Abstract
Cystic fibrosis is a prominent genetic disease caused by mutations of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Among the many disease-causing alterations are pre-mRNA splicing defects that can hamper mandatory exon inclusion. CFTR exon 9 splicing depends in part on a polymorphic UG(m)U(n) sequence at the end of intron 8, which can be bound by TDP-43, leading to partial exon 9 skipping. CELF proteins, like CUG-BP1 and ETR-3, can also bind UG repeats and regulate splicing. We show here that ETR-3, but not CUG-BP1, strongly stimulates exon 9 skipping, although both proteins bind efficiently to the same RNA motif as TDP-43 and with higher affinity. We further show that the skipping of this exon may be due to the functional antagonism between U2AF65 and ETR-3 binding onto the polymorphic U or UG stretch, respectively. Importantly, we demonstrate that the divergent domain of ETR-3 is critical for CFTR exon 9 skipping, as shown by deletion and domain-swapping experiments. We propose a model whereby several RNA-binding events account for the complex regulation of CFTR exon 9 inclusion, with strikingly distinct activities of ETR-3 and CUG-BP1, related to the structure of their divergent domain.
INTRODUCTION
Cystic fibrosis is a common genetic disease caused by defects in the multidomain cystic fibrosis transmembrane conductance regulator (CFTR) protein. More than 1500 distinct mutations have been recorded, among which 200 or more are believed to result in pre-mRNA splicing defects (http://www.genet.sickkids.on.ca/cftr/app). Such anomalies were shown to lead to dysfunction of several organs in CF patients. One such case is exemplified by the congenital bilateral absence of vas deferens (CBAVD) syndrome (1–4), a mild form of CF, due to the production of an inactive CFTR protein devoid of exon 9-encoded sequence, which prevents incorporation of a mandatory domain of the protein (5,6). Among these, exon 9 inclusion has been shown to be highly variable, depending on several cis-elements and proteins (7). Tia-1 binds to a sequence immediately downstream of exon 9, called PCE (proximal control element) and stimulates exon inclusion, while SF2/ASF and SRp40 bind further downstream in intron 9, which favours exon exclusion (8). The third control element is a polymorphic UG(m)U(n) sequence at the end of intron 8. Strikingly, UG(m)U5 alleles are often associated with low UG numbers in healthy subjects, but with high numbers in affected individuals (1,9–14). TDP-43 (HIV-1 TAR DNA-binding protein) was shown to bind to this UG(m)U(n) sequence, leading to moderate exon skipping (15,16). This effect was further reinforced, in the case of UG11U5, by over-expressed SF2/ASF (7). Importantly, the ability of splicing factors to control exon 9 splicing was shown to be strongly dependent on the number of repeated elements, underlying the natural effects of the polymorphisms (17). Hence, in vitro TDP-43 binding was observed if at least 6UGs were present, its affinity increasing with the number of UG repeats (16). However, its effect was strongly reduced with U7 or U9 alleles. Furthermore, TDP-43 could interact with hnRNP A2/B1, hnRNP A1, hnRNP C1/C2 and hnRNP A3, all reported to act as splicing inhibitory proteins (18), an effect that could promote exon 9 skipping. Finally, suppression of TDP-43 rescued exon 9 splicing in live cells (19).
Another set of splice proteins, including CUG-binding protein 1 (CUG-BP1) (20) and embryonic lethal abnormal vision-type RNA-binding protein 3 (ETR-3)-like (CELF) protein family members, plays key roles in pre-mRNA splicing (for a review see (21)). These two proteins share a high degree of overall sequence similarity (76%). They have three RNA recognition motifs (RRMs) that are at least 90% identical between the two proteins. By contrast, a large portion of the proteins between RRM2 and RRM3, known as the divergent domain (DD), is poorly conserved (29% similarity). Nevertheless, both CUG-BP1 and ETR-3 regulate several RNA targets in a similar way (21). A systematic evolution of ligands by exponential enrichment (SELEX) approach has led to the identification of the optimal RNA targets for the ETR-3 protein. It was found that ETR-3 could bind avidly to UG repeats and control splicing events in live cells (22). In addition, CUG-BP1 could also bind to the identified UG-repeats to which ETR-3 could bind. This binding was proposed recently to depend on the activity of RRM3, which was shown to bear strong preference for UG sequences (23). When tested in the context of a specific minigene that had incorporated the polymorphic UG(m)U(n) element from CFTR intron 8, ETR-3 was able to control the splicing event occurring downstream. This was not the case if the site was mutated, demonstrating that ETR-3 could indeed play a role in CFTR splicing through this sequence (22).
The aims of the present study were to determine the potential of ETR-3 and CUG-BP1 to regulate CFTR exon 9 splicing in the context of CFTR minigenes carrying TG(11–13)T(5–9) repeated elements from intron 8. Our results show that ETR-3 and CUG-BP1 could bind to this element with higher affinity than TDP43, and with an absolute requirement for UG repeats. Importantly, ETR-3, but not CUG-BP1, strongly suppressed exon 9 inclusion in a U5 context, regardless of the length of the UG repeat, suggesting that the level and/or activity of ETR-3 may play a major role in the control of expression of a functional CFTR protein. Furthermore, ETR-3 promoted CFTR exon 9 skipping also in the endogenous gene. Finally replacing CUG-BP1 divergent domain with that of ETR-3 fully converted the activity of CUG-BP1 into that of ETR-3 on this splicing event. These results extend the roles of CELF proteins to the control of CFTR gene expression in a polymorphism- and CELF protein type-dependent fashion, opening the possibility that—at least one of—these proteins might also play a role in cystic fibrosis.
MATERIAL AND METHODS
Plasmids
CFTR hybrid minigenes, carrying the TGmTn (m = 11, 12, 13 or 0; n = 5, 7 or 9), have been described previously (17,24). The pRc-CMV-TDP-43 was previously described (16). The pcDNA3.1-nV5-ETR-3 and pcDNA3.1-nV5-CUG-BP1 were constructed by inserting ETR3 and CUG-BP1 cDNAs in a gateway entry vector (pENTRD, Invitrogen), and next inserted by recombination into the pcDNA3.1-nV5 destination vector. The pet28a-His-CUG-BP1 and pet28a-His-ETR-3 plasmids were described before (25–28). The pcDNA3.1-TDP-43-V5-His was produced using the pcDNA3.1/V5-His©TOPO®TA Expression kit (Invitrogen) according to the manufacturer’s instructions, using the full-length coding region of TDP-43 produced using the forward primer: 5′-ATGTCTGAATATATTCGGGTAACCGA-3′ and the reverse primer: 5′-CATTCCCCAGCCAGAAGACTTAGAA-3′ and the sequence was verified by sequencing. The GFPcETR3vL, GFPcETR3vL-DD.1, GFPcETR3vL-DD.2, GFPcETR3vL-DD.3 have been previously described (29), as the GST-U2AF65 and pEGFP-C3-U2AF65 (30,31). The chimera CUG-BP1-DD.ETR-3 protein was produced by gene synthesis (GenScript Corporation). The cTnT minigene (RTB300 minigene vector, centred on cTnT exon 5) was a gift of Dr T. Cooper.
Cell culture and transfection
HGT-1 gastric cells, HCT116 colon cancer cells and HeLa cervix carcinoma cells were grown in DMEM containing 4.5 g of glucose (Lonza) and 10% fetal bovine serum (Gibco) at 37°C in a humidified incubator in the presence of 5% CO2. They were plated at a density of 4.105 cells in 6-well plates at 70% confluency 24 h before transfection. Transfection was performed using Lipofectamine 2000 (Invitrogen) with CFTR hybrid minigenes plasmids (500 ng) and with TDP-43 (2.5 µg), ETR-3 (500 ng), CUG-BP1 (500 ng) expression vectors or with the empty vector (a total of 3µg of DNA), and harvested 16 h later. For siRNA experiments, cells were plated at 1.105 cells in 6-well plates, 24 h before transfection (Lipofectamine 2000). Cells were transfected with siNeg (Eurogentec), siTDP-43: 5′-GCAAAGCCAAGAUGAGCCU-3′, siETR-3: 5′-GGGUGAUGUUCUCUCCAUU-3′, siCUG-BP1: 5′-GUUACGACAAUCCUGUUUC-3′, siSF3b49: 5′-GAAGUUGCUUUAUGAUACU-3′ (Sigma) or siSF3b145 : 5′-GUAUGUGACUGAAGAACCU-3′ (Sigma) at a final concentration of 20 nM. After 72 h of transfection, CFTR hybrid minigene (500 ng) and the same amounts of siRNAs were transfected again and cells were harvested 16 h later.
RNA extraction and RT–PCR
RNAs were extracted using Tri-Reagent (Ambion). RNAs were quantified by spectrophotometry and the quality was verified by electrophoresis. RT–PCR primers were as follows:
CFTR minigene primers:
forward primer 5′-CAACTTCAAGCTCCTAAGCCACTGC-3′,
reverse primer 5′-TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA-3′;
CFTR endogenous gene primers:
forward primer hCFTR E8: 5′-TAACAGCCTTCTGGGAGGAG-3′,
reverse primer hCFTR E10: 5′-CCAGGCATAATCCAGGAAAA-3′;
Caspase-2 endogenous gene primers:
forward primer hCasp-2 E8: 5′-GTTACCTGCACACCGAGTCACG-3′
reverse primer hCasp-2 E10: 5′-GCGTGGTTCTTTCCATCTTGTTGGTCA-3′;
RTB300 minigene primers:
forward primer: 5′-CATTCACCACATTGGTGTGC-3′
reverse primer: 5′-GGTGCTGCCGCCGGGCGGTGGCTG-3′.
Two hundred nanograms of RNA (minigene condition) or 1 µg RNA (endogenous mRNA) were reverse transcribed and amplified with the QIAGEN OneStep reverse transcriptase PCR (RT–PCR) kit according to the manufacturer’s instructions.
Recombinant proteins
His-ETR-3, His-CUG-BP1, His-CUG-BP1-DD.ETR3 and His-TDP-43 were purified using the Protino Ni-TED 2000 system (Macherey-Nagel) according to the manufacturer’s instructions. GST-U2AF65 was purified as previously described (30).
In vitro transcription and gel retardation assays
To generate RNA probes covering a portion of the 3′ region of CFTR intron 8, PCR was performed on the hybrid minigene carrying the TGmT5 polymorphisms, using the following oligos: 5′-TAATACGACTCACTATAGGGACTCATCTTTTATTTTTGA-3′ (sense) (containing the T7 promoter sequence) and 5′-TTGCTTTCTCAAATAATTCCCCAAATCC-3′ (antisense). The PCR product was purified using NucleoSpin Extract II (Macherey-Nagel). In vitro transcription was performed using the MEGAshortscriptTM kit (Ambion) according to the manufacturer’s instructions.
For ElectroMobility Shift Assay (EMSA), 1 pmol (10 000 cpm) of labelled RNA was incubated with various quantities of recombinant proteins in 25 mM HEPES-KOH pH 7.5, 0.3 mM MgCl2, 200 mM potassium glutamate, 0.2 mM EDTA pH 8, 10% glycerol, 0.5 mM DTT, 0.1 mg/mL BSA, 0.1 mg/mL Heparin and 2 mM ATP for 10 min at 30°C. The samples were loaded onto 6% native acrylamide gels and run for 3 h at 12 mA. The gels were fixed 20 min and dried before autoradiography.
The Scatchard plot analysis was conducted by measuring the ratios between protein-bound and protein-free RNA signals.
UV crosslinking
UV crosslinking of recombinant proteins was performed using 30 pmol of [α-32]P-UTP radiolabelled transcript in the presence of various quantities of recombinant proteins. Proteins and RNA were incubated at 30°C for 10 min with a mixture containing 0.3 mM MgCl2, 2 mM ATP, 25 mM HEPES (pH 7.5) 200 mM potassium glutamate, 0.2 mM EDTA, 0.5 mM DTT, 0.1 mg/ml BSA, 0.1 mg/ml Heparin and 10% glycerol. Reaction mixtures were put on ice and irradiated 4 cm from a Philips G15T8 germicidal lamp for 10 min. Samples were digested with 1 μg each of RNase A and RNase T1 (Sigma) at 37°C for 20 min. An equal volume of protein-loading buffer was added, and samples were denatured at 90°C and run on SDS–10% polyacrylamide gels. The gels were fixed 20 min and dried before autoradiography.
For U2AF65 immunoprecipitation experiments, 50 µl of anti-U2AF65 monoclonal antibody, MC3 (32), was added to the crosslinked material and added to a solution containing 20 mM HEPES pH 8, 0.5 mM EDTA, 20% glycerol, 1 mM DTT, 0.05% NP-40 to a final volume of 120 µl. Antibody and antigen were allowed to react on ice for 1 h with periodic swirling. After the addition of 50 µl of a slurry of Protein A + Protein G Sepharose 4 Fast Flow (at 1:1 proportion; Amersham) in a low salt buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40), samples were incubated at 4°C with rotation for 1 h. Beads were washed three times with a high-salt buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 1% NP-40), three times with a low-salt buffer and resuspended in 10 µl of SDS loading dye and boiled for 5 min. After centrifugation, the supernatant was run on SDS–10% polyacrylamide gels, fixed, dried and submitted to autoradiography.
Western blotting analysis
Whole cell extracts were prepared as previously described (33). Nuclear cell extracts were prepared using the Nuclear co-IP kit (Active-Motif), following the manufacturer’s instructions. Fifty micrograms of total proteins or 30 µg of nuclear proteins were boiled in the Laemmli buffer for 5 min before separation by SDS–PAGE. Proteins were then electro-blotted onto polyvinyl difluoride membranes (Millipore), and non-specific binding sites were blocked for 1 h at room temperature by 5% (wt/vol) fat-free milk before an overnight incubation at 4°C with specific antibodies: mouse monoclonal anti-CUG-BP1 (1/1000, Sigma, clone HL 1190), mouse monoclonal anti-ETR-3 (1/1000, Santa-Cruz, clone 1H2), mouse monoclonal anti-TDP-43 (1/1000, Abcam), mouse monoclonal anti-V5 tag (1/1000, Invitrogen), mouse monoclonal anti-hnRNPA1 (Abcam) and mouse monoclonal anti-GFP (Roche). Mouse monoclonal or rabbit polyclonal anti-Hsc70 (1/8000, Abcam) were used as a loading control for total protein, whereas rabbit polyclonal anti-Topoisomerase II β (1/1000, Santa-Cruz) was used for nuclear proteins. Primary antibodies were detected with horseradish peroxidase-conjugated goat antimouse or goat anti-rabbit IgGs (1/10 000; Jackson ImmunoResearch Laboratories). Blots were revealed using an enhanced chemiluminescence detection method.
Densitometry analysis
Ethidium bromide signals obtained following gel electrophoresis of RT–PCR products were integrated under unsaturated conditions (Gel Doc software, Bio-Rad) and used to derive band intensities.
RESULTS
ETR-3 strongly stimulates exon 9 skipping
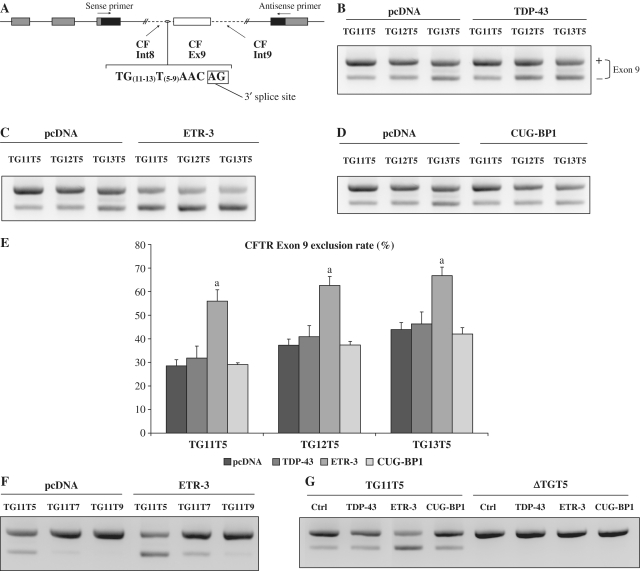
We have previously characterized extensively the mechanism through which TDP-43 binds to the polymorphic intron 8 UG(m)U(n) stretch and inhibits CFTR exon 9 splicing (16). We sought to analyse the possible effect of other UG-binding proteins, ETR-3 and CUG-BP1, two members of the CELF family of splice proteins. We used minigenes harbouring various polymorphic versions of the TG(m)T(n) repeated element, as they occur in CF patients (17), in combination with expression vectors for splice proteins or an empty vector. Plasmid vectors encoding TDP-43, ETR-3 and CUG-BP1 allowed similar levels of protein production in HGT-1 cells (Supplementary Figure S1a). Following transient cell transfections, minigene-encoded transcripts were analysed by gel electrophoresis after reverse transcription and PCR with primers from both sides of exon 9 (Figure 1A). The ratios of exon 9-lacking to exon 9-containing transcripts were then determined by densitometry analysis. As shown in Figure 1B and E, TDP-43 weakly stimulated exon 9 skipping, as previously reported, for all lengths of the TG (11–13) repeat, although the effect did not reach statistical significance. Strikingly, ETR-3 had a much stronger effect, ranging between 2-fold and 1.5-fold the exclusion levels obtained in control, for TG11 to TG13 (Figures 1C and E). In addition, the effect of ETR-3 decreased as the T length increased (Figure 1F). Finally, a CFTR minigene lacking TG sequences showed full exon 9 inclusion, regardless of the addition of a splice protein, demonstrating the absolute requirement for TG in this negative control mechanism (Figure 1G). Hence, these results show that, more than TDP-43, ETR-3 could stimulate exon 9 exclusion, with a somewhat smaller effect as the length of the TG stretch increased, cases where basal exclusion levels were raised. Strikingly, over-expressed CUG-BP1 had no effect on CFTR exon 9 exclusion levels (Figures 1D and E), although over-expressed CUG-BP1, like ETR-3, was able to suppress efficiently cTnT exon 5 exclusion in the control minigene (Supplementary Figure S1b). Hence, although CUG-BP1 and ETR-3 are highly similar, especially in their RRMs, and recognize similar sequences (28), they behaved very differently on CFTR exon 9 splicing.
Figure 1.
ETR-3 induces CFTR exon 9 skipping. (A) Schematic representation of the hybrid minigenes containing CFTR exon 9 and its flanking introns, carrying the TGmTn polymorphism. (B) CFTR hybrid minigenes were co-transfected with TDP-43 or with the empty vector. RT–PCR products were resolved by agarose gels electrophoresis. The exon 9 exclusion rate was determined by quantification using Bio-Rad Chemidoc XR software. (C) Same experiments as in (B) with ETR-3 expression plasmid. (D) Same experiments as in (B) with CUG-BP1 expression plasmid. The results are representative of six experiments with similar results. (E) Histogram recapitulating the results. The exclusion levels are expressed as percent. The standard deviation of the mean is shown (n = 6); aSignificantly different from the pcDNA conditions (Student’s t-test, P < 0.05). (F) CFTR hybrid minigenes carrying TG11-T9, -T7 or -T5 polymorphisms were co-transfected with ETR-3 or the empty vector, (G) CFTR hybrid minigenes carrying 11 or no TG repeats were co-transfected with TDP-43, ETR-3, CUG-BP1 vectors or the empty vector.
To extend these observations to cells that express ETR-3 at various levels, we made use of HCT116 cells that express high levels of ETR-3 (34) and HeLa cells that do not express ETR-3 (N. Charlet-Berguerand, unpublished). As can be shown in Supplementary Figure S2a, these cell lines, like HGT-1, expressed high levels of TDP-43, reinforcing the notion that this protein is widely and strongly expressed (E. Buratti, unpublished results). By contrast, ETR-3 was barely detectable in HGT-1 cells and no signal was observed for HeLa cells, even with up to 100 µg nuclear protein amounts. As expected, a strong signal was observed in HCT116 cells. In minigene assays, ETR-3 strongly induced exon 9 skipping in HCT116 and HeLa cell lines, but it had a smaller effect in HCT116 cells, probably because these showed a higher constitutive exon exclusion level (Supplementary Figure S2b). Similarly to HGT-1 cells, over-expressed CUG-BP1 and TDP-43 had no effect.
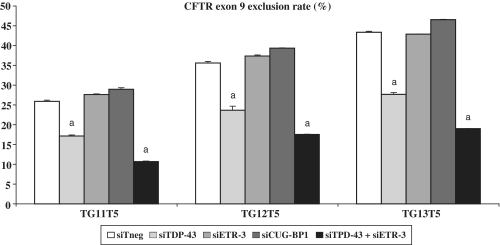
To explore further the roles of ETR-3 and CUG-BP1, in comparison with TDP-43, we used siRNAs. As shown in Figure 2, a siRNA directed against TDP-43 strongly reduced exon 9 exclusion levels, as previously reported (19). By contrast, a siRNA against ETR-3 had no effect (Figure 2), despite a good suppressing activity of the siRNA (Supplementary Figure S3a). This was likely due to the low expression of ETR-3 in the recipient cells used (Supplementary Figure S3b). Nevertheless, the extinction of both TDP-43 and ETR-3 induced a stronger effect on exon 9 exclusion, suggesting that suppression of TDP-43 unmasked the weak basal exon 9 exclusion activity of ETR-3. In HeLa cells, which do not express ETR-3 but express TDP-43, over-expression of ETR-3 induced a much stronger effect when TDP-43 was silenced (7-fold to 21-fold, Supplementary Figure S4). Silencing CUG-BP1 had no effect on exon 9 exclusion and the extinction of both TDP-43 and CUG-BP1 did not restore exon 9 inclusion any better than the extinction of TDP-43 alone (data not shown).
Figure 2.
Effect of siRNA-mediated silencing of TDP-43, ETR-3 or CUG-BP1 on CFTR exon 9 splicing. After 72 h of siRNA transfection, hybrid minigenes were co-transfected with the same amount of siRNAs for 16 h. RT–PCR products were resolved by agarose gels electrophoresis. Histogram recapitulating the results. The exclusion levels are expressed as percent. The standard deviation of the mean is shown (n = 5). aSignificantly different from the siTneg condition (Student’s t-test, P < 0.05).
ETR-3 and CUG-BP1 proteins bind to the UG repeat from CFTR intron 8
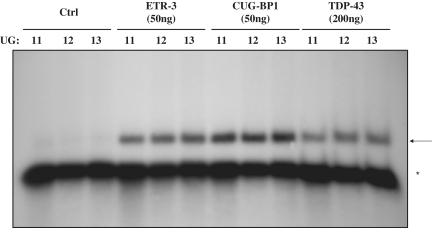
The results obtained so far indicated that these proteins may interact with the UG(m)U(5) polymorphic stretch from intron 8. In order to analyse directly their binding onto this sequence, we performed RNA gel retardation assays (EMSA) with purified tagged proteins, using in vitro transcribed RNAs as labelled probes. As shown in Figure 3, the three proteins could interact with the probe, with a small increase in signal being apparent with increasing UG numbers. His-ETR-3 and His-CUG-BP1 seemed to have a comparable affinity with one another and a stronger affinity than His-TDP-43, as at least four times more of the TDP-43 protein was necessary to give a comparable binding signal as those obtained for ETR-3 and CUG-BP1 (Figure 3).
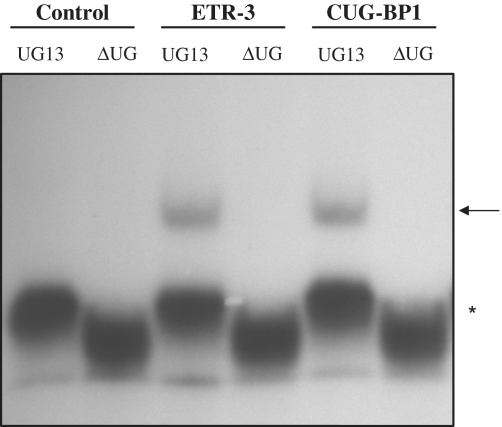
Figure 3.
Electromobility shift assay using recombinant TDP-43, ETR-3 and CUG-BP1. Using a primer containing the T7 promoter sequence, an in vitro transcription assay was performed, allowing the synthesis of a 100 nt radiolabelled RNA containing the CFTR intron 8 polymorphic sequences (UG11, UG12 or UG13 followed by the U5 tract). Recombinant His-TDP-43, His-CUG-BP1 and His-ETR-3 were incubated for 20 min at 30°C with 32P-labelled RNA (1pmole). Native polyacrylamide gels (6%) were used for separation. Ctrl represents the His-tag alone. The results are representative of five experiments with similar results. More TDP-43 protein (200 ng) than ETR-3 or CUG-BP1 (50 ng) was used to compensate for its lower RNA-binding activity. The arrow points to the protein-RNA complexes. The asterisk symbol points to the free RNA probe.
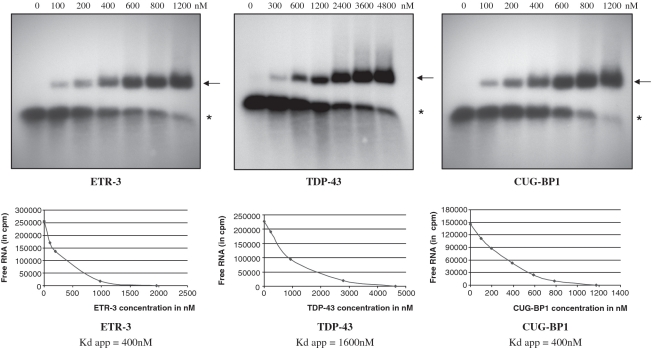
To measure the in vitro binding affinity of the proteins for the RNA motif, we prepared Scatchard plots following EMSA analyses. As shown in Figure 4, the apparent Kds were 400 nM and 1600 nM for ETR-3 and TDP-43, respectively. Hence, ETR-3 had a 4-fold higher affinity for the RNA probe than TDP-43. CUG-BP1 bound to the sequence with the same affinity as ETR-3 (400 nM). Finally, ETR-3 and CUG-BP1 had an absolute requirement for UG sequences, which deletion from the RNA probe abrogated binding (Figure 5). This last result fully agrees with the lack of effect of the proteins in functional assays when the UG sequences were removed from the minigene (Figure 1G).
Figure 4.
Scatchard plot analysis of TDP-43, ETR-3 and CUG-BP1 RNA binding. Gel retardation analyses were performed with the same RNA (UG11U5) probe type and quantity for each protein (see the legend to Figure 3). The results are representative of three experiments with similar results. The arrow points to the protein-RNA complexes. The asterisk symbol points to the free RNA probe.
Figure 5.
ETR-3 and CUG-BP1 bind onto CFTR intron 8 UG repeats. Gel retardation analyses were performed with an RNA probe containing 13 or no UG repeats (see the legend to Figure 3). The results are representative of four experiments with similar results. The arrow points to the protein-RNA complexes. The asterisk symbol points to the free RNA probe.
U2AF65 binding to the CFTR intronic sequence can be displaced by ETR-3
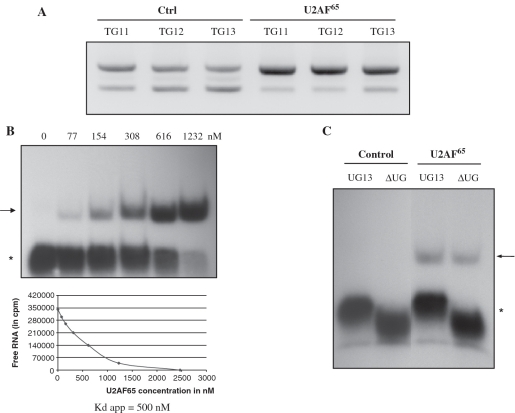
In a functional assay, U2AF65 could strongly stimulate exon 9 inclusion, indicating that it could bind to, and act through, the intronic sequence (Figure 6A). As shown in Figure 6B, U2AF65 could indeed bind to the probe with a slightly lower affinity than ETR-3 (Kd app = 500 nM). However, contrary to ETR-3 or CUG-BP1, its binding was independent of the presence of UG repeats, suggesting that it could bind elsewhere, presumably onto the U repeats (Figure 6C). In order to determine the sequence requirement for U2AF65 binding, we performed UV crosslinking experiments with cell nuclear extracts, followed by U2AF65 IP. As shown in Figure 7A, a dose-dependent increase in protein binding was observed if at least 5Us were present on the probe. In the presence of only 3Us, barely any binding was detectable. In addition, this binding was independent of the presence of UGs (Figure 7B). These results show that U2AF65 binding does not require the same sequences as ETR-3, but the protein can bind to an element containing both UGs and Us through selective interaction with U residues. This suggests that U2AF65 could stimulate spliceosome assembly and agrees to the fact that over-expression of U2AF65 strongly increased CFTR exon 9 inclusion (Figure 6C).
Figure 6.
U2AF65 binds CFTR intron 8 and can be displaced by ETR-3. (A) Exon 9 splicing analysis in the presence of over-expressed U2AF65. CFTR hybrid minigenes were co-transfected with the U2AF65 vector or the empty vector. RT–PCR products were resolved by agarose gels electrophoresis. (B) Determination of U2AF65 apparent Kd for CFTR intron 8 sequence by the Scatchard plot analysis. The same amount of RNA (UG11U5) probe as in Figure 4 was used. The results are representative of three experiments with similar results. The arrow points to the protein-RNA complexes. The asterisk symbol points to the free RNA probe. (C) RNA binding analysis of U2AF65. Gel retardation analyses were performed with an RNA probe containing 13 or no UG repeats (see the legend to Figure 3). The results are representative of three experiments with similar results. The arrow points to the protein-RNA complexes. The asterisk symbol points to the free RNA probe.
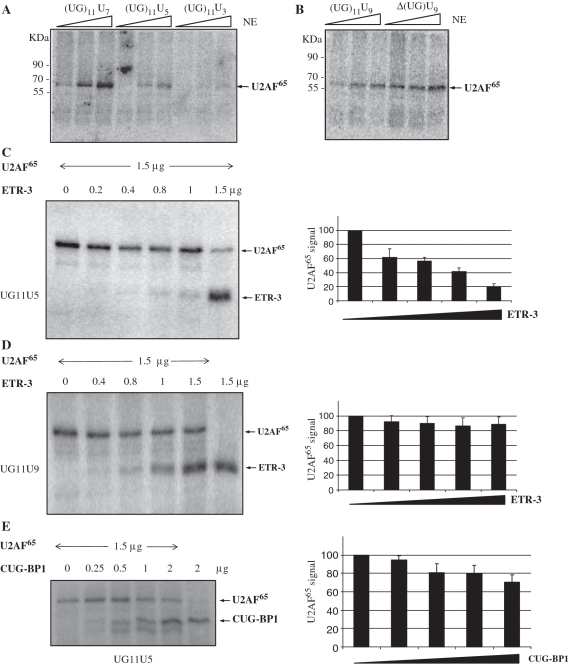
Figure 7.
U2AF65 binds onto CFTR U-repeat and it is displaced by ETR-3 but not by CUG-BP1. After UV crosslinking and RNAse treatment, a U2AF65 immunoprecipitation was performed. (A) Using UG11-U7, -U5 and -U3, (B) using the same length of U-repeat (U9) but with 11 or 0 UG-repeat. (C) UV crosslinking experiment using 30 pmoles of labelled UG11U5 RNA probe and the indicating amounts of U2AF65 and ETR-3. A histogram with the means and standard deviations from triplicate experiments is shown on the right part of the figure. (D) Same as (C) except with a labelled UG11U9 RNA probe. A histogram with the means and standard deviations from triplicate experiments is shown on the right part of the figure. (E) UV crosslinking experiment using 30 pmoles of labelled UG11U5 RNA probe and the indicating amounts of U2AF65 and CUG-BP1. A histogram with the means and standard deviations from triplicate experiments is shown on the right of the figure.
As the length of the polypyrimidine tract following the UG repeat seemed to be very important for the effect of ETR-3 (Figure 1F), we postulated that ETR-3 could induce exon 9 skipping through displacement of U2AF65, which would be expected to lead to a lower efficiency of 3′ splice site recognition (35). We, therefore, wished to determine if ETR-3 and U2AF65 proteins might bind RNA in a mutually exclusive manner. To this end, we performed additional UV crosslinking experiments. As shown in Figure 7C, ETR-3 could largely displace U2AF65 from the probe (UG11U5) when equal amounts (1.5 µg) of each protein were added. The last lane showed an RNA-binding signal for both proteins, indicating that they could bind at the same time to the probe, albeit with a strong preference for ETR-3. When a UG11U9 probe was used, the best ETR-3 binding obtained (1.5 µg protein) was associated with still a good binding of U2AF65, showing that both proteins could bind simultaneously to the probe and with comparable efficiency, provided the U stretch was long enough (Figure 7D). These RNA-binding data agree to the fact that the longest 9U stretch was less responsive to ETR-3 for the skipping of CFTR exon 9 than the 5U stretch in the UG11 context (Figure 1F). Taken together, these results suggest that the skipping induced by ETR-3 was associated with displacement of U2AF65 from the RNA motif, and was inversely related to the length of the U stretch under the UG11 context. Nevertheless, we cannot fully rule out that the binding of the two proteins may be mutually exclusive, with ETR-3 binding to some oligonucleotide probes and U2AF65 binding to a smaller subset of oligo probes in proportion to their relative affinities.
In order to address the functional difference between ETR-3 and CUG-BP1, we analysed the ability of increasing amounts of CUG-BP1 to bind to the RNA probe in the presence of a fixed amount of U2AF65. As shown in Figure 7E, 2 µg CUG-BP1 did not displace 1.5 µg U2AF65 (compare lanes 1 and 5). Hence, contrary to ETR-3 (Figure 7C), CUG-BP1 did not displace U2AF65 from the UG11U5 probe, although it demonstrated strong binding to the probe. This observation agrees with the absence of effect of over-expressed CUG-BP1 on exon 9 skipping (Figure 1D), the ability of U2AF65 to promote inclusion being preserved, even in the UG11U5 context.
The divergent domain of ETR-3 is essential for its suppressive effect
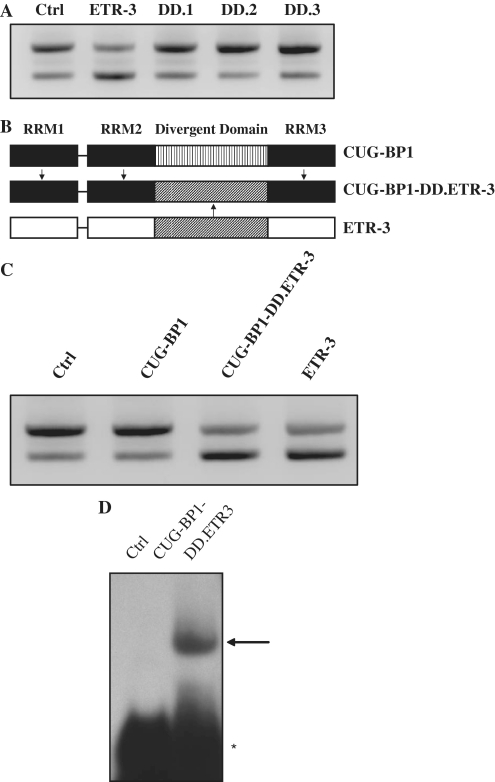
To try to understand the difference observed between CUG-BP1 and ETR-3 on CFTR exon 9 splicing, we hypothesized that the divergent domain might account for the selectivity of ETR-3, as the RRMs were highly similar between the two proteins. Using deletion mutants of ETR-3 (Supplementary Figure S5) (29), we observed that its DD was necessary for the increase in exon 9 skipping (Figure 8A). Indeed, disruption of the DD by deletion in DD.1, DD.2 or DD.3 mutants, which did not modify ETR-3 nuclear localization (data not shown), as previously reported (29), abolished CFTR exon 9 skipping. In fact, this skipping was abrogated every time the DD was altered, as compared to the control, regardless of the particular deletion. In order to look for the RNA-binding ability of the various ETR-3 proteins to the RNA sequence, we transfected expression vectors for these proteins and prepared nuclear protein extracts to conduct EMSAs with the UG11U5-labelled RNA probe. As shown in Supplementary Figure S6, wt ETR-3 and its deletion derivatives strongly bound to the RNA sequence. These results show that all DD deletions could still localize to the nucleus and bind RNA, yet they did not induce exon 9 skipping. This suggests that a defective RNA binding alone cannot explain their lack of effect on splicing. We, therefore, hypothesized that replacing the CUG-BP1 DD by that of ETR-3 might confer exon 9 splicing inhibitory activity. To address this possibility, we engineered a CUG-BP1 expression vector in which the DD of ETR-3 had replaced that of CUG-BP1 (Figure 8B). Strikingly, after transient co-transfection of the TG11T5 minigene and the vector encoding the chimera, we observed that the hybrid protein behaved strictly like ETR-3, strongly inducing exon 9 skipping (Figure 8C). As shown by EMSA, the chimera was able to bind efficiently to the UG11U5 probe (Figure 8D). These data demonstrate that exon 9 skipping was strongly, if not solely, controlled by ETR-3 DD.
Figure 8.
Role of ETR-3 divergent domain on CFTR exon 9 splicing. (A) The TG11T5 CFTR was co-transfected with ETR-3, ETR-3 DD mutants or with the empty vector. RT–PCR products were resolved by agarose gel electrophoresis. The results are representative of three experiments with similar results. (B) Schematic representation of ETR-3, CUG-BP1 and the CUG-BP1-DD.ETR-3 chimeric protein. (C) Same experiment as in (A), with ETR-3, CUG-BP1, CUG-BP1-DD.ETR-3 or the empty vector. The results are representative of five experiments with similar results. (D) Gel retardation analysis of the recombinant CUG-BP1-DD.ETR-3 protein with the UG11U5 labelled RNA probe (see the legend to Figure 3).
The spliceosomal SF3b49/145 components control the functional activity of ETR-3
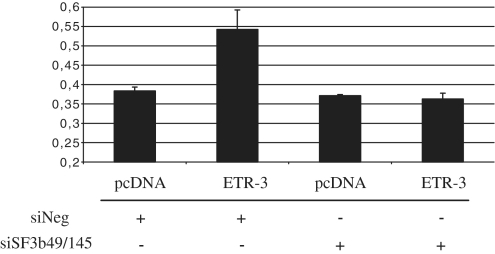
A recent study from the Cooper lab has shown, by a proteomic analysis, that ETR-3 and CUG-BP1 could be distinguished by their ability to bind U2 snRNP-associated spliceosome components. ETR-3, but not CUG-BP1, was shown to interact with SF3b49 and SF3b145, suggesting that some splicing control events could be distinctive between these two CELF proteins (36). In order to look for a potential role of these proteins, we silenced these two proteins and analysed the effect of ETR-3 on CFTR exon 9 exclusion. As shown in Figure 9, no more response to ETR-3 was observed, suggesting that indeed at least part of the effect of ETR-3 on this splicing event could be driven by a functional interaction of ETR-3 and SF3b49/SF3b145.
Figure 9.
Effect of siSF3b49 and siSF3b145 on ETR-3-mediated CFTR exon 9 exclusion. After 72 h of siRNA transfection, cells were co-transfected with pcDNA3.1-nV5-ETR-3 or the empty vector with added siRNAs for another 16 h. RT–PCR products were resolved by agarose gels electrophoresis. Histogram summarizing the level of exon 9 exclusion from triplicate experiments is shown. The mean and the standard deviation are shown.
DISCUSSION
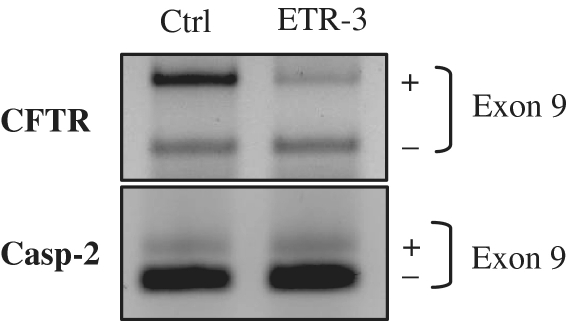
CFTR pre-mRNA splicing plays an important role in the CF condition, and molecular approaches have identified splice proteins that take part in the regulation of CFTR exons assembly. The polymorphic sequence element from intron 8 of the gene has been shown to bind TDP-43, which resulted in a small but consistent level of exon 9 skipping, and hence was proposed to be involved in the allele-dependent effect in CBAVD patients (16). In the present study, we show that one additional protein, ETR-3, could strongly increase the level of CFTR exon 9 skipping, especially in the TG11T5 context, much more efficiently than TDP-43. This was associated with its ability to bind avidly to the polymorphic sequence in vitro, with an absolute requirement for UG sequences. Furthermore, this effect was also observed upon over-expression of ETR-3 on splicing of the endogenous gene (Figure 10), suggesting that such a control may also occur in tissues. By contrast, CUG-BP1 had no effect despite a comparable affinity for the RNA sequence, which was four times that of TDP-43. As previously reported, the longer TG stretches led to increased basal exon 9 exclusion levels. Silencing the abundantly expressed TDP-43 protein led to a strong increase in exon 9 inclusion in the T5 context, for all lengths of TGs, whereas a siRNA against ETR-3 had no effect. This indicates that TDP-43 may be constitutively active in the control of minigene exon 9 splicing, whereas ETR-3, which was expressed at low levels in the recipient cells used, had almost no effect unless its expression was increased by transfection. In addition, silencing of TDP-43, coupled with over-expression of ETR-3 led to a larger increase in exon 9 skipping as compared to over-expression of ETR-3 alone (HeLa cells, Supplementary Figure S4). Silencing CUG-BP1 had no effect. Finally, silencing TDP-43 and ETR-3 at the same time produced the largest splicing suppression effect. That a siRNA against ETR-3 had no effect (presumably because of the very low expression level in the recipient cell), unless it was used together with a siRNA against TDP-43, may be best explained by either one of the following possibilities: (i) ETR-3 could help TDP-43 binding to the target sequence or (ii) the two proteins could bind to distinct sequence subsets, such that dual silencing of the proteins would have a stronger effect than silencing of TDP-43 only. We favour the second possibility since the two proteins bind to the same target sequence. More studies will be needed to investigate this aspect further.
Figure 10.
Effect of ETR-3 over-expression on endogenous CFTR exon 9 splicing. HeLa cells were transfected with the ETR-3 vector or the empty vector. After RNA extraction, RT–PCR was performed using a forward primer located in CFTR exon 8 and a reverse primer in CFTR exon 10. A control RT–PCR experiment was performed to assay the Caspase-2 exon 9 splicing event.
We report here a novel role for two CELF proteins in the regulation of CFTR exon 9 splicing. We confirm that they bind to UG repeats, as previously reported (22). In vitro RNA-binding assays showed that indeed these proteins could recognize the polymorphic sequence much more efficiently than TDP-43. According to studies from the Cooper lab, CUG-BP1 and ETR-3 had comparable abilities to control inclusion of cTNT exon 5, which was clearly not the case for the CFTR minigene used here, where only ETR-3 was active in splicing inhibition (28). In addition, the same group has shown that ETR-3 could induce cTNT exon 5 inclusion through activation of the polymorphic sequence from CFTR intron 8 (22), which is strikingly different from our results. We can only speculate that the structural differences between the minigenes would likely explain these differences. In the case of Cooper’s minigene, the CFTR intron 8 was not associated with CFTR exon 9 and part of intron 9, but with unrelated sequences. In addition, we are not aware of the effect of CUG-BP1 in the context of the Cooper minigene. If CUG-BP1 had the same effect as ETR-3 with the Cooper minigene, then it could be proposed that the CFTR polymorphism would only allow recruitment of the proteins, but the resulting functional activity of the proteins would be more strictly dependent on the overall sequence context and the availability of interacting proteins. Moreover, ETR-3 also triggered CFTR exon 9 skipping from the endogenous gene in the present study, pointing to a putative physiological relevance of our findings. The differences in the functional activity of ETR-3 and CUG-BP1, as observed in the present study, were most likely due to the strong dissimilarity of the DD protein region that localizes outside of the RRMs (28). Consistent with this view, over-expression of ETR-3 deletion mutants lacking parts of the DD, although they showed proper nuclear localization (data not shown), as reported previously (29), triggered no splicing inhibition. Remarkably, replacing the DD of CUG-BP1 by that of ETR-3 fully converted CUG-BP1 into an ETR-3-like protein with respect to exon 9 splicing inhibition. To our knowledge, this is the first report to show that the DD of these two CELF proteins can be functionally quite distinct in its ability to control pre-mRNA splicing events, whereas the CFTR RNA-binding activity and affinity were very similar between CUG-BP1 and ETR-3. As stated by Ladd et al. (28), this DD contains no known protein–protein, protein–RNA or protein–DNA interaction motifs, targeting signals, or predicted secondary structure, making it unique for each CELF protein. However, deletion experiments conducted later had shown that ESE-specific ETR-3 binding and activation of splicing within the cTNT gene could be mediated by either RRM1 and RRM2 plus 70 residues of the adjacent downstream DD or RRM3 plus 119 residues of the adjacent upstream DD (37). In the present study, the DD swapping experiment demonstrated that there was no requirement for the ETR-3 RRMs to convert CUG-BP1 into an ETR-3-like protein, pointing to the major role of ETR-3 DD in the control of CFTR exon 9 splicing. Looking at the amino-acid composition of this domain, it appeared that the number of hydrophobic residues was low, when compared to that of the RRMs, indicating that it could have a somewhat loose conformation, by opposition with the RRMs (I. Callebaut, personal communication). In addition, the DD could be involved in protein–protein interactions or be the target for specific phosphorylations, which would be distinctive between ETR-3 and CUG-BP1, accounting for the strong differences in activities of the proteins with respect to CFTR exon 9 splicing. In any case, the fact that all of the particular DD deletions tested in this study triggered full exon inclusion suggests that the DD overall structure and/or size may be the major determinant of the splicing control activity. Because U2 snRNP binding onto the branch point is functionally linked to the recruitment of U2AF65/U2AF35 onto the 3′ splice site, it is tempting to speculate that the binding of ETR-3, which would antagonize that of U2AF65, could reduce proper exon 9 recognition. The DD of ETR-3, but not that of CUG-BP1, would thus play a major role in these effects.
Preliminary experiments have shown that ETR-3, but not CUG-BP1, could interact with U2AF65 either directly or indirectly, as shown by co-immunoprecipitation of nuclear extracts containing over-expressed GFP-U2AF65 and V5-ETR-3 or V5-CUG-BP1 (data not shown). Such experiments, although showing that U2AF65 and ETR-3 were part of the same protein complex, did not demonstrate that the two proteins interacted directly with each other. In addition, we have not been able to detect, either by native gel retardation or UV crosslinking assays, the occurrence of large protein complexes that may contain the two proteins. Hence, although these two proteins have the ability to co-immunoprecipitate, more studies remain to be performed to see whether they interact directly or not.
As shown by the Cooper lab, ETR-3, but not CUG-BP1, could interact with SF3b49 and SF3b145 spliceosomal components. Our study shows that this interaction is likely to have a functional role on coupling the activity of ETR-3 to the spliceosomal proteins that, altogether, trigger CFTR exon 9 exclusion. It is tempting to speculate that this interaction may involve the DD of ETR-3, but not that of CUG-BP1, which had no effect on splicing CFTR exon 9. In addition, it could be proposed that SF3b49/145 proteins could bridge ETR-3 and U2AF65, which would bind to the intron 9 branch point site, according to the Cooper model (36). Further analyses will be required to address this possibility.
According to the results presented here and previously, it is very clear that the polymorphic UG(m)U(n) stretch is a major determinant of exon 9 splicing. In fact, we have obtained the proof of concept, through in vitro splicing experiments, that addition of a UG(12) oligonucleotide to sequester TDP-43 resulted in increased recognition of the CFTR exon 9 acceptor site (38). Regardless of the particular negatively acting protein that may bind to the UG repeats, it appears that masking the site might thus relieve the inhibitory effect.
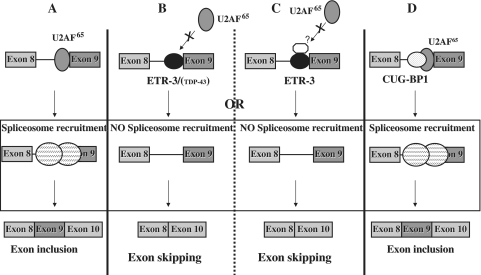
A model that summarizes our observations is presented in Figure 11. In the absence of recruitment of ETR-3 and/or TDP-43, or in the absence of any UG repeat, the spliceosome is correctly assembled, and exon 9 is correctly inserted (part A). In the presence of bound ETR-3 and/or TDP-43, exon 9 skipping is stimulated through hampering the recruitment of U2AF65 onto the polypyrimidine tract, which decreases the 3′-splice site recognition (part B), possibly through additional intermediate proteins that would, for example, interact with the DD of ETR-3 (part C). We suggest that the relative amount and nuclear addressing level of TDP-43 and ETR-3 will dictate which of them—or if both—will bind to the UG sequence. This possibility is substantiated by the fact that in cells that contain higher amounts of ETR-3 than the HGT-1 cells used here, like HCT-116 colon carcinoma cells, silencing TDP-43 had no effect on exon 9 splicing (data not shown). In any case, binding of ETR-3 led to a drop in the efficiency of U2AF65 recruitment. Finally, CUG-BP1 binding allowed spliceosome recruitment and splicing to proceed (part D). As a whole, the abundance, together with the relative binding affinity and sub-cellular localization of CUG-BP1, TDP-43, ETR-3 and U2AF65, among other spliceosomal components, in the living cell, might determine the level of correctly spliced, exon 9-containing, CFTR mRNA. Although the exact relevance to the CF condition of our findings remains to be established, a recent study suggests that the lack of expression of the CFTR gene in muscles—where ETR-3 is strongly expressed—may be linked with muscle alteration. Indeed, CFTR KO mice show specific defects in muscles, and it can be postulated that, depending on the polymorphic status of intron 8 in CF patients, the defect in muscles could be due to the activity of ETR-3 (39).
Figure 11.
Model of CFTR exon 9 splicing regulation by TDP-43 and the CELF proteins, ETR-3 and CUG-BP1. Large circles with dashed lines represent the spliceosomal complex, other circles represent splicing proteins. See the ‘Discussion’ section. (A) Exon 9 inclusion through U2AF65 interaction. (B) ETR-3 (TDP-43) prevents U2AF65 interaction, which blocks exon 9 inclusion. (C) Intermediate protein may help ETR-3 prevent U2AF65 interaction and exon 9 inclusion. (D) CUG-BP1 binding does not hamper U2AF65 interaction or exon 9 inclusion.
In conclusion, this study has identified ETR-3 as a novel CFTR splice repressor protein that, depending on its level and type of intron 8 polymorphism, may impose a strong control of the amount of correctly spliced CFTR transcript in the region of exon 9. This is also the first demonstration of a major functional divergence between ETR-3 and CUG-BP1 CELF proteins.
FUNDING
The Italian association against Cystic Fibrosis and INSERM, the French association against Cystic Fibrosis (‘Vaincre la Mucoviscidose’), the University of Brest, the Federal Institute for Research (IFR ScInBioS, no. 148) and the Brittany Region. G.D. was supported by a Grant from the French Ministry of Education and Research and by a fellowship from Vaincre la Mucoviscidose. Funding for open access charge: ECLA-INSERM U613, Faculty of Medicine, Brest, France.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Dr TA Cooper for expression plasmids encoding ETR-3 and DD mutants and the RTB300 minigene vector, Dr J. Valcarcel for U2AF65 expression plasmid, Dr Ludovic Guigou for his help with Scatchard analysis of EMSA experiments, Dr I. Callebaut for her in silico structural analysis of ETR-3 and CUG-BP1 proteins, Dr Ghislaine Henneke for her help with the Typhoon system, Drs M.C. De Cian, B. Chabot and G. Friocourt for thoughtful discussions, A. François and members of the ECLA team for suggestions.
REFERENCES
- 1.Chillon M, Casals T, Mercier B, Bassas L, Lissens W, Silber S, Romey MC, Ruiz-Romero J, Verlingue C, Claustres M, et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. New Engl. J. Med. 1995;332:1475–1480. doi: 10.1056/NEJM199506013322204. [DOI] [PubMed] [Google Scholar]
- 2.Dork T, Dworniczak B, Aulehla-Scholz C, Wieczorek D, Bohm I, Mayerova A, Seydewitz HH, Nieschlag E, Meschede D, Horst J, et al. Distinct spectrum of CFTR gene mutations in congenital absence of vas deferens. Hum. Genet. 1997;100:365–377. doi: 10.1007/s004390050518. [DOI] [PubMed] [Google Scholar]
- 3.De Braekeleer M, Ferec C. Mutations in the cystic fibrosis gene in men with congenital bilateral absence of the vas deferens. Mol. Hum. Reprod. 1996;2:669–677. doi: 10.1093/molehr/2.9.669. [DOI] [PubMed] [Google Scholar]
- 4.Claustres M. Molecular pathology of the CFTR locus in male infertility. Reprod. Biomed. Online. 2005;10:14–41. doi: 10.1016/s1472-6483(10)60801-2. [DOI] [PubMed] [Google Scholar]
- 5.Delaney SJ, Rich DP, Thomson SA, Hargrave MR, Lovelock PK, Welsh MJ, Wainwright BJ. Cystic fibrosis transmembrane conductance regulator splice variants are not conserved and fail to produce chloride channels. Nat. Genet. 1993;4:426–431. doi: 10.1038/ng0893-426. [DOI] [PubMed] [Google Scholar]
- 6.Strong TV, Wilkinson DJ, Mansoura MK, Devor DC, Henze K, Yang Y, Wilson JM, Cohn JA, Dawson DC, Frizzell RA, et al. Expression of an abundant alternatively spliced form of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is not associated with a cAMP-activated chloride conductance. Hum. Mol. Genet. 1993;2:225–230. doi: 10.1093/hmg/2.3.225. [DOI] [PubMed] [Google Scholar]
- 7.Pagani F, Buratti E, Stuani C, Romano M, Zuccato E, Niksic M, Giglio L, Faraguna D, Baralle FE. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- 8.Buratti E, Stuani C, De Prato G, Baralle FE. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat. Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 10.Mak V, Jarvi KA, Zielenski J, Durie P, Tsui LC. Higher proportion of intact exon 9 CFTR mRNA in nasal epithelium compared with vas deferens. Hum. Mol. Genet. 1997;6:2099–2107. doi: 10.1093/hmg/6.12.2099. [DOI] [PubMed] [Google Scholar]
- 11.Rave-Harel N, Kerem E, Nissim-Rafinia M, Madjar I, Goshen R, Augarten A, Rahat A, Hurwitz A, Darvasi A, Kerem B. The molecular basis of partial penetrance of splicing mutations in cystic fibrosis. Am. J. Hum. Genet. 1997;60:87–94. [PMC free article] [PubMed] [Google Scholar]
- 12.Teng H, Jorissen M, Van Poppel H, Legius E, Cassiman JJ, Cuppens H. Increased proportion of exon 9 alternatively spliced CFTR transcripts in vas deferens compared with nasal epithelial cells. Hum. Mol. Genet. 1997;6:85–90. doi: 10.1093/hmg/6.1.85. [DOI] [PubMed] [Google Scholar]
- 13.Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, Goossens M, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J. Clin. Invest. 1998;101:487–496. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larriba S, Bassas L, Gimenez J, Ramos MD, Segura A, Nunes V, Estivill X, Casals T. Testicular CFTR splice variants in patients with congenital absence of the vas deferens. Hum. Mol. Genet. 1998;7:1739–1743. doi: 10.1093/hmg/7.11.1739. [DOI] [PubMed] [Google Scholar]
- 15.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. Embo J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niksic M, Romano M, Buratti E, Pagani F, Baralle FE. Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet. 1999;8:2339–2349. doi: 10.1093/hmg/8.13.2339. [DOI] [PubMed] [Google Scholar]
- 18.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 19.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 20.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell. Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 26.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell. Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 27.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 28.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladd AN, Cooper TA. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J. Cell. Sci. 2004;117:3519–3529. doi: 10.1242/jcs.01194. [DOI] [PubMed] [Google Scholar]
- 30.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 31.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 32.Gama-Carvalho M, Krauss RD, Chiang L, Valcarcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell. Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logette E, Le Jossic-Corcos C, Masson D, Solier S, Sequeira-Legrand A, Dugail I, Lemaire-Ewing S, Desoche L, Solary E, Corcos L. Caspase-2, a novel lipid sensor under the control of sterol regulatory element binding protein 2. Mol. Cell. Biol. 2005;25:9621–9631. doi: 10.1128/MCB.25.21.9621-9631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramalingam S, Natarajan G, Schafer C, Subramaniam D, May R, Ramachandran I, Queimado L, Houchen CW, Anant S. Novel intestinal splice variants of RNA-binding protein CUGBP2: isoform-specific effects on mitotic catastrophe. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G971–G981. doi: 10.1152/ajpgi.00540.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 36.Goo YH, Cooper TA. CUGBP2 directly interacts with U2 17S snRNP components and promotes U2 snRNA binding to cardiac troponin T pre-mRNA. Nucleic Acids Res. 2009;37:4275–4286. doi: 10.1093/nar/gkp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh G, Charlet BN, Han J, Cooper TA. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 2004;32:1232–1241. doi: 10.1093/nar/gkh275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Divangahi M, Balghi H, Danialou G, Comtois AS, Demoule A, Ernest S, Haston C, Robert R, Hanrahan JW, Radzioch D, et al. Lack of CFTR in skeletal muscle predisposes to muscle wasting and diaphragm muscle pump failure in cystic fibrosis mice. PLoS Genet. 2009;5:e1000586. doi: 10.1371/journal.pgen.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.