Abstract
The physiological and behavioral circadian rhythms of most creatures are controlled by a harmony of functional relationships between clock genes. In mammals, several core clock genes show rhythmic profiles of their mRNA and protein expression. Among them, Rev-erb α functions as a transcriptional repressor, affecting expression patterns of other clock genes. For the continuous and robust oscillation of the molecular clock system, the levels of Rev-erb α protein are expected to be tightly regulated with the correct timing. Here, we demonstrate that Rev-erb α has an internal ribosomal entry site (IRES) in its 5′ untranslated region. Furthermore, we demonstrate that heterogeneous nuclear ribonucleoprotein Q and polypyrimidine tract-binding protein (PTB) modulate the IRES-mediated translation of Rev-erb α. We suggest that the rhythmic binding affinity of hnRNP Q to the Rev-erb α IRES and the change in PTB cytosolic levels lead to maintenance of the oscillation profile of the Rev-erb α protein.
INTRODUCTION
Almost all living organisms have daily physiological and behavioral rhythms due to the rotation of the earth over a nearly 24-h period. Even though the existence of the day and night cycle is the main factor governing the circadian rhythm, living creatures from cyanobacteria to humans have a circadian oscillation control system (1,2). This endogenous system is composed of an autoregulatory transcription–translation feedback loop (TTFL) composed of several clock genes (3–5). The Bmal1 and Clock heterodimer is an active transcription complex that binds to promoter E box elements (CACGTG) upstream of three Period (Per) genes, two Cryptochrome (Cry) genes, Rev-erb α, Ror α and many other clock-controlled genes (6). After Per and Cry proteins are translated, they form a heterodimer in the cytosol and translocate to the nucleus. This complex binds to a BMAL1/CLOCK dimer and directly inhibits its transcriptional activity (7,8). The suprachiasmatic nucleus (SCN) of the anterior hypothalamus is in charge of master clock that synchronizes the biological rhythms of peripheral tissues so that they function in a defined manner (9).
Among the core clock genes, Rev-erb α, also known as Nr1d1, was identified as a regulator of lipid metabolism. It transcriptionally represses the apoA1 and apoCβ genes, which reduce free cholesterol accumulation and constitute the high-density lipoprotein and very low-density lipoprotein, respectively (10). In conjunction with these reports, Rev-erb α−/− mice have dyslipidemic characteristics (11). In addition, increasing evidence has shown that Rev-erb α plays an important role in maintenance of circadian timing in brain and liver tissue (12,13). Because Rev-erb α is a well-known transcriptional repressor in the positive limb of circadian transcription, the amplitude, period length and phase of the mRNA oscillation pattern of several clock genes such as Bmal1, Clock and Cry1 are largely altered in Rev-erb α−/− mice (14,15).
Until now, the generation of mRNA and protein oscillation profiles has been mainly explained by transcriptional regulation by TTFL. Because proteins are the primary effectors within cells, many scientific researches in the field of circadian rhythm have focused on protein–protein interactions, protein localization, signaling cascades and post-translational modifications (16,17). However, mRNA quantity and quality controls should be set forth beforehand to encode proteins properly (18,19). Among them, regulation of translation initiation is the most important step with clock genes in particular, in order to accurately regulate protein oscillation. Here, we suggest that internal ribosomal entry site (IRES)-mediated translation may be a mechanism that controls the mouse Rev-erb α (mRev-erb α) protein oscillation pattern in NIH3T3 mouse embryo fibroblast cells, which are a good model for studying the molecular mechanism of the mammalian circadian clock system (20,21). Even though the IRES was first discovered in viral genes (22), many studies have shown that mammalian cells utilize an IRES-mediated translation mechanism for rapid adaptation to certain environments such as chemotoxic stress (23), mitosis (24) and apoptosis (25). For IRES-mediated translation, proteins known as IRES trans-acting factors (ITAFs) must recognize IRES elements in a structure or sequence-dependent manner. In this context, we suggest that polypyrimidine tract-binding protein (PTB), also called heterogeneous nuclear ribonucleoprotein (hnRNP) I, and hnRNP Q are ITAFs for the IRES-mediated translation of mRev-erb α, and that they are necessary for maintenance of the circadian profile of mRev-erb α protein by enhancing its translation, which is dependent on the circadian phase.
MATERIALS AND METHODS
Plasmids
To generate pRF Rev1–489, the mouse Rev-erb α (accession no. AY336125) 5′-UTR was amplified with the forward primer 5′-AAGTCGACAGAGTGAAATATTACTGCT-3′ and the reverse primer: 5′-AACCCGGGGTCTTCACCAGCTGAAAGC-3′ using Pfu polymerase (Solgent) and was confirmed by sequencing. The PCR product was digested with SalI and SmaI and inserted into the intercistronic region of a pRF dicistronic vector (26). To generate the inverse construct, the mRev-erb α 5′-UTR was amplified with the forward primer 5′-AACCCGGGAGAGTGAAATATTACTGCT-3′ and the reverse primer 5′-AAGTCGACGTCTTCACCAGCTGAAAGC-3′, digested with SmaI and SalI, and cloned into the intercistronic region of the pRF vector. To generate serial deletion constructs, mRev-erb α 5′-UTR fragments were amplified with forward primers 5′-AAGTCGACGCTTCTCTTCCTTTGGGAC-3′ for pRF100–489, 5′-AAGTCGACTCCGGTGCACTGCAGAGAC-3′ for pRF180–489, 5′-AAGTCGACCACTCTCTGCTCTTCCCAT-3′ for pRF 256–489, and the reverse primer 5′-AACCCGGGGTCTTCACCAGCTGAAAGC-3′ for all deletion constructs. To construct the pRF reporter lacking a promoter, the CMV promoter was removed from pRF Rev1–489 by digestion with BglII/NheI, and then was self-ligated. To create a hairpin-harboring construct, a palindrome was inserted at the NheI site upstream of pRF Rev1–489.
For in vitro binding followed by UV crosslinking, full-length and deletion fragments of the mRev-erb α 5′-UTR were amplified as described earlier. The PCR products were digested with EcoRI and XbaI, and then inserted into the pSK′ vector (27), yielding pSK′ Rev1–489, pSK′ Rev100–489, pSK′ Rev180–489 and pSK′ Rev256–489. To generate the dicistronic mRNA reporter for mRNA transfection, full-length and deletion fragments of the mRev-erb α 5′-UTR were amplified and ligated into the intercistronic region of the pCY2-RF vector (26), yielding pCY2-RF Rev1–489 and pCY2-RF Rev256–489.
Cell culture and dexamethasone shock
NIH3T3 cells were maintained in DMEM (Welgene) supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin–streptomycin (Welgene) in a humidified atmosphere containing 5% CO2 at 37°C. The dexamethasone shock was performed as described previously (19). In brief, ∼1.5 × 105 cells were seeded in each well of a 12-well plate. When the cells reached confluence, the medium was exchanged with medium containing 100 nM dexamethasone. After 2 h, this medium was replaced with complete medium. Cells were harvested at the indicated times for further experiments.
In vitro RNA synthesis and luciferase assay
For mRNA transfection, pCY2-RF Rev1–489 and pCY2-RF Rev256–489 were linearized by digesting with EcoRI. This plasmid contains a 20-nt long poly (A) sequence upstream of the EcoRI site. Reporter mRNA was in vitro transcribed with SP6 RNA polymerase (Roche) in the presence of a cap analog. Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. IRES activity was determined as the ratio of Firefly to Renilla activity.
Transient transfection
For plasmid transfection, NIH3T3 cells were seeded in 24-well plates at a density of 1 × 105 cells per well 12 h before transfection. Transfection was carried out using Metafectene (Biontex) according to the manufacturer’s instructions. After incubation for 48 h, cells were harvested for further experiments. For mRNA transfection, NIH3T3 cells were seeded in 24-well plates at a density of 1 × 105 cells per well 12 h prior to synchronization. Two micrograms of capped reporter mRNA were transfected into NIH3T3 cells at the indicated time points using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After incubation for 6 h, cells were harvested for further experiments.
Quantitative real-time RT–PCR
Quantitative real-time RT–PCR was performed as described previously (19). In brief, total RNA was isolated using TRI Reagent (Molecular Research Center). RNA was reverse transcribed using the ImProm-IITM Reverse Transcription System (Promega) according to the manufacturer’s instructions. For detection and quantification, the MyiQTM real-time PCR detection system (Bio-Rad) was used. The sequences of the forward and reverse primers were as follows: endogenous mRev-erb α, 5′-CTGGAGGGCTGCAGTATAGC-3′ and 5′-GTCCAGGGTCGTCATGTCTT-3′; ribosomal protein L32 (mRPL32), 5′-AACCCAGAGGCATTGACAAC-3′ and 5′-CACCTCCAGCTCCTTGACAT-3′.
In vitro binding assay and immunoprecipitation
In vitro binding assay was performed as described earlier (26). In brief, XbaI-linearized pSK′-5′-UTR constructs were transcribed using T7 RNA polymerase (Promega) in the presence of [α-32P] UTP. Twenty micrograms of nuclear extracts or 40 μg of cytosolic extracts were incubated with labeled RNAs at 30°C. After 30 min of incubation, the mixtures were UV-irradiated on ice for 15 min with a CL-1000 UV-crosslinker (UVP). The samples were detected with autoradiography after SDS–PAGE. To confirm the identity of the UV cross-linked protein, 3 μg of a polyclonal antibody against hnRNP Q (anti-SYNCRIP-N antibody; provided by A. Mizutani, University of Tokyo, Japan) or polyclonal anti-PTB, or pre-immune serum were added to RNase-digested reaction mixtures. After 16 h, Protein G agarose beads (Amersham bioscience) were added. After a further incubation for 3 h, precipitates were detected with autoradiography after SDS–PAGE.
Protein preparation and immunoblot analysis
For immunoblotting, cells were disrupted with complete protein solubilizing buffer containing 1% SDS and 2 M urea in PBS, followed by sonication. Fractionation of NIH3T3 cells into cytosolic and nuclear extracts was performed as described (28). Immunoblot analyses were performed with polyclonal anti-PTB, monoclonal anti-14-3-3ξ (Santa Cruz), polyclonal antibody against hnRNP Q (Sigma-Aldrich), polyclonal anti-mRev-erb α (Cell Signaling), polyclonal phosphor-S6 ribosomal protein (Ser 235/236) antibody (Cell Signaling), polyclonal anti-phospho-eIF4E-BP (Cell Signaling) and polyclonal anti-histone H2B (Upstate). HRP-conjugated mouse, rabbit (KPL) or rat (Santa Cruz) secondary antibodies were detected with SUPEX ECL reagent (Neuronex) and a LAS-4000 system (FUJI FILM), according to the manufacturer’s instructions.
RNA interference
The sequences of synthesized siRNAs were as follows. siCon: 5′-UUCUCCGAACGUGUCACGUTT-3′ (Samchully Pharm.), siPTB: 5′-ACACCUGUGCCUAGCAAUATT-3′ (Samchully Pharm.), sihnRNP Q: 5′-AGACAGUGAUCUCUCUCAUTT-3′ (Dharmacon Research). For siRNA transfection into NIH3T3 cell lines, a microporator (Digital-Bio) was used, according to the manufacturer’s instructions.
RESULTS
Inhibition of cap-dependent translation does not alter the kinetics of mRev-erb α protein expression
mTOR is a serine/threonine protein kinase that regulates several cellular physiological events such as cell growth, cell proliferation, cell survival and protein synthesis(29). p70-S6 Kinase 1 (S6K1) and the eukaryotic initiation factor 4E (eIF4E) binding proteins (4E-BPs) are well-characterized targets of mTOR. Active S6K1 can stimulate the protein synthesis by phosphorylating S6 ribosomal proteins (30). 4E-BP is another important regulator of cellular translation levels. When 4E-BP is hypophosphoryated, it impairs recruitment of the 40S ribosomal subunit to the cap structure of mRNAs by binding to eIF4E. However, when 4E-BP is hyperphosphoryated, it is released from eIF4E and translation efficiency is increased. Rapamycin inhibits the function of mTOR, leading to inhibition of the cap-dependent translation by interfering both phosphorylation of S6 ribosomal proteins and 4E-BPs (29,31). Indeed, rapamycin changed the global translation level in NIH3T3 fibroblasts (Supplementary Figure S1).
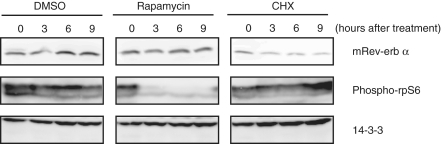
To explore the regulation of translational initiation of mRev-erb α, we suppressed protein synthesis in NIH3T3 mouse fibroblasts by treating the cells with rapamycin or cycloheximide. Compared to vehicle control, the general translation elongation inhibitor cycloheximide caused a rapid reduction of mRev-erb α protein 3 h after treatment. Interestingly, another translation inhibitor rapamycin did not alter the mRev-erb α protein level, even at 9 h after treatment (Figure 1). We confirmed the activity of rapamycin by measuring the phosphorylation status of S6 ribosomal proteins. This result could be explained by three possibilities. First, mRev-erb α protein could be translated in a cap-independent manner. Second, certain proteins that protect the mRev-erb α protein from degradation could be synthesized in a cap-independent mechanism. Third, some factors that promote the degradation of mRev-erb α protein could have disappeared when the mammalian target of rapamycin pathway was inhibited by rapamycin.
Figure 1.
mRev-erb α protein is not degraded by rapamycin treatment. The levels of mRev-erb α protein were analyzed by western blotting. mRev-erb α protein was rapidly decreased following cycloheximide treatment, but was insensitive to rapamycin. Phospho-rpS6 is a phosphorylated S6 ribosomal protein. 14-3-3 was used as a loading control. DMSO, dimethyl sulfoxide; CHX, cycloheximide.
mRev-erb α mRNA possesses IRES sequences in its 5′-UTR
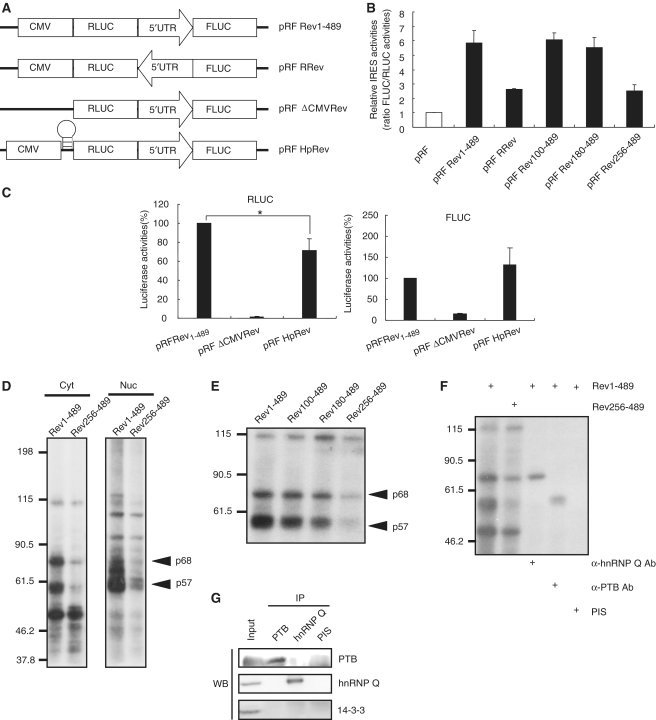
To identify the existence of an IRES in the mRev-erb α transcript, we utilized a dicistronic pRF vector system (26,32). The 5′ untranslated region (UTR) of the mRev-erb α mRNA was inserted between the Renilla luciferase (Rluc) and firefly luciferase (Fluc) coding sequences (pRF Rev1–489), because increasing evidence has shown that the 5′-UTR is mainly responsible for translational regulation. Because Rluc is translated in a cap-dependent manner, and the translation of Fluc is induced in a cap-independent manner by the sequences inserted between the Rluc stop codon and the Fluc start codon in a cap-independent manner, the ratio of Fluc to Rluc indicates the IRES-mediated translation efficiency. We also generated another pRF-based construct to which the 5′-UTR of the mRev-erb α mRNA was inserted in the reverse orientation to confirm the sequence specificity of the IRES elements (pRF RRev) (Figure 2A). We conducted dual luciferase assays 48 h after transfection into NIH3T3 cells. Interestingly, the 5′-UTR of the mRev-erb α mRNA enhanced the Fluc level ∼6-fold compared to the negative control that was transfected with pRF. On the other hand, the reverse-orientated mRNA induced Fluc translation <3-fold. To determine the cis-acting region of IRES-mediated translation, we performed dual luciferase assays with deletion constructs. While there was little reduction in the Fluc level when the 5′ proximal 99 and 179 nt within the 5′-UTR of the mRev-erb α mRNA were deleted (pRF Rev100–489 and pRF Rev 180–489, respectively), additional deletion of 76 nt (pRF Rev 256–489) failed to induce Fluc synthesis as much as control vector (Figure 2B).
Figure 2.
Determination of a cis-acting element and trans-acting factors in IRES-mediated translation of mRev-erb α. (A) Schematic diagram of the dicistronic vector. The sequence orientation of the mRev-erb α 5′-UTR is indicated by the direction of the arrow. The location of the hairpin structure is shown. (B) The efficiency of IRES-mediated translation of each construct is shown. pRF Rev 100–489, pRF Rev 180–489 and pRF Rev 256–489 are deletion constructs of pRF Rev 1–489. The numbers indicate the remaining nucleotides in the mRev-erb α 5′-UTR. The IRES activity of the pRF mock vector was set as 1. (C) The activities of RLUC (left) and FLUC (right) are shown. The level of each protein from pRF Rev 1–489 construct-transfected cells was set as 100. All results are presented as the mean ± SD of at least three independent experiments. The significance of differences of RLUC levels between two constructs was determined by one-way analysis of variance (ANOVA). *P < 0.005 (D) Cellular proteins that bound to the wild-type or truncated forms of the mRev-erb α 5′-UTR were analyzed with the in vitro binding followed by UV-crosslinking assay. Cytoplasmic (Cyt) and nuclear (Nuc) lysates were extracted from NIH3T3 cells. The sizes of proteins are indicated to the left of the panels. (E) The binding patterns of cytosolic proteins to serially deleted mRev-erb α 5′-UTR. In (D) and (E), arrows indicate the candidate ITAF proteins (p68 and p57). (F) Identification of p68 and p57 as hnRNP Q and PTB, respectively, using UV crosslinking followed by immunoprecipitation. (G) Confirmation of existence of hnRNP Q and PTB in immunoprecipitates with hnRNP Q antibody and PTB antibody, respectively. PIS, pre-immune serum.
To exclude the possibility that the enhancement of Fluc levels came from additionally created Fluc transcripts driven by a cryptic promoter within the 5′-UTR of the mRev-erb α mRNA, we deleted the CMV promoter in pRF Rev (pRF ΔCMVRev in Figure 2A). As expected, both Rluc and Fluc expression were barely detected (Figure 2C). This result indicated that there is no promoter activity in the 5′-UTR of the mRev-erb α mRNA. To exclude the possibility that ribosomes released from the Rluc stop codon could re-associate with the start codon of Fluc to maintain synthesis of the downstream Fluc cistron, we designed a modified pRF Rev construct that harbors a hairpin loop upstream of Rluc (pRF HpRev in Figure 2A). Although this loop blocked cap-dependent translation of Rluc by ∼30%, Fluc activity still remained (Figure 2C). These data showed that Fluc translation was not driven by ribosome reinitiation. There was no alternative splicing or unexpected mRNA decay as confirmed by northern blot (Supplementary Figure S2).
hnRNP Q and PTB modulate IRES-mediated translation of mRev-erb α
To determine the ITAFs that modulate IRES-mediated translation of mRev-erb α, we conducted in vitro binding followed by UV crosslinking. Interestingly, both a 68-kDa protein (p68) and a 57-kDa protein (p57) showed strikingly higher binding affinities to the full-length 5′-UTR of the mRev-erb α mRNA (Rev1–489) in both cytosolic and nuclear fractions, but not to the deletion transcript, which displayed only mild Fluc activity (Figure 2D). As anticipated, binding of these two proteins remained until the 5′ proximal 179 nt were deleted (Figure 2E). This strong correlation between the IRES activity of the serial deletion reporters and the binding pattern of the two proteins to the deletion constructs suggested that p68 and p57 are strong candidates for ITAFs.
We previously reported that hnRNP Q enhances IRES-mediated translation of AANAT in rat pinealocytes (26). Moreover, several lines of evidence have demonstrated that PTB can function as an ITAF at viral or cellular IRES (25,33). Notably, the molecular weights of hnRNP Q and PTB are ∼68 and 57 kDa, respectively. Therefore, we determined whether hnRNP Q and PTB bound to the 5′-UTR of the mRev-erb α mRNA with UV crosslinking followed by immunoprecipitation with antibodies against hnRNP Q and PTB.
Both the 68 kDa and the 57 kDa bands, which showed dramatically reduced binding to the Rev256–489 construct, were clearly detected in precipitates using anti-hnRNP Q and anti-PTB, respectively. In contrast, we did not observe any detectable bands in precipitates using pre-immune serum (Figure 2F). In addition, we tested whether both antibodies against hnRNP Q and PTB interact with their targets specifically in UV cross-linking followed by immunoprecipitation experiment that we performed (Figure 2G). PTB was detected in immunoprecipitate with PTB antibody, but absent in other precipitates. hnRNP Q was also only found in immunoprecipitate with hnRNP Q antibody. 14-3-3 was not detectable in any precipitates. Therefore, we confirmed that the 68 kDa and the 57 kDa bands were hnRNP Q and PTB, respectively.
Knockdown of hnRNP Q and PTB induces rapamycin sensitivity in the mRev-erb α protein
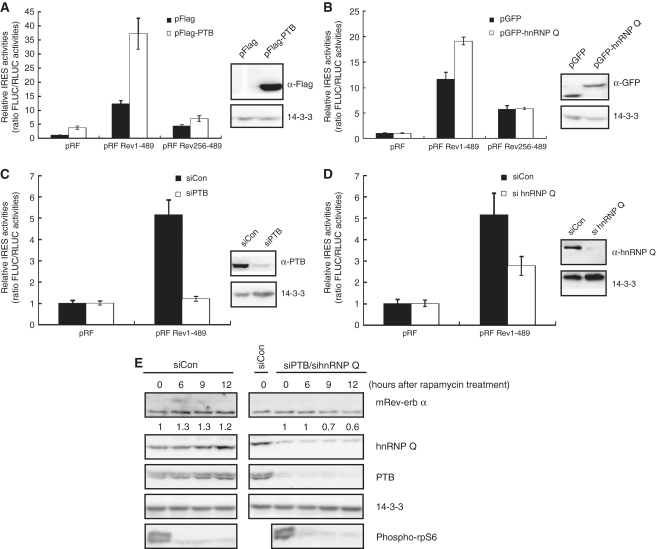
To confirm whether hnRNP Q and PTB modulate the IRES activity of mRev-erb α, overexpression and knockdown experiments were performed. Overexpression of hnRNP Q and PTB augmented the IRES activity of mRev-erb α by about 2- and 3-fold, respectively. The negative control pRF mock vector and the deletion construct pRF Rev256–489 were barely affected by the overexpression of hnRNP Q and PTB (Figure 3A and B). As expected, reduction of hnRNP Q and PTB reduced the IRES activity of mRev-erb α to about 60 and 20%, respectively, compared to the control treated with scrambled siRNA. The negative control was not influenced by the above knockdown experiments. This result suggested that hnRNP Q and PTB may modulate the IRES activity of mRev-erb α (Figure 3C and D).
Figure 3.
hnRNP Q and PTB modulate IRES-mediated translation of mRev-erb α in combination. (A and B) The enhancement of IRES activity of mRev-erb α was shown in hnRNP Q (A) and PTB (B) overexpression experiments. pRF mock and mRev-erb α 5′-UTR-truncated forms were negative controls. All results are representative of at least three independent experiments. The error bars represent the mean ± S.E.M. of duplicate measurements (left panels). Overexpression of hnRNP Q (A) and PTB (B) was confirmed by western blotting (right panels). (C and D) The suppression of IRES activity of mRev-erb α is shown in the hnRNP Q (C) and PTB (D) knockdown experiments. pRF mock is the negative control. Downregulation of hnRNP Q (C) and PTB (D) was confirmed by western blotting (right panels). (A–D) All results are representative of at least three independent experiments. The error bars represent the mean ± SEM of duplicate measurements (left panels). (E) Kinetics of mRev-erb α protein was analyzed by western blotting when both hnRNP Q and PTB were downregulated, in which mRev-erb α protein became sensitive to rapamycin treatment. In contrast, mRev-erb α protein was still insensitive to rapamycin treatment in control siRNA transfected cells. The relative band intensities of mRev-erb α proteins are shown below the bands. The intensities at 0 h were arbitrarily set as 1. siCon, control siRNA-transfected extract. Phospho-rpS6 was used as a marker of rapamycin treatment. Knockdown efficiencies of hnRNP Q and PTB were identified by western blotting. 14-3-3 was used as a loading control. siCon, control siRNA-transfected extract.
In light of the results that insensitivity of mRev-erb α to rapamycin is derived from IRES-mediated translation of mRev-erb α by ITAFs, reduction of hnRNP Q and PTB should diminish the level of mRev-erb α when cap-dependent translation is blocked by rapamycin. To test this hypothesis, we treated vehicle or rapamycin to NIH3T3 mouse fibroblasts in which hnRNP Q and/or PTB level is lessened. When hnRNP Q or PTB was downregulated independently, the level of the mRev-erb α protein still remained high 9 h after rapamycin treatment (Supplementary Figure S3A and B). However, when both hnRNP Q and PTB were downregulated, mRev-erb α protein was clearly diminished 9 h after treatment with rapamycin (Figure 3E). We therefore suggest that hnRNP Q and PTB modulate IRES-mediated translation of mRev-erb α in a combinatorial mode.
Knockdown of hnRNP Q and PTB disrupts the rhythmic oscillation of the mRev-erb α protein
Because mRev-erb α has its own molecular circadian rhythm, we hypothesized that alteration in IRES-mediated translation would lead to an abnormal oscillation pattern of the mRev-erb α protein. To analyze the rhythmic profiles of the mRev-erb α mRNA and protein, we harvested NIH3T3 fibroblasts every 6 h after phase-synchronization with dexamethasone treatment. Using real-time quantitative RT–PCR, we identified the first peak between 18 and 24 h after synchronization in the endogenous mRev-erb α mRNA profile. The second peak was observed at nearly 42 h after dexamethasone treatment (Figure 4A). This result indicates that mRev-erb α mRNA oscillates with about a 24-h period in NIH3T3 cells. We further confirmed that the mRev-erb α protein also oscillates with approximately a 24-h period, even though the peak time was delayed by ∼6 h compared to the mRNA oscillation pattern (Figure 4B).
Figure 4.
Co-knockdown of hnRNP Q and PTB disrupts the protein oscillation profiles of mRev-erb α. (A and C) The endogenous mRev-erb α mRNA levels were determined using quantitative real-time RT–PCR and normalized to RPL32 mRNA levels at the indicated time points in phase-synchronized control (A) and siRNA (against hnRNP Q and PTB)-transfected (C) NIH3T3 cells. Initial levels of mRev-erb α mRNA were arbitrarily set as 1.0. These results represent the mean ± SD of three experiments. (B) The levels of endogenous mRev-erb α proteins were analyzed by western blotting at the indicated time points in phase-synchronized NIH3T3 fibroblasts. Histone H2B was used as loading controls. (D and E) The oscillation patterns of endogenous mRev-erb α proteins were analyzed by western blotting at the indicated time points in phase-synchronized, control siRNA-transfected (D) and siRNAs against both hnRNP Q and PTB-transfected (E) NIH3T3 cells. The relative band intensities of mRev-erb α proteins are shown below the bands. The intensities at 0 h were arbitrarily set as 1. siCon, control siRNA-transfected extract. 14-3-3 was used as a loading control. Knockdown of hnRNP Q and PTB was confirmed by western blotting in (E).
To test whether a deficiency in hnRNP Q and PTB affects the mRNA and protein profiles of mRev-erb α, we transfected siRNAs against hnRNP Q and PTB into NIH3T3 cells. Interestingly, the mRev-erb α protein did not oscillate robustly in hnRNP Q and PTB siRNA-cotransfected NIH3T3 fibroblasts (Figure 4E). mRev-erb α protein oscillation was not altered in control siRNA-transfected cells (Figure 4D). On the other hand, mRNA oscillation showed a similar pattern between hnRNP Q and PTB siRNA-cotransfected NIH3T3 cells and control siRNA transfected NIH3T3 cells (Figure 4C). This result suggests that IRES-mediated translation by hnRNP Q and PTB is necessary to generate the oscillation of the mRev-erb α protein without requiring a significant difference in the mRNA oscillation pattern.
IRES-mediated translation occurs in a phase-dependent manner
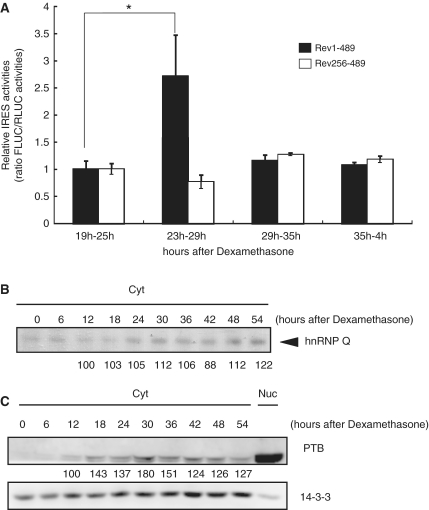
It should be noted that the oscillation may be generated when synthesis and degradation are dynamically regulated. In other words, during the rising phase, the synthesis rate would exceed the decay rate, and this would occur reversely during the declining phase. In this context, we hypothesized that IRES-mediated translation may be regulated in a phase-dependent manner in which IRES-activity would be the highest when the mRev-erb α protein is approaching its peak level. Two mRNA reporters were generated from in vitro transcription of the pRF Rev 1–489 and pRF Rev 256–489 constructs (Rev1–489 and Rev256–489, respectively). These two mRNA reporters were transiently transfected into NIH3T3 fibroblasts at 19, 23, 29 and 35 h after phase-synchronization with dexamethasone. Six hours after transfection, we analyzed the IRES activity of each mRNA reporter in the different phases. Interestingly, the IRES activity of the Rev 1–489 mRNA reporter was greatly increased during the 29- to 35-h time period, compared to the other time points (Figure 5A). Remarkably, this time period correlated well to the peak time of mRev-erb α protein oscillation. At other time points, the IRES activity of the Rev 256–489 mRNA reporter was not significantly changed.
Figure 5.
IRES activity of mRev-erb α is dynamically regulated and phase dependent. (A) The IRES activity of mRev-erb α was differentially regulated and phase dependent, and was analyzed with mRNA transfection. The IRES activity of the truncated form was nearly constant. This result is representative of at least three independent experiments. The error bars represent the mean ± SEM of duplicate measurements. The labels on the x axis indicate mRNA transfection time-harvest time pairs of each mRNA reporter after dexamethasone synchronization. The significance of differences of IRES activities between different phases was determined by one-way analysis of variance (ANOVA). *P < 0.005 (B) Dynamics of the binding affinity of hnRNP Q to mRev-erb α 5′-UTR mRNA after synchronization was analyzed with the UV crosslinking assay. The loading amounts of cytosolic extracts were confirmed by western blotting of the 14-3-3 level in (C). The normalized band intensities are shown below the bands. The intensity at 12 h after synchronization was arbitrarily set as 100. (C) Dynamics of the levels of cytosolic PTB after synchronization as analyzed by western blotting. 14-3-3 protein was used as a loading control. The normalized band intensities are shown below the bands. The intensity at 12 h after synchronization was arbitrarily set as 100.
Because phase-dependent IRES activity may be modulated by phase-dependent ITAF association with IRES, we analyzed the binding dynamics between the 5′-UTR of the mRev-erb α mRNA and hnRNP Q and PTB using UV crosslinking. As expected, hnRNP Q bound to the 5′-UTR of the mRev-erb α mRNA more strongly during both the 24- to 30-h time period and the 48- to 54-h time period after synchronization, in agreement with the mRev-erb α protein profile (Figure 5B). For PTB, we previously reported that PTB differentially translocates to the cytoplasm, depending on the phase (18). We confirmed that the cytosolic PTB level was the highest 30 h after synchronization, even though the total level of PTB was constant (Figure 5C). Collectively, our data suggest that IRES-mediated translation of mRev-erb α is regulated in a phase-dependent manner through the rhythmic binding affinity of hnRNP Q and cytosolic PTB proteins to their 5′-UTR s.
DISCUSSION
Our results provide several evidences suggesting that the mRev-erb α mRNA is translated to protein in a cap-independent manner as well as cap-dependent manner and that both hnRNP Q and PTB modulate the IRES activity of mRev-erb α. The importance of hnRNP Q and PTB in maintaining the rhythmicity of mRev-erb α protein oscillation was verified by knockdown experiments in phase-synchronized NIH3T3 mouse fibroblasts. Furthermore, we suggest that the phase-dependent IRES-mediated translation by hnRNP Q and PTB is a regulatory mechanism for the rhythmic control of the mRev-erb α protein.
Nevertheless, we could not totally exclude the possibility that hnRNP Q and PTB also regulate mRev-erb α protein oscillation by affecting other factors that modulate the mRev-erb α protein level. Because it has been reported that both hnRNP Q and PTB can mediate cap-independent translations of several genes, there still remains the possibility that certain proteins which could be synthesized in a cap-independent mechanism by hnRNP Q or PTB can affect the mRev-erb α protein level in some extent. In our results, downregulation of hnRNP Q and PTB does not affect the mRNA stability of mRev-erb α (Supplementary Figure S4B and C). However, as we previously reported that PTB could control the mRNA stability of mPer2 and mPer2 proteins can inhibit the transcription of mRev-erb α mRNA, PTB and hnRNP Q can regulate mRev-erb α level indirectly. It is also possible that hnRNP Q and PTB post-translationally influence mRev-erb α protein activity, perhaps by affecting protein degradation kinetics. Indeed, we could not observe the full synergistic reduction of Fluc level when both of hnRNP Q and PTB were perturbated in dicistronic pRF vector transient transfection (Supplementary Figure S4A). Therefore, it would be important to explore whether other kinds of modulation of mRev-erb α by hnRNP Q and PTB also exist.
We observed an ∼6-fold increase in IRES activity of mRev-erb α in our DNA transient transfection experiment compared to the negative control, but we observed less than a 2-fold increase in Fluc translation in the mRNA transfection test (unpublished data). This may be explained by two reasons. First, it takes time for ITAFs to recruit several different types of translation initiation machineries to fully induce IRES-mediated translation. Indeed, we analyzed luciferase activity only 6 h after transfection of mRNA, compared to 36 h after DNA transfection. The more likely possibility is that nuclear experience is important for sufficient translation via an IRES. Even though IRES-mediated translation occurs in the cytoplasm, increasing evidence has shown that gene expression steps are interconnected from transcription to translation (34). Therefore, a number of hnRNPs and RNA-binding factors that are involved in transcription, splicing, mRNA export, mRNA decay and translation associate with their target mRNAs in a combinatorial and/or serial manner. Meanwhile, they recruit other factors that are needed for the next stage. In this context, some proteins, including hnRNP Q and PTB, which bound to the IRES element of mRev-erb α in the nucleus may enhance the IRES activity by facilitating recruitment of other unidentified ITAFs or translation initiation machineries. Accordingly, the in vitro transcribed mRNA reporter did not exhibit full IRES activity because it remained only in the cytoplasm. In addition, because the level of hnRNP Q and PTB is usually much higher in the nucleus than in the cytoplasm, in vitro transcribed mRNA reporters may interact with a limited amount of hnRNP Q and PTB.
Because mRev-erb α is a main component of the autoregulatory transcription–translation feedback loop in the mammalian circadian clock system, perturbation of mRev-erb α protein oscillation may affect the transcription efficiency, mRNA level and oscillation pattern of mRNAs and proteins of other clock genes. Indeed, we observed alteration of the mRNA levels of several clock genes following knockdown of hnRNP Q and PTB (unpublished data). Although the variation in the oscillatory pattern of other clock molecules remains to be determined, our results suggest that post-transcriptional regulation by hnRNP Q and PTB is necessary to maintain the feedback loop.
It will be necessary to test whether IRES-mediated translation of mRev-erb α occurs in vivo. In particular, because the SCN functions as the master clock for regulating the peripheral clock, the molecular study of clock genes in the SCN is a prerequisite for understanding animal physiology. If the IRES-mediated translation of Rev-erb α is also present in the SCN and is regulated by both hnRNP Q and PTB, dysfunction of hnRNP Q and PTB may lead to perturbations in animal circadian behaviors such as sleep regulation or physiological homeostasis. In addition, because Rev-erb α can function during adipocyte differentiation and the immune response, the role of post-transcriptional regulation including IRES-mediated translation in various physiological conditions should be investigated in the future.
Studies of cellular IRES-mediated translation have continued since the viral IRES was discovered. Increasing evidence suggests that IRES-mediated translation is involved in various physiological activities such as mitosis, apoptosis and stress conditions. More rapid translation via the IRES is likely beneficial for cells to quickly adapt to certain situations without changing the efficiency of transcription, thus saving time and energy. In the case of circadian rhythms, it is possible that many clock-controlled genes utilize IRES-mediated translation so that they are properly and robustly induced at the right time. Our results shed light on the molecular regulatory mechanism of the mammalian circadian oscillation system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) grants (No. 20090063547, No. 20090081464 and WCU program R31-2008-000-10105-0); Korean Ministry of Education, Science and Technology, Regional Core Research Program/Anti-aging and Well-being Research Center and Brain Korea 21program. Funding for open access charge: National Research Foundation of Korea (NRF) grants (No. 20090063547, No. 20090081464); Korean Ministry of Education, Science and Technology, Regional Core Research Program/Anti-aging and Well-being Research Center and Brain Korea 21 program.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu. Rev. Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 2.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 3.Mirsky HP, Liu AC, Welsh DK, Kay SA, Doyle FJ., 3rd A model of the cell-autonomous mammalian circadian clock. Proc. Natl Acad. Sci. USA. 2009;106:11107–11112. doi: 10.1073/pnas.0904837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Florez JC, Takahashi JS. The circadian clock: from molecules to behaviour. Ann Med. 1995;27:481–490. doi: 10.3109/07853899709002457. [DOI] [PubMed] [Google Scholar]
- 5.Barinaga M. Circadian rhythms. Two feedback loops run mammalian clock. Science. 2000;288:943–944. doi: 10.1126/science.288.5468.943a. [DOI] [PubMed] [Google Scholar]
- 6.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 7.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 8.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 10.Coste H, Rodriguez JC. Orphan nuclear hormone receptor Rev-erbalpha regulates the human apolipoprotein CIII promoter. J. Biol. Chem. 2002;277:27120–27129. doi: 10.1074/jbc.M203421200. [DOI] [PubMed] [Google Scholar]
- 11.Chomez P, Neveu I, Mansen A, Kiesler E, Larsson L, Vennstrom B, Arenas E. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor. Development. 2000;127:1489–1498. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- 12.Teboul M, Guillaumond F, Grechez-Cassiau A, Delaunay F. The nuclear hormone receptor family round the clock. Mol. Endocrinol. 2008;22:2573–2582. doi: 10.1210/me.2007-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in –mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 14.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan SN, Muscat GE. The orphan Rev-erb nuclear receptors: a link between metabolism, circadian rhythm and inflammation? Nucl. Recept. Signal. 2006;4:e009. doi: 10.1621/nrs.04009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak E, Kim TD, Kim KT. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- 20.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 21.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 22.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 23.Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ, de Moor CH, Bushell M, Willis AE. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27:1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, Cornelis S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganguly S, Coon SL, Klein DC. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–137. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- 28.Hahm B, Cho OH, Kim JE, Kim YK, Kim JH, Oh YL, Jang SK. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 29.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 30.Shah OJ, Anthony JC, Kimball SR, Jefferson LS. 4E-BP1 and S6K1: translational integration sites for nutritional and hormonal information in muscle. Am. J. Physiol. Endocrinol. Metab. 2000;279:E715–E729. doi: 10.1152/ajpendo.2000.279.4.E715. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Bjornsti MA, Houghton PJ. Rapamycins: mechanism of action and cellular resistance. Cancer Biol. Ther. 2003;2:222–232. doi: 10.4161/cbt.2.3.360. [DOI] [PubMed] [Google Scholar]
- 32.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kafasla P, Morgner N, Poyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol.Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.