present on pES1.
To determine the approximate distance and relative orientation of the bspRIR and bspRIM genes, four PCR reactions were performed using B. sphaericus DNA as template and the following combinations of primers: AK108 + AK112, AK106 + AK112, AK106 + AK111 and AK108 + AK111 (Figure 1B). AK111 and AK112 were designed on basis of the previously determined sequence flanking the bspRIM gene (43). Only the AK106 + AK111 combination yielded a PCR product (∼850 bp). The same result was obtained when pES1 plasmid DNA was used as template. These results showed that the genes of the BspRI R-M system are closely located in tandem arrangement with the REase gene being upstream (Figure 1).
Another conclusion following from this observation was that the entire bspRIR gene must be on the BamHI fragment cloned in pES1. This was surprising because of the lack of endonuclease activity in the clone. It seemed possible that the methods used (endonuclease assay in crude extracts and phage restriction) were not sensitive enough to detect low BspRI activity. This question was addressed by testing how inactivation of the MTase would affect viability of the clone. To obtain an m− r+ plasmid, the small SalI fragment carrying the 3′-terminal half of the bspRIM gene in pES1 (Figure 1A) was deleted. Escherichia coli cells carrying the resulting plasmid were perfectly viable. Taking into account the large number of BspRI sites in the E. coli genome and that restriction cuts producing blunt ends are likely to be highly damaging due to the absence of DNA–ligase-mediated repair (44), the viability of the m− r+ clone convincingly showed that BspRI expression from pES1 in E. coli was undetectable.
The nucleotide sequence of a 1028 bp segment preceding and that of a 535 bp segment following the published sequence (43) was determined (accession number: X15758). Comparison of the deduced amino acid sequence with the peptide sequences determined by Edman sequencing and MS analysis (Figures 1 and 2) unequivocally identified the ORF encoding R.BspRI. A great majority of the peptides detected in the unfractionated tryptic digest (29/34) fit to the amino acid sequence derived from the DNA sequence (Figure 2). This ORF starts with TTG at 168 and ends with TAG at 1082 defining a 304 amino acid protein. TTG is not an unusual start codon in B. sphaericus. Approximately 8% of the genes of another B. sphaericus strain (C3–41), whose sequence has recently been published (45), have TTG initiation codon (Xiaomin Hu, personal commumication). The stop codon of the REase and the ATG start codon of the MTase are separated by 77 bp. Re-sequencing part of the MTase gene identified an error in the published sequence: the correct amino acid at position 394 is Ala rather than Thr.
Figure 2.
BspRI endonuclease sequence coverage by mass spectrometry. Upper panel, MALDI–TOF mass spectrum of BspRI unfractionated tryptic digest. Numbers in brackets indicate sequence positions of the peptides. Lower panel, amino acid sequence of BspRI restriction endonuclease. Sequences identified from MS/MS data (italic) (Table 1) or from mass only are underlined.
The G + C content of the sequenced region (36.6%) corresponds well to the G + C content of the genome of B. sphaericus C3–41 (37.29%) (45). The region encompassing the BspRI R-M system (3178 bp) is devoid of BspRI recognition sites.
The nucleotide sequence offered an explanation for the lack of expression of R.BspRI in E. coli. First, TTG is an inefficient translational initiatior in E. coli (46). Second, there is only a weak Shine–Dalgarno sequence preceding the start codon. Third, there is no typical E. coli promoter upstream of the initiation codon: a TATAAT sequence is present, but the −35 sequence is missing (Figure 1B).
To test whether the lack of expression of R.BspRI in E. coli was due to the lack of proper transcriptional and translational signals, the plasmid pBAD-Bsp3 was constructed as described in ‘Materials and Methods’ section. In pBAD-Bsp3 the bspRIR gene has an ATG start codon and the vector provides a strong Shine–Dalgarno sequence as well as an inducible E. coli promoter (araBAD). Arabinose induction led to high expression of R.BspRI indicating that the lack of expression of the native gene in E. coli was caused by the absence of proper transcriptional and translational signals.
Sequence analysis of BspRI endonuclease
The DNA sequence defines a protein with a calculated Mr of 34 278 Da and a theoretical isoelectric point of 8.76. The results of Edman-sequencing showed that the mature protein does not contain the N-terminal formyl-Met.
A comparison with protein sequences in the GenBank database identified nine proteins (all annotated as hypothetical proteins) displaying relatively high sequence similarity to R.BspRI (Table 2). Seven of the predicted proteins are very similar in size to R.BspRI, the number of amino acids falling between 293 and 302. The genes of eight proteins are located adjacently to genes, whose predicted translation products show the signature motifs of C5-MTases, and share strong sequence similarity with M.BspRI (Table 2). The significant amino acid sequence similarity, the similar size and the co-localization with a C5-MTase gene strongly suggests that all eight proteins are restriction endonucleases (Table 2). As a result of evolutionary self-defence, R-M genes are typically characterized by the absence or paucity of the recognition site specific for the system (1). A search of the DNA regions encompassing the ORFs of the predicted REases and the counterpart C5-MTases revealed a striking scarcity of BspRI (GGCC) sites (Table 2), suggesting that these R-M systems might be functional and might have GGCC specificity, or if they are inactive, they probably have lost activity only recently. The nineth protein sharing amino acid sequence similarity with R.BspRI is a predicted protein of a strain of Streptococcus thermophilus. The protein is much shorter than R.BspRI, and the similarity extends for only the N-terminal half of the enzyme. In this case, the BLAST search did not find a gene encoding a MTase-like protein, instead it identified a gene whose translational product shows similarity with proteins playing a role in regulation of REase in some R-M systems. No significant similarity was found between R.BspRI and its isoschizomer R.BsuRI, which contrasts sharply with the very high sequence identity (67%) found between M.BspRI and its closest homolog in the database, M.BsuRI. Interestingly, the genes of the putative BspRI-like R-M system in the Bacteroides sp. 3_1_33FAA genome (Table 2) are located next to the genes of another putative R-M system annotated as ‘DNA cytosine MTase’ (ZP_06087300) and ‘type II restriction enzyme HaeIII’ (ZP_06087301), suggesting that this bacterium might contain two R-M systems of identical specificity. Whereas the amino acid sequences of the two predicted C5-MTases are highly similar, the REases of the two systems do not show significant similarity (not shown).
Table 2.
Putative R-M systemsa showing the highest sequence similarity to R.BspRI
| Organism | Putative REase |
Putative MTase |
Frequency of GGCC sitesb | ||||
|---|---|---|---|---|---|---|---|
| Protein | Length (aa) | E-value to R.BspRI | Protein | Length (aa) | E-value to M.BspRI | ||
| Acinetobacter haemolyticus ATCC 19194 | Conserved hypothetical protein ZP_06729282 | 302 | 5e−51 | ZP_06729281 (M.AhaBGORF3490P) | 336 | 3e−53 | 2/1900 |
| Gardnerella vaginalis ATCC 14019 | Conserved hypothetical protein ZP_03936874 | 298 | 3e−44 | ZP_03936873 (M.GvaORF417P) | 333 | 1e−52 | 1/1950 |
| Roseburia intestinalis L1-82 | Hypothetical protein RintL_00030 ZP_04741872 | 293 | 6e−41 | ZP_04741871 (M.RinLORF5004P) | 432 | 3e−149 | 0/2191 |
| Bacteroides sp. 3_1_33FAA | Conserved hypothetical protein ZP_06087297 | 297 | 7e−40 | ZP_06087299 (M.BspFAAORF965P) | 466 | 2e−49 | 0/2585 |
| Bacteroides ovatus SD CMC 3f | Conserved hypothetical protein ZP_06618190c | 298 | 3e−36 | ZP_06618187(M1.BovSDORF2192P) | 459 | 8e−51 | 3/4846 |
| ZP_06618188 (M2.BovSDORF2192P) | 337 | 8e−51 | |||||
| Lysinibacillus sphaericus C3-41 | Hypothetical protein Bsph_0498 YP_001696253 | 249 | 7e−32 | YP_001696252 (M.LspCORF497P) | 426 | 2e−148 | 1/2132 |
| Providencia alcalifaciens DSM 30120 | Hypothetical protein PROVALCAL_01484 ZP_03318550 | 295 | 7e−30 | ZP_03318551 (M.PalDORF1485P) | 330 | 2e−52 | 2/1927 |
| Uncultured marine crenarchaeote HF4000_APKG3B16 | Hypothetical protein ALOHA_HF4000APKG3B16ctg1g5 ABZ08412 | 300 | 2e−26 | ABZ08413 (M.UcrHFORF6P) | 377 | 2e−48 | 1/2026 |
| Streptococcus thermophilus CNRZ1066 | Hypothetical protein str0690 YP_141100d | 188 | 9e−15 | None | |||
MTase names (in parentheses) are from REBASE (1).
aIdentified by BLAST (blastp) search of the GenBank non-redundant protein database.
bNumber of BspRI recognition sites in the DNA regions (in base pairs) encompassing the ORFs of the predicted REases and the counterpart C5-MTases.
cOne of the flanking genes encodes a putative Vsr DNA mismatch endonuclease.
dOne of the flanking genes encodes a putative regulatory protein of an R-M system.
Based on crystallographic data and on structure predictions, most Type II REases are classified into five superfamilies (47). R.BspRI as well as the other R.BspRI-like proteins identified in the BLAST search (Table 2) appear to belong to the largest superfamily characterized by the PD-(D/E)XK motif forming the active site. This assignment is supported by results of secondary-structure predictions, which identified the αβββαβ core typical for PD-(D/E)XK nucleases (Supplementary Figure S1), and, in all but two proteins, the essential charged residues at characteristic positions (D in the βII chain and EXK in the βIII chain) (48) (Figure 3). In two of the R.BspRI-like proteins (those of Lysinibacillus sphaericus C3–41 and Providencia alcalifaciens), parts of the predicted active site motif are missing (Figure 3), suggesting that these proteins are inactive.
Figure 3.
Alignment of the predicted active site motifs of BspRI and the putative REases (Table 2) sharing the highest amino acid sequence similarity with BspRI. Numbers at the beginning of the sequences designate the position of the first amino acid shown. The numbers of residues between the shown blocks of sequences are given in parentheses. Predicted α-helices and β-strands (41), are indicated at the top. The convention of numbering, with Roman numerals, the secondary structural elements of the PD-(D/E)XK conserved fold is adopted from (48). Conserved residues of the putative PD-(D/E)XK nuclease fold (47,48) are highlighted with colored background: red, acidic; grey, lysine; yellow, uncharged.
BspRI cleaves the two DNA strands sequentially
Previous results showed that R.BspRI is a monomer in free state (13). This raised questions about the mode of substrate recognition and cleavage. If BspRI is a monomer with one active site, how does it cleave the two strands of the symmetrical recognition sequence? The most likely mechanism appeared to be cutting the DNA in two steps: first making a nick, then, in a second binding event, cleaving the other strand. To test this model, a 2910 bp plasmid (pC194) having a single BspRI site was digested under steady state conditions with His-tagged BspRI, and conversion of the supercoiled plasmid DNA into the nicked and linear forms was followed as described in ‘Materials and Methods’ section. During the digestion, the nicked DNA accumulated before being converted to the linear form (Figure 4A), showing that the enzyme first cleaved just one strand. The amount of DNA in the different forms was quantitated by densitometry and the kinetics of the cleavage reactions were analyzed in the framework of the proposed reaction scheme shown in Figure 4B. The fit of the derived curves to the experimental data (Figure 4C) shows that the proposed reaction scheme is consistent with the results. Although the available experimental data did not allow determination of the full set of elementary rate constants shown in the reaction scheme, several of their pairwise respective ratios proved to be stable in the framework of the present model. For example we could establish that the rate constants of the first and second cleavage do not differ within the error of the data (∼25%), suggesting independency of these processes. Accumulation of a significant amount of nicked DNA during the reaction assumes dissociation of the majority of the DNA•E complex before the final cutting process takes place. This implies that BspRI cuts one strand at a time, and the second cut occurs after binding of the enzyme to the other strand. Introducing an alternative pathway characterized by flipping of the enzyme from the cut to the uncut strand (Figure 4B, step 5) improved the fit only slightly, suggesting that double cutting may occur without formal dissociation of the monomer, but with a significantly lower probability (<20%). More complicated models, including enzyme dimerization on the DNA, did not further improve the fit, whereas the existence of sequential cutting steps in the reaction scheme remained obligatory.
Cleavage by untagged BspRI was analyzed less thoroughly, but it displayed similar kinetics as the His-tagged enzyme (not shown). Accumulation of nicked intermadiate was not dependent on specific reaction conditions, it was observed in all buffers tested (Supplementary Figure S2).
The isoschizomer BsuRI (GG/CC), another REase reported to be a monomer (15) produced a similar accumulation of the open circular form before reaching complete cleavage (Figure 4A). Thus R.BsuRI, which does not share sequence similarity with R.BspRI and is a considerably larger protein (576 versus 304 aa) (49), also appears to cut the double-stranded substrate in two consecutive reactions.
Of the Type IIP REases that have been shown by crystallographic evidence to be monomeric, only HinP1I has been analyzed with regard to the cleavage mechanism. It was shown that cleavage of supercoiled pUC19 DNA went through a nicked intermediate, but because of the 17 HinP1I sites in this plasmid, the time-course of the second-strand cleavage could not be reliably assessed (23). Since pC194 contains a single site also for HinP1I, we could test the cleavage kinetics of HinP1I with this more informative substrate. Conversion of supercoiled pC194 into the linear form by HinP1I was accompanied by the appearance of the open circular intermediate in a very similar fashion as for BspRI (Figure 4A), which is consistent with both enzymes acting as a monomer.
DISCUSSION
Amino acid sequence information obtained by sequencing the purified enzyme was used to identify the gene of the BspRI endonuclease, which was not expressed in the cloning host E. coli. The TTG start codon, the poor Shine–Dalgarno sequence and the lack of an E. coli promoter may explain why the bspRIR gene, in its native form, was silent in E. coli. Of these factors, the effect of the suboptimal initiation codon appears to be the most important. This can be concluded from the phenotype of the plasmid pTZ-Bsp5, which was an intermediate in the process of constructing pBAD-Bsp3. In pTZ-Bsp5 the original TTG initiation codon is already replaced with ATG, but even the rudimentary AGG Shine–Dalgarno sequence present in the native bspRIR gene is missing, and the orientation of the bspRIRM genes is opposite to the lac transcription starting from the pTZ57R vector. Nevertheless, E. coli cells harboring pTZ-Bsp5 produce BspRI endonuclease, suggesting that the increased expression was predominantly due to the replacement of the TTG initiation codon. Although TTG is much less efficient as start codon in E. coli than ATG (46), the observed dramatic difference in BspRI expression is surprising, and may indicate interactions between the start codon and downstream sequences of the mRNA that might be formed differently in B. sphaericus and E. coli (50).
The different start codons (TTG for R.BspRI and ATG for M.BspRI) suggest more efficient translation initiation for the MTase than for the REase, which can be important for ensuring safe protection of the host DNA against the cognate REase, especially when R-M genes enter a new cell. A regulatory mechanism based on the different efficiencies of translation signals was suggested to operate also in some other R-M systems (18,49). Such a mechanism could be an alternative to transcriptional control coordinating REase and MTase expression (51).
Putative R-M systems are typically identified in new genome seqences on basis of the signature sequence motifs characterizing DNA MTases. In most cases, due to their great variety, REase genes cannot be recognized directly (1). In this work, sequence similarity to a REase led to the discovery of putative R-M systems in the database. The sequence conservation characterizing both enzymes suggests common evolutionary origin for these R-M systems, which is puzzling for such diverse group of organisms (Table 2). Sequence analysis suggests that R.BspRI and its nine putative homologs are typical PD-(D/E)XK nucleases. Sequence conservation between the enzymes is highest in the region encompassing the predicted active site (Figure 3 and Supplementary Figure S1).
One of the goals of this work was to determine whether BspRI, which was a monomer in free state (13), acts on its target sequence as a monomer or dimerizes before the cleavage reaction takes place. For example, SalI and Eco29kI were also shown to exist predominantly as monomers (52,53), but assemble on the target sequence to act as dimers (54,55). In spite of using a wide range of conditions, no specific complex of BspRI with cognate DNA could be detected by gel electrophoretic mobility shift assay (our unpublished results). As an alternative approach, we chose to characterize the stoichiometry of the interaction using a plasmid containing a single BspRI site. Since its introduction (56) the use of plasmid DNA substrates containing a single target site has been a very productive method in studying the cleavage mechanism of REases because it allows separate kinetic analysis of single and double strand cleavage (57). This technique has been applied under steady-state as well as single-turnover conditions, e.g. (58–62). However, with few exceptions (61), the requirement for a single-site plasmid tended to restrict such studies to enzymes with longer recognition sequences. Finding a plasmid containing just a single GGCC site allowed us to analyze kinetically the first- and second-strand cleavage reactions by BspRI. The results of this analysis were consistent with the model of BspRI cleaving the two strands in two consecutive binding reactions. Qualitatively similar cleavage kinetics were observed in sub-optimal buffers (Supplementary Figure S2) indicating that sequential cleavage of the two strands is an inherent property of the enzyme. The simplest interpretation of these data is that BspRI acts as a monomer. This assignment is supported by the similar time-course of cleavage detected for HinP1I, an enzyme shown by structural evidence to act as a monomer (Figure 4A).
Interestingly, all Type IIP REases that have been shown, by biochemical or structural evidence, to exist as monomers, recognize short sequences (4–5 bp; ‘Introduction’ section). Further work will determine whether this is just a coincidence or reflects an inherent property of the recognition mechanism.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hungarian Scientific Research Fund (T038343 to A.K.); National Science Foundation (DMB 8217553.000 to R.J.R.); Exxon Corporation (grant to R.J.R.); National Institutes of Health (NCRR RR015804 and P41RR001614 to the UCSF Mass Spectrometry Facility, Director A.L. Burlingame, to K.F.M., in part). Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Dubnau and Lise Raleigh for the strains BD364(pC194) and ER1821, respectively, and Ibolya Anton for the technical assistance.
REFERENCES
- 1.Roberts RJ, Vincze T, Pósfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modrich P, Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J. Biol. Chem. 1976;251:5866–5874. [PubMed] [Google Scholar]
- 3.Bingham AH, Atkinson T, Sciaky D, Roberts RJ. A specific endonuclease from Bacillus caldolyticus. Nucleic Acids Res. 1978;5:3457–3467. doi: 10.1093/nar/5.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke CM, Hartley BS. Purification, properties and specificity of the restriction endonuclease from Bacillus stearothermophilus. Biochem. J. 1979;177:49–62. doi: 10.1042/bj1770049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith LA, Chirikjian JG. Purification and characterization of the sequence-specific endonuclease BamHI. J. Biol. Chem. 1979;254:1003–1006. [PubMed] [Google Scholar]
- 6.Kelly TJ, Jr, Smith HO. A restriction enzyme from Hemophilus influenzae. II. Base sequence of the recognition site. J. Mol. Biol. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 7.Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of EcoRI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 8.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 10.Athanasiadis A, Vlassi M, Kotsifaki D, Tucker PA, Wilson KS, Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nat. Struct. Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Balendiran K, Schildkraut I, Anderson JE. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 13.Koncz C, Kiss A, Venetianer P. Biochemical characterization of the restriction-modification system of Bacillus sphaericus. Eur. J. Biochem. 1978;89:523–529. doi: 10.1111/j.1432-1033.1978.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 14.Kiss A, Sain B, Csordás-Tóth É, Venetianer P. A new sequence-specific endonuclease (Bsp) from Bacillus sphaericus. Gene. 1977;1:323–329. doi: 10.1016/0378-1119(77)90037-3. [DOI] [PubMed] [Google Scholar]
- 15.Bron S, Hörz W. Purification and properties of the Bsu endonuclease. Methods Enzymol. 1980;65:112–132. doi: 10.1016/s0076-6879(80)65017-4. [DOI] [PubMed] [Google Scholar]
- 16.Petrusyte M, Janulaitis A. Isolation and some properties of the restriction endonuclease BcnI from Bacillus centrosporus. Eur. J. Biochem. 1982;121:377–381. doi: 10.1111/j.1432-1033.1982.tb05797.x. [DOI] [PubMed] [Google Scholar]
- 17.de la Campa AG, Springhorn SS, Kale P, Lacks SA. Proteins encoded by the DpnI restriction gene cassette. Hyperproduction and characterization of the DpnI endonuclease. J. Biol. Chem. 1988;263:14696–14702. [PubMed] [Google Scholar]
- 18.Szilák L, Venetianer P, Kiss A. Cloning and nucleotide sequence of the genes coding for the Sau96I restriction and modification enzymes. Nucleic Acids Res. 1990;18:4659–4664. doi: 10.1093/nar/18.16.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozidis C, Vlatakis G, Bouriotis V. Sequence-specific DNA affinity chromatography: application of a group-specific adsorbent for the isolation of restriction endonucleases. J. Chromatogr. 1993;630:151–157. doi: 10.1016/0021-9673(93)80451-d. [DOI] [PubMed] [Google Scholar]
- 20.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Xu QS, Roberts RJ, Guo HC. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005;14:2590–2600. doi: 10.1110/ps.051565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Horton JR, Maunus R, Wilson GG, Roberts RJ, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaus-Drobek M, Czapinska H, Sokolowska M, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007;35:2035–2046. doi: 10.1093/nar/gkm064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007;369:722–734. doi: 10.1016/j.jmb.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Szomolányi É, Kiss A, Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980;10:219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed I, Yokota A, Yamazoe A, Fujiwara T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb.nov. Int. J. Syst. Evol. Microbiol. 2007;57:1117–1125. doi: 10.1099/ijs.0.63867-0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BP, Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 31.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 32.Iordanescu S, Surdeanu M, Della Latta P, Novick R. Incompatibility and molecular relationships between small Staphylococcal plasmids carrying the same resistance marker. Plasmid. 1978;1:468–479. doi: 10.1016/0147-619x(78)90005-7. [DOI] [PubMed] [Google Scholar]
- 33.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Hunkapiller MW, Hewick RM, Dreyer WJ, Hood LE. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Kalb SR, Cotter RJ. Improved procedures for N-terminal sulfonation of peptides for matrix-assisted laser desorption/ionization post-source decay peptide sequencing. Rapid Commun. Mass Spectrom. 2004;18:96–102. doi: 10.1002/rcm.1289. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Nucleic Acids Res. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin FH, Castro MM, Aboul-ela F, Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985;13:8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pósfai G, Kiss A, Erdei S, Pósfai J, Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J. Mol. Biol. 1983;170:597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- 44.Tímár E, Venetianer P, Kiss A. In vivo DNA protection by relaxed-specificity SinI DNA methyltransferase variants. J. Bacteriol. 2008;190:8003–8008. doi: 10.1128/JB.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu X, Fan W, Han B, Liu H, Zheng D, Li Q, Dong W, Yan J, Gao M, Berry C, et al. Complete genome sequence of the mosquitocidal bacterium Bacillus sphaericus C3-41 and comparison with those of closely related Bacillus species. J. Bacteriol. 2008;190:2892–2902. doi: 10.1128/JB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 47.Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552–3569. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knizewski L, Kinch L, Grishin N, Rychlewski L, Ginalski K. Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches. BMC Struct. Biol. 2007;7:40. doi: 10.1186/1472-6807-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiss A, Pósfai G, Keller CC, Venetianer P, Roberts RJ. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985;13:6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenstrom CM, Holmgren E, Isaksson LA. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 51.Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxwell A, Halford SE. The SalGI restriction endonuclease. Purification and properties. Biochem. J. 1982;203:77–84. doi: 10.1042/bj2030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pertzev AV, Kravetz AN, Mayorov SG, Zakharova MV, Solonin AS. Isolation of a strain overproducing endonuclease Eco29kI: purification and characterization of the homogeneous enzyme. Biokhimiia. 1997;62:732–741. [PubMed] [Google Scholar]
- 54.Maxwell A, Halford SE. The SalGI restriction endonuclease. Mechanism of DNA cleavage. Biochem. J. 1982;203:85–92. doi: 10.1042/bj2030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibryashkina EM, Sasnauskas G, Solonin AS, Zakharova MV, Siksnys V. Oligomeric structure diversity within the GIY-YIG nuclease family. J. Mol. Biol. 2009;387:10–16. doi: 10.1016/j.jmb.2009.01.048. [DOI] [PubMed] [Google Scholar]
- 56.Greene PJ, Poonian MS, Nussbaum AL, Tobias L, Garfin DE, Boyer HW, Goodman HM. Restriction and modification of a self-complementary octanucleotide containing the EcoRI substrate. J. Mol. Biol. 1975;99:237–261. doi: 10.1016/s0022-2836(75)80143-4. [DOI] [PubMed] [Google Scholar]
- 57.Bennett SP, Halford SE. Recognition of DNA by Type II restriction enzymes. Curr. Top. Cell. Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- 58.Rubin RA, Modrich P. Substrate dependence of the mechanism of EcoRI endonuclease. Nucleic Acids Res. 1978;5:2991–2997. doi: 10.1093/nar/5.8.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halford SE, Johnson NP, Grinsted J. The reactions of the EcoRI and other restriction endonucleases. Biochem. J. 1979;179:353–365. doi: 10.1042/bj1790353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halford SE, Johnson NP. Single turnovers of the EcoRI restriction endonuclease. Biochem. J. 1983;211:405–415. doi: 10.1042/bj2110405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zebala JA, Choi J, Barany F. Characterization of steady state, single-turnover, and binding kinetics of the TaqI restriction endonuclease. J. Biol. Chem. 1992;267:8097–8105. [PubMed] [Google Scholar]
- 62.Sasnauskas G, Jeltsch A, Pingoud A, Siksnys V. Plasmid DNA cleavage by MunI restriction enzyme: single-turnover and steady-state kinetic analysis. Biochemistry. 1999;38:4028–4036. doi: 10.1021/bi982456n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.