Abstract
miRNAs play key roles in the nervous system, where they mark distinct developmental stages. Accordingly, dysregulation of miRNA expression may have profound effects on neuronal physiology and pathology, including cancer. Among the neuronal miRNAs, miR-9 was shown to be upregulated during in vitro neuronal differentiation and downregulated in 50% of primary neuroblastoma tumors, suggesting a potential function as an oncosuppressor gene. In this study we characterized the promoter and the transcriptional regulation of the miR-9-2 gene during neuronal differentiation. We found that, despite its localization inside an exon of a putative host-gene, miR-9-2 is expressed as an independent unit with the promoter located in the upstream intron. By promoter fusion and mutational analyses, together with RNAi and Chromatin immunoprecipitation assays, we demonstrated that the concerted action of the master transcriptional factors RE1-silencing transcription factor (REST) and cAMP-response element binding protein (CREB) on miR-9-2 promoter induces miRNA expression during differentiation. We showed that the repressor REST inhibits the activity of the miR-9-2 promoter in undifferentiated neuroblastoma cells, whereas REST dismissal and phosphorylation of CREB trigger transcription in differentiating cells. Finally, a regulatory feed-back mechanism, in which the reciprocal action of miR-9 and REST may be relevant for the maintenance of the neuronal differentiation program, is shown.
INTRODUCTION
The mammalian nervous system is the anatomic district including the highest cellular diversity, with each cell type displaying a specific gene expression profile that dynamically changes in response to synaptic activity. Such flexibility requires a highly orchestrated coordination of gene expression programs, which is achieved through integrated levels of gene regulation at all stages of neuronal development.
A significant portion of such regulation is provided by microRNAs, ‘fine-tuning’ modulators of gene expression at the post-transcriptional level (1) that, through their pleiotropic action, have a relevant role in neuronal development as well as in maintenance and plasticity of neural connections (2).
Mammalian miRNA genes are organized either in gene clusters, which are transcribed in a coordinate manner from a single polycistronic transcription unit, or as monocistronic genes (3). Such genomic assortment is further complicated by intronic and exonic localization. About 62% of miRNAs are located within the introns of either non-protein-coding or protein-coding transcription units, whereas only 10% are encoded inside exons. A further 27% displays intergenic organization (4,5).
miRNA expression in different cell types and organisms is a spatio-temporal regulated process (6,7). Notably, miRNA mis-expression is often associated with human diseases, such as cancer (8,9), making relevant the study of the mechanisms governing their expression.
Evidence is emerging for the importance of post-transcriptional control of miRNAs (10); nevertheless, transcriptional control, which is the major level of regulation, is still poorly understood, since only a handful of miRNA promoters have been functionally characterized (11).
In this article, we present structural and functional characterization of the promoter region of the human miR-9-2 gene, encoding for an evolutionary conserved miRNA, expressed almost exclusively in the brain (12,13). miR-9 has been implicated in nervous system development, physiology and pathology in several organisms (14,15). We recently highlighted its property as an onco-suppressor miRNA, with a crucial role in repressing cell proliferation of human neuroblastoma and medulloblastoma cell lines (16,17).
We demonstrate that the transcription of the miR-9-2 gene, which displays an unusual exonic localization, is independent from that of its host gene, whereas it is directed by a previously unidentified promoter.
We identify a relevant function for the transcription factors RE1-silencing transcription factor (REST) and cAMP-response element binding protein (CREB) in controlling miR-9-2 gene expression. The repressor REST is crucial for maintaining low levels of miR-9-2 in undifferentiated cells, whereas REST dismissal and simultaneous phosphorylation of the already bound neuronal activator CREB trigger miR-9-2 transcription, ensuring high levels of miR-9 during in vitro neuronal differentiation.
Finally, our data directly demonstrates the existence of a feedback mechanism in which the reciprocal action of miR-9 and REST, previously described as one of the miR-9 targets (18), may be responsible for the maintenance of the neuronal differentiation program.
MATERIALS AND METHODS
Cell cultures and treatments
SK-N-BE cells were cultured in RPMI medium 1640 (Gibco, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (EU Standard, South American Origin, Gibco), 1× l-glutamine, and penicillin/streptomycin, and induced to differentiation by 10 μM all-trans-RA (Sigma-Aldrich, St Louis, MO, USA). Plasmids, siRNAs and LNA oligonucleotides were transfected into SK-N-BE cells by Lipofectamine and Plus Reagent (Invitrogen, Carlsbad, CA, USA).
RNA extraction and cDNA synthesis
Total RNA was extracted from confluent plates of untreated or RA-treated SK-N-BE cells, using TRIzol (Invitrogen) as recommended by the manufacturer’s protocol. Prior to cDNA synthesis, 5 µg of RNA samples were treated with 1 U of DNase (Promega, Madison, WI, USA). cDNA was generated using SuperScript III (Invitrogen).
Rapid amplification of 5' complementary DNA ends (5′RACE)
miR-9-2 Transcriptional Start Site (TSS) was mapped by 5′RACE System (Invitrogen). DNA-free RNA was retro-transcribed using a specific primer (5′-CTCGGCTGTAGTCTTTCATTCTCACACGC-3′) that anneals downstream of miR-9-2 stem-loop structure; the generated cDNA was amplified utilizing the oligonucleotide 5′-CCAAAGATAACAACTCGCTTCCC-3′ as the specific primer.
Primer extension
The position of the miR-9-2 TSS was further confirmed by primer extension. An amount of 20 µg of DNA-free RNA were reverse-transcribed with the γ-32P-labeled oligonucleotide 5′-CCTCTGTCTTCCTCTTGCCAGACTCC-3′. Reaction products were fractionated by denaturing PAGE together with the sequencing reaction, obtained from M13 single strand DNA, by using the Sequenase Version 2.0 DNA Sequencing Kit (GE Healthcare).
Promoter analysis
Conserved DNA fragments located upstream of miR-9-2 TSS were PCR-amplified employing the oligonucleotides reported in Supplementary Table S1, and inserted into pGL3-Basic vector (Promega). Single plasmids were co-transfected into SK-N-BE cells together with the control plasmid (pRL-TK). After 24–72 h of incubation in the presence or absence of RA, cells were assayed with the Dual-Luciferase Assay (Promega). The ΔCREB mutant derivative, lacking CREB binding site, was obtained from the wild-type construct by inverse PCR, utilizing the primers reported in Supplementary Table S1.
Protein extraction and western blot assay
Whole-cell protein extracts were prepared from SK-N-BE cells lysed in RIPA buffer. Extracts were separated by electrophoresis on 4–12% poly-acrylamide gel (Invitrogen) and electroblotted onto nitrocellulose membrane (Protran, S&S, Drammen, Norway); immunoblots were incubated overnight with polyclonal antibodies anti-REST 07-579, anti-CREB 06-863, anti-phospho-CREB (Ser133) 06-519 (Millipore, Billerica, MA, USA), and for 1 h with monoclonal antibody against GAPDH (6C5) sc-32233 (Santa Cruz Biotechnology, Inc.) as a loading control.
Analysis of RNA expression by RT–PCR and Quantitative Real Time RT–PCR
An amount of 1–5 µg of DNA-free RNA were reverse-transcribed using a mixture of poly-dT oligonucleotides and random examers (Invitrogen). In order to quantify the expression of miR-9 host gene transcripts, 50 ng of each generated cDNA were amplified in triplicate by Quantitative Real Time RT–PCR (qRT–PCR), in the presence of Power SYBR Green PCR Master Mix (Applied Biosystems) and 3 pmol of each sequence-specific primer (Supplementary Table S2). qRT–PCR was performed, data were collected and analyzed using 7500 Fast Real-Time PCR System (Applied Biosystems). Thermal cycle conditions were the following: 2 min at 50°C for activation and 10 min of initial setup at 95°C, followed by 40 cycles at 95°C for 15 s and at 60°C for 60 s. Data were calculated with 7500 Software v2.0.3 (Applied Biosystems) by the ΔΔCT method and expressed as relative quantities (RQs) after GAPDH normalization.
The same cDNA was utilized to determine pri-miR-9-1, pri-miR-9-2 and pri-miR-9-3 expression levels through semi-quantitative PCR (25–30 cycles at 94°C for 30 s, at 55°C for 45 s and at 72°C for 30 s), performed using specific oligonucleotides listed in Supplementary Table S2. PCR fragments were separated by 2.5% agarose gel electrophoresis.
Analysis of REST mRNA 3′UTR
RNA was reverse-transcribed as described earlier. The 3′UTR was analyzed by semi-quantitative PCR using the oligonucleotides reported in Supplementary Table S2. PCR fragments were separated by 2% agarose gel electrophoresis.
Northern blot analysis
An optimized northern blot protocol for small RNA analysis was employed. Total RNA extracted from SK-N-BE cells was fractionated on 10% poly-acrylamide gel in MOPS–NaOH (pH 7), 7 M Urea and transferred onto Amersham Hybond-NX nylon membrane (GE Healthcare). RNA cross-linking was performed in 0.16 M N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and 0.13 M 1-methylimidazole (Sigma-Aldrich) at pH 8, for 2 h at 60°C (19). DNA oligonucleotides complementary to the sequence of mature miR-9 (17) and to 5S-rRNA (5′-TGAACGCGCCCGATCTCGTCT-3′) were 32P-labeled and used as probes.
Chromatin immuno-precipitation assay
Untreated or 1 day and 6 days RA-treated SK-N-BE cells, grown to confluence in 10-cm dishes, were cross-linked with PBS in the presence of formaldehyde at a final concentration of 1% at room temperature for 10 min. Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (5), with minor modifications. Before immunoprecipitation, 30 µl of supernatant were saved as input (IN). Chromatins were immunoprecipitated (IP) with 5–10 µg of specific antibodies, anti-REST 07-579, anti-CREB 06-863, anti-phospho-CREB (Ser133) 06-519, anti-acetyl-Histone H3 06-599 (Millipore), at 4°C overnight. No addition of antibody was used as a negative control (NA). After incubating 30 µl of Protein A Agarose/Salmon Sperm DNA with IP and NA samples at 4°C for 1 h, beads were sequentially washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.0 and 150 mM NaCl), high-salt buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA, 20 mM Tris–HCl, pH 8.0 and 0.5 M NaCl), LiCl buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris–HCl, pH 8.0), and TE buffer (10 mM Tris–HCl, pH 8.0 and 1 mM EDTA). The DNA–protein complexes were eluted by elution buffer (1% SDS and 0.1 M sodium bicarbonate) at room temperature. Reverse-cross-linking was performed by incubating the eluted DNA–protein complexes with 300 mM NaCl at 65°C overnight and treating with Proteinase K (Roche) at 45°C for 1 h. Purified DNA fragments from IP, IN and NA samples were amplified by multiplex semi-quantitative PCR, employing Immolase (Bioline) in the presence of [α-32P]-dATP (PerkinElmer) and a mixture of specific primers, spanning across CR599257 gene, and primers targeting tRNAGlu gene transcriptional unit, for the unrelated control (Supplementary Table S3). Thermal cycle conditions were: 30 cycles of 94°C for 30 s, 60°C for 60 s and 72°C for 40 s. PCR fragments were separated by 8% polyacrylamide gel electrophoresis. Gels were exposed onto Storage Phosphor Screen (GE Healthcare) and data were collected by densitometric analysis performed with Typhoon Trio (GE Healthcare). The specific immuno-precipitation enrichment was calculated as the ratio between IP and IN, after subtracting NA from IP, and normalized versus the unrelated region (tRNAGlu gene).
miR-9 ectopic expression
Overexpression of miR-9 was carried out by transfecting SK-N-BE cells with a plasmid in which the miR-9 coding region was cloned between the polII promoter and termination region of the U1 snRNA gene. As control, a plasmid producing an unrelated 21-nt long RNA, bearing no homology to any known miRNA or mRNA sequence in human was employed (17). miR-9 over-expression was assessed by northern blot analysis.
CREB and REST knock-down
CREB expression was silenced by RNAi. SK-N-BE cells were transfected with a specific siRNA against CREB mRNA at a final concentration of 20 nM (Hs CREB1 5 SI00299894, Qiagen) or, as control, with an unrelated siRNA (AllStars Neg. Control siRNA, Qiagen), and treated with RA for 3 days. The efficiency of CREB knock down was evaluated by western blot analysis.
REST expression was silenced both by over-expressing miR-9 and by RNAi. SK-N-BE cells were transfected in absence of RA with the plasmid expressing human miR-9 (17), or with a mixture of specific siRNAs against REST mRNA at a final concentration of 20 nM (Hs REST 1 SI00701407, Hs REST 5 SI04153765; Hs REST 7 SI04333588, Qiagen); as controls, cells were transfected with a plasmid expressing an unrelated 21-nt long RNA (17), or with an unrelated siRNA (AllStars Neg. Control siRNA, Qiagen). The efficiency of REST knock-down was evaluated by western blot analysis.
miR-9 knock down
FITC-labeled LNA oligonucleotide (Exiqon,Vedbaek, Denmark) against miR-9 was transfected into SK-N-BE cells overexpressing miR-9 at a final concentration of 50 nM.
RESULTS
Expression profiling of miR-9 and host genes during neuronal differentiation
According to the UCSC genome browser (http://genome.ucsc.edu/), human miR-9 is encoded by three distinct genomic loci, miR-9-1, miR-9-2 and miR-9-3, generating three mature miR-9 species with identical sequences. These loci are located on chromosome 1, 5 and 15, respectively and each one is associated with annotated UCSC cDNAs (Supplementary Figure S1). In particular, miR-9-1 is located in the second intron of the C1orf61 gene encoding for the CROC4 protein, miR-9-2 is positioned in the last exon of the annotated overlapping BC036480, AX746931 and CR599257 non-coding genes and miR-9-3 is embedded within the first intron of the non coding CR612213 gene.
To correlate the expression of the putative host genes with those of the individual miRNAs, we analyzed the expression profiles of both the individual genes and the pri-miR-9 species in proliferating versus differentiating cells. As model system, we utilized the human neuroblastoma-derived SK-N-BE cell line that can be induced to neuronal differentiation by treatment with retinoic acid (RA).
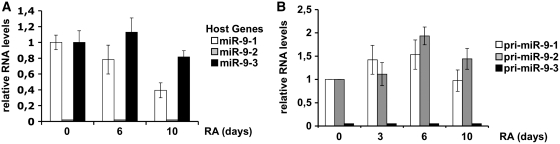
The expression profile of miR-9 host genes was analyzed by qRT–PCR, utilizing oligonucleotides specific for putative exonic regions. In the case of miR-9-1 and miR-9-2 host genes the oligos were selected in regions common to all the annotated transcripts (Supplementary Figure S1). Figure 1A shows that the expression of the miR-9-1 host gene was high in proliferating cells and decreased upon RA treatment; the miR-9-3 host transcript was instead expressed both in untreated and in RA-treated cells, whereas the miR-9-2 host transcripts were not expressed both in undifferentiated and in differentiating cells. The identification of the latter transcripts in NB4 cells indicated the suitability of the oligos utilized for transcript identification (Supplementary Figure S2).
Figure 1.
Expression profiles of miR-9 transcripts. (A) Expression profiling of miR-9-1, miR-9-2 and miR-9-3 host gene transcripts by qRT–PCR. (B) Expression profiling of miR-9 primary transcripts (pri-miR-9) by RT–PCR. In both (A) and (B) panels, the expression levels were evaluated at specific time-points (days, indicated below each bar) of RA treatment and normalized against GAPDH mRNA levels. White, grey and black bars refer to miR-9-1, miR-9-2 and miR-9-3 genes, respectively. Error bars in the histograms represent the standard deviation obtained from three independent experiments.
The analysis of pri-miR-9 species was carried out by RT–PCR utilizing oligonucleotides matching each one of the pri-miRNA-9 species. Figure 1B shows that the expression of both pri-miR-9-1 and pri-miR-9-2 increased up to 6 days of RA-treatment to drop off later; pri-miR-9-3 expression was, instead, not detectable both in untreated and RA-treated cells. These results are in agreement with the reported exclusive expression of miR-9-1 and miR-9-2 in mammalian brain (20).
Altogether, these results indicate that there was no correlation between the expression of pri-miR-9 species and that of the corresponding host genes, suggesting that they are expressed by independent pathways. In particular, we focused our studies on miR-9-2 for its peculiar exonic localization.
Identification of miR-9-2 TSS
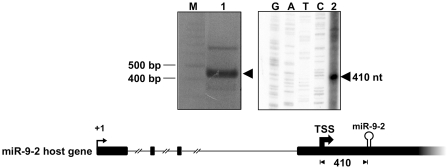
To investigate the possibility that miR-9-2 could be independently transcribed from its host gene, its TSS was firstly determined. Primer extension and 5′RACE assays on total RNA from RA-treated SK-N-BE cells were carried out using primers designed to anneal the stem structure of the pre-miRNA (Figure 2). The TSS mapped 410-nt upstream of the 5′ end of the pre-miRNA, in a position not matching any of the UCSC reported TSS of the miR-9-2 host transcripts. This result suggests that miR-9-2 transcription relies on an independent promoter.
Figure 2.
Identification of miR-9-2 TSS by 5′RACE and primer extension analyses. Left panel: the product of 5′RACE (lane 1) was fractionated along with a DNA ladder (lane M). The arrowhead points to the amplified product. Right panel: the primer extension reaction (lane 2) was run in parallel with the sequencing reaction (lanes G, A, T and C). The arrowhead indicates the extended product. In the underneath schematic representation, the 5′ end of the reported miR-9-2 host transcript (+1) is pointed out by a thin arrow. The thick arrow points to the miR-9-2 TSS; the distance of the TSS from miR-9-2 stem loop structure is reported as nucleotide length.
Identification of miR-9-2 promoter
Several histone modifications have been described as chromatin signatures for promoter identification in human cells (21–23). Among them, acetylation of histone H3 lysine 9 and 14 (H3K9,14Ac) was shown to be diagnostic for transcriptionally active promoters (21). To identify the promoter region driving miR-9-2 expression, we initially investigated the H3 lysine 9 and 14 acetylation level on the chromatin of the presumptive host gene promoter and of the hypothetical regulative region upstream of miR-9-2 TSS.
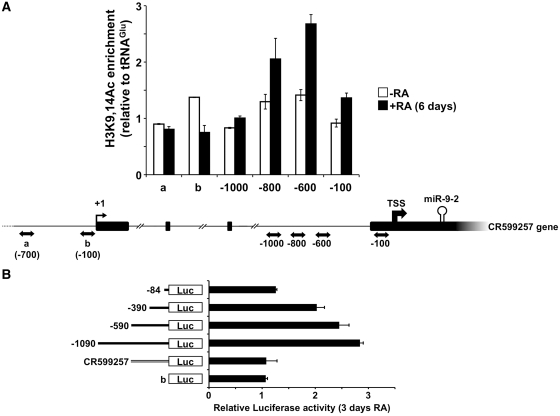
ChIP assays using antibodies against H3K9,14Ac were performed on untreated and 6 days RA-treated SK-N-BE cells. The regions analyzed were selected according to their evolutionary conservation, derived from the UCSC genome browser. This search highlighted three intronic highly conserved regions, spanning ∼1-kb upstream of miR-9-2 TSS. As control, two conserved regions at ∼100- and 700-bp upstream of the CR599257 gene TSS were analyzed. The results of Figure 3A show that acetylated H3 histone is specifically associated with the intronic regions (−600 and −800) upstream of the miR-9-2 TSS after cell treatment with RA for 6 days, whereas background interaction was detected in the absence of RA and in the presumptive promoter region of the host gene. These results point to the intronic region upstream of miR-9-2 as the one competent for miRNA transcriptional activity upon RA-treatment.
Figure 3.
Identification of miR-9-2 promoter. (A) Analysis of histone acetylation on miR-9-2 and CR599257 gene upstream regions. ChIP assays were performed on chromatin samples from 6 days-RA treated (black bars) or untreated (white bars) cells, immunoprecipitated with antibodies against H3K9,14Ac. PCR-amplified regions are reported below each bar of the histogram and are indicated by double-headed arrows in the underneath schematic representation. Immunoprecipitation levels are expressed as fold enrichment with respect to an unrelated region (tRNAGlu). Error bars in the histogram represent the standard deviation obtained from three independent experiments. (B) Analysis of CR599257 and miR-9-2 promoter activity by luciferase assay. The luciferase-based reporter construct (−1090), containing the 1-kb region upstream of the miR-9-2 TSS, and its deletion mutant derivatives thereof were tested together with the construct containing the conserved 500-bp region upstream of the CR599257 gene TSS. For each reporter construct, schematized on the left, the histogram shows luciferase expression levels, evaluated as the ratio between the values measured in 3 days RA-treated versus untreated cells. The promoter-less reporter construct was tested as control (b). Error bars in the histogram represent the standard deviation obtained from three independent experiments.
Based on the chromatin signature, we focused on the 1-kb region upstream of miR-9-2 TSS to characterize the sequences promoting miR-9-2 transcription during neuronal differentiation. We generated a first construct in which the 1-kb region was cloned in a promoter-less reporter vector, pGL3-Basic, upstream of the FLuc reporter gene (−1090 construct) (Figure 3B). According to the sequence conservation data, three additional constructs containing shorter upstream sequences of 590, 390 and 84 bp (constructs −590, −390 and −84, respectively) were derived. As control, the most conserved 500-bp region upstream of CR599257 gene TSS was cloned in the same reporter vector (CR599257 construct). These five different constructs were transfected into untreated or RA-treated SK-N-BE cells, and Fluc activity was measured as the ratio between the values obtained in RA-treated versus untreated cells. Among the different constructs, the −1090 displayed the strongest Fluc induction upon RA treatment. This result indicated that the 1-kb region is the minimal promoter triggering miR-9-2 transcription during in vitro neuronal differentiation.
Identification of the transcription factors controlling miR-9-2 gene expression
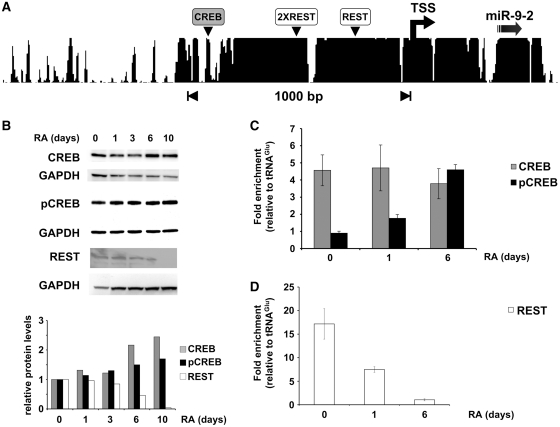
To identify the transcription factors involved in the regulation of miR-9-2 expression, an algorithmic prediction of potential transfactor binding sites within the identified promoter was carried out using the Chip-Mapper Web Server (http://mapper.chip.org/). Among the others, putative binding sites for the neuro-restrictive silencer factor REST and the cAMP responsive element binding protein CREB were identified in very conserved regions (Figure 4A). To investigate the involvement of these transcription factors in miR-9-2 gene regulation during neuronal differentiation, we first examined their expression profile in RA-treated SK-N-BE cells by western blot (Figure 4B). We utilized antibodies against total CREB (lanes CREB), recognizing both the unphosphorylated and the phosphorylated forms of the transcriptional factor, and antibodies specific for the phosphorylated form (lanes pCREB); we found no marked modulation either for CREB and for its active phosphorylated form. On the contrary, the level of REST significantly decreased along the differentiation process.
Figure 4.
Association of transcriptional factors with miR-9-2 promoter chromatin. (A) Conservation of the region upstream of miR-9-2 stem loop structure in mammals, according to UCSC genome browser. The conserved binding sites for CREB and REST are indicated by grey and white boxes, respectively, according to the Chip-Mapper Web Server. (B) Analysis of total CREB (CREB), phospho-CREB (pCREB) and REST (REST) protein levels. Upper panels: SK-N-BE cells were treated with RA for the times indicated above each lane and total protein extracts were analyzed by western blot. Signals were normalized for GAPDH levels. Lower panel: densitometric analyses of western blots. Results are normalized against GAPDH and expressed as RQs versus untreated cells (lane 0). (C) and (D) ChIP analysis of total CREB and phospho-CREB (CREB and pCREB respectively) and REST occupancy at the miR-9-2 promoter. Chromatin samples were obtained by cells treated with RA for the times (days) indicated below each bar. Immunoprecipitation levels are expressed as fold enrichment with respect to the tRNAGlu gene. Error bars in the histograms represent the standard deviation obtained from three independent experiments.
As a second step, we examined the promoter occupancy of the selected factors by ChIP assays (Figure 4C and D and Supplementary Figure S3). Chromatin from untreated and RA-treated SK-N-BE cells was immunoprecipitated with antibodies against total CREB, pCREB, and REST. Figure 4C shows that CREB is already bound to the promoter region at time 0 and persists up to 6 days of RA treatment. Notably, the association of pCREB on the promoter region, very low in proliferating cells, increased in RA-treated cells, reaching the highest level at 6 days upon RA-treatment (Figure 4C). From these results it appears that CREB is already bound to the promoter in undifferentiated cells, but it is the phosphorylated form that is present during differentiation. The analysis of the association of the repressor REST with the promoter region revealed that it was at maximum level in undifferentiated cells, dropping off at 6 days upon RA-treatment (Figure 4D).
Altogether, these results indicate that pCREB and REST are associated to the miR-9-2 promoter region in vivo, in a differentiation-controlled fashion; in particular, with the progression of differentiation, the occupancy of phosphorylated CREB parallels REST dismissal.
Functional analysis of CREB and REST transcriptional regulation
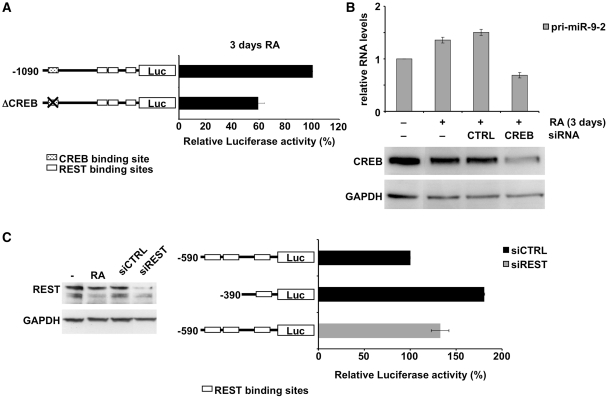
To assess the functional contribution of CREB to miR-9-2 transcriptional control, a deletion mutant lacking the CREB binding site (ΔCREB) was derived from the −1090 construct (Figure 5A). The wild-type −1090 construct and the ΔCREB mutant were transfected in SK-N-BE treated with RA for 3 days and in control untreated cells. Luciferase expression from both constructs was evaluated as the ratio between the values observed in RA-treated versus untreated cells. In comparison to the wild-type, the luciferase expression from ΔCREB construct was 40% lower, suggesting that CREB is a critical element for inducing transcriptional activation during RA-induced differentiation (Figure 5A).
Figure 5.
Functional analysis of transcriptional factor activities. (A) Functional analysis of CREB by luciferase assay. Cells were transfected with the constructs, represented aside each histogram bar, and grown in the presence or in the absence of RA. Values for each construct were calculated as the ratio of luciferase activities in 3 days RA-treated versus untreated cells, normalized against control cells transfected with the −1090 construct, and expressed as percentage. (B) Upper panel: functional analysis of CREB by knock down experiment. Cells treated with RA for 3 days (+RA) were transfected with siRNAs against CREB (CREB siRNA) or with an unrelated siRNA (CTRL siRNA). Results are based on qRT–PCR analysis of pri-miR-9-2 levels. Error bars in the histograms represent the standard deviation obtained from three independent experiments. Lower panel: western blot analysis of CREB levels in untreated (lane −RA), RA-treated (lanes +RA), and CREB-interfered cells (lane CREB siRNA). The anti-CREB RNAi caused a 60% decrease in protein level as evaluated from three independent experiments. As control, cells transfected with an unrelated siRNA were utilized (lane CTRL siRNA). GAPDH was employed as a loading control. (C) Left panel: western blot analysis of REST levels in untreated (lane −), 3 days RA-treated (lane RA), and REST-interfered cells (lane siREST). The anti-REST RNAi caused a 80% decrease in protein level as evaluated from three independent experiments. As control, cells transfected with an unrelated siRNA were utilized (lane siCTRL). GAPDH was employed as a loading control. Right panel: functional analysis of REST. Luciferase-based constructs, represented aside each histogram bar, were co-transfected with an unrelated siRNA (black bars) or with siRNAs-interfering for REST expression (grey bar) in proliferating cells. Luciferase activity was normalized against control cells transfected with the −590 construct and the control siRNA, and expressed as percentage. Error bars in the histograms represent the standard deviation obtained from three independent experiments. In both (A) and (C) panels, the binding sites for CREB and REST are depicted as dotted and white boxes, respectively.
To confirm this result, CREB knock down experiment was performed in RA-treated SK-N-BE cells and the levels of the endogenous pri-miR-9-2 evaluated. Figure 5B shows that specific siRNA-mediated knockdown of CREB (lower panel) resulted in a decrease of ∼60% of pri-miR-9-2 level (upper panel).
To determine the role played by REST, we analyzed luciferase expression in proliferating cells (a condition in which REST is bound to the promoter region) from the −590 construct, containing the three binding sites for REST, and from the −390 construct, a deletion mutant lacking two REST binding sites. Compared to the expression level of the −590 construct, that from the −390 was 70% higher (Figure 5C, right panel). Moreover, a loss-of-function approach was carried out and both the luciferase expression of the reporter construct (-590) and the endogenous level of pri-miR-9-2 transcript were evaluated. siRNA-mediated knockdown of REST resulted in upregulation of ∼40% of luciferase expression from −590 construct (Figure 5C), whereas the level of the endogenous pri-miR-9-2 was unaffected (data not shown). To verify whether the latter result could be due to the residual REST still present on the miR-9-2 promoter, following the siRNA-mediated knockdown, ChIP experiment was performed in REST-interfered cells. The results indicate that no REST is associated to miR-9-2 promoter (Supplementary Figure S4).
Overall, the luciferase assays demonstrate the repressor activity of REST on the reporter gene. On the other hand, the analysis of the endogenous transcript reveals that dismissal of REST is not sufficient for transcriptional activation.
Since miR-9-1 gene expression pattern was similar to that of miR-9-2 gene during in vitro differentiation (Figure 1B), we asked whether the REST/CREB regulation was a common mechanism for the two loci. Preliminary in silico analysis about the occurrence of REST and CREB sites on the miR-9-1 promoter described by Ko et al. (24), showed the presence of putative binding sites at −130-nt and at −1232-nt upstream of miR-9-1 stem loop structure for CREB and REST, respectively. To demonstrate the involvement of these factors in miR-9-1 transcription, the levels of the endogenous pri-miR-9-1 were evaluated in the cells knocked down for REST and CREB, as described for miR-9-2 analysis. We found that the knock down of the transcriptional activator CREB in RA-treated cells caused a decrease of ∼40% of pri-miR-9-1 levels (Supplementary Figure S5). On the contrary, the knock down of REST in undifferentiated cells did not affect pri-miR-9-1 transcription (data not shown).
miR-9 controls REST expression during neuronal differentiation
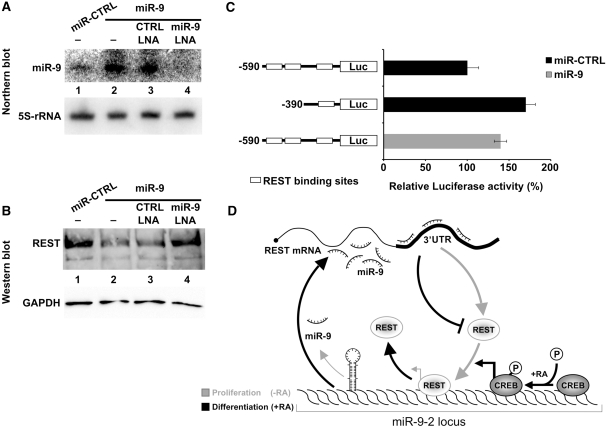
It was previously shown (18) that miR-9 targets REST mRNA in HEK 293 cells and in RA-induced NT-2 cells. Notably, miR-9, which is upregulated during RA-induced differentiation of SK-N-BE cells (17), inversely correlates with the amount of REST, which undergoes downregulation after RA treatment. This suggests that REST expression might be modulated by miR-9 during neuronal differentiation. To confirm this hypothesis, we analyzed in a preliminary study the 3′UTR of REST mRNA in proliferating versus differentiating SK-N-BE cells. In fact, it has been reported that some transcripts undergo 3′UTR shortening, resulting from alternative cleavage and polyadenylation, during neuronal activation (25) or embryonic development (26). Evaluation of the 3′UTR of REST mRNA was performed by RT–PCR, and revealed that the ∼3.7-kb long region, containing the three binding sites for miR-9 (18), was not altered during in vitro differentiation of SK-N-BE cells (Supplementary Figure S4). This result indicates that REST mRNA may be targeted by miR-9 during neuronal differentiation. To validate this hypothesis, miR-9 was overexpressed (∼10-fold) in neuroblastoma cells (Figure 6A) and REST levels were evaluated by western blot. Figure 6B shows that the ectopic expression of miR-9 caused ∼70% decrease of REST (lane 2). Moreover, treatment of neuroblastoma cells overexpressing miR-9 with an anti-miR-9 LNA oligonucleotide restored the endogenous level of REST (Figure 6B, compare lane 4 with lane 1), further demonstrating the specific action of miR-9 on REST gene expression.
Figure 6.
Feed-back regulatory loop between miR-9 and REST. (A) The ectopic expression of miR-9 was verified by northern blot (lanes 2–4) and normalized with respect to 5S-rRNA expression. SK-N-BE cells were transfected with the plasmid overexpressing miR-9 in the absence of LNA (lane 2), or together with a control LNA (lane 3), or with the anti-miR-9 LNA (lane 4). RNA from cells transfected with the control plasmid expressing an unrelated 21-nt long RNA was analyzed in lane 1. (B) Western blot analysis of REST protein levels. SK-N-BE cells were transfected with the plasmid overexpressing miR-9 in the absence of LNA (lane 2), or together with a control LNA (lane 3), or with the anti-miR-9 LNA (lane 4). The overexpression of miR-9 caused a 70% decrease in protein level (lane 2) as evaluated from three independent experiments. Proteins from cells transfected with the control plasmid expressing an unrelated 21-nt long RNA were analyzed in lane 1. GAPDH protein levels were used as a loading control. (C) Effect of miR-9 over-expression on REST activity. Reporter luciferase-based constructs (indicated on the left) were co-transfected with the control plasmid expressing an unrelated 21-nt long RNA (black bars) or with the plasmid over-expressing miR-9 (grey bar) in proliferating cells. Luciferase activity was normalized against cells transfected with the −590 construct and with the unrelated 21-nt long RNA, and expressed as percentage. Binding sites for REST are depicted as white boxes. Error bars in the histogram represent the standard deviation obtained from three independent experiments. (D) Minicircuitry illustrating the functional interplay between miR-9, REST and CREB. In proliferating SK-N-BE cells (grey arrows), REST occupies miR-9-2 promoter and represses miR-9 expression at the transcriptional level. Upon RA treatment (black arrows) the dismissal of REST and the phosphorylation of CREB, already bound to the promoter, induce miR-9 transcription, which in turn represses REST expression, contributing to the maintenance of the differentiation program.
To correlate miR-9-dependent REST downregulation with transcriptional induction, luciferase expression from the −590 construct, containing the three REST binding sites, was measured in SK-N-BE cells overexpressing miR-9 or an unrelated 21-nt long RNA. Figure 6C shows that miR-9 ectopic expression (grey bar) increased of ∼40% luciferase expression with respect to the control cells transfected with the same −590 construct and an unrelated 21-nt long RNA. Notably, such an induction was comparable to that observed upon specific REST knockdown performed by RNAi (Figure 5C).
DISCUSSION
miR-9 has been proposed as one of the crucial regulators of neuronal development in several organisms. In Drosophila, it is involved in cell fate specification of developing sensory organs (12); in zebrafish, it participates in midbrain-hindbrain boundary definition (14); in mouse cortical development, it is involved in differentiation of Cajal Retzius cells (15). In mammals, miR-9 is induced upon embryonic stem cell neuronal differentiation and blocking its function during this process decreases neuronal differentiation at the expense of astrocytes (27). Alteration of miR-9 expression has been reported in Alzheimer’s disease and its downregulation has been observed in Huntington’s disease, suggesting its further implication in neurodegeneration (18,28). We previously showed that miR-9 is upregulated during in vitro neuronal differentiation, whereas it is downregulated in 50% of primary neuroblastoma (NB) tumors (17). However, miR-9 altered expression is not genetically determined by chromosome aberrations, since no one of the three distinct loci encoding for human miR-9 are positioned within the chromosomal regions rearranged in neuroblastomas (29,30). It transpires that miR-9 dys-regulation in these tumors could be due to alteration in gene expression.
Analyses aimed at profiling the expression of the three individual miR-9 host genes and the pri-miR-9 species, in proliferating versus differentiating NB cells, underlined a non correlative expression between them. This suggests that the transcription of both the intronic and the exonic miR-9 genes is independent from that of the host genes, and is driven by their own promoters. Notably, this conclusion is in accordance with the recent finding highlighted by several research groups that, combining bioinformatic strategies with experimental approaches, found many intronic miRNAs, the expression of which is controlled by upstream regulatory elements, independent of the host genes (22,31,32).
Further analyses were aimed at unveiling the transcriptional regulation of miR-9-2 gene, that displays a less common exonic localization. We showed that the TSS of the pri-miRNA maps at 410-nt upstream of the pre-miRNA, and that the promoter region is located in the upstream intron, which displays histone modifications typical for an active chromatin domain. These results demonstrated that miR-9-2 is expressed by a transcription unit, different from that of the host gene, depending on a dedicated promoter, and that, therefore, miR-9-2 and its putative host gene are physically overlapped but functionally independent. Such conclusions are in line with genome scale computational studies of intergenic miRNA gene promoters, which claimed that putative promoters of ∼85% of these genes reside between −500- and −1000-bp upstream of the pre-miRNA. This implies that real core promoters of most intergenic miRNA genes are relatively close to the pre-miRNA structure (33). Accordingly, we found that the region encompassing ∼1-kb upstream of miR-9-2 TSS was sufficient for transcriptional induction during in vitro neuronal differentiation, thus representing the miR-9-2 minimal promoter.
Search for transcription factors involved in miR-9-2 gene regulation allowed us to highlight two antagonistic molecules, the repressor REST and the activator CREB, which function in an opposite and temporally regulated manner. REST is a well known suppressor of neuronal genes in non neuronal cells; it functions by binding to a conserved element (RE1) in neuronal loci and recruiting histone deacetylases by corepressors, associated with its repressor domains (34). The downregulation of this master regulator of neuronal genes is imperative during neuronal differentiation to allow neuronal genes to be expressed. On the other side, the activator CREB plays a pivotal role in regulating a wide range of neuronal functions, as neuronal survival, neuronal proliferation and differentiation, and is crucial for mediating long-term memory and synaptic plasticity (35). Furthermore, CREB was demonstrated to regulate transcription of miR-132, which has a crucial role in neuronal morphogenesis (36). It has been reported that the binding of this factor to its target genes does not require its phosphorylated status, whereas the crucial event in the activator function of CREB is the phosphorylation at Ser 133 (37). The involvement of these factors in miR-9-2 gene expression control was previously suggested by computational studies (38) and by experimental data that identified a REST binding site 45-kb upstream of miR-9-2 stem loop precursor (39). However, no functional studies on the promoter activity of that region were performed, missing the identification of the proximal promoter here characterized, that is instead necessary and sufficient to reconstitute miR-9-2 correct temporal expression.
In this study, we found that in proliferating cells, where it is highly expressed, REST is bound to the miR-9-2 promoter preventing transcriptional activity. Upon RA treatment, REST dismissal and the concomitant phosphorylation of CREB, already bound to the promoter, activate transcription. In the light of these data, we propose that the opposite and temporally coordinated action of these factors would ensure the low level of miR-9 in undifferentiated cells and its induction during differentiation. Notably, the functional analyses of the involvement of REST and CREB in the transcriptional regulation of the miR-9-1 gene, showed that miR-9-1 expression is unaffected by REST knockdown, whereas it is impaired by CREB depletion. These results strongly suggest that REST/CREB regulation is a common mechanism shared by miR-9-1 and miR-9-2 loci, both contributing to miR-9 production during neuronal differentiation.
Noteworthy, we also demonstrated that REST mRNA is targeted by miR-9 during in vitro neuronal differentiation of SK-N-BE cells. This finding strongly supports the earlier evidence, provided by Packer and colleagues (18), of a regulative double negative feed-back loop between REST and miR-9, which may contribute to the maintenance of differentiation programs (Figure 6D).
In conclusion, our data add new insight into the definition of cell-specific mechanisms underlying miRNA biosynthesis, and support the general conclusion that a complex network of transcription activators and repressors acting together with miRNAs may be responsible for coordinating neuronal gene expression during neuronal development (38).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union project SIlencing Rnas: Organizers and Coordinators of Complexity in eukaryotic Organisms (SIROCCO) (LSHG-CT-2006-037900); European Science Foundation project NuRNASu; Associazione Italiana per la Ricerca sul Cancro; Progetti di Ricerca di Interesse Nazionale; Istituto Italiano di Tecnologia SEED-project; and Centro di Eccellenza Biologia e Medicina Molecolare. Fondazione Italiana per la Ricerca sul Cancro (Fellowship to U.G.). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Ballarino and E. Girardi for advice in setting up ChIP experiments, P. Filetici for providing anti-H3K9,14Ac antibodies, M. Arceci and M. Marchioni for technical help and M. Morlando for helpful discussion.
REFERENCES
- 1.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 3.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Kim K, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr. Opin. Genet. Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am. J. Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 11.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 15.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, et al. MicroRNA profiling in human medulloblastoma. Int. J. Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 17.Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc. Natl Acad. Sci. USA. 2007;104:7957–7962. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 20.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 21.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 24.Ko MH, Kim S, Hwang do W, Ko HY, Kim YH, Lee DS. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2008;275:2605–2616. doi: 10.1111/j.1742-4658.2008.06408.x. [DOI] [PubMed] [Google Scholar]
- 25.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J. Inorg. Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonini GP, Romani M. Genetic and epigenetic alterations in neuroblastoma. Cancer Lett. 2003;197:69–73. doi: 10.1016/s0304-3835(03)00081-8. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Xuan Z, Zhao X, Li Y, Zhang MQ. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res. 2009;19:266–275. doi: 10.1101/gr.081638.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput. Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 35.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 36.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl Acad. Sci. USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl Acad. Sci. USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.