Abstract
Precise regulation of gene expression is crucial to myogenesis and is thought to require the cooperation of various transcription factors. On the basis of a bioinformatic analysis of gene regulatory sequences, we hypothesized that myogenic regulatory factors (MRFs), key regulators of skeletal myogenesis, cooperate with members of the SIX family of transcription factors, known to play important roles during embryonic skeletal myogenesis. To this day little is known regarding the exact molecular mechanism by which SIX factors regulate muscle development. We have conducted a functional genomic study of the role played by SIX1 and SIX4 during the differentiation of skeletal myoblasts, a model of adult muscle regeneration. We report that SIX factors cooperate with the members of the MRF family to activate gene expression during myogenic differentiation, and that their function is essential to this process. Our findings also support a model where SIX factors function not only ‘upstream’ of the MRFs during embryogenesis, but also ‘in parallel’ to them during myoblast differentiation. We have identified new essential nodes that depend on SIX factor function, in the myogenesis regulatory network, and have uncovered a novel way by which MRF function is modulated during differentiation.
BACKGROUND
Skeletal muscle development is a complex process regulated in large part by sequence-specific transcription factors (TFs). They govern the ordered expression of a multitude of genes along the successive steps of myogenesis, from the engagement of mesodermal cells in the muscle lineage, to the differentiation of somitic cells, to the terminal differentiation of myocytes into myofibers [reviewed in refs. (1–3)].
In terms of gene regulation, the differentiation of muscle is a system where complexity manifests itself both in the precise location and timing of gene expression patterns. Accurate gene transcription is made possible in part by the convergence of regulatory inputs from different TFs on single genes. For example, during fruit fly larva mesoderm development, several factors converge to drive in a combinatorial fashion the expression of genes to specific mesodermal compartments and during precise time periods (4).
In vertebrates, the making of muscle is coordinated in part by the action of TFs of the myogenic regulatory factor family (MRFs), which counts four members: MYOD, MYF5, MYOG (myogenin) and MRF4 (5). These basic helix–loop–helix TFs bind DNA as dimers with E-proteins to a DNA sequence element termed the E-box (consensus CANNTG). MRFs function in a cascade fashion and may regulate each other’s expression. For example, MYOD regulates the expression of Myog, and MYOG can activate the expression of Mrf4 (6). At the same time, both MYOD and MYOG have the ability to enhance their own expression (7).
It is clear that the function of the MRFs in regulating target gene expression is coupled to that of other TFs. Logically, it is difficult to envision a regulatory scheme by which these four TFs could establish by themselves the complex gene expression profiles that pattern muscle. For instance, Molkentin et al. (8) have shown that transcriptional activation by the MRFs is enhanced by the function of MEF2 TFs. However, as key regulators, MRFs’ cooperation with other TFs in regulating precise gene expression in myogenesis deserves to be further studied. Previous work by us and others has revealed the identity of a large number of direct transcriptional targets of the MRFs (9–11). We have demonstrated that certain TF binding sites are specifically enriched among the regulatory regions of this large set of MRF targets, suggesting the identity of partners in combinatorial regulation. The A/T rich element recognized by MEF2 was among those discovered, and indeed we and others have confirmed the recruitment of MEF2 to several of these sites. Nevertheless, cooperation of MRFs and their co-factors is far from being fully understood, and cooperation of MRFs with not one but a variety of TFs, for example at different time points or on different target gene promoters, is likely to occur.
Another motif we discovered as specifically enriched among MRF target promoters was the MEF3 element (consensus TCAGGTTTC) (9). The MEF3 sequence was found to be most abundant for MRF target genes whose expression is significantly induced during differentiation. This sequence element is known to be recognized by TFs of the SIX family, which led us to hypothesize that MRF function during myogenic differentiation is modulated by combinatorial regulation with the SIX factors.
The SIX proteins belong to a family of six homeodomain TFs, from SIX1 to SIX6, which are involved in controlling the development of a number of tissue types [reviewed in refs. (12,13)]. Importantly, the functions of SIX1 and SIX4 are tied to skeletal muscle development: Six1-null mice die at birth from respiratory failure, with grave defects in trunk musculature due in part to impaired primary myogenesis (14–16). While Six4-null mice appear normal (17), compound mutant Six1;Six4-null mice display a more pronounced impairment in myogenesis than Six1-null animals (18). Because loss of Six1 leads to perinatal lethality, comparatively less attention has been given to the role it plays after birth, in adult muscle and during muscle repair. It is known however that in addition to their role in the early phase of muscle development, SIX1 has an influence on establishing muscle fiber type: enhancing the activity of SIX1 leads to an increase in the number of fast-twitch (glycolytic) muscle fibers, and to higher expression of genes stereotypical of this class of myofibers (19). Expression profiling data from embryonic tissue of Six1;Six4 double knock-out embryonic tissue also support the notion that SIX factors are required to activate in the myotome the expression of fast-type muscle genes (20). In line with this, in zebrafish, loss of function of the six1a gene causes abnormal fast-twitch muscle formation (21). Taken together, these data indicate that SIX1 and SIX4 are critical for embryonic muscle development.
Despite its clear role in regulating the formation of muscle, and other tissues, few genes have been shown to be under the direct transcriptional control of SIX1 during myogenesis. Over-expression approaches followed by gene expression profiling have been used, leading to the identification of potential SIX1 and SIX4 target genes (22). On the basis of their deregulation in Six1 knock-out animals, a number of targets have been identified or are thought to represent direct targets (16,18). Importantly, among direct SIX1 target genes are two MRFs, Myog and Myf5 (23,24). The expression levels of Pax3, Myod1, MRF4 and Lbx1, all encoding TFs playing critical roles in embryonic myogenesis, have also been shown to be affected by the loss of Six1, but it is unknown in these cases if the regulation is direct or mediated by the product of one or more genes regulated by SIX1. Considering the essential roles of MRFs and SIX factors for muscle differentiation, we hypothesize that MRFs and SIX factors function in a combinatorial manner.
In order to verify our hypothesis that MRFs may cooperate with SIX factors during myogenesis, we have used a combination of genomic approaches to analyze SIX1 AND SIX4 function during myoblast differentiation. We found that as predicted by our bioinformatic analyses, MRFs’ function is potentiated by SIX factors. These two families of TFs together play a pervasive role in establishing the proper gene expression profiles that enable differentiation of muscle precursor cells, by directly regulating the expression of genes that control not only the development of muscle but also its function once terminally differentiated.
MATERIALS AND METHODS
Cell culture and RNA interference
C2C12 myoblasts were acquired from the American Type Culture Collection and cultured as described earlier (9). For all ChIP-on-chip and expression profiling experiments performed on differentiated myotubes (MT), the cells were separated from undifferentiated ‘reserve’ cells using differential trypsinization (25). Human embryonic kidney carcinoma cells HEK293T cells were obtained from the ATCC and grown in DMEM supplemented with 10% fetal bovine serum, glutamine and antibiotics. For RNA interference experiments, C2C12 cells were seeded at 100 000 cells in 6-well plates (Corning) and were transfected with siRNA (Dharmacon) using Lipofectamine 2000 and the recommended lipofection protocol (Invitrogen). siRNA oligo sequences are given in Supplementary Table S6.
Antibodies
Recombinant mouse SIX1 (full-length protein) or mouse SIX4 (amino acids 414–775) fused to N-terminal hexa-histidine tags and purified from Escherichia coli on Ni–NTA beads (GE Healthcare) were used to immunize rabbits (Open Biosystems). Rabbit sera were purified by affinity to the cognate recombinant proteins, purified from E. coli as N-terminal glutathione-S-transferase fusion proteins on glutathione agarose (Pierce Chemicals). Western blots on C2C12 whole lysates using anti-SIX1 or anti-SIX4 each revealed only one band, and chromatin immuno-precipitations followed by immuno-blotting confirmed antibody specificity (data not shown, and Supplementary Figure S6). Other antibodies used include anti-myosin heavy chain (MHC, MF20 clone, obtained from the Developmental Studies hybridoma Bank, DSHB), anti-myogenin (clone F5D, DSHB, contributed by W. Wright), anti-β-tubulin (clone E7, DSHB). Secondary antibodies against mouse or rabbit IgG, coupled to horseradish peroxidase (Pierce Chemicals), were used in western blotting experiments. The secondary antibody used in immuno-fluorescence experiments is anti-mouse coupled to FITC (Jackson Immunoresearch).
ChIP assays and ChIP-on-chip
Chromatin immunoprecipitations were performed as described in ref. (9), using 2 μg of antibody and 25 μg of chromatin DNA. Normal rabbit IgG (Pierce Chemicals) was used as a negative control antibody. After clean-up, the immunoprecipitated DNA was quantitated using qPCR employing the SYBR green chemistry on a MX3005p instrument (Stratagene). Each sample was used in duplicate PCR reactions. For each time point and each biological replicate, an aliquot of the ‘input’ starting chromatin material was used to create a dilution curve in order to express target DNA abundance in the IP samples as a percentage of that in the starting material (percent of input values). The Hoxd10 promoter was used as a negative control locus, with no recruitment of the TFs being studied. See Supplementary Table S6 for a list of primers used in ChIP assays. A stringent multi-criteria method was used to evaluate factor binding by qChIP. To be considered a true binding event, each IP sample had to pass three tests: giving higher enrichment than with the control antibody (at the same time point and same locus, one-tailed t-test paired with P < 0.05), higher enrichment than at the HoxD10 locus (at the same time point and with same antibody, one-tailed t-test unpaired with P < 0.05), and average enrichment levels at least 10 times higher than with the control antibody. Details of ChIP-on-chip experiments and microarray design are provided in Supplementary Methods. The normalized ChIP-on-chip dataset has been deposited in the GEO database, under accession number GSE20150.
Quantitative reverse-transcription PCR
Total RNA was isolated from cells using the Absolutely RNA Miniprep Kit (Stratagene) following the manufacturer protocol including DNase I treatment. RT–PCR was performed using the SuperScript® First-Strand Synthesis System for RT–PCR (Invitrogen) using random hexamers to initiate the reaction. qRT–PCR was performed with the MX3000P platform (Stratagene) using the Syber Green quantification method and ROX normalization. Relative quantification was performed using the efficiency-corrected method. Genes were normalized using the Rps26 gene.
Gene expression profiling
For the time course of C2C12 differentiation, cells were handled as described in ref. (9). For profiling following RNAi, cells were transfected subconfluent in growth medium. The next day, differentiation was induced. Cells were harvested one day later. Expression profiling was done using the One-Color Microarray Gene Expression Platform and the 4x44k Whole Mouse Genome Oligo microarray (Design ID #014868) from Agilent Technologies. Details are given in Supplementary Methods. Microarray data can be downloaded from the Gene Expression Omnibus, under accession number GSE19988.
Gene expression data from the mouse GeneAtlas V3 [GNF3, (26)] were downloaded from GEO (GSE10246) and analyzed using the dChip program, with quantiles normalization (27). Genes were further normalized to a median of 0 and a SD of 1, across all tissues. Affymetrix microarray probes were matched to target genes or to Agilent expression profiling probes using matching GeneID numbers.
k-means clustering of expression profiling microarray data was performed with the Cluster 3 program using Euclidean metrics, and visualized using Java Treeview (28,29). When performed on the differentiation time-course experiments, clustering was done on all probes on the expression microarray and genes were assigned to one of five clusters. After clustering, only SIX1-bound genes were retained.
Immuno-fluorescence
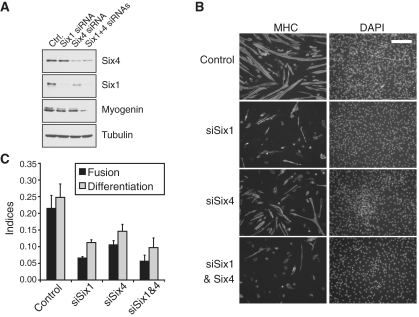
C2C12 myoblasts were grown in slide flasks (Nunc) and transfected with siRNA when they reached a confluence of 75–80%. Differentiation was induced the next day, and the cells were transfected again with the same siRNA molecules 24 h later to maintain the knock-down of target genes. The cells were fixed at 72 h after start of differentiation, and processed for immuno-fluorescent detection as described in ref. (30). Images were taken on a Leica DMI6000 B immuno-fluorescence microscope at a magnification of 10×. Images were acquired with the Velocity software version 5.0.2 (Improvision). Ten random fields were taken on each slide. The TIFF images from each color channel were merged and intensity adjusted using the Levels tool in Adobe Photoshop version 13.0. Fusion and differentiation indices were calculated; they are, respectively, defined as the fraction of MT with at least three nuclei over the total number of nuclei, and the fraction of nuclei within MHC-positive cytoplasms over the total number of nuclei.
Reporter constructs
All luciferase reporter constructs used in this study were made in the pGL3-Basic backbone (Promega). The pGL3-TATA-Luc construct, containing the TATA box from the adenoviral E1B gene, has been described before (31). The 6xMEF3-TATA-Luc construct was made by annealing complementary oligos (sequence given in Supplementary Table S6) containing the MEF3 site from the myogenin promoter (TCAGGTTTC), which were then ligated together to form concatamers and cloned into pGL3-TATA-Luc. The 4xEbox-TATA-Luc was constructed by excising the four E-box copy insert from 4R-tk-luc construct (32) and cloning into pGL3-TATA-Luc. The 5xE2F-TATA-Luc construct contains five copies of the E2F binding site sequence upstream of the E1B TATA box and has been described before (31). Constructs containing various combination of these TF binding sites were made using similar strategies. The myogenin gene promoter was cloned by PCR amplification from C2C12 genomic DNA. A mutation in the MEF3 site was incorporated by changing the sequence of this element from GGGCTCAGGTTTCTGT to GGGgagagagagaTGT.
Expression plasmids
The mouse SIX1 coding sequence cloned into the p3xFlag-myc plasmid (Sigma) was a kind gift from G. Merlino (33). The mouse SIX4, MYOD, MYOG and Heb-beta coding sequences were amplified from cDNA prepared from C2C12 total RNA and cloned into p3xFlag-myc. The human E2F1 and DP1 expression plasmids, with a pcDNA3 backbone, have been described previously (31).
Promoter reporter assays
Cells (C2C12 or 293T) were seeded in 24-well tissue culture plates, and transfected the following day with 1 μg of DNA using TurboFect (MBI Fermentas) according to the manufacturer’s protocol. The amount of DNA typically included 250 ng of reporter DNA and 5 ng of pRL-CMV (Promega), expressing renilla luciferase under the control of the CMV promoter. The cells were harvested 24 hours after transfection by washing in phosphate-buffered saline (PBS) and lysing in passive lysis buffer (Promega). Firefly and renilla luciferase activities were then assayed using the Dual-luciferase assay system (Promega) on a GloMax luminometer (Promega), and firefly enzymatic activities were divided by those of the renilla enzyme. Data were normalized by the intensities obtained with the empty, pGL3-Basic plasmid, or by those obtained with an empty expression plasmid. We quantified transcriptional synergy as the ratio of the transcriptional activation obtained with two TFs over the sum of activations obtained with each factor alone, as reported in ref. (34). In experiments where differentiation of C2C12 myoblasts was induced, the cells were transfected at the myoblast stage, and harvested either one day later, in their undifferentiated, sub-confluent state in growth medium, or allowed to grow to confluence, switched to differentiation medium, and harvested 4 days later. In these experiments, renilla activity drops several folds during the time course of differentiation and becomes an unreliable internal control. To circumvent this problem, firefly luciferase activity values were instead normalized by the total protein concentrations of the lysates, measured using the bicinchoninic acid protein assay (BCA, Pierce Chemicals).
RESULTS
Genome-wide binding profile of SIX1
In order to study the role of SIX factors in relation with the MRFs, we used the well characterized C2C12 mouse myoblast cell line, which represents a model of activated satellite cells undergoing terminal differentiation. Western blot on lysates of C2C12 cells at different time points of the differentiation program indicates that SIX1 is expressed in this myogenesis model system (Figure 1A). It is expressed in myoblasts, its levels increase at the onset of differentiation, and then decline in differentiated MT. In comparison, the expression of myogenin occurs after 24 h of differentiation (T24h) and is maintained in MT.
Figure 1.
SIX1 is functional in C2C12 myoblasts. (A) Western blot quantitation of SIX1 expression in differentiating C2C12 cells. Total cell lysates were used, and membranes were probed for SIX1 and myogenin expression. T0 h, confluent myoblasts at the onset of differentiation induction. T24 h, myoblasts induced to differentiate for 24 h. Beta-tubulin is given as a loading control. (B) Binding of SIX1 to the endogenous myogenin promoter, measured by qChIP. The chromatin from C2C12 cells at different stages of differentiation was immunoprecipitated with antibodies against SIX1 or MYOG, or control rabbit IgG. Enrichment of the ChIP samples for the myogenin promoter sequence and a negative control region (Hoxd10), were measured by qPCR. The binding signal expressed as a proportion of the starting material recovered in the ChIP reaction (percent of input). Asterisks indicate that the enrichment passes three significance tests, as indicated in ‘Materials and Methods’ section. The data represent the average of at least three biological replicates. Error bars: standard error on the mean (SEM). (C) Venn diagram showing the overlap in regions bound by SIX1 in myoblasts, at 24 h of differentiation, and in MT. The numbers given are the number of bound regions, and those in parentheses represent the number of distinct genes bound. (D) Validation of SIX1 ChIP-on-chip findings using qChIP. ChIP and data analysis were performed as described in Figure 1B. Average of at least three biological replicates. Error bars: SEM. Asterisks indicates significant binding event, as in B.
We next wanted to verify that SIX1 is functional in C2C12 cells. For this, chromatin immuno-precipitation assays followed by qRT–PCR (qChIP) were performed to detect binding of Six1 on the myogenin proximal promoter, a known target of SIX factors during embryogenesis (23). We could detect a specific binding of SIX1 at this promoter, but not at the negative control locus, the promoter of the HOXD10 gene, which is not expressed in C2C12 cells (Figure1B). Interestingly, the profile of binding of SIX1 changes over time, during differentiation: while we detected significant recruitment of SIX1 in both myoblasts and MT, it is in MT that we detected the strongest binding signal of SIX1 to the myogenin promoter. MYOG binding to its own promoter is only reliably detected in differentiated cells, as expected.
The phenotype of Six1-null mouse embryos suggests a substantial deregulation of the myogenic gene expression program in these animals, since several genes are affected by SIX1 loss. Further, our bioinformatic analyses suggest a widespread role for SIX1 in myogenesis. We reasoned that SIX1 must play a global, genome-wide role in controlling the differentiation of myoblasts into MT, and therefore decided to perform ChIP-on-chip analyses to determine the genomic binding profile of SIX1 in C2C12 cells. Because SIX1 expression is modulated during this process (Figure 1A) and SIX1 can be recruited on Myog promoter region (Figure1B), we performed ChIP-on-chip at three different time points: in proliferating myoblasts, in myoblasts after 24 h of differentiation induction (T24h) and in fully differentiated MT. We used oligonucleotide tiling DNA microarrays containing a total of 2.9-million probes representing ∼17% of the mouse genome, at an average density of more than five probes per kilobase (kb) (see Supplementary Material and methods for details).
We identified with high confidence 1019 binding sites of SIX1 in myoblasts, 739 at T24h, and 1852 in MT, for a total of 2624 individual genomic binding sites (Figure 1C and Supplementary Table S1). Representative genomic binding profiles are shown in Supplementary Figure S2. We used a multi-factorial algorithm to assign target genes to these binding events (‘Materials and Methods’ section), allowing us to determine that SIX1 targets 750 genes in myoblasts, 568 genes at T24, and 1313 genes in MT, for a total of 1793 genes. This reduction from number of binding sites to number of genes is explained in part by the facts that in certain instances SIX1 binds at more than one site near or within a gene, and conversely that some binding events are extremely far from any known transcribed region. Interestingly, the genome binding profile of SIX1 appears to be modulated during myogenesis, as this protein binds only partially overlapping sets of target loci at the three time points investigated. Despite being the least abundant in MT, it is in these differentiated cells that Six1 appeared to bind the largest number of loci. We stress however that due to the stringent cut-off we established in data analysis, a number of loci appearing as ‘condition-specific’ may in fact be bound by SIX1 at more than one time point (we privileged a low rate of false positives at the expense of reporting more false negatives). Figure 1D represents validatory qChIP on selected targets. Additional qChIP confirmatory data are shown in Supplementary Table S2. For these validations, we also used an antibody directed against SIX4, since it is expressed in C2C12 cells just like SIX1 and it is expected to bind a DNA sequence similar to that of SIX1(23,24). Interestingly, we found that SIX4 binding was indeed detected to most SIX1 target loci we tested, suggesting that SIX1 and SIX4 are recruited to similar loci in muscle cells, and that perhaps both factors operate together.
SIX1 binds to genes belonging to several functional categories
We used the gene ontology classification (35) to identify biological processes that would be over-represented among the SIX1-bound genes (Table 1). Genes participating in developmental processes are the most abundant and represent the most highly enriched functional category among SIX1 targets. Not surprisingly, the category of muscle development is a process whose abundance is well above random expectations. For example, and as expected, our genome-wide analysis identified the myogenin locus as bound by SIX1. Characterization of the role of SIX1 in regulating this category of target genes is currently ongoing and will be reported later. Interestingly, genes participating in neuronal development also constitute a large fraction of genes bound by SIX1. This may explain the appearance of a cluster of SIX1 target genes with predominant expression in nervous system tissues (Supplementary Figure S1). An interesting observation we made is that SIX factors engage in auto-regulation within their family: we detected binding of SIX1 to the Six1, Six4 and Six5 regulatory regions (Supplementary Table S2). This has important implications as it represents a putative mechanism for functional compensation, which is hypothesized to occur in the Six1 or Six4 knock-out mice (14,17,18,36). The Eya4 gene is also targeted by SIX1, suggesting a further layer of auto-regulation, since this protein functions as transcriptional co-regulator of SIX factor function, and interacts directly with SIX1 (37).
Table 1.
Selected gene ontology biological processes significantly enriched among genes targeted by SIX1
| Category | Targets: total (Mb; T24 h; Mt) | P-valuea | Example gene products |
|---|---|---|---|
| System development | 260 (124; 91; 183) | 2.40E-15 | PBX1, MEIS2, FOXA2, GSK3B, HOXC6, INHBB |
| Nervous system development | 99 (48; 35; 68) | 1.70E-04 | NEUROD6, GDNF, DPYSL2, FGF5, HES6, NRXN3 |
| Reg. of transcription from RNA pol. II promoter | 86 (39; 33; 65) | 5.51E-07 | HDAC4, ARNTL, E2F6, ELF1, KLF13, MEIS1 |
| Muscle development | 58 (21; 20; 48) | 7.35E-15 | MYF5, MYOD1, MYOG, VGLL2, MEF2C, PITX2 |
| Cell motility | 65 (30; 23; 48) | 3.14E-04 | ITGA6, ITGB3, STMN1, MTSS1, DCC, TNS1 |
| Regulation of cell proliferation | 60 (28; 25; 41) | 2.01E-03 | CDKN1A, E2F7, GAS1, KIFAP3, MYC, RB1 |
| Vasculature development | 56 (26; 20; 38) | 4.05E-06 | ADAMTS1, ANGPT1, ANXA2, CYR61, EMCN, PDGFA |
| Heart development | 41 (15; 15; 27) | 1.09E-05 | ACVR1, PITX2, NFATC4, BMP4, CSRP3, MEF2C |
| Muscle contraction | 33 (10; 7; 30) | 9.66E-10 | TTN, TNNI3, RYR1, MYH7, TPM3, ACTN3 |
| Kidney development | 17 (10; 7; 12) | 1.48E-02 | GLI2, BDNF, GZF1, NPNT, GREM1, SPRY1 |
aThe P-values given are using Fisher’s exact test, with correction for multiple hypothesis testing by the algorithm of Benjamini and Hochberg. The background set of genes used in these calculations was the entire mouse genome.
Interestingly, the function of SIX1 is not limited to the regulation of genes that enable the early developmental phases of myogenesis: we note that several genes encoding components of the contractile apparatus are also targeted by SIX1, such as titin, various troponin (Tnni1, Tnni2, Tnni3) and myosin (Myh7, Myh13, Myl1, Myl4) isoforms and ATP-dependent sodium–potassium channels (Atp1a1, Atp1a2, Atp2a1). These genes tend to have an expression pattern highest in muscle tissues, as compared to other adult mouse tissues (Supplementary Figure S1 and Table S3). We note that genes characteristic of both fast and slow muscle fiber types are bound by SIX1, for example Tnnc1, Tnni1 and Tnnt1 (slow) and Atp2a1, Tnni2 and Tnnt3 (fast).
Coordinate genomic binding of SIX factors and the MRFs
DNA sequence analysis of the genomic regions bound by SIX1 revealed that the E-box element, recognized by MYOD and MYOG, is significantly enriched among these sequences, as compared to randomly selected sequences surveyed in our ChIP-on-chip experiments (data not shown). Cao et al. (11) recently published the global mapping of MYOD binding sites using ChIP-seq in C2C12 cells. Strikingly, we found that, regardless of the condition, 39% of SIX1-bound loci identified herein overlap with the MYOD-bound regions reported by Cao et al. (400 out of 1021 in myoblasts, and 741 out of 1853 in MT, with overlap of at least 1 bp) This significant overlap (P < 1 × 10−15 by hypergeometric distribution test) strongly suggests a concerted mode of action of SIX factors and the MRFs.
SIX1 and SIX4 are required for myoblast differentiation
Considering our finding that SIX factors bind to a large number of genes playing important roles in myogenesis, we used short interfering RNA duplexes (siRNAs) to knock-down their expression from myoblasts, in order to determine if SIX1 and SIX4 functions are required for myoblast differentiation. We transfected C2C12 myoblasts with siRNAs targeting either the Six1 mRNA, the Six4 mRNA, both together or a control non-targeting RNA duplex. One day later, the cells were induced to differentiate and were harvested after 2 or 4 days in differentiation medium, for immuno-blotting or for immuno-fluorescent scoring of their differentiation, respectively. Western blots (Figure 2A) indicate that the knock-downs of SIX1 and SIX4 were substantial. Interestingly, the expression of myogenin was markedly decreased after knock-down of either Six factor, an effect in line with the role of SIX factors on MYOG induction during embryonic myogenesis. The cells were stained for their expression of myosin heavy-chain (MHC), a terminal myogenic differentiation marker, and their DNA was counterstained with DAPI, allowing us to measure their differentiation and fusion indices (Figure 2B and C). We found a remarkable statistically significant impairment of both myoblast differentiation and fusion when either SIX factors were knocked-down, indicating that these factors are essential not only for induction of myogenin expression, but also for the proper course of differentiation. Similar effects were obtained when knock-down was performed for SIX1 or SIX4 using pools of three siRNA duplexes (Supplementary Figure S3) which exclude the sequences used in Figure 2, making it unlikely that the failure to differentiate caused by knock-down of SIX proteins is due to off-target effects. Together, these results show that SIX1 and SIX4 are functional in C2C12 cells and that they are likely to directly regulate the induction of myogenin expression. Our results also indicate that because they are required for myoblast differentiation, SIX factors function extend beyond the early phase of embryonic muscle development.
Figure 2.
Six1 required for proper myogenesis and cell differentiation. (A) Protein levels of SIX1, SIX4 and MYOG after the knock-down of SIX1 and/or SIX4 from C2C12 cells, after 48 h of differentiation. The levels of beta-tubulin are shown as a loading control. (B) Fusion and differentiation indices of C2C12 transfected with siRNAs against SIX1 or a control non-targeting siRNA, and induced to differentiate for 72 hours. Asterisks indicate a paired t-test P < 0.05 in comparison with the control siRNA conditions. (C) Representative microscopic fields from each condition tested in panel B. MHC: myosin heavy chain. Scale bar: 200 microns. The same magnification was used for all frames shown.
A pervasive role for SIX proteins: activating gene expression during myogenesis
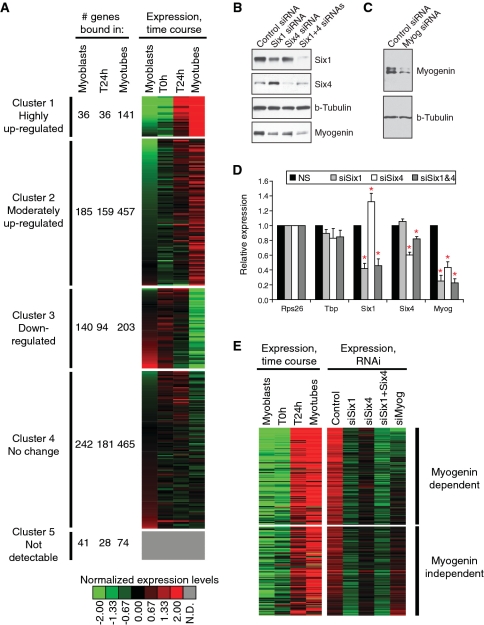
The genome-wide binding profile of SIX1 suggests that several genes important for muscle formation and activity are directly regulated by SIX factors. In order to establish a correlation between gene regulation and SIX function, we performed expression profiling of C2C12 cells undergoing differentiation (Supplementary Table S4). Figure 3A represents as a heat-map the expression profiles of direct SIX1 targets as myoblasts differentiate into MT, clustered by a k-means algorithm. We note that SIX1 target genes have a variety of expression profiles during differentiation, with substantial numbers falling into each of five clusters, including genes with no detectable expression in C2C12 cells (cluster #5). Interestingly however, a large proportion of target genes are either highly or moderately induced, during differentiation (clusters #1 and #2). This analysis reveals that binding by SIX1 in MT is strongly associated to target gene activation: for example, of the 21 493 genes profiled using our expression microarrays, 446 fall into cluster #1 (highly induced), and 141 of those are bound by SIX1 in MT. The chance for this association to occur randomly is exceedingly small (P < 10−20 for the cumulative hypergeometric probability). Since this analysis shows that nearly one-third of the most induced genes during myoblast differentiation are targeted by SIX1, we conclude that this factor must play a very important role in activating gene expression during myogenesis.
Figure 3.
SIX1 binding is associated to gene activation during myoblast differentiation. (A) Heat-map representing the expression profiles of SIX1 target genes identified in our ChIP-on-chip, grouped in clusters formed using all genes represented on the expression profiling microarrays. A legend giving the color code for normalized gene expression values is given at the bottom. The color grey is assigned to genes with undetectable (N.D.) expression in C2C12 cells. For each of five clusters, the number of targets bound in each condition (myoblasts, T24 h or MT) is given. SIX1 target genes not represented on expression arrays were omitted from this analysis. (B) Protein expression levels of SIX1 and SIX4 after their knock-down using siRNA duplexes. Myogenin levels are shown as well. Beta-tubulin levels are shown as a loading control. (C) Expression levels of myogenin after its knock-down by siRNA. Beta-tubulin levels are shown as a loading control. (D) mRNA expression levels of Six1 or Six4 after their knock-down, as measured by qRT-PCR. Data were normalized relative to the Rps26 gene. The levels of Tbp (TATA-binding protein) mRNA serve as an additional invariant control. The data represent the average of at least three biological replicates. Error bars: SEM. Asterisks indicate a Student’s paired t-test P-value lower than 0.05, when compared with the control siRNA duplex. (E) Heat-map representing the expression levels of SIX1 target genes affected by knock-down of SIX1 (2-fold change or more) during normal differentiation (left-hand side) or after the knock-down of SIX1, SIX4, SIX1 and SIX4 or MYOG (right-hand side). The color code is the same used in panel A. Genes are ranked from top to bottom in decreasing order of down-regulation after myogenin knock-down. Those for which the knock-down of MYOG gave a 50% or more reduction in expression are represented in the top portion (MYOG-dependent), otherwise they lie in the bottom portion (MYOG-independent).
In order to establish a more direct link between the role of SIX proteins and gene activation during differentiation, and to verify whether the function of these proteins is required for the efficiency of myogenesis, we examined the effects of loss of SIX expression caused by RNA interference. As for Figure 2A, we transfected C2C12 myoblasts targeting either the SIX1, SIX4 or both, or a control non-targeting RNA duplex, and induced the cells to differentiate one day later. For comparison, we also used an siRNA duplex against myogenin. The cells were allowed to differentiate for one day, after which we harvested their RNA and performed gene expression profiling. This early time point was chosen to minimize secondary effects, which may plague this sort of assay. Western blots and quantitative reverse-transcription PCR (qRT–PCR) indicate that substantial knock-downs were achieved (Figure 3B–D). Here again, the knock-down of either SIX1 or SIX4 led to a significant decrease in the levels of MYOG protein and mRNA. By microarray expression profiling, we found 380 genes to be down-regulated by more than 50% after SIX1 loss of function; among those, 165 are direct targets as they are bound by SIX1 (Supplementary Table S5 and Supplementary Figure S3). We lined up the expression profiles of these deregulated SIX1 targets with their normal profiles during differentiation (Figure 3E). Strikingly, we found that the vast majority are induced during myogenesis, and belong to cluster 1 (highly induced, 112 genes) or cluster 2 (moderately induced, 48 genes), in the differentiation time course expression profiles shown in Figure 3A. Interestingly, the knock-down of SIX4 also caused the down-regulation of several of these SIX1 target genes (Supplementary Table S5), further reinforcing the notion that SIX1 and SIX4 may exert their functions at least partially together. Gene expression information after the knock-down of MYOG allows us to discern two categories of SIX1 targets: those that also depend on MYOG function for efficient induction during differentiation, and the MYOG-independent SIX1 targets, which are unaffected by the suppression of this MRF. MYOG-dependent targets therefore reflect the requirement of these genes for both SIX and MYOG in the induction process. In addition to demonstrating an essential regulatory role for SIX1 and SIX4 in enabling gene expression induction and differentiation of myoblasts, these results provide further support to the notion of cooperation between the MRF and SIX classes of transcriptional regulators.
Transcriptional synergy between SIX and MRFs
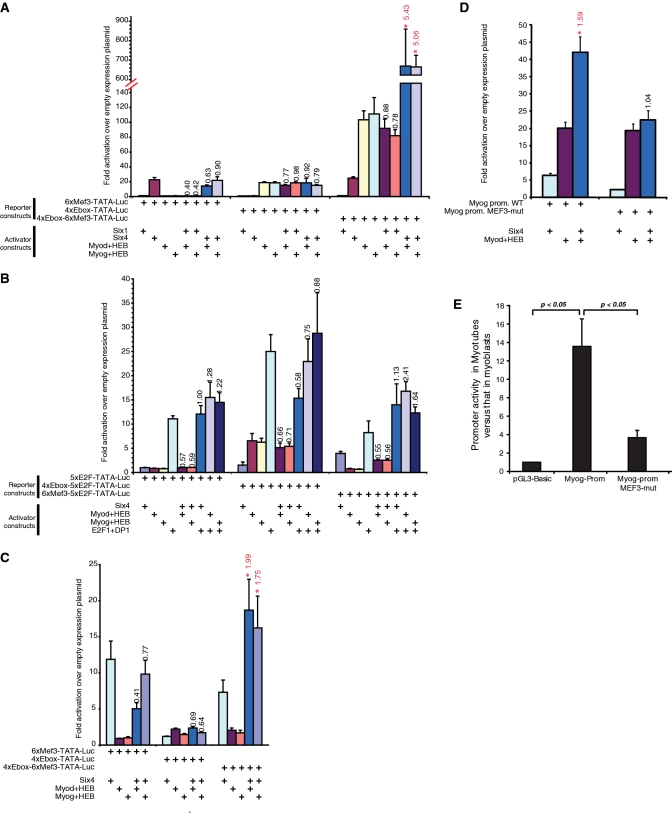
Having identified a group of genes depending on SIX1, SIX4 and MYOG expression for appropriate induction during differentiation, we finally set out to determine if these TFs can regulate gene expression in a cooperative manner. For this, we used luciferase assays where reporter constructs contained tandem copies of the MEF3 or the E-box elements, or both types of elements together, upstream of a TATA box and the firefly luciferase gene. These constructs were co-transfected in 293T cells along with various combinations of expression plasmids for SIX1, SIX4, and MYOD or MYOG together with the E-protein HEB-β. This cell line was chosen for its lack of expression of the MRFs, giving lower background signal in these assays. We found that SIX4, but not SIX1, activates constructs containing MEF3 sites (Figure 4A). This is perhaps due to the facts that SIX1 requires additional co-activators to be present which might be in limiting amounts (38), or that it functions in certain contexts as a repressor (39). Both MYOD and MYOG activated transcription from the E-box-containing constructs. Interestingly, there was a significant and strong, well above additive effect of combined SIX4 and MYOD or MYOG, indicating that the two classes of TFs indeed have the ability to synergize in regulating gene expression. The extent of synergy is 5.43 for SIX4 and MYOD, and 5.06 for SIX4 and MYOG. We note that the effect of MYOD and MYOG are stronger on the construct containing both MEF3 and E-box elements, perhaps due to the fact that 293T cells express certain members of the SIX family, including SIX4 (data not shown). The synergy between SIX4 and the MRFs depends on the presence of both cognate binding sites; therefore it is unlikely that it relies only on protein–protein interactions. This fact also rules out the possibility that the abundance of one TF impacts transcription indirectly, for example by enhancing the expression of the other factor.
Figure 4.
Transcriptional synergism between SIX and MRF transcription factors. (A) 293T cells were transfected with the indicated firefly luciferase reporter constructs and various combinations of expression plasmids for transcription factors, as indicated. A plasmid driving the expression of renilla luciferase was used as internal control, for normalization purposes. Normalized firefly luciferase activities were divided by those obtained in cells transfected with the empty expression plasmid, giving values of fold activation over empty plasmid. Each experiment was performed a minimum of three times. Histogram values represent the average ± SEM of all replicates. Note the axis break and change of scale. For conditions with a combination of transcriptional activators, the synergy value is indicated above the histogram bar. Synergy values that are statistically significant (P < 0.05 by one-tailed paired t-test) are marked in red with an asterisk. (B) The experiment was performed as in panel A, but using different combinations of reporter constructs and transcriptional activators. Synergy is indicated as in panel A. (C) Transcriptional activity of SIX factors and the MRFs in C2C12 myoblasts. Cells were transfected with the indicated firefly luciferase reporter constructs and various combinations of expression plasmids for transcription factors, as indicated. Samples and data were processed as in panel A. (D) Transcriptional activity of SIX4 and MYOD in 293T cells transfected with a reporter construct consisting of the myogenin proximal promoter fused to the luciferase gene, or with a similar construct where the MEF3 site has been mutated. Samples and data were processed as in panel A. (E) Luciferase activity in cells transfected with the wild-type myogenin promoter reporter construct or a version with its MEF3 site mutated, or the empty luciferase plasmid (pGL3-basic). The results show the ratio of luciferase activity in MT divided by that in myoblasts.
In order to further validate the specificity of this transcriptional synergy, we generated reporter constructs containing multiple copies of the E2F binding site, alone or with tandem copies of the MEF3 or E-box sequence elements (Figure 4B). E2F sites are bound by the E2F1-DP1 pair, which is involved in regulating progression through the cell cycle (40). No significant transcriptional cooperation was observed between E2F1-DP1 and either SIX4 or the MRFs. This supports the idea that the synergy between SIX4 and the MRFs is specific to these two classes of transcriptional activators, and that it does not reflect a general propensity for these factors to cooperate with other regulators indiscriminately.
We also verified that transcriptional synergy between the MRFs and SIX factors can occur in myoblasts, in addition to 293T cells. We therefore repeated these luciferase reporter assays in C2C12 cells, and found that indeed SIX4 can cooperate with MYOD or MYOG to activate transcription in this setting (Figure 4C).
Finally, because the myogenin gene promoter has been shown to be under the control of MYOD (41) and Six factors (23) during development, we asked whether members of each family could synergize together. SIX4 and MYOD activated the reporter gene luciferase fused to the myogenin promoter in a synergistic fashion (Figure 4D). The synergy depends entirely on the presence of a MEF3 site since it is completely abrogated when a reporter construct with a mutation in the MEF3 site is used. Here again, the MEF3 site requirement rules out the possibility that the synergy merely relies on protein–protein interactions between SIX factors and the MRFs. To conclude, we analyzed the luciferase activity from the wild-type or MEF3-mutated myogenin reporter construct, in C2C12 cells that are undergoing differentiation. As expected, luciferase activity is much higher in MT than in myoblasts, when the wild-type construct is compared with the empty reporter plasmid pGL3-Basic (Figure 4E). When a mutation is introduced in the MEF3 site, this induction is strikingly reduced by 70%, as compared to the wild-type construct. These results strongly suggest that cooperation between SIX and MRF factors is required for proper induction of the expression of target genes such as myogenin.
Together, our data indicate a likely mechanism by which the transcriptional function of MRFs may be modulated during myogenesis, that is by cooperation with SIX TFs.
DISCUSSION
The work presented here provides evidence that MRFs and SIX family TFs function in a cooperative manner to regulate gene expression during skeletal myogenesis. Additionally, the data contained herein offers interesting insight to the question of the molecular basis of the developmental defects in Six1-null animals.
Gene transcription is rarely regulated by single TFs in isolation, and combinatorial regulation offers a means by which precise expression profiles can be established. This occurs when the combined action of two transcriptional regulators together generates a transcriptional output that is different from that obtained with either factor alone. The timing, amplitude or duration of the transcriptional response, or the selection of target sites, may be modulated in a combinatorial manner (42). These considerations are often discussed in theoretical terms, and while the scientific literature abounds with tools to predict instances of combinatorial regulation [e.g. (43,44)], much fewer studies have gone beyond reporting the co-occurrence of TF binding sites. The work presented herein represents a step further in explaining fine-tuning of gene expression within the transcriptional regulatory network that oversees myogenesis.
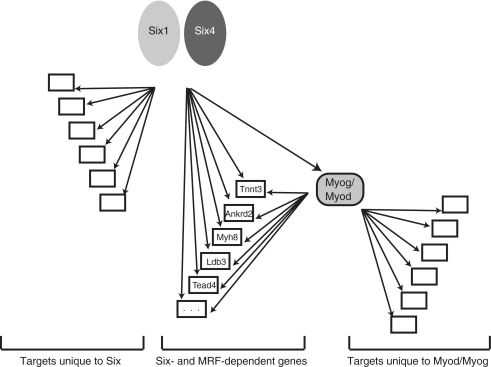
Using bioinformatic analysis of DNA sequences, we have uncovered a novel way by which MRF function might be modulated, that is by cooperation with SIX family TFs. In our previous work, a high abundance of MEF3 sites, the DNA sequence recognized by SIX factors, was found at MRF target genes most induced during differentiation, but not at MRF targets with modest or no induction (9). Conversely, our current findings demonstrate the specific enrichment of the E-box motif among SIX1-bound genomic regions. Moreover, our ChIP-on-chip and expression profiling experiments confirmed that the function of SIX and MRF factors are in several cases tightly connected: we uncovered a substantial group of direct SIX1 target genes which are also direct targets of MYOD or MYOG (9–11). This novel notion of cooperation between SIX and MRFs is reinforced by the results of our promoter reporter assays: the two families of TFs activate transcription in a synergistic fashion. Our data suggest that the function of SIX factors in combinatorial regulation with the MRFs would be to allow maximum target gene activation by these myogenic regulators. Our findings evoke a model (Figure 5) whereby SIX1 function is not only upstream of the MRFs, during embryonic development, but also parallel to them, during terminal differentiation of myoblasts. At the same time, SIX factors also function independently of MRF activity to induce during myogenic differentiation the expression of a distinct set of target genes, possibly in concert with different TFs.
Figure 5.
Model of SIX and MRF function during myoblast differentiation. See text for details.
The exact mechanism by which the synergy between SIX4 and the MRFs is made possible is not perfectly clear at this stage. Logically, one would expect the two classes of TFs to enhance distinct steps of the transcription process, such as initiation, elongation, or the establishment of a permissive chromatin structure at the proximal promoter (45). The MRFs are able to recruit certain transcriptional co-activators to target promoters. For instance, MYOD can recruit the PCAF histone acetyltransferase, via an interaction with P300, which enhances its transcriptional activity (46,47). It can also interact with TAF3 (TATA-binding protein Associated Factor 3), thereby enhancing activation of target genes (48). MYOG can interact with RBP3, a core subunit of the RNA polymerase II complex (49), an interaction which is also thought to facilitate target gene transcription. SIX factors, on the other hand, are best characterized with regards to their interactions with the EYA and DACH families of transcriptional co-regulators (50). How EYA co-factors enhance RNA pol II-dependent transcription is currently unclear, but is thought to involve their enzymatic activity as protein phosphatases, and they may act at least partly by reversing the repressive effect of DACH proteins on SIX function (16). This is thought to involve the CREB-binding protein CBP, a protein related to P300 (51). Combinatorial regulation of transcription may in certain instances involve interaction between cooperating TFs. However, we believe this not to be the case with SIX and MRF proteins. In numerous attempts, we have failed to detect an interaction between members of these two families, in co-immunoprecipitation assays. Furthermore, our results with luciferase reporter assays clearly indicate that cooperative function of the two families of TFs requires the binding of each factor to their respective binding site. Even if direct contacts between these proteins are made, they are unlikely to be sufficient for synergy to occur. Finally, two recent reports shed light on possible mechanisms by which SIX factors could regulate target gene expression and potentiate the function of MRFs. A recent report demonstrates that recruitment of SIX4 and the histone demethylase UTX at the Myog and Ckm (muscle creatine kinase) promoters is required for induction of these genes at the onset of myoblast differentiation (52). UTX functions by removing methyl groups from histone H3 lysine 27, a covalent chromatin mark which is associated with transcriptional repression (53). A second recent study has shown that the myogenin locus, which depends on MRFs and SIX factors for proper expression, is methylated on DNA in undifferentiated cells, and that loss of CpG methylation coincides with its activation; this removal or loss of DNA methylation depends on SIX1 function (54). These two studies therefore reveal the possibility that SIX factors recruit enzymes, or enzyme complexes, which render the chromatin template more conducive to transcription and to MRF function, explaining synergistic function. Another non-mutually exclusive scenario is that binding of MRFs to their target loci is facilitated by the chromatin reorganization mediated by SIX factors. More work will be required to elucidate the mechanistic details of these possibilities and to determine if these phenomena are widespread or limited to only a subgroup of SIX and MRF target loci.
Our luciferase assays demonstrated a strong activity of SIX4, but not SIX1, as transcriptional activator on the constructs tested. It is possible that levels of transcriptional co-regulators required to interact with over-expressed SIX1 in order for an activation response to be generated are suboptimal in the system we used. Both cell lines used in luciferase assays (C2C12 and 293T) endogenously express SIX1 and SIX4, but endogenous SIX4 may be the only one in limiting amounts. It is also possible that SIX1 is required for SIX4 transcriptional function, and that the synergy with the MRFs requires the transcriptional activation domain of SIX4 [the SIX1 protein lacking a similar domain (12)]. This would not be an isolated case, since the cooperative function between SIX4 or SIX5 and their EYA co-factors indeed requires these C-terminal activation domains (55). In further support of the notion that the functions of SIX1 and SIX4 are tied together are the following three facts: (i) a marked number of SIX1 targets we tested in gene-specific qChIP assays also proved to be bound by SIX4, (ii) our knock-down experiments indicate that a very substantial number of these SIX1/SIX4 targets depend on wild-type levels of both proteins for their induction during differentiation and (iii) terminal differentiation is greatly lowered upon SIX4 or SIX1 knock-down. This is not due to decreased SIX1 expression after SIX4 knock-down, since in fact Six1 mRNA levels are slightly but significantly higher after SIX4 knock-down (Supplementary Figure S4).
A clear requirement for SIX1 during embryonic muscle development has been established before (14–16). However, the molecular basis of the developmental defects in Six1-null animals remains to be firmly established. Using ChIP-on-chip, we have identified a large number of new direct target genes of the SIX1 TF, which significantly extend its previously known target repertoire. Interestingly, we have discovered binding sites for SIX1 in the vicinity of genes that play critical roles in skeletal myogenesis, and which have been shown to be deregulated in SIX1 knock-out embryos. For example, the Pax3, Myf5, Myod1, Myog and Pbx1 gene were found to represent direct SIX1 and SIX4 targets. The relevance of binding of these genes by SIX factors in C2C12 myoblasts, which do not express certain of these target genes at any detectable level, and in the developing embryonic muscle, where a clear requirement for these genes has been established, deserves further scrutiny.
A related observation we made is that numerous SIX1 targets identified herein are well known to control the function or development of other tissues affected by the loss of SIX1 in mice. For instance, SIX1 is essential for the development of various sensory tissues, such as the otic and olfactory epithelia, as well as the kidney, and we found SIX1 to bind several genes involved in these processes (e.g. Gdnf, Bdnf, Pbx1, Spry1, Grm1, Mef2c, Hes6) (56–61). It is certainly a surprise to find SIX1 binding to such genes in a myoblast cell line, which is presumably restricted to a mostly myogenic fate. It will be interesting to determine whether these genes also participate in muscle development and/or function, and if their deregulation in Six1-null animals contributes to the muscle phenotype, since most of these genes are in fact expressed, and therefore potentially functional, in myoblasts. Mef2c and Hes6 are two examples of developmental regulators which definitely support this hypothesis, as they are involved in both neuronal and muscular processes (62–66), and their expression in differentiating myoblasts depends on SIX factors (Supplementary Table S6 and Supplementary Figure S5). Likewise, Pbx1 is a gene involved in both myogenesis and nephrogenesis (67,68) and could work downstream of SIX1 in both muscle and kidneys.
Using RNA interference of SIX factors, we discovered that SIX4 is essential to terminal differentiation of myoblasts. This reinforces the notion that the synergistic function SIX4 carries out with the MRFs is crucial for myogenesis. However, this observation is seemingly at odds with the absence of a clear muscle phenotype in Six4 knock-out mice (17). We suggest that the compensatory mechanisms which mask the effects of loss of SIX4 expression during embryonic development do not function in our myoblast culture model. SIX5 is the only other SIX family member with an extended C-terminal activation domain similar to that of SIX4. It has been hypothesized that SIX5 levels in embryonic myogenic structures are sufficiently high to compensate for the absence of SIX4 (17,69). In contrast, SIX5 is expressed at only very low levels in C2C12 cells (Supplementary Figure S5), and its levels are unaffected by SIX1 or SIX4 knock-down (Supplementary Table S4). Therefore, it may not function in a compensatory manner as it does in the embryo. Moreover, the time scales of events in these two loss-of-function systems (RNA interference in cultured cells, compared with developing knock-out embryo) are different, which may also explain differences in the emergence of compensatory responses, or lack thereof. Finally, this SIX4 ablation phenotype specific to adult myoblasts suggests that SIX4 function may be required primarily for myogenic regeneration. In this context, it will be of particular interest to study the impact of conditional loss of function of SIX factors during the repair of adult muscle tissue.
FUNDING
Ontario Graduate Scholarship award (to Y.L.); National Science and Engineering Research Council of Canada (NSERC) scholarship (to I.C.); University of Ottawa and research grants from the Muscular Dystrophy Association and the NSERC (to A.B.). Funding for open access charge: Muscular Dystrophy Association (to A.B.).
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Glen Merlino (National Institutes of Health) for the gift of murine SIX1 expression plasmid; Heide Ford (U. Colorado in Denver) for the gift of anti-human SIX1 antibodies in the early phase of the project; Geneviève Pelletier and Galen Chen for technical assistance; Ilona Skerjanc, Jean-François Couture and Jeffrey Dilworth for advice and critical review of our manuscript.
REFERENCES
- 1.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 3.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 5.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 6.Naidu PS, Ludolph DC, To RQ, Hinterberger TJ, Konieczny SF. Myogenin and MEF2 function synergistically to activate the MRF4 promoter during myogenesis. Mol. Cell Biol. 1995;15:2707–2718. doi: 10.1128/mcb.15.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl Acad. Sci. USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 13.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol. Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- 15.Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech. Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 17.Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, Okamura HO, Kitamura K, Muto S, Kotaki H, et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 18.Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 19.Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti JE, Kawakami K, Xu PX, Kelly R, Petrof BJ, et al. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell Biol. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niro C, Demignon J, Vincent S, Liu Y, Giordani J, Sgarioto N, Favier M, Guillet-Deniau I, Blais A, Maire P. Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev. Biol. 2009;338:168–182. doi: 10.1016/j.ydbio.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 21.Bessarab DA, Chong SW, Srinivas BP, Korzh V. Six1a is required for the onset of fast muscle differentiation in zebrafish. Dev. Biol. 2008;323:216–228. doi: 10.1016/j.ydbio.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Ando Z, Sato S, Ikeda K, Kawakami K. Slc12a2 is a direct target of two closely related homeobox proteins, Six1 and Six4. FEBS J. 2005;272:3026–3041. doi: 10.1111/j.1742-4658.2005.04716.x. [DOI] [PubMed] [Google Scholar]
- 23.Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl Acad. Sci. USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc. Natl Acad. Sci USA. 2007;104:11310–11315. doi: 10.1073/pnas.0611299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carnac G, Fajas L, L'Honore A, Sardet C, Lamb NJ, Fernandez A. The retinoblastoma-like protein p130 is involved in the determination of reserve cells in differentiating myoblasts. Curr. Biol. 2000;10:543–546. doi: 10.1016/s0960-9822(00)00471-1. [DOI] [PubMed] [Google Scholar]
- 26.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blais A, Monte D, Pouliot F, Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J. Biol. Chem. 2002;277:31679–31693. doi: 10.1074/jbc.M204554200. [DOI] [PubMed] [Google Scholar]
- 32.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc. Natl Acad. Sci. USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66:1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 34.Markus M, Du Z, Benezra R. Enhancer-specific modulation of E protein activity. J. Biol. Chem. 2002;277:6469–6477. doi: 10.1074/jbc.M110659200. [DOI] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klesert TR, Cho DH, Clark JI, Maylie J, Adelman J, Snider L, Yuen EC, Soriano P, Tapscott SJ. Mice deficient in Six5 develop cataracts: implications for myotonic dystrophy. Nat. Genet. 2000;25:105–109. doi: 10.1038/75490. [DOI] [PubMed] [Google Scholar]
- 37.Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 38.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl Acad. Sci. USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- 40.Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 42.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das D, Zhang MQ. Predictive models of gene regulation: application of regression methods to microarray data. Methods Mol. Biol. 2007;377:95–110. doi: 10.1007/978-1-59745-390-5_5. [DOI] [PubMed] [Google Scholar]
- 44.Babu MM. Computational approaches to study transcriptional regulation. Biochem. Soc. Trans. 2008;36:758–765. doi: 10.1042/BST0360758. [DOI] [PubMed] [Google Scholar]
- 45.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 48.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corbi N, Di Padova M, De Angelis R, Bruno T, Libri V, Iezzi S, Floridi A, Fanciulli M, Passananti C. The alpha-like RNA polymerase II core subunit 3 (RPB3) is involved in tissue-specific transcription and muscle differentiation via interaction with the myogenic factor myogenin. FASEB J. 2002;16:1639–1641. doi: 10.1096/fj.02-0123fje. [DOI] [PubMed] [Google Scholar]
- 50.Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu. Rev. Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- 51.Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 54.Palacios D, Summerbell D, Rigby PW, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse myogenin gene. Mol. Cell Biol. 2010 doi: 10.1128/MCB.00050-10. doi:10.1128/MCB.00050-10 [24 May 2010, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bricaud O, Collazo A. The transcription factor six1 inhibits neuronal and promotes hair cell fate in the developing zebrafish (Danio rerio) inner ear. J. Neurosci. 2006;26:10438–10451. doi: 10.1523/JNEUROSCI.1025-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christophorou NA, Bailey AP, Hanson S, Streit A. Activation of Six1 target genes is required for sensory placode formation. Dev. Biol. 2009;336:327–336. doi: 10.1016/j.ydbio.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 59.Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, et al. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda K, Ookawara S, Sato S, Ando Z, Kageyama R, Kawakami K. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev. Biol. 2007;311:53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 64.Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl Acad. Sci. USA. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–2943. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- 66.Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH. Hes6 regulates myogenic differentiation. Development. 2002;129:2195–2207. doi: 10.1242/dev.129.9.2195. [DOI] [PubMed] [Google Scholar]
- 67.Schnabel CA, Godin RE, Cleary ML. Pbx1 regulates nephrogenesis and ureteric branching in the developing kidney. Dev. Biol. 2003;254:262–276. doi: 10.1016/s0012-1606(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 68.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 69.Fougerousse F, Durand M, Lopez S, Suel L, Demignon J, Thornton C, Ozaki H, Kawakami K, Barbet P, Beckmann JS, et al. Six and Eya expression during human somitogenesis and MyoD gene family activation. J. Muscle Res. Cell Motil. 2002;23:255–264. doi: 10.1023/a:1020990825644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.