Abstract
Mitomycin C induces both MC-mono-dG and cross-linked dG-adducts in vivo. Interstrand cross-linked (ICL) dG-MC-dG-DNA adducts can prevent strand separation. In Escherichia coli cells, UvrABC repairs ICL lesions that cause DNA bending. The mechanisms and consequences of NER of ICL dG-MC-dG lesions that do not induce DNA bending remain unclear. Using DNA fragments containing a MC-mono-dG or an ICL dG-MC-dG adduct, we found (i) UvrABC incises only at the strand containing MC-mono-dG adducts; (ii) UvrABC makes three types of incisions on an ICL dG-MC-dG adduct: type 1, a single 5′ incision on 1 strand and a 3′ incision on the other; type 2, dual incisions on 1 strand and a single incision on the other; and type 3, dual incisions on both strands; and (iii) the cutting kinetics of type 3 is significantly faster than type 1 and type 2, and all of 3 types of cutting result in producing DSB. We found that UvrA, UvrA + UvrB and UvrA + UvrB + UvrC bind to MC-modified DNA specifically, and we did not detect any UvrB- and UvrB + UvrC–DNA complexes. Our findings challenge the current UvrABC incision model. We propose that DSBs resulted from NER of ICL dG-MC-dG adducts contribute to MC antitumor activity and mutations.
INTRODUCTION
Mitomycin C (MC) is a potent antitumor drug. Although this drug was discovered decades ago, it is still being actively used clinically, in combination with other antitumor drugs, for the treatment of advanced cancers (1–4). Upon entering cells MC is chemically reduced to a form that can react with deoxyguanosine (dG) residues in DNA to form a MC-mono-dG adduct. Mitomycin C can further form an intrastrand biadduct at –GG- sites and an interstrand cross-linked (ICL) dG-MC-dG lesion at a –CG- site (5). If not repaired, these DNA adducts, particularly ICL lesions, can block transcription and DNA replication and cause cell death (6). The antitumor activity of MC is generally believed to be derived from these interactions with DNA (7,8).
Both eukaryotes and prokaryotes have the capacity to repair MC-DNA adducts. The MC-mono-dG adducts are repaired by the nucleotide excision repair (NER) mechanism, which is similar to what occurs with other bulky DNA adducts, such as benzo(a)pyrene diol epoxide-dG adducts and cyclobutane pyrimidine dimers (CPD) photoproducts (9,10). We previously have shown that UvrABC nuclease, the NER enzyme complex in Escherichia coli cells, makes an incision 7–8 nt 5′ to and 3–4 nt 3′ to an MC-mono-dG adduct (10). It is likely that the intrastrand biadduct at –GG- sites is repaired in the same fashion as CPD and MC-mono-dG adducts, but the repair of ICL dG-MC-dG lesion is less clear. The current understanding of ICL lesions is derived primarily from results of studying UvrABC nuclease incision on ICL thymidine-psoralen-thymidine (dT-psoralen-dT) lesions (11,12). However, results from studies on UvrABC recognition and repair of ICL dT-psoralen-dT lesions are inconsistent; while van Houten et al. (11,12) have shown that the UvrABC nuclease makes dual incisions only at the DNA strand with the furan side of ICL dT-psoralen-dT lesion, Ramaswamy and Yeung (13) have demonstrated that the UvrABC nuclease can make incisions on both strands of this type of lesion. Sladek et al. (14,15) have demonstrated that the ICL dT-psoralen-dT adduct in a circular double-stranded DNA can be removed and repaired by the concerted actions of UvrABC incision, recA-mediated DNA strand invasion and DNA polymerase I-mediated repair synthesis.
An ICL dG-MC-dG has two distinct structural differences from an ICL dT-psoralen-dT: 1, dG residues are cross-linked instead of dT residues, and 2, the MC moiety sticks out of the minor groove and does not induce DNA bending (16), while the psoralen moiety is hidden within the base stacks and induces a 46.5° kink in the DNA helix (17). Similar to the ICL dG-MC-dG adduct, it has been shown that an MC-mono-dG adduct does not induce DNA bending (16). If an ICL dG-MC-dG adduct does not cause DNA bending then what are the signals for UvrABC to recognize this adduct as a DNA lesion? Since the 2 dGs in both DNA strands in this lesion are covalently bonded with the same MC molecule, how does UvrABC incise an ICL dG-MC-dG adduct?
To address these questions, we determined the recognition and incision of two types of MC–DNA lesions by UvrABC nuclease, using substrates of 61-bp DNA fragments containing either a site-specific ICL dG-MC-dG or MC-mono-dG adduct at the same site.
MATERIALS AND METHODS
Materials
The restriction enzymes SmaI and SnaBI were obtained from Roche. T4 polynucleotide kinase was obtained from DuPont New England Biolabs. Acrylamide, bis-acrylamide, APS and yeast tRNA were purchased from the Sigma Chemical Company. The γ-32P-ATP was purchased from Perkin Elmer.
Preparation of 61-mer containing a site-specific MC-mono-dG or an ICL dG-MC-dG adduct
15-mer containing a MC-mono-adduct
The 15-mer oligonucleotide [d(TCTATACGTAGAATT)] was annealed to a 9-mer [d(TAC5-MeGTATAG)] and then reacted with MC (4 μmol/ml) using a previously described method (18,19). The adducted duplexes were separated by high-performance liquid chromatography (HPLC).
61-mer duplex containing a MC-mono-dG
The 61-mer containing a site-specific MC-mono-dG adduct was constructed by ligating the MC-mono-adducted 15-mer, the phosphorylated 23-mer, [d(CCCATAGGAGAATTCCTACCCTA)], unphosphorylated 23-mer, [d(GCTCGGTACCCGGGGATATCCTC)] and a 39-mer scaffold oligonucleotide, [d(TTCTCCTATGGGAATTCTACGTATAGAGAGGATATCCCC)]. The ligated 61-mer was isolated by gel electrophoresis and stored at −20°C.
MC-cross-linked 61-mer duplex
The 61-bp fragments containing a site-specific ICL dG-MC-dG adduct were generated by hybridizing the MC-mono-adduct–containing 61-mer with its complementary 61-mer, and the duplexes underwent further MC cross-linking using previously described reaction conditions (18,19). The N2-dG-MC-N2-dG interstrand cross-link–containing 61-mer duplex was purified by PAGE.
Preparation of 32P-labeled DNA fragments
The single-stranded (SS) 61-mer fragment containing an MC-mono-dG adduct or its complementary SS 61-mer was dissolved in 1 × TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA), 5′-end labeled with γ-32P-ATP by T4 polynucleotide kinase (New England, BioLab), hybridized to each other, and then separated in a nondenaturing 8% polyacrylamide gel. The DS 61-bp fragments containing a site-specific ICL dG-MC-dG were 5′-end-labeled with γ-32P-ATP by T4 polynucleotide kinase at both ends; the DNA fragments were then digested with SmaI to generate single 5′-end 32P-labeled fragments.
UvrABC nuclease reactions
The UvrA, UvrB and UvrC proteins were purified as previously described (9). An aliquot of the 32P-labeled 61-bp DNA fragments (0.6–2.0 nM) was reacted with the UvrA, UvrB and UvrC proteins (15 nM each) in UvrABC reaction buffer (50 mM Tris–HCI, pH 7.5, 10 mM MgC12, 100 mM KC1, 1 mM ATP and 1 mM DTT) at 37°C for different time periods.
Kinetics of UvrABC incision
The 32P-labeled DS DNA fragments containing a site-specific MC adduct were pre-incubated with 15 nM UvrA and 15 nM UvrB at 37°C in UvrABC reaction buffer and the reaction was then started with the addition of UvrC (15 nM). The UvrABC reaction mixtures were sampled at different time intervals and the resultant DNAs were purified and separated in a 12% polyacrylamide denaturing or non-denaturing gel. The DS DNA markers (15, 30 and 46 bp) with and without an ICL dG-MC-dG lesion were generated by digestion of the 5′-end 32P-labeled 61-bp DNA fragments with EcoRV and SnaBI restriction enzymes. The intensity of the electrophoresis-separated bands was determined by scanning with a Packard CycloneTM Storage Phosphor System.
Detection of Uvr–DNA binding by gel mobility shift assay
The 61-bp DNA fragments with a site-specific MC-mono-dG or an ICL dG-MC-dG adduct were 32P 5′-end-labeled and reacted with Uvr proteins under three different conditions: (i) UvrA (15 nM) only for 5 min; (ii) UvrA and UvrB (15 nM each) for 5 min; and (iii) the same as in (ii) and then UvrC (15 nM) was added. The DNA amount for each reaction was 0.6–2.0 nM. The reactions were carried out in the same UvrABC reaction buffer used in the UvrABC incision reactions except bovine serum albumin (BSA) (0.1 mg/ml) was added to maintain the stability of the Uvr–DNA complexes. The presence of BSA did not affect the UvrABC incision activity. The mixtures were incubated at 37°C for 20 min and then separated in a 4.5% polyacrylamide gel containing 10 mM MgCl2 and 2 mM ATP by electrophoresis in 0.5 × TBE buffer the same as described by Delagoutte et al. (20) except that the electrophoresis was conducted at 2°C. After 1 h of electrophoresis, the gel was exposed to phosphor screen (30 min) and then rerun for 1 more hour under the same conditions. The DNA and protein–DNA complexes were analyzed using a Packard CycloneTM Starage Phosphor System. All gel retardation experiments were repeated at least three times.
Determination of the composition of Uvr-MC–modified DNA complexes
The Uvr–DNA complex bands separated by gel electrophoresis were cut, smashed and heated at 95°C for 5 min in the presence of 2% SDS to dissociate the Uvr proteins from the complexes. The resultant mixtures were then electrophoresed in an 8% SDS PAGE and stained with silver stain.
RESULTS
Construction of 61-bp DNA fragments containing a site-specific MC-mono-adduct or ICL dG-MC-dG adduct
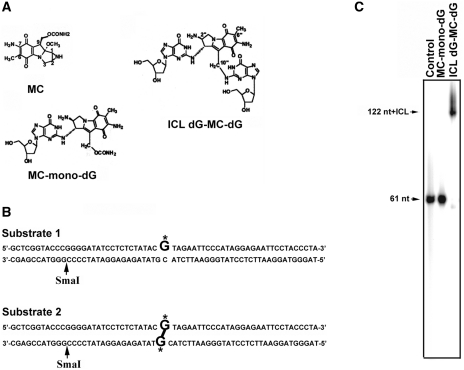
In order to obtain definitive UvrABC–DNA adduct interactions 61-bp DNA fragments containing a site-specific MC-mono-dG adduct (substrate 1) or an ICL dG-MC-dG adduct were constructed (substrate 2) (Figure 1B). The presence of an MC-mono-dG adduct and ICL-dG-MC-dG adduct in these DNA fragments was confirmed by enzymatic digestion followed by HPLC separation and by LC-ESI-MS analyses (18) (Supplements 1–3). The interstrand crosslink was further characterized by denatured electrophoresis. Results in Figure 1C show that the denatured 61-bp fragments that contain an ICL dG-MC-dG migrate significantly slower than the denatured 61-bp fragments that contain an MC-mono-dG, indicating that the two DNA strands in the former are covalently linked.
Figure 1.
(A) Chemical structures of MC, MC-mono-dG, and ICL dG-MC-dG DNA adducts. (B) The DNA sequence of the 61-bp DNA fragment containing a site-specific MC-mono-dG (substrate 1) or an ICL dG-MC-dG adduct (substrate 2). G* represents MC-modified deoxyguanosine. Restriction enzyme SmaI site is indicated. (C) Substrate 1, substrate 2 and control 61-bp DNA fragments were 5′-end 32P labeled, denatured and then separated by electrophoresis in a denaturing gel.
Recognition and incision of MC-mono-dG and ICL dG-MC-dG adducts by UvrABC
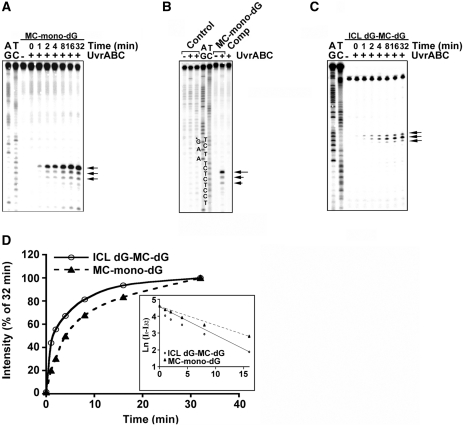
Previously, using DNA fragments modified with MC, we have shown that the NER enzyme complex UvrABC nuclease is able to recognize MC-mono-dG adducts and make dual incisions 7–8 nt 5′ to and 4–5 nt 3′ to the adduct (10,21). Since dG residues in both strands of these DNA fragments were modified with MC, our previous results did not rule out the possibility that MC-mono-dG adducts may induce UvrABC incision on the DNA strand opposite to the MC-mono-dG adduct. To test this possibility the 61-bp DNA fragments containing a site-specific MC-mono-dG adduct were 5′-end-32P labeled at either the MC-adduct strands (substrate 1) or the non-adducted strands as shown in Figure 1B, and then reacted with UvrABC nuclease for different time periods. The results in Figure 2A show that UvrABC incises the MC-mono-dG-containing strands as a function of incubation time and incisions mainly occur at 7 nt with minor incisions 8 and 9 nt, 5′ to the MC-dG; these results are consistent with previous findings (10,21). In contrast, UvrABC does not incise the complementary strands that do not contain an MC-mono-dG adduct even after a 60-min incubation period (Figure 2B).
Figure 2.
Kinetics of UvrABC incision of MC-mono-dG and ICL dG-MC-dG adducts. In (A) the MC-mono-dG–containing strand of substrate 1 was 5′-32P-end labeled. In (B) the MC-mono-dG–containing strand of substrate 1, the non-modified strand (Comp) of substrate 1, and control 61-bp fragments were 5′-32P-single-end labeled. In (C) substrate 2 was 32P-labeled at the 5′ end on both strands and digested with SmaI to generate single 5′-end-32P-labeled DNA fragments. The DNA fragments were incubated with UvrABC for different time periods (A and C), or for 60 min (B), and the resultant DNAs were separated by electrophoresis in a 12% polyacrylamide denaturing gel. (A–C) represent typical autoradiographs, and (D) represents the quantitations. The band intensities at different times (It) were normalized to the intensity at 32 min (I32), which encompasses 90% of total activity. The inset shows a semilogarithmic plot; the slopes yield two apparent first-order rate constants, Kobs,ABC, mono = 0.111 min−1 and Kobs,ABC, ICL = 0.169 min−1. Symbols: AG and TC represent Maxam and Gilbert sequencing reaction products (42); G*, MC-modified guanine; the arrows indicate the UvrABC incision bands; MC-mono-dG (filled triangle with dashed line); and ICL dG-MC-dG adducts (open circle with continous line).
In order to determine whether the NER mechanism can recognize and repair ICL dG-MC-dG adducts, the 61-bp DNA fragments that contain a site-specific ICL dG-MC-dG (substrate 2) were 32P-labeled on both 5′ ends, digested with SmaI to generate a single 5′-end 32P-labeled fragment and then incubated with UvrABC nucleases for different time periods. The results in Figure 2C show that these reactions generate a major 32P band that corresponds to the incision at 9 nt and also to minor incisions 8 and 10 nt 5′ to the lesion, and the incision is a function of the incubation time. These results indicate that UvrABC is able to recognize the ICL dG-MC-dG adduct and makes incisions mainly 9 nt 5′ to this lesion. To determine the cutting efficiency of UvrABC toward these MC-mono-dG and ICL dG-MC-dG adducts, we determined the kinetics of UvrABC incision on these two types of lesions. Results in Figure 2D show that UvrABC incises these two types of adducts with similar pseudo-first order kinetics and rate constants. Although the initial slopes of the two curves are different, we interpret these results as indicating that the two types of adducts are recognized and incised by the UvrABC nuclease with similar efficiencies.
UvrABC incision of an ICL dG-MC-dG adduct results in double-stranded DNA break
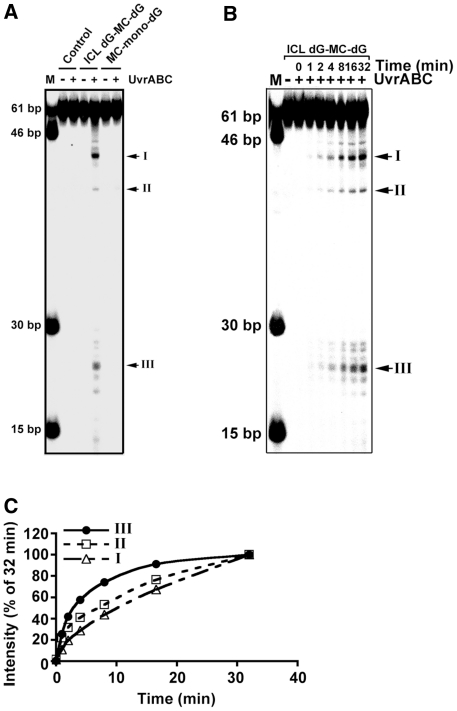
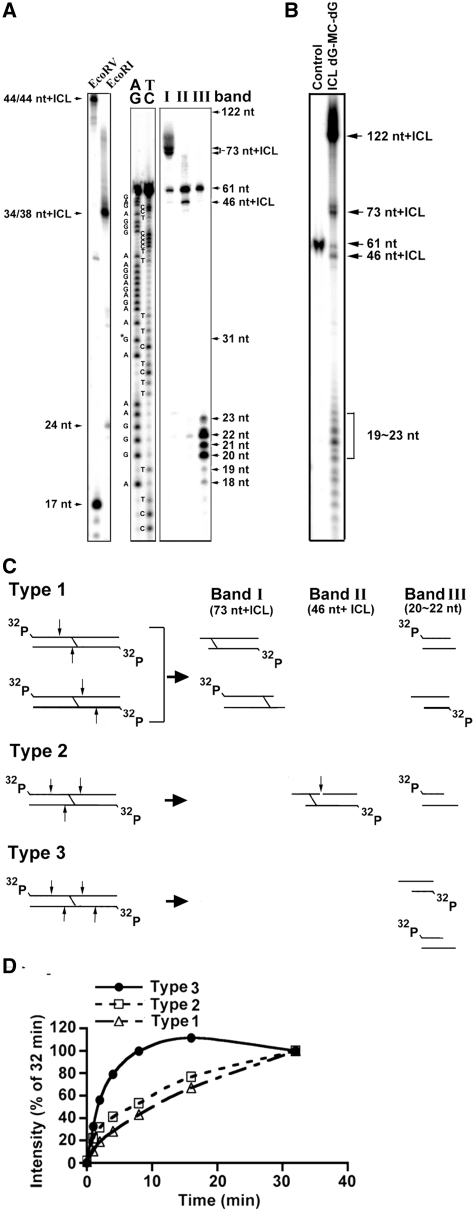
The results from LC-MS/MS analysis (Supplement 3) and gel electrophoresis (Figure 1C) indicate at the ICL dG-MC-dG lesion the two dGs at the two DNA strands are indeed covalently bonded by the same MC molecule. If UvrABC makes a dual incision at 5′ and 3′ of these two MC-bonded dG’s, similar to the dual incision seen with the MC-mono-dG adduct, then this type of incision should produce a DSB. To test this possibility, the 61-bp DNA fragments containing a site-specific ICL dG-MC-dG or a MC-mono-dG were labeled with 32P at both 5′ ends, incubated with UvrABC nuclease for different times, and the resultant DNAs were separated by electrophoresis in a non-denaturing gel. The results in Figure 3A show that while UvrABC incision of MC-mono-dG fragments does not produce any shorter DNA fragments than the original 61-bp DNA fragments, UvrABC incision of ICL dG-MC-dG fragments produces three major DNA fragments I (42 bp), II (40 bp) and III (22–24 bp). Results in Figure 3B show that the formation of these three bands is a function of incubation time and no intermediate products were produced besides the end products observed after very short as well as after very long periods of incubation. Kinetic analysis shows that the rate constant of band III formation is significantly larger than Band I and Band II. These results indicate that once the UvrABC–ICL dG-MC-dG complex is formed the pattern of UvrABC incision is determined. To identify the nature of these three bands, DNA in each was isolated, denatured and separated by electrophoresis in a 12% denaturing gel. The results in Figure 4A show that the size of band I is ∼73 nt + ICL, band II is ∼46 nt + ICL and band III is 20–23 nt. It should be noted that all three bands contain a band with a size that corresponds to a single-stranded 61-nt fragment. We believe this single-stranded 61-nt fragment is a contaminant originating from the double-stranded DNA construction. To exclude the trivial possibility that the DNA isolation process may generate different size of DNA fragments, UvrABC incised ICL dG-MC-dG fragments were denatured and separated by electrophoresis in a denatured gel. Results in Figure 4B show the same four DNA fragments (73 nt + ICL, 61 nt, 46 nt + ICL and 20–23 nt) were observed indicating that these DNA fragments were indeed the UvrABC incision products.
Figure 3.
Double-stranded DNA break formation resulted from UvrABC incision of an ICL dG-MC-dG adduct. (A) 32P-labeled substrate 1, substrate 2 and control 61-bp DNA fragments were reacted with UvrABC for 60 min and then separated in a nondenaturing polyacrylamide gel. (B) The kinetics of DSB formation resulted from UvrABC incision of an ICL dG-MC-dG DNA adduct. The 5′-end-32P-labeled 61-bp DNA fragments containing a site-specific ICL dG-MC-dG–DNA adduct were incubated with UvrABC nucleases as described in Figure 2. At different time periods of incubation, the resultant DNAs were separated in a nondenaturing polyacrylamide gel. The double-stranded DNA fragments standards are shown in lane M. (C) Quantitations. The band intensities at different times (It) shown in (B) were normalized to the intensity at 32 min (I32), which encompasses 90% of total activity. The data were plotted the same as in Figure 2.
Figure 4.
Identification of the products resulting from UvrABC incision of the 61-bp DNA fragments containing an ICL dG-MC-dG–DNA adduct. (A) The three major bands (I, II and III) as shown in Figure 3 were extracted and separated by electrophoresis in a 12% polyacrylamide denaturing gel as described in Figure 2. Note: the DNA fragments in band I resulted in a band corresponding to size of ∼73 nt + ICL, the DNA fragments in band II resulted in a band corresponding size of ∼46 nt + ICL, and the DNA fragments in band III resulted a band of size ∼22-nt bands. The size standards were generated using the 61– bp DNA fragment containing an ICL dG-MC-dG adduct cut with EcoRI or EcoRV. The 61-nt band results from an unreacted 61-mer contamination during the strand construction. AG and TC represent Maxam and Gilbert sequencing reaction products (42). (B) 32P-labeled 61-bp DNA fragments containing an ICL dG-MC-dG DNA adduct were incubated with UvrABC for 32 min and the resultant DNAs were separated by electrophoresis the same as in (A). (C) The three possible types (1, 2 and 3) of UvrABC incision on the double-stranded 61-bp fragments containing a site-specific ICL dG-MC-dG DNA adduct that would result in generating fragments of the following approximate sizes: 73 nt + ICL, 46 nt + ICL and 22 nt. The arrows indicate the UvrABC cutting sites. For clarity, only the major incision positions are shown. It should be noted that type 1, 2 and 3 incisions resulted in producing DSB. (D) The kinetics of the three types of UvrABC cutting on the ICL dG-MC-dG lesion. The calculations were based on the band intensity shown in Figure 3B and the cutting models presented in Figure 4C.
The pattern of UvrABC incision on of 61-bp DNA fragments containing a site-specific ICL dG-MC-dG adducts
Based on the results shown in Figures 2–4B, we constructed the possible patterns of UvrABC incision on the double-end 32P-labeled 61-bp DNA fragments containing a site-specific ICL dG-MC-dG (Figure 4C). It appears that UvrABC makes three types of incisions on this type of adduct: type 1, a single incision at 8–10 nt 5′ to the ICL dG-MC-dG on 1 strand and 4 nt 3′ to the other strand; type 2, dual incisions on 1 strand and a single incision 3′ to the ICL dG-MC-dG; and type 3, dual incisions at 8–10 nt 5′ to and 4–5 nt 3′ to the ICL dG-MC-dG on both strands. The type 1 cut generates two 32P-labeled DNA duplexes: a 24-/27-nt and a 34-/39-nt, with the latter containing the ICL dG-MC-dG adduct. The type 2 cut generates two 32P-labeled DNA duplexes: a 24-/27-nt and a 34-/39-nt, and the latter contains the ICL dG-MC-dG adduct, just as with type 1. However with the added incision site, a single-strand break is generated, resulting in a 46-nt fragment containing the adduct. The type 3 cut generates one kind of 32P-labeled DNA duplex: 24-/27-nt. All three types of incision result in generation of double-stranded DNA break (DSB). Based on the band intensity shown in Figure 3B and the models of cutting presented in Figure 4C we calculated the kinetics and the distribution of the three types of UvrABC cutting. The results indicate: (i) the rate constant of type 3 cutting is significantly higher than type 1 and type 2 cutting (Figure 4D), and (ii) type 1 and type 3 cuttings (40% each) are more frequent than type 2 cutting (20%).
Binding of MC-mono-dG and ICL dG-MC-dG adducts by Uvr proteins
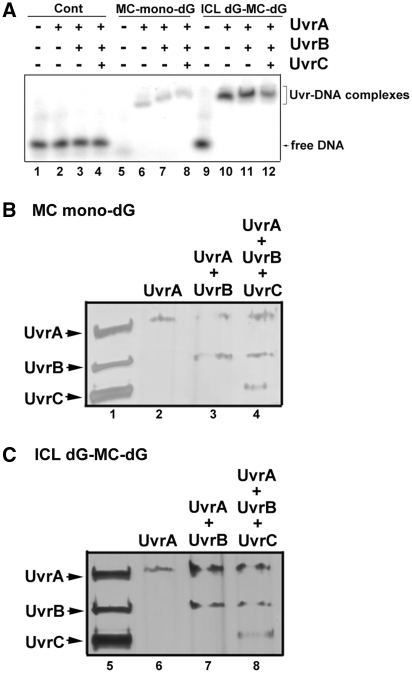
Using a site-specific cholesterol-dG as a substrate it has been found that the sequential steps of binding and incision of the lesion by the three Uvr proteins are as follows: (i) a dimerized UvrA binds to the lesion to form a (UvrA)2–DNA lesion complex; (ii) the complex attracts a UvrB, which binds to form a (UvrA)2(UvrB)–lesion complex; (iii) UvrA is released from the complex and a UvrC joins in to form a (UvrB)(UvrC)–lesion complex and triggers dual incisions at both the 5′ and 3′ sides of the DNA lesion (22–25). While this Uvr–DNA interaction model can account for the dual UvrABC incision on a MC-mono-dG adduct, it cannot account for the type 2 and type 3 UvrABC incisions on an ICL dG-MC-dG adduct. The type 2 and type 3 UvrABC incisions on an ICL dG-MC-dG adduct raise the possibility that UvrABC may form a complex or complexes with this type of adduct that is different from the ones that are formed with the MC-mono-dG adduct. To test this possibility, we determined the binding pattern of Uvr proteins UvrA, UvrA + UvrB and UvrA + UvrB + UvrC with the 61-bp DNA fragments containing a MC-mono-dG adduct or an ICL dG-MC-dG under the incision reaction conditions. The results in Figure 5A show that: (i) Uvr proteins do not bind to undamaged control DNA fragments significantly; this result is distinctly different from other reports that show a significant amount of Uvr binding to undamaged DNA fragments (26,27). (ii) UvrA binds to MC-mono-dG, resulting in a single band, which is indicative of the formation of a (UvrA)2(MC-mono-dG) complex since it is well established that UvrA dimerizes in UvrABC reaction buffer (22–25,28). (iii) Addition of UvrB results in a different, slower moving band, representing the formation of a (UvrA)2(UvrB)(MC-mono-dG) complex. (iv) Addition of UvrC protein results in the generation of a further retarded band, indicating the formation of a (UvrA)2(UvrB)(UvrC)(MC-mono-dG) complex.
Figure 5.
The binding pattern of Uvr proteins with 32P-labeled DS 61-bp DNA fragments with and without a site-specific MC-mono-dG or an ICL dG-MC-dG adduct. In (A) the DNA fragments were incubated with UvrA, UvrA + UvrB and UvrA + UvrB + UvrC for 20 min and the resultant complexes were separated by electrophoresis in a 4.5% non-denaturing polyacrylamide gel containing 50 mM MgCl2 and 1 mM ATP at 2°C in 0.5 × TBE buffer for 2 h (lanes 1–12). The Uvr–DNA complexes are: (UvrA)2–DNA (lane 6); (UvrA)2-(UvrB)–DNA (lane 7); (UvrA)2-(UvrB)-(UvrC)–DNA (lane 8); (UvrA)4–DNA (lane 10); (UvrA)4-(UvrB)2–DNA (lane 11) and (UvrA)4-(UvrB)2-(UvrC)2–DNA (lane 12). To determine the Uvr protein composition in the Uvr–DNA complexes resulting from Uvr-bound DS 61-bp DNA fragments containing a site-specific (B) MC-mono-dG or (C) ICL dG-MC-dG adduct, bands isolated from Figure 5A were denatured by heat (95°C in 2% SDS for 5 min), separated in an 8% SDS-PAGE, and stained with silver stain. The Uvr proteins added to the DNA fragments that resulted in the different retarded bands are shown on the top of the panel. In (B) lane 2 is from lane 6 of Figure 5A; lane 3 is from lane 7 of Figure 5A; lane 4 is from lane 8 of Figure 5A In (C) lane 6 is from lane 10 of Figure 5A; lane 7 is from lane 11 of Figure 5A; lane 8 is from lane 12 of Figure 5A. Lanes 1 and 5 are the UvrA, UvrB and UvrC standards. Note: The complexes in each band contain all the added Uvr protein (i.e. UvrA was found in the MC-modified DNA added with UvrA; UvrA and UvrB were found in MC-modified DNA added with these two proteins; and UvrA, UvrB and UvrC were found in the MC-modified DNA added with these three proteins).
We found that all Uvr proteins bound to the 61-bp DNA fragments containing an ICL dG-MC-dG adduct, and all the resultant complexes moved more slowly than the corresponding Uvr proteins bound to 61-bp fragments containing a MC-mono-dG adduct (Figure 5A). The mobility of the ICL dG-MC-dG 61-bp DNA fragments bound to UvrA is significantly slower than the mobility of the corresponding MC-mono-dG DNA fragment bound to UvrA; we interpret this to mean that two dimerized UvrA proteins bind to the ICL dG-MC-dG lesion [(UvrA)2-(UvrA)2-ICL dG-MC-dG] while only 1 dimerized UvrA protein binds to the MC-mono-dG lesions [(UvrA)2-MC-mono-dG]. Addition of UvrB slightly retarded mobility further, suggesting the formation of a (UvrA)2(UvrB)-(UvrA)2(UvrB)-ICL dG-MC-dG complex. Addition of UvrC resulted in further mobility retardation of the complex, and we interpret this to being due to the formation of a (UvrA)2(UvrB)(UvrC)-(UvrA)2(UvrB)(UvrC)-ICL dG-MC-dG complex.
To determine the composition of the Uvr-MC-mono-dG and Uvr-ICL-dG-MC-dG complexes the bands in Figure 5A were isolated and the Uvr proteins in the bands were further separated by SDS-PAGE. The results in Figure 5B and C show that adding UvrA, UvrA + UvrB and UvrA + UvrB + UvrC to the MC-modified DNA, the Uvr proteins remain binding at damaged DNA. These results indicate that UvrABC incisions of MC-mono-dG and ICL-dG-MC-dG adducts require the interactions and presence of all three Uvr proteins.
DISCUSSION
The results presented here are consistent with a model in which UvrABC incision of an ICL dG-MC-dG leads to DSB formation because UvrABC makes single and dual incisions 5′ and 3′ of an ICL dG-MC-dG adduct on both DNA strands. Our results indicate that a significant amount of UvrABC incisions of ICL dG-MC-dG result in DSB. The underlying mechanisms of NER are conserved from prokaryotic cells to human cells (29–31). It has been found that the initial incision step of NER in human cells is similar to NER in E. coli cells, except that in human cells 28–32 nt are excised, including the damaged base(s) (31). If the NER mechanism in human cells for ICL dG-MC-dG is similar to NER in E. coli cells, then NER should produce DSB in human cells and NER of MC-induced DNA damage therefore would contribute to MC antitumor activity since an unrepaired DSB is a potential lethal event.
Our results suggest that the action of the UvrABC complex on the ICL dG-MC-dG substrate results in excision of a 12–13-bp DNA fragment. In human cells, we expect NER of ICL dG-MC-dG will result in the excision of a 24–32-bp DNA segment. If this type of DSB is repaired by the end-to-end joining mechanism in situ then it would result in a small deletion. If the DSB causes a chromosome translocation, it would also result in a deletion in the translocated chromosome. In either case, an ICL dG-MC-dG–induced, NER-mediated, end-to-end joining event is mutagenic and this event probably contributes to the secondary tumorigenesis that is very often found in patients after long-term use of MC (32).
In contrast with our finding that UvrABC incision of the ICL dG-MC-dG adduct results in DSB, using DNA fragments containing a site-specific ICL dT-psoralen-dT lesion van Houten et al. (11,12) found that UvrABC makes a dual incision only on the strand that interacts with the furan moiety of psoralen and does not cause a DSB. Moreover, while Pitte and Hearst (33) found that a dT-psoralen-dT ICL causes a 46.5° kink, Rink et al. (16) found that both MC-mono-dG and ICL dG-MC-dG lesions do not cause DNA bending. Given these contrasting results, it is likely that the differing effects on DNA secondary structure caused by these two different ICL lesions determines the mode of UvrABC incision. While our work was in progress Peng et al. (34) and Sczepanski et al. (35) reported that UvrABC incision of ICL dT-dA results in both single-stranded DNA breaks and DSBs.
Two distinct models of how UvrABC recognizes and incises DNA damage have been proposed (22–25,28). Works from Grossman’s laboratory (28) supported the model that suggests dimerized UvrA form a (UvrA)2UvrB complex that localizes and binds to the DNA lesion; UvrC then joins the complex and triggers dual incisions. On the other hand, works from Sancar (22), Van Houten and McCullough (23), Moolenaar et al.(24) and Fuchs and Seeberg (36), support the model that suggest UvrA locates the damaged region and attracts UvrB binding. After UvrB binding, UvrA is released from the damaged region (20,22–25). The reason for the disparate results from these laboratories that lead to two models to account for UvrABC incision is unclear. One possibility is due that the DNA lesions used as UvrABC substrates and the method to assess Uvr–DNA lesion complex formation by Grossman’s laboratory (28) are different from Sancar (22), Van Houten and McCullough (23), Moolenaar et al. (24) and Fuchs and Seeberg (36). Our current results using MC–DNA adducts as UvrABC substrate are consistent with Grossman and Yeung’s model (28). There is evidence that DNA wraps around UvrB and causes strand separation in the damaged region (37,38). UvrC then joins the UvrB–DNA damage complex and triggers strand incision (23–25,29). The role of strand separation for UvrB and UvrC incision is clear for mono-DNA adduct lesions but is unclear for ICL lesions. The dual incision on the outward furan-linked strand containing an ICL dT-psoralen-dT lesion suggests that helix kinking of the DNA wrapped around UvrB plays a crucial role. If this is the case, then it is expected that the UvrABC incision of an ICL dG-MC-dG lesion will be different since this type of lesion does not cause DNA kinking or bending and either DNA strand can wrap around UvrB molecules. The signal for UvrA binding at this ICL lesion probably comes from the changing helix fluidity caused by this type of lesion (39). Our gel retardation results indicate that UvrA remains bound to the MC-modified DNA fragments after addition of UvrB and UvrC proteins; this differs from the results of van Houten et al. (11,12), who showed that upon UvrB binding UvrA was released from the complex. In addition and in contrast to their results, we did not observe a significant amount of UvrA, (UvrA + UvrB) or (UvrA + UvrB + UvrC) binding on control DNA, indicating that the Uvr-MC modified DNA complexes we observed are productive complexes that eventually lead to UvrABC incisions.
It appears that the recognition and binding of the ICL dG-MC-dG site by Uvr proteins do not follow a strict sequence; if they did, a single type of UvrABC excision would be observed, as we found for interaction of UvrABC with MC-mono-dG adducts. Instead, we observed three 32P-labeled fragments resulting from UvrABC incision of an ICL dG-MC-dG adduct. The possible models to account for these incisions are many. However, the gel retardation results in Figure 5A show that the UvrA-dG-MC-dG complex band moves more slowly than the UvrA-MC-mono-dG complex band. These results suggest that more than 1 dimerized UvrA binds at ICL dG-MC-dG–adducted DNA, assuming that 1 dimerized UvrA binds at UvrA-MC-mono-dG–adducted DNA. We propose that two dimerized UvrA proteins bind at each strand of an ICL dG-MC-dG-adducted DNA with the same polarity relative to the lesion. These two dimerized UvrA–DNA complexes can attract two sets of UvrB and UvrC, and three types of incision could then be triggered: (i) dual 5′ and 3′ incisions on both strands (type 3); (ii) dual incisions on 1 strand and 3′ single incision on the other strand (type 2); and (iii) a single 5′ incision on 1 strand and a single 3′ incision on the other (type 1), as depicted in Figure 4. Although, in general, UvrABC makes dual incisions 5′ and 3′ to bulky DNA damage, uncoupled incision 5′ to or 3′ to DNA damage has also been observed (40,41).
Our results suggest that the type 1 and type 3 incisions for ICL dG-MC-dG are the most common UvrABC repair mechanisms for ICL lesions, which result in the generation of either 1 or 2 DSB, respectively. A significant fraction of UvrABC repair is through type 2 incisions, which also generate 1 DSB. It is unclear what factors determine what type of incision occurs. Since we only have observed these three types of incision bands and not intermediate bands, we suggest that once the Uvr protein–ICL-dG-MC-dG complexes are formed they are not interchangeable, and once a particular form of the complex is formed at the ICL lesion it precludes formation of other forms of the complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (CA114541, ES014641, CA99007 and ES00260); National Institute of Environmental Health (ES09127 and ES013324); National Cancer Institute (CA101864). Funding for open access charge: National Institutes of Health (CA114541 and ES00260).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Michael Patrick and Yen-Yee Nydam for critical review of this manuscript. We thank Yong Jiang (University of California, Riverside) for MS analyses.
REFERENCES
- 1.Kusenda Z, Kerger J, Awada A, Geurs F, Van Vreckem A, Habboubi N, Piccart MJ. Mitomycin C and vinblastine in combination with amifostine in metastatic breast cancer. A feasibility study of the EORTC–Investigational Drug Branch for Breast Cancer (IDBBC) Support Care Cancer. 1997;5:414–416. doi: 10.1007/s005200050101. [DOI] [PubMed] [Google Scholar]
- 2.Lenaz L. Mitomycin C in advanced breast cancer. Cancer Treat. Rev. 1985;12:235–249. doi: 10.1016/0305-7372(85)90007-6. [DOI] [PubMed] [Google Scholar]
- 3.Bolenz C, Cao Y, Arancibia MF, Trojan L, Alken P, Michel MS. Intravesical mitomycin C for superficial transitional cell carcinoma. Expert Rev. Anticancer Ther. 2006;6:1273–1282. doi: 10.1586/14737140.6.8.1273. [DOI] [PubMed] [Google Scholar]
- 4.Snegovoy A, Manzuk L, Artamonova E. Mitomycin plus capecitabine in metastatic breast cancer. 2008 Breast Cancer Symposium. 2008 General Poster Session E, Abstract No: 264. [Google Scholar]
- 5.Bizanek R, McGuinness BF, Nakanishi K, Tomasz M. Isolation and structure of an intrastrand cross-link adduct of mitomycin C and DNA. Biochemistry. 1992;31:3084–3091. doi: 10.1021/bi00127a008. [DOI] [PubMed] [Google Scholar]
- 6.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 7.Iyer VN, Szybalski W. Mitomycins and porfiromycin: chemical mechanism of activation and cross-linking of DNA. Science. 1964;145:55–58. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- 8.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation of DNA as the molecular basis of their activity. Pharmacol. Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 9.Tang M-s. Mapping and quantification of bulky chemical-induced DNA damage using UvrABC nucleases. Technol. Detect. DNA Damage Mutat. 1996;Chapter 11:139–152. [Google Scholar]
- 10.Li VS, Reed M, Zheng Y, Kohn H, Tang M-s. C5 cytosine methylation at CpG sites enhances sequence selectivity of mitomycin C-DNA bonding. Biochemistry. 2000;39:2612–2618. doi: 10.1021/bi991307h. [DOI] [PubMed] [Google Scholar]
- 11.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl Acad. Sci. USA. 1986a;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Houten B, Gamper H, Hearst JE, Sancar A. Construction of DNA substrates modified with psoralen at a unique site and study of the action mechanism of ABC excinuclease on these uniformly modified substrates. J. Biol. Chem. 1986b;261:14135–14141. [PubMed] [Google Scholar]
- 13.Ramaswamy M, Yeung AT. Sequence-specific interactions of UvrABC endonuclease with psoralen interstrand cross-links. J. Biol. Chem. 1994;269:485–492. [PubMed] [Google Scholar]
- 14.Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 1989a;264:6755–6765. [PubMed] [Google Scholar]
- 15.Sladek FM, Melian A, Howard-Flanders P. Incision by UvrABC excinuclease is a step in the path to mutagenesis by psoralen crosslinks in Escherichia coli. Proc. Natl Acad. Sci. USA. 1989b;86:3982–3986. doi: 10.1073/pnas.86.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rink SM, Lipman R, Alley SC, Hopkins PB, Tomasz M. Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem. Res. Toxicol. 1996;9:382–389. doi: 10.1021/tx950156q. [DOI] [PubMed] [Google Scholar]
- 17.Pearlman DA, Holbrook SR, Pirkle DH, Kim SH. Molecular models for DNA damaged by photoreaction. Science. 1985;227:1304–1308. doi: 10.1126/science.3975615. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. Structure elucidation of DNA interstrand cross-link by a combination of nuclease P1 digestion with mass spectrometry. Anal. Chem. 2003;75:6306–6313. doi: 10.1021/ac034683n. [DOI] [PubMed] [Google Scholar]
- 19.Borowy-Borowski H, Lipman R, Chowdary D, Tomasz M. Duplex oligodeoxyribonucleotides cross-linked by mitomycin C at a single site: synthesis, properties, and cross-link reversibility. Biochemistry. 1990;29:2992–2999. doi: 10.1021/bi00464a015. [DOI] [PubMed] [Google Scholar]
- 20.Delagoutte E, Bertrand-Burggraf E, Lambert IB, Fuchs RP. Binding and incision activities of UvrABC excinuclease on slipped DNA intermediates that generate frameshift mutations. J. Mol. Biol. 1996;257:970–976. doi: 10.1006/jmbi.1996.0216. [DOI] [PubMed] [Google Scholar]
- 21.Kohn H, Li VS, Tang M-S. Recognition of mitomycin C-DNA monoadducts by UVRABC nuclease. J. Am. Chem. Soc. 1992;114:5501–5509. [Google Scholar]
- 22.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 23.Van Houten B, McCullough A. Nucleotide excision repair in E. coli. Ann. NY Acad. Sci. 1994;726:236–251. doi: 10.1111/j.1749-6632.1994.tb52822.x. [DOI] [PubMed] [Google Scholar]
- 24.Moolenaar GF, Herron MF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N. The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J. Biol. Chem. 2000;275:8044–8050. doi: 10.1074/jbc.275.11.8044. [DOI] [PubMed] [Google Scholar]
- 25.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 26.Gordienko I, Rupp WD. A specific 3' exonuclease activity of UvrABC. EMBO J. 1998;17:626–633. doi: 10.1093/emboj/17.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caron PR, Grossman L. Incision of damaged versus nondamaged DNA by the Escherichia coli UvrABC proteins. Nucleic Acids Res. 1988;16:7855–7865. doi: 10.1093/nar/16.16.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman L, Yeung AT. The UvrABC endonuclease system of Escherichia coli–a view from Baltimore. Mutat. Res. 1990;236:213–221. doi: 10.1016/0921-8777(90)90006-q. [DOI] [PubMed] [Google Scholar]
- 29.Friedberg EC, Walker GC, Siede W. Nucleotide excision repair in prokaryotes. DNA Repair Mutagen. 1995;Chapter 5:191–225. [Google Scholar]
- 30.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 31.Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 32.Verweij J, Pinedo HM. Mitomycin C: mechanism of action, usefulness and limitations. Anticancer Drugs. 1990;1:5–13. [PubMed] [Google Scholar]
- 33.Piette J, Hearst J. Sites of termination of in vitro DNA synthesis on psoralen phototreated single-stranded templates. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1985;48:381–388. doi: 10.1080/09553008514551381. [DOI] [PubMed] [Google Scholar]
- 34.Peng X, Ghosh AK, Van Houten B, Greenberg MM. Nucleotide excision repair of a DNA interstrand cross-link produces single- and double-strand breaks. Biochemistry. 2010;49:11–19. doi: 10.1021/bi901603h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Double-strand break formation during nucleotide excision repair of a DNA interstrand cross-link. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs RP, Seeberg E. pBR322 plasmid DNA modified with 2-acetylaminofluorene derivatives: transforming activity and in vitro strand cleavage by the Escherichia coli uvrABC endonuclease. EMBO J. 1984;3:757–760. doi: 10.1002/j.1460-2075.1984.tb01880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhoeven EE, Wyman C, Moolenaar GF, Hoeijmakers JH, Goosen N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 2001;20:601–611. doi: 10.1093/emboj/20.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Lu M, Tang M-s, Van Houten B, Ross JB, Weinfeld M, Le XC. DNA wrapping is required for DNA damage recognition in the Escherichia coli DNA nucleotide excision repair pathway. Proc. Natl Acad. Sci. USA. 2009;106:12849–12854. doi: 10.1073/pnas.0902281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter RB, Pierce J, Case R, Tang M-s. Recognition of the DNA helix stabilizing anthramycin-N2 guanine adduct by UVRABC nuclease. J. Mol. Biol. 1988;203:939–947. doi: 10.1016/0022-2836(88)90119-2. [DOI] [PubMed] [Google Scholar]
- 40.Tang M-s, Lee CS, Doisy R, Ross L, Needham-VanDevanter DR, Hurley LH. Recognition and repair of the CC-1065-(N3-adenine)-DNA adduct by the UVRABC nucleases. Biochemistry. 1988;27:893–901. doi: 10.1021/bi00403a009. [DOI] [PubMed] [Google Scholar]
- 41.Nazimiec M, Lee CS, Tang YL, Ye X, Case R, Tang M-s. Sequence-dependent interactions of two forms of UvrC with DNA helix-stabilizing CC-1065-N3-adenine adducts. Biochemistry. 2001;40:11073–11081. doi: 10.1021/bi010953p. [DOI] [PubMed] [Google Scholar]
- 42.Maxam AM. Sequencing the DNA of recombinant chromosomes. Fed. Proc. 1980;39:2830–2836. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.