Abstract
Solid-phase hybridization, i.e. the process of recognition between DNA probes immobilized on a solid surface and complementary targets in a solution is a central process in DNA microarray and biosensor technologies. In this work, we investigate the simultaneous effect of monovalent and divalent cations on the hybridization of fully complementary or partly mismatched DNA targets to DNA probes immobilized on the surface of a surface plasmon resonance sensor. Our results demonstrate that the hybridization process is substantially influenced by the cation shielding effect and that this effect differs substantially for solid-phase hybridization, due to the high surface density of negatively charged probes, and hybridization in a solution. In our study divalent magnesium is found to be much more efficient in duplex stabilization than monovalent sodium (15 mM Mg2+ in buffer led to significantly higher hybridization than even 1 M Na+). This trend is opposite to that established for oligonucleotides in a solution. It is also shown that solid-phase duplex destabilization substantially increases with the length of the involved oligonucleotides. Moreover, it is demonstrated that the use of a buffer with the appropriate cation composition can improve the discrimination of complementary and point mismatched DNA targets.
INTRODUCTION
Solid-phase hybridization, in which nucleic acid strands tethered to a solid support (DNA oligonucleotide ‘probes’) bind DNA molecules from a solution (‘targets’), represents a key process in DNA microarray and biosensor technologies. In the last two decades, these technologies have made great advances and have been applied in a number of important areas such as genotyping, gene expression profiling and biological detection (1–3). Although the underlying hybridization process is fairly well understood in bulk solution, the understanding of solid-phase hybridization is much less developed and is therefore highly desired (1,4).
The solid–liquid interfacial environment is substantially different from the bulk solution. Typically, DNA probe layers are characterized by a surface density of 1012–1013 probes/cm2 and a layer thickness of several nanometers (1,5). These values correspond to a local concentration of oligonucleotides of ∼3–30 mM, which is much higher than the typical levels used in hybridization experiments in a solution. As the oligonucleotides are negatively charged, not only steric but also electrostatic effects play an important role in the solid-phase hybridization processes. Therefore, it is not surprising that the results of solid-phase hybridization experiments often deviate substantially from the models established for hybridization in a solution (1,6–8).
One of the key factors affecting the electrostatic field of the surface with the immobilized negatively-charged probes, and thus solid-phase hybridization, is the presence of cations. In general, cations compensate for the negative charge of the oligonucleotide backbone and stabilize the oligonucleotide complexes. It is anticipated that the shielding effect of cations will be more pronounced in solid-phase hybridization in comparison with hybridization in a solution due to the high surface density of negatively charged probes in the layer arrangement. Due to the need for cation shielding to compensate for the negative charge of closely spaced probes, DNA duplexes have been shown to exhibit lower stability in solid-phase than in solutions with the same cation content (1,7,8). Several experimental and theoretical works have demonstrated repulsion of the immobilized probes and its destabilizing effect on duplexes and related them with the cation shielding (1,9–11). Peterson et al. (11) and Yu et al. (9) reported that the hybridization efficiency (HE) decreases with an increase of the surface probe density. Peterson et al. (11) showed that the yield of the probe immobilization on the surface increases with an increase of the concentration of monovalent sodium in the immobilization buffer. The dependence of the HE on sodium concentration in the range of 0–500 mM was studied in the work of Okahata et al. (10). In agreement with the electrostatic theory, it was determined that the HE increases with an increase of the sodium concentration. Recently Cho et al. (12) used a surface plasmon resonance (SPR) sensor to demonstrate that the key factor influencing the probe density and subsequent hybridization is the total concentration of sodium, whereas the type of buffer had only a minor effect. Vainrub and Pettitt (13) developed a macroscopic theoretical model considering the repulsion between the charged immobilized probe layer and the target as the main factor influencing the hybridization yield and the binding kinetics on a DNA chip. This model was subsequently improved by Halperin et al. (14) who treated the immobilized probes as a 3D layer. In summary, the experimental and theoretical works dealing with the cation effect on solid-phase hybridization published so far have focused more on probe density, while the role of different types and concentrations of cations has been addressed to a lesser extent.
In contrast to the case of solid-phase hybridization, numerous models and empirical formulas have been developed to describe oligonucleotide hybridization in a solution and used to determine thermodynamic parameters of oligonucleotide hybridization in the presence of monovalent and divalent cations (15–20). SantaLucia et al. (18,19) employed a nearest-neighbor model to predict a duplex stability for a wide range of sodium concentrations. Recently Owczarzy et al. (17), based on the analysis of a large set of UV absorption spectra, proposed an empirical formula for oligonucleotide duplexes enabling the prediction of melting temperatures, transition enthalpies, entropies and free energies in buffers containing magnesium and monovalent cations. The most advanced theoretical model, referred to as the tightly bound ion (TBI) model, was proposed by Tan and Chen to describe cation–oligonucleotide interactions in a solution (20,21). This model employs separate treatments for tightly bound ions and for diffuse ions surrounding DNA molecules.
In this work, we present for the first time the study of the simultaneous effect of sodium (monovalent) and magnesium (divalent) ions on the solid-phase hybridization of fully complementary and partly mismatched DNA targets to densely immobilized probes on a solid surface. The shielding effect of cations on hybridization is investigated with respect to both the cation type and concentration using an SPR sensor and the results are compared with the results obtained in a solution using UV absorption spectroscopy.
MATERIALS AND METHODS
Oligonucleotides
A biotinylated 23-mer DNA oligonucleotide (BdO23) of the sequence biotin-(TEG)2-5′-d(CAG TGT GGA AAA TCT CTA GCA GT)-3′ was used as a DNA oligonucleotide probe. This base sequence plays a crucial role in the replication of the human immunodeficiency virus (HIV) (22). The following DNA oligonucleotides were used as targets: (i) the fully matched target 5′-d(ACT GCT AGA GAT TTT CCA CAC TG)-3′ (CdO23), (ii) the partly mismatched target containing three purine–pyrimidine mismatches (the mismatched bases are underlined) 5′-d(ACT GCC AGA GAT CTT CCA TAC TG)-3′ (Mism3), (iii) the partly mismatched target containing four purine–pyrimidine mismatches 5′-d(ACT GCC AGA AAT TCT CCA TAC TG)-3′ (Mism4). The oligonucleotides were synthesized in LMFR Masaryk University in Brno, Czech Republic. The thiolated DNA probe S1 [SH-(MC6-D)-5′-d(CCT AAA AGG TTA CTC CAC CGG CT)-3′] and the DNA complementary target S1C 5′-d(AGC CGG TGG AGT AAC CTT TTA GG)-3′ were purchased from Integrated DNA Technologies, Inc., USA, and from LMFR Masaryk University, Czech Republic, respectively. All the oligonucleotides were HPLC purified.
Reagents
All the buffers were prepared using ultrapure water (18 MΩ/cm resistance, Direct-Q from Millipore). Sodium acetate, KH2PO4, Na2HPO4, KCl, NaCl, MgCl2, HEPES (C8H18N2O4S), sodium cacodylate [(CH3)2AsONa.3H2O], Na2HPO4, Tris (C4H11NO3) and TSTU [N,N,N′,N′-Tetramethyl-O-(N-succinimidyl) uronium tetrafluoroborate] were purchased from Sigma Aldrich, USA, in mol. biol. grade or higher. Streptavidin from Streptomyces Avidinii was obtained from Sigma Aldrich, USA. Carboxylic (HS-C15-COOH) and hydroxylic [HS-C11-(EG)2-OH] alkanethiols were purchased from Prochimia, Poland. Ethanol for spectroscopy (purity ≥99.9%) was purchased from Merck, USA.
The composition of the phosphate buffer (PB) was 1.4 mM KH2PO4, 8 mM Na2HPO4, 2.7 mM KCl, pH 7.4 at 25°C. The PBS buffer had the same composition as the PB buffer with 137 mM NaCl, pH 7.4 at 25°C. The immobilization and hybridization were measured in 10 mM Tris (in some cases in 10 mM PB, 10 mM HEPES or 10 mM cacodylate) containing a selected combination of mixed 0, 50, 150, 500 and 1000 mM NaCl and 0, 1.5, 5 and 15 mM MgCl2, pH 7.4 at 25°C.
The SPR biosensor
A laboratory SPR sensor (PLASMON IV) developed at the Institute of Photonics and Electronics, Prague, Czech Republic, was used for the purpose of this study (23). In brief, the sensor is based on wavelength spectroscopy of surface plasmons in four independent sensing channels and uses a flow-through flow-cell to confine the liquid sample during the experiments. In terms of performance, the sensor is comparable with the best commercial SPR sensor systems. A detailed description of the sensor system is available elsewhere (23).
SPR experiments
In most SPR experiments reported herein, a conventional immobilization method utilizing streptavidin–biotin interaction was used for the attachment of DNA probes to the sensor surface (gold layer). The immobilization procedure is described in detail elsewhere (23). In brief, the buffer (10 mM Tris, pH 7.4 at 25°C) was allowed to flow along the streptavidin-modified sensor surface for 10 min. Then 100 nM solution of the probe (biotinylated oligonucleotide BdO23) was introduced to the buffer and injected in the flow-cell. The immobilization process was monitored using the SPR sensor. After about 35 min, when a stable level of the sensor response to the probe immobilization was reached, the running buffer was injected in the flow-cell to wash the functionalized surface. The hybridization experiments were performed immediately after the immobilization of probes was completed. The solution of the target (100 nM) was introduced into the running buffer and allowed to flow through the flow-cell until the equilibrium response was reached (about 35 minutes). Both the immobilization and the hybridization steps were performed in the same running buffers. The temperature and the flow rate were strictly maintained at 25°C and 30 µl/min, respectively.
In control experiments reported in this work, thiolated oligonucleotide probes (S1) were directly attached to the SPR sensor surface. The clean SPR chip was rinsed with deionized water and immersed in a solution of 2 μM probe (S1) in PBS for 2 h. Prior to the SPR measurements, the functionalized chip was rinsed with deionized water, dried with a stream of nitrogen and mounted into the SPR instrument. The solid-phase hybridization of the target (S1C, 100 nM) with S1 probe was performed in 10 mM Tris with 15 mM magnesium and 1000 mM sodium, respectively. The hybridization process was carried out for 20 min followed by washing with Tris containing the selected cation content. Regeneration of the SPR chip was performed by flowing 3% HCOOH along the sensor surface for 5 min. The experiment was carried out at a flow rate of 30 μl/min and a temperature of 25°C.
Analysis of SPR measurements
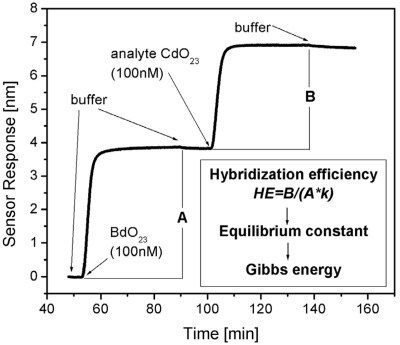
SPR measurements provided the sensor response (proportional to the mass of captured molecules) as a function of time. A typical kinetic curve containing sensor response to both the probe immobilization and the target hybridization process is shown in Figure 1. The equilibrium levels corresponding to the yields of the immobilization A and of the hybridization B were determined. The total surface density of the immobilized probes D was calculated according to the formula 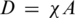 , where χ = 1.42 × 1012 oligonucleotide/cm2/nm is the calibration coefficient for BdO23. This calibration coefficient χ was calculated for the used SPR sensor system taking into account the molecular weight of the probe. Details of the calculation are provided in Ref. (24). The HE, i.e. the ratio of formed duplexes to immobilized oligonucleotide probes, was calculated as
, where χ = 1.42 × 1012 oligonucleotide/cm2/nm is the calibration coefficient for BdO23. This calibration coefficient χ was calculated for the used SPR sensor system taking into account the molecular weight of the probe. Details of the calculation are provided in Ref. (24). The HE, i.e. the ratio of formed duplexes to immobilized oligonucleotide probes, was calculated as 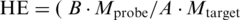 ), where Mprobe and Mtarget correspond to the molecular weight of the probe and target, respectively.
), where Mprobe and Mtarget correspond to the molecular weight of the probe and target, respectively.
Figure 1.
Design of the SPR experiment. The oligonucleotide immobilization giving the immobilization level A was followed with the hybridization process to get the final amount of formed complexes (level B). Both processes were monitored for 35 min in a buffer with the desired Na+/Mg2+ composition at 25°C and a flow rate of 30 μl/min. The inset shows the scheme of data processing. k in the equation for the HE represents the molecular-weight ratio of the target and the probe.
Considering the duplex formation as a pseudo first-order process, its equilibrium association constant K is given by HE according to the formula 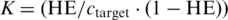 , where ctarget is the target concentration (ctarget = 100 nM in our experiments). The decrement of Gibbs energy associated with the duplex formation was determined using the van’t Hoff equation:
, where ctarget is the target concentration (ctarget = 100 nM in our experiments). The decrement of Gibbs energy associated with the duplex formation was determined using the van’t Hoff equation: 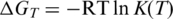 .
.
UV absorption measurements
The oligonucleotide solutions (BdO23 and CdO23) for UV absorption measurements were prepared in the buffers with a cation content corresponding to the SPR experiments (i.e. 10 mM Tris with 1000 mM NaCl and 10 mM Tris with 15 mM MgCl2). The equimolar solutions of mixed oligonucleotides with a total strand concentration of 0.5, 1, 2 and 5 μM, and with a total strand concentration of 0.3, 0.6, 2 and 6 μM, were prepared in Tris containing sodium and magnesium cations, respectively, and placed in cuvettes with a 1-cm path length. The absorbance at the wavelength of 260 nm was measured with the UV–VIS absorption spectrometer Varian 4000. The heating cycles were performed at the temperature range of 30–90°C with a heating rate of 1°C/min. Each sample was measured in duplicate. The melting temperatures were calculated by the standard derivative method. The resulting ΔG298 values were calculated according to the prediction formula based on the nearest-neighbor model described by SantaLucia (18).
RESULTS
Hybridization of fully matched targets
The main set of experiments was performed with a model system—a DNA 23-mer target (CdO23) containing a base sequence fully complementary to the sequence of the immobilized probe (BdO23). Prior to the detailed study of the cation effect, the influence of the buffer being used was investigated. HE was measured in four common buffers, namely in the Tris, HEPES, phosphate and cacodylate buffers. The HE was measured five times in each buffer at 10 mM concentration, with the addition in each case of one of the five combinations of sodium/magnesium: 0/0 mM, 0/15 mM, 50/1.5 mM, 150/5 mM, 1000/0 mM and 1000/15 mM. The obtained results did not display any noticeable differences between the buffers (Supplementary Data), which is in agreement with the observations made by Cho et al. (12). The only exception was the case when no cations were added to the buffer, i.e. for the ‘0/0 mM’ combination of sodium/magnesium concentration. In this case, no sensor response to the target hybridization was observed in Tris, while a very low response to hybridization was observed in the other measured buffers. This was due to the fact that only in the case of Tris, no small cations were present in a solvent after adjusting to the required pH (pH 7.4 at 25°C). To ensure that the solid-phase hybridization experiments are conditioned solely by the presence of free cations coming from the added salts (NaCl and MgCl2), Tris was selected as a hybridization buffer for all the SPR experiments and the melting study. Tris was used despite the temperature sensitivity of its pH (17) as it did not contain any additional cations and was optimal for reported measurements with low concentrations of sodium and magnesium cations.
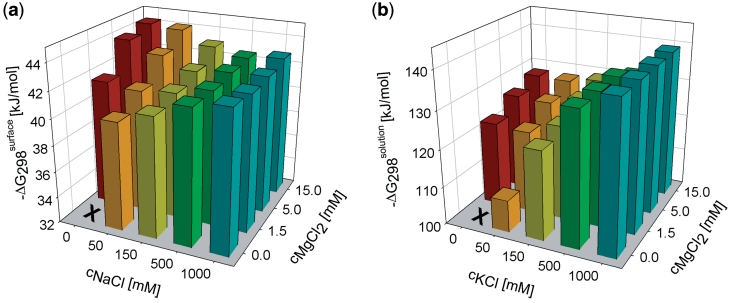
Gibbs energy values (ΔG298) corresponding to the hybridization of the 23-mer target (CdO23) with the immobilized probe (BdO23) were derived from the hybridization efficiencies as described in the ‘Materials and Methods’ section (Figure 1). The results are displayed in Figure 2a as a 2D function of sodium and magnesium concentration (the values are presented in the form of tables in Supplementary Data for all the 2D graphs). The cation concentration ranges were 0–1 M and 0–15 mM for sodium and magnesium, respectively. The comparative Gibbs energy values for duplex hybridization in a solution are shown in Figure 2b. These values were calculated using the standard empirical prediction formulas as described in Owczarzy et al. (17).
Figure 2.
Gibbs energies ΔG298 of 23-mer fully matched duplex formation in the presence of various monovalent and divalent cation concentrations. (a) Gibbs energy values obtained using the SPR sensor. The value for zero content of small cations is not shown, because no sensor response to hybridization was observed. (b) The comparative Gibbs energy values for hybridization in a solution calculated using the empirical formulas described by Owczarzy et al. (17). The unknown parameter of Equation 23 (17) 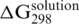 was determined to be equal to −139 ± 7 kJ/mol using UV absorption measurement at a sodium concentration of 1 M [this experimental value agrees with that predicted by SantaLucia’s model (18) in a solution]. Monovalent potassium is considered as equivalent with sodium cation in the stabilization of duplexes in a solution (16,17).
was determined to be equal to −139 ± 7 kJ/mol using UV absorption measurement at a sodium concentration of 1 M [this experimental value agrees with that predicted by SantaLucia’s model (18) in a solution]. Monovalent potassium is considered as equivalent with sodium cation in the stabilization of duplexes in a solution (16,17).
Figure 2a demonstrates the expected overall tendency of increased duplex stability with an increase of the cation content. However, several obvious features differ considerably from the results usually obtained in solutions: (i) magnesium is more efficient in the duplex stabilization than even a much higher concentration of sodium, (ii) the absolute values of ΔG298 are substantially lower than those reported in a solution with the same cation content, (iii) local stability minimum can be observed when the sodium concentration is gradually increased while the magnesium level is kept constant.
The observation that for solid-phase hybridization, the 15 mM magnesium (divalent) cation is more efficient in the duplex stabilization than even a very high concentration of monovalent sodium (1 M NaCl) is rather surprising. For oligonucleotide complexes in a solution (present typically at micromolar concentrations), the opposite trend in cation efficiency has been described. Owczarzy et al. (17) observed that 1 M monovalent potassium, which is equivalent with sodium ions in the stabilization of duplexes in a solution (16,17), is more efficient in stabilization of the oligonucleotide duplexes than any concentration of divalent magnesium. Clearly, the trend in monovalent/divalent ion efficiency obtained by applying Owczarzy’s formula (17) to our buffer parameters (Figure 2b) is opposite to our experimental results.
To further confirm these observations and to address the potential issue of differences in the experimental conditions, a comparative UV absorption experiment was performed with the same oligonucleotide system (BdO23 and CdO23 oligonucleotides) as used in the SPR experiments. The selected cation content was 1000 mM sodium (without magnesium) and 15 mM magnesium (without sodium). The melting temperatures of 75.3 ± 0.5°C and 70.1 ± 0.5°C corresponding to Gibbs energies ΔG298 of −139 ± 7 kJ/mol and −130 ± 6 kJ/mol for 1000 mM sodium and 15 mM magnesium, respectively, were achieved (for a detailed data analysis, see Supplementary Data). A comparison of these results with the ΔG298 values for solid-phase hybridization obtained with the SPR sensor, i.e. −42.6 ± 0.8 kJ/mol for 1000 mM sodium and −44.3 ± 0.8 kJ/mol for 15 mM magnesium confirms the opposite ranking of the sodium and magnesium efficiency.
A substantial difference in the sodium effect on the duplex stability observed in solid-phase hybridization and hybridization in a solution can be further illustrated by a comparison of our results with the published works. For hybridization in a solution, a quantitative description of the effect of sodium concentration on duplex stability has been derived (18,25). Based on these studies both the experimental data (18) and the TBI model (25) suggest a linear dependence of ΔG on the logarithm of sodium concentration in the concentration range of 50–1000 mM, i.e. 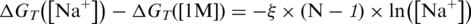 where (N–1) is the number of phosphates in a single DNA strand. For our experimental conditions (N–1 = 22, T = 298 K) the values of the coefficient ξ were determined to be 0.46 kJ/mol (18) and 0.28 kJ/mol (25). By fitting the formula to our solid-phase hybridization results in the used sodium concentration range (Supplementary Data), a linear trend was found as described for hybridization in a solution. However, the corresponding value of the coefficient ξ was only 0.036 kJ/mol, which is about one tenth of the values for hybridization in a solution. This result is consistent with the observation made by Gong and Levicky (26) using shorter (18-mer) oligonucleotides. This demonstrates that the sodium concentration (at a concentration range of 50–1000 mM) influences duplex stability much less in solid-phase hybridization than in hybridization in a solution.
where (N–1) is the number of phosphates in a single DNA strand. For our experimental conditions (N–1 = 22, T = 298 K) the values of the coefficient ξ were determined to be 0.46 kJ/mol (18) and 0.28 kJ/mol (25). By fitting the formula to our solid-phase hybridization results in the used sodium concentration range (Supplementary Data), a linear trend was found as described for hybridization in a solution. However, the corresponding value of the coefficient ξ was only 0.036 kJ/mol, which is about one tenth of the values for hybridization in a solution. This result is consistent with the observation made by Gong and Levicky (26) using shorter (18-mer) oligonucleotides. This demonstrates that the sodium concentration (at a concentration range of 50–1000 mM) influences duplex stability much less in solid-phase hybridization than in hybridization in a solution.
As follows from Figure 2a, the duplex stability dependence on the sodium concentration (at a fixed concentration of magnesium) shows local minima. Specifically, those minima were found for 50 mM sodium at 1.5 mM magnesium, 150 mM sodium at 5 mM magnesium, and 500 mM sodium at 15 mM magnesium. All of these combinations correspond to an ∼30-fold excess of sodium over magnesium. These local minima are believed to be caused by the competition of sodium and magnesium cations in their binding to DNA duplexes (17,27,28). For example, the magnesium as well as sodium ions are known to bind in both minor and major groove in DNA duplex (29). However, due to variable hydration shell, the monovalent ions have larger variability of the binding sites within the DNA duplex. Detailed discussion of sodium magnesium competition can be found in Ref. (17,27,28). A competition effect of monovalent and divalent cations has been recently observed using the UV absorption method (17) and predicted by the TBI model (25) for oligonucleotides in a solution.
To determine the reproducibility of Gibbs energy values, a series of independent hybridization experiments (using 22 measuring channels on eight different chips) was performed at the conditions corresponding to the highest achieved HE (i.e. 15 mM magnesium and 0 mM sodium). The obtained ΔG298 values were found to be highly reproducible with a standard deviation of <2%.
In all the SPR experiments described above, the probes were immobilized to the sensor surface via a streptavidin linker. To rule out the potential effect of cation–streptavidin interaction on the studied cation–oligonucleotide interactions, SPR measurements employing another probe immobilization strategy were carried out. In those experiments the thiol-derivatized oligonucleotide probes were attached directly to the SPR sensor surface (without the use of streptavidin). The hybridization with complementary targets in buffer with selected cation content was monitored (see Supplementary Data for more details). The results clearly demonstrate the same trend in cation efficiency as was observed for the streptavidin-based immobilization method. In particular, the sensor response to the complementary target binding was considerably higher for the analyte solvent containing only magnesium in comparison with that containing only sodium (Supplementary Data). This confirms our assumption that the presence of streptavidin does not influence the conclusions derived from the hybridization experiments utilizing the streptavidin-based immobilization method.
Our results suggest that when formulating assay protocols for DNA microarrays or biosensors, it is advantageous to use a hybridization buffer containing a relatively high concentration of magnesium (∼15 mM) and a minimum amount of sodium.
Hybridization of targets containing mismatched bases
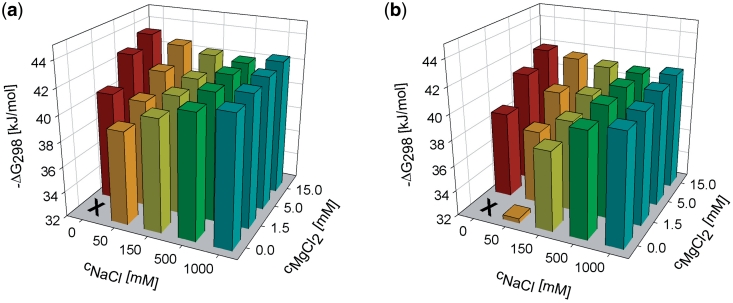
To determine the cation effect on stability of targets containing internal base mismatches, a series of SPR experiments analogous to the previous experiments were carried out with the targets containing three (Mism3) or four (Mism4) purine–pyrimidine mismatched base pairs as a model system. These point mismatched bases were distributed uniformly along the target strand. Figure 3 shows ΔG298 values in dependence on sodium and magnesium concentrations. The overall trends of the duplex stability as a function of sodium and magnesium concentrations for Mism3 and Mism4 are similar to those obtained for the fully matched targets. These include, in particular, the higher efficiency of magnesium over sodium and the stability minima when the sodium concentration is gradually varied and the magnesium concentration is fixed.
Figure 3.
Gibbs energies ΔG298 of partly matched duplex formation at the SPR sensor in the presence of various sodium and magnesium concentrations. The left graph (a) corresponds to three and the right graph (b) to four purine–pyrimidine mismatches distributed regularly within the 23-mer chain. In the case of no added cations, no hybridization was observed.
In agreement with the previous DNA biosensor (30) and microarray (6) studies, total ΔG298 values were reduced due to the presence of mismatched base pairs. The destabilizing effect was not proportional to the number of mismatches. For instance, in the case of 15 mM magnesium without sodium, the difference for three mismatches ΔG298(Mism3) – ΔG298(Match) = 0.84 kJ/mol is three times less than that for four mismatches ΔG298(Mism4) – ΔG298(Match) = 2.45 kJ/mol.
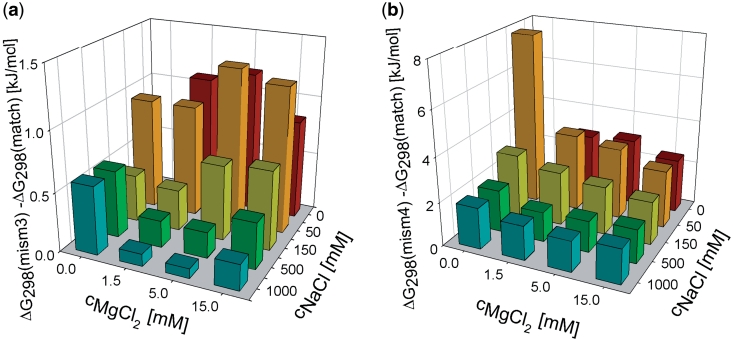
To investigate whether it is possible to improve the level of discrimination of mismatched and fully complementary targets by optimizing the combination of cations in the hybridization buffer by type and cation concentration, the levels of destabilization dependent on cation content were plotted out. Figure 4 shows the levels of destabilization determined as the difference between Gibbs energies for three and four base mismatched targets and those for the fully matched target with the same cation content. The values of the calculated destabilization levels were relatively small and therefore these values were subject to a relatively high uncertainty. However, the general trend of the sodium and magnesium effect is obvious. There is no clear dependence of the mismatched duplex destabilization on the magnesium concentration except for the case of 50/0 mM, in which, however, the stability of the used fully matched duplex is also substantially reduced. It is evident that at high sodium concentrations (higher than 100 mM) the destabilization of mismatched duplexes is lower in comparison with a low concentration of sodium (50 mM or less). In the optimization of the cation content for the discrimination of fully complementary and mismatched targets, the need for sufficiently high HE must be taken into account as well. The optimized cation composition yielding the highest level of discrimination between fully complementary and mismatched targets while maintaining a high hybridization signal is clearly the combination of the following cation concentrations, sodium: 0–50 mM and magnesium: 1.5–15 mM. It should be noted that the results were obtained for the model oligonucleotide system containing point purine–pyrimidine mismatches. Extension of the conclusions of our work to general oligonucleotide systems with different lengths and mismatch types would require further study which will be a subject of our future work.
Figure 4.
Differences between the Gibbs energies of duplexes containing three (a) and four (b) mismatched base pairs and the Gibbs energy of fully matched duplex for various sodium and magnesium concentrations. The horizontal axes are rotated for better visibility. The value for zero content of small cations is not shown, because no SPR signal to hybridization was observed.
DISCUSSION
The molecular model describing the observed shielding effect of sodium and magnesium cations on solid-phase hybridization is presented below. All the interactions of probes with targets occur within an active layer with a thickness of ∼10 nm. This thickness was estimated from the characteristic dimensions of 23-mer oligonucleotide and streptavidin molecules. Considering the observed values of probe coverage (∼5 × 1012 molecules/cm2) and hybridization efficiencies of BdO23/CdO23 duplexes (∼75%), the molar concentration of duplexes in the active layer was 5 mM for 1000 mM sodium. This indicates that oligonucleotide duplexes occupy about 25% of the layer volume. Similarly it can be concluded that in the solution containing 15 mM magnesium and no sodium, as much as 35% of the active layer is filled with duplexes. Considering that 75% of the active layer is freely accessible for cations and assuming that DNA duplexes form cylindrical polyanions with a radius of 10 Å (28,31) it is estimated that only a 6–8 Å thick shell surrounding each duplex is available on average for solvent with cations compensating for the negative charge of the DNA duplex. Using the Poisson–Boltzmann theory, Misra and Draper (31) determined concentration profiles for sodium and magnesium cations surrounding an isolated DNA duplex. These calculations showed that both sodium and magnesium cations effectively occupy an area up to 10 Å from the DNA duplex surface, which exceeds our estimated free zone. This suggests a spatial stress restricting the ability of cations to completely shield the negative charges on deoxyribophosphate backbones, which may explain the decreased stability of highly packed duplexes in comparison with low concentrations of oligonucleotides in a solution. It has been shown that the distribution of magnesium around an isolated DNA duplex is more compact than that of sodium (31). Therefore, the total sum of positive charges is higher for magnesium under the restricted spatial conditions. This can explain the stronger stabilization effect of magnesium over sodium on the duplex stability for solid-phase hybridization as well as the higher importance of the sodium–magnesium competition.
It is important to mention that we cannot completely exclude the effect of other magnesium binding mechanisms to DNA probes, which would also support its relatively high stabilization efficiency. For instance, the limited space between duplexes may evoke the idea of a possible parallelism with the known role of multivalent cations in the process of DNA condensation and redissolution (32–34). Magnesium is also believed to specifically interact with the oligonucleotide bases (35). All these interactions may result in solid-phase hybridization of non-complementary target. However, for our solid-phase hybridization experiments, no considerable SPR signal was observed when a non-complementary target with a high concentration (2 µM) was injected over the surface of the immobilized probes even for the highest used concentration of magnesium (data not shown here). Therefore, it is not likely that magnesium-mediated interchain binding (other than duplex formation) takes place in the solid-phase hybridization processes. On the other hand, the interactions between closely packed probes may affect the target affinity to probes (especially for buffers with high cation concentrations) (26,36,37). For example, the studies of Gong and Levicky (26) and Matveeva et al. (36) confirmed that mutual probe interactions may result in lower target affinity to probes.
Duplex destabilization for solid-phase hybridization (in comparison with hybridization in a solution with the same cation content) was observed in this work (Figure 2). This trend can be also found in several other DNA sensor and microarray studies (1,7–9,11,12). Due to the different experimental conditions used in these works, it is difficult to form a meaningful quantitative comparison among our results and the published data. However, for selected experimental conditions, the differences between Gibbs energy values for solid-phase hybridization and hybridization in a solution (i.e. 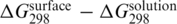 ), which should be independent of particular base sequence (7), can be compared. The selected conditions include a comparable probe density (1012–1013 probes/cm2) and the hybridization of fully complementary targets in buffer containing 1000 mM sodium without magnesium. In the cases when solution hybridization data were not published,
), which should be independent of particular base sequence (7), can be compared. The selected conditions include a comparable probe density (1012–1013 probes/cm2) and the hybridization of fully complementary targets in buffer containing 1000 mM sodium without magnesium. In the cases when solution hybridization data were not published, 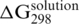 values were estimated using the nearest-neighbor prediction method by SantaLucia (18). The obtained
values were estimated using the nearest-neighbor prediction method by SantaLucia (18). The obtained 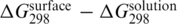 differences varied significantly among the reported data (Supplementary Data for details). However, the correlation with the length of the DNA strand was obvious. In particular, the level of duplex destabilization (expressed as the
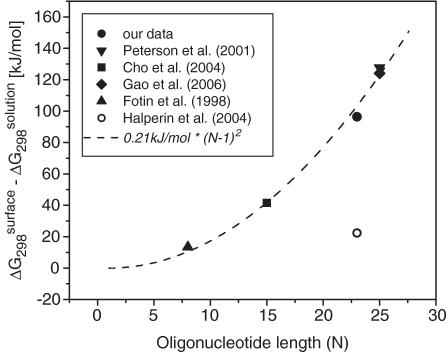
differences varied significantly among the reported data (Supplementary Data for details). However, the correlation with the length of the DNA strand was obvious. In particular, the level of duplex destabilization (expressed as the 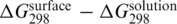 difference) was directly proportional to the squared number of negatively charged phosphate units in the oligonucleotide chain (Figure 5). This dependence is in qualitative agreement with the theoretical prediction of Halperin et al. (14), who treated the immobilized probes as a 3D layer. On the other hand, the predicted duplex destabilization is significantly lower than observed in our experiments (Figure 5). It suggests that the cation shielding for solid-phase hybridization is probably overestimated by the theory (14). In addition to the electrostatic repulsion, mutual interactions between probes may be also involved in duplex destabilization (26).
difference) was directly proportional to the squared number of negatively charged phosphate units in the oligonucleotide chain (Figure 5). This dependence is in qualitative agreement with the theoretical prediction of Halperin et al. (14), who treated the immobilized probes as a 3D layer. On the other hand, the predicted duplex destabilization is significantly lower than observed in our experiments (Figure 5). It suggests that the cation shielding for solid-phase hybridization is probably overestimated by the theory (14). In addition to the electrostatic repulsion, mutual interactions between probes may be also involved in duplex destabilization (26).
Figure 5.
Differences between the duplex stability in the case of solid-phase hybridization and that for hybridization in a solution, expressed in ΔG298 values at standard ionic content of 1 M sodium without magnesium. Filled circle: our result, streptavidin–biotin immobilization, probe surface density 5 × 1012 probes/cm2. Triangle down: Peterson et al. (11), immobilization via thiolate group, 5.2 × 1012 probes/cm2. Square: Cho et al. (12), immobilization via thiolate group, 4.3 × 1012 probes/cm2. Diamond: Gao et al. (8), immobilization via thiolate group, 4.5 × 1012 probes/cm2. Triangle up: Fotin et al. (7), gel immobilization, probe density not determined, five unpairing nucleotides added to the target. Opened circle: theoretical prediction by Halperin et al. for our system (14). Dashed line: squared number of phosphate units in a single strand multiplied by a coefficient fitted to the experimental data obtained for known probe densities.
Figure 5 suggests a good agreement among our results and the published data despite the different immobilization methods used. This confirms our assumption that the immobilization method as well as the detection technique does not influence the general properties of cation–oligonucleotide interactions. Therefore, one can conclude that the trends in the shielding effect of combined monovalent and divalent cations on DNA solid-phase hybridization established in this work could be expanded to any solid-phase hybridization utilizing similar surface density and oligonucleotide length.
CONCLUSIONS
In this work, the simultaneous effect of monovalent and divalent cations on solid-phase hybridization for closely packed oligonucleotides on a biosensor chip was investigated using SPR biosensor technology. The results were compared with the cation efficiency trends established for oligonucleotides in a solution as well as with the relevant published data on cation effect in solid-phase hybridization.
Our study shows that both the type and concentration of cations play an important role in the compensation for the negative charge of densely packed oligonucleotides on solid surfaces and thus in solid-phase hybridization. It is also shown that the efficiency with which cations stabilize duplexes is greatly affected by the restricted surface conditions and thus may substantially differ from that observed in a solution. Specifically, divalent magnesium was found to be much more efficient in stabilization of duplexes than monovalent sodium. This finding sharply contrasts with the results in solution where the trend is the opposite. Moreover, the duplex stability was found to be influenced much less by the concentration of monovalent sodium (when no magnesium was present) in contrast with the relatively high sodium concentration effect on stability observed in a solution.
In our experiments, the remarkable destabilization of duplexes in solid-phase hybridization in comparison with duplexes in a solution was observed. This result was in agreement with previously published works. The level of duplex destabilization was found to be strongly dependent on the oligonucleotide length. In particular, the trend of increasing duplex destabilization with an increase of the oligonucleotide length was demonstrated. Our results indicate that for solid-phase hybridization, the cation composition is critical for accurate determination of the duplex stability. Stability estimates based solely on comparable oligonucleotide and cation systems in a solution are inaccurate, in particular, when oligonucleotides longer than ∼10 nt are involved.
The obtained results have a number of important consequences for the development of the technology of DNA microarrays and biosensors, especially in terms of formulating assays protocols, developing data analysis tools and optimizing the device design. It is clearly demonstrated that the HE can be controlled by optimizing the cation composition in the running buffer. Investigation of model oligonucleotides with point mismatches suggests that discrimination of fully matched and partially mismatched targets can be improved by optimizing the cation composition in the hybridization buffer. This finding is of great interest in many practical applications. In this work, it was demonstrated that a several millimolar concentration of magnesium and a low concentration of sodium (<50 mM) provide favorable experimental conditions offering an improved discrimination of mismatched and fully complementary targets and a high sensitivity of hybridization detection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation under contract # 202/09/0193; Academy of Sciences of the Czech Republic under contract KAN200670701. Funding for open access charge: National Science Foundation of the Czech Republic.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Levicky R, Horgan A. Physicochemical perspectives on DNA microarray and biosensor technologies. Trends Biotechnol. 2005;23:143–149. doi: 10.1016/j.tibtech.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Mir KU, Southern EM. Sequence variation in genes and genomic DNA: methods for large-scale analysis. Ann. Rev. Genomics Hum. Genet. 2000;1:329–360. doi: 10.1146/annurev.genom.1.1.329. [DOI] [PubMed] [Google Scholar]
- 3.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Wu CL, Carta R, Zhao HT. Free energy of DNA duplex formation on short oligonucleotide microarrays. Nucleic Acids Res. 2007;35:e18. doi: 10.1093/nar/gkl1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piliarik M, Párova L, Homola J. High-throughput SPR sensor for food safety. Biosens. Bioelectron. 2009;24:1399–1404. doi: 10.1016/j.bios.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Fish DJ, Horne MT, Brewood GP, Goodarzi JP, Alemayehu S, Bhandiwad A, Searles RP, Benight AS. DNA multiplex hybridization on microarrays and thermodynamic stability in solution: a direct comparison. Nucleic Acids Res. 2007;35:7197–7208. doi: 10.1093/nar/gkm865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fotin AV, Drobyshev AL, Proudnikov DY, Perov AN, MIrzabekov AD. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 1998;26:1515–1521. doi: 10.1093/nar/26.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y, Wolf LK, Georgiadis RM. Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F, Yao DF, Knoll W. Oligonucleotide hybridization studied by a surface plasmon diffraction sensor (SPDS) Nucleic Acids Res. 2004;32:e75. doi: 10.1093/nar/gnh067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okahata Y, Kawase M, Niikura K, Ohtake F, Furusawa H, Ebara Y. Kinetic measurements of DNA hybridisation an an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal. Chem. 1998;70:1288–1296. doi: 10.1021/ac970584w. [DOI] [PubMed] [Google Scholar]
- 11.Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho YK, Kim S, Kim YA, Lim HK, Lee K, Yoon DS, Lim G, Pak YE, Ha TH, Kim K. Characterization of DNA immobilization and subsequent hybridization using in situ quartz crystal microbalance, fluorescence spectroscopy, and surface plasmon resonance. J. Colloid Inter. Sci. 2004;278:44–52. doi: 10.1016/j.jcis.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Vainrub A, Pettitt BM. Coulomb blockage of hybridization in two-dimensional DNA arrays. Phys. Rev. E. 2002;66 doi: 10.1103/PhysRevE.66.041905. art. no. 041905. [DOI] [PubMed] [Google Scholar]
- 14.Halperin A, Buhot A, Zhulina EB. Sensitivity, specificity, and the hybridization isotherms of DNA chips. Biophys. J. 2004;86:718–730. doi: 10.1016/S0006-3495(04)74150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano S, Fujimoto M, Hara H, Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 18.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. 3Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SantaLucia J, Allawi HT, Seneviratne A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 20.Tan ZJ, Chen SJ. Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length. Biophys. J. 2006;90:1175–1190. doi: 10.1529/biophysj.105.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan ZJ, Chen SJ. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J. Chem. Phys. 2005;122 doi: 10.1063/1.1842059. art. no. 044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaisocherová H, Snášel J, Špringer T, Šípová H, Rosenberg I, Štěpánek J, Homola J. Surface plasmon resonance study on HIV-1 integrase strand transfer activity. Anal. Bioanal. Chem. 2009;393:1165–1172. doi: 10.1007/s00216-008-2485-y. [DOI] [PubMed] [Google Scholar]
- 23.Vaisocherová H, Zítová A, Lachmanová M, Štěpánek J, Králíková S, Liboška R, Rejman D, Rosenberg I, Homola J. Investigating oligonucleotide hybridization at subnanomolar level by surface plasmon resonance biosensor method. Biopolymers. 2006;82:394–398. doi: 10.1002/bip.20433. [DOI] [PubMed] [Google Scholar]
- 24.Homola J. Surface Plasmon Resonance Based Sensors. Germany, Berlin: Springer-Verlag; 2006. [Google Scholar]
- 25.Tan ZJ, Chen SJ. RNA helix stability in mixed Na+/Mg2+ solution. Biophys. J. 2007:227A–227A. doi: 10.1529/biophysj.106.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong P, Levicky R. DNA surface hybridization regimes. Proc. Natl Acad. Sci. USA. 2008;105:5301–5306. doi: 10.1073/pnas.0709416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Record MT. Effects of Na+ and Mg++ ions on helix-coil transition of DNA. Biopolymers. 1975;14:2137–2158. [Google Scholar]
- 28.Korolev N, Lyubartsev AP, Nordenskiold L. Application of polyelectrolyte theories for analysis of DNA melting in the presence of Na+ and Mg2+ ions. Biophys. J. 1998;75:3041–3056. doi: 10.1016/S0006-3495(98)77745-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egli M. DNA-cation interactions: quo vadis? Chem. Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 30.Peterson AW, Wolf LK, Georgiadis RM. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 31.Misra VK, Draper DE. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J. Mol. Biol. 1999;294:1135–1147. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 32.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Raspaud E, Chaperon I, Leforestier A, Livolant F. Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J. 1999;77:1547–1555. doi: 10.1016/S0006-3495(99)77002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar T, Conwell CC, Harvey LC, Santai CT, Hud NV. Condensation of oligonucleotides assembled into nicked and gapped duplexes: potential structures for oligonucleotide delivery. Nucleic Acids Res. 2005;33:143–151. doi: 10.1093/nar/gki156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rincon E, Jaque P, Toro-Labbe A. Reaction force analysis of the effect of Mg(II) on the 1,3 intramolecular hydrogen transfer in thymine. J. Phys. Chem. A. 2006;110:9478–9485. doi: 10.1021/jp062870u. [DOI] [PubMed] [Google Scholar]
- 36.Matveeva OV, Shabalina SA, Nemtsov VA, Tsodikov AD, Gesteland RF, Atkins JF. Thermodynamic calculations and statistical correlations for oligo-probes design. Nucleic Acids Res. 2003;31:4211–4217. doi: 10.1093/nar/gkg476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray J, Manning GS. Effect of counterion valence and polymer charge density on the pair potential of two polyions. Macromolecules. 1997;30:5739–5744. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.