Abstract
The molecular basis of the genetic code relies on the specific ligation of amino acids to their cognate tRNA molecules. However, two pathways exist for the formation of Gln-tRNAGln. The evolutionarily older indirect route utilizes a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) that can form both Glu-tRNAGlu and Glu-tRNAGln. The Glu-tRNAGln is then converted to Gln-tRNAGln by an amidotransferase. Since the well-characterized bacterial ND-GluRS enzymes recognize tRNAGlu and tRNAGln with an unrelated α-helical cage domain in contrast to the β-barrel anticodon-binding domain in archaeal and eukaryotic GluRSs, the mode of tRNAGlu/tRNAGln discrimination in archaea and eukaryotes was unknown. Here, we present the crystal structure of the Methanothermobacter thermautotrophicus ND-GluRS, which is the evolutionary predecessor of both the glutaminyl-tRNA synthetase (GlnRS) and the eukaryotic discriminating GluRS. Comparison with the previously solved structure of the Escherichia coli GlnRS-tRNAGln complex reveals the structural determinants responsible for specific tRNAGln recognition by GlnRS compared to promiscuous recognition of both tRNAs by the ND-GluRS. The structure also shows the amino acid recognition pocket of GluRS is more variable than that found in GlnRS. Phylogenetic analysis is used to reconstruct the key events in the evolution from indirect to direct genetic encoding of glutamine.
INTRODUCTION
In a catalytic step that defines the molecular basis of the genetic code, most amino acids are ligated directly to their cognate tRNAs by a specific aminoacyl-tRNA synthetase (aaRS). A few amino acids, including Gln, Asn, Cys and Sec, can be or are only biosynthesized on their cognate tRNA [reviewed in (1,2)]. In archaea (3), most bacteria (4,5), and eukaryotic organelles (6–9) a special GluRS, the so-called non-discriminating GluRS (ND-GluRS), can attach glutamate not only to tRNAGlu but also to tRNAGln (5). The misacylated Glu-tRNAGln generated by ND-GluRS is converted to Gln-tRNAGln by the action of Glu-tRNA-dependent amidotransferase (Glu-AdT) (1).
Phylogenetic evidence clearly indicates that this indirect aminoacylation pathway was present at the time of the last common ancestor of all life (10) while a direct pathway for Gln-tRNAGln formation was absent from the biosphere until glutaminyl-tRNA synthetase (GlnRS) evolved in early eukaryotes by a gene duplication of the eukaryotic GluRS (11). Phylogenetic analysis presented here and elsewhere (11–14) shows that GlnRS was then vertically inherited in eukaryotes, but the bacteria received GlnRS in several independent horizontal gene transfer events. The bacterial GluRS responded to the presence of the GlnRS in these cases by evolving a discriminating GluRS (D-GluRS) from a ND-GluRS.
The tRNA substrates of ND-GluRS differ at the third position of the anticodon as tRNAGln has the 34YUG36 anticodon and tRNAGlu the 34YUC36 anticodon. In bacterial ND-GluRSs, which have been studied in detail, discrimination of these tRNA species hinges on recognition of the nucleotide base at position 36. Mutation of a single arginine residue (Arg358 in Thermus thermophilus GluRS) to glutamine was sufficient to convert the D-GluRS from T. thermophilus to a ND-GluRS (15). Furthermore, in Helicobacter pylori one of the organism’s two copies of GluRS could be converted from an enzyme with a strong preference for tRNAGln to a tRNAGlu specific enzyme by means of two-point mutations (Glu334Arg/Gly417Thr) in the anticodon-binding domain (16). The bacterial GluRS has a α-helix cage anticodon recognition domain, where these residues are located. This portion of the bacterial GluRS is homologous to the anticodon-binding domain of class I LysRS (17,18) (Supplementary Figure S1), yet unrelated to the β-barrel shaped anticodon-binding domain of the archaeal and eukaryotic GluRSs and GlnRS (12). Discrimination, therefore, evolved differently in the bacterial GluRSs compared to their eukaryotic counterparts.
The mode of discrimination by the bacterial GluRS has been structurally and biochemically well characterized (15,19), yet a lack of crystallographic data has left open the question of how the archaeal ND-GluRS is able to efficiently aminoacylate both tRNAGlu and tRNAGln. Here, we present the ND-GluRS crystal structure from the archaeon Methanothermobacter thermautotrophicus. Comparison with the GlnRS and D-GluRS structures (15,20,21) shows that the GluRS amino acid recognition pocket tolerates more variability than the GlnRS active site, and reveals regions of the GlnRS structure, which are absent in the ND-GluRS, that likely contribute to the relaxed tRNA specificity required by the ND-GluRS. Finally, comparison of ND-GluRS with related aaRS structures and sequenced-based phylogeny were used to re-construct the key events underlying the evolutionary progression from indirect to direct genetic coding of glutamine.
MATERIALS AND METHODS
Protein purification
Methanothermobacter thermautotrophicus ND-GluRS protein was overexpressed in Escherichia coli. The ND-GluRS gene was subcloned into the Nde I and Xho I sites of pET28a, and the E. coli BL21(DE3) strain was transformed with this vector. The transformed E. coli cells were grown at 37°C at O.D.600 = 0.5, and the gene expression was induced with 1 mM IPTG, followed by cultivation at 37°C for 12 h. The harvested cells were resuspended in the buffer containing 50 mM Tris–HCl (pH 8.0), 0.2 mM EDTA, 10 mM KCl and 5 mM β-mercaptoethanol, and then were disrupted by sonication. The supernatant after centrifugation was heat-treated at 67°C for 30 min, and then was recentrifuged to exclude the denatured E. coli proteins. The supernatant was loaded onto a Q Sepharose FF column (Amersham Bioscience), and the proteins were eluted with an NaCl gradient from 10 mM to 1 M. The eluted sample was dialyzed against 50 mM sodium phosphate buffer (pH 7.5) with 5 mM β-mercaptoethanol. Then, 4 M ammonium sulfate was added to a final concentration of 0.8 M, and the sample was loaded onto a Resource Phe column (Amersham Bioscience). The ND-GluRS was eluted by reverse gradient of 0.8–0 M ammonium sulfate. The eluted sample was dialyzed against 10 mM Tris–HCl buffer (pH 7.0) containing 10 mM MgCl2 and 5 mM β-mercaptoethanol, and was concentrated with Amicon Ultra (M.W.C.O., 30 000) for crystallization.
Crystallization and data collection
The crystallized protein represents the full-length ND-GluRS encompassing residues 1–552, including four cysteines and 18 methionine residues, with a molecular weight of 63 kDa. Crystals with average dimensions of 300 × 300 × 100 µm were grown at 293 K within 5 days using the hanging-drop method by mixing 1.0 µl of reservoir buffer containing 50 mM sodium cacodylate (pH 7.0), 50 mM calcium chloride and 8% PEG6000, and 1.0 µl of protein solution at 20 mg/ml. For data collection, crystals were sequentially soaked for 1 min in 50 µl drops containing reservoir buffer supplemented with 30% glycerol and were flash-cooled directly in the nitrogen cryostream at the beamline.
The data sets of the native crystal were collected at stations BL41XU at SPring-8 (Harima, Japan). The native crystal belongs to the orthorhombic space group P212121, with unit-cell parameters a = 54.07 Å, b = 99.90 Å, c = 105.4 Å. The data were processed, scaled, and merged with the program HKL2000 (22).
For the sulfur SAD phasing experiment using Cr Kα radiation (λ = 2.29 Å), diffraction data were collected to a resolution of 2.4 Å using an in-house X-ray source (Rigaku FR-E SuperBright with a Cu/Cr dual target; 40 kV, 40 mA for Cr) and a Rigaku R-AXIS VII imaging-plate detector. In order to eliminate X-ray absorption, the cryo-buffer around the protein crystal was removed before data collection. A total of 720 images with 0.5 oscillation were collected with a crystal-to-detector distance of 80 mm. The resulting average redundancy was about 15 with separate Friedel pairs. The collected intensities were indexed, integrated, corrected for absorption, scaled using HKL2000 (22) with the ‘scale anomalous’ flag to keep Bijvoet pairs separate. The resulting data were merged and the substructure structure factors were calculated using SHELXC (23).
Structure determination and refinement
The positions of anomalous scatterers were located using SHELXD (23,24), and thereby the initial phases were estimated by SOLVE (25,26) with these sites. The phase improvement by density modification and automatic model building were performed using RESOLVE (25,26). After the manual investigation and modification by O (27) and Coot (28), the built model was refined against the synchrotron dataset using reflections to 1.65 angstrom resolution by CNS (29) and phenix.refine (30). The structure factors and coordinates have been deposited in the Protein Data Bank (accession code 3AII).
Phylogenetic analysis
GluRS, GlnRS, CysRS, class I LysRS, Glu-QRS and MetRS sequences were downloaded from the National Center for Biotechnology Information non-redundant database and structures from the Protein Data Bank (31). Structure-based alignments were completed by using the Multiseq 2.0 module in VMD 1.8.5 (32). The structural alignments were then used to guide sequence alignments in Geneious 4.7.4 (http://www.geneious.com/). The insertion domain specific to the CysRS and GlxRS catalytic domains were removed and the alignments manually refined in Geneious 4.7.4 prior to phylogenetic calculations. Phylogenetic analyses were as described (33) using Phyml (34). Briefly, starting trees were generated with BioNJ and tree space was searched by the SPR algorithm followed by optimization using the NNI algorithm with likelihood parameters initially estimated from the alignments.
RESULTS AND DISCUSSION
Overall structure
The crystal structure of M. thermautotrophicus ND-GluRS (552 residues, Mr of 63 237) was determined by the single-wavelength anomalous dispersion (SAD) method, using sulfur anomalous dispersion (Table 1). The final model contains residues 90–292, 298–324 and 332–552, and has an R-factor of 20.5% (free R-factor of 24.2%) against the diffraction data of the native crystal up to 1.8 Å resolution (Table 1). The crystal structure of GluRS contains one monomer per asymmetric unit, and is bound with one zinc ion and a putative calcium ion (Figure 1A). The GluRS structure consists of a structurally-disordered N-terminal YQEY domain (residues 1–85), Rossmann fold catalytic domain (residues 86–170 and 288–331), connective polypeptide (CP) insertion domain (residues 171–287), stem-contact-fold (SCL) domain (residues 354–399), proximal β-barrel (pβ) domain (residues 400–408 and 500–552) and C-terminal distal β-barrel (dβ) domain (residues 409–499) (Figure 1A).
Table 1.
Data collection and refinement statistics
| Data set | ND-GluRS |
|---|---|
| Data collection statistics | |
| Beamline | SPring-8BL41XU |
| Wavelength (Å) | 1.0000 |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 54.07, 99.90, 105.4 |
| Resolution (Å) | 50.0–1.65 (1.71–1.65) |
| Unique reflections | 67 808 (5552) |
| Redundancy | 10.5 (3.7) |
| Completeness (%) | 97.6 (81.3) |
| I/s(I) | 42.9 (2.5) |
| Rsym | 0.063 (0.312) |
| Refinement statistics | |
| Resolution (Å) | 50.0–1.65 |
| Rwork (%) | 19.3 |
| Rfree (%) | 21.7 |
| σA coordinate error (Å) | 0.19 |
| No. of | |
| Protein atoms | 3,751 |
| Ligands/ions | 10 |
| Waters | 445 |
| Average B-factor (Å2) | |
| Protein | 27.8 |
| Ligand/ion | 28.8 |
| Water | 39.2 |
| RMSD from ideal values | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.041 |
| Ramachandran plot (%) | |
| Preferred | 91.4 |
| Allowed | 8.6 |
| Outliers | 0 |
Rwork = Σ|Fo – Fc|/ΣFo for reflections of work set.
Rfree = Σ|Fo – Fc|/ΣFo for reflections of test set.
Figure 1.
Overall structure of the ND-GluRS from M. thermautrophicus. (A) Crystal structure of M. thermautrophicus ND-GluRS. Two metal ions bound to ND-GluRS are colored yellow. (B) The structure of E. coli GlnRS·tRNAGln complex (20). The same color code as in panel A is used. The protruding loop in the GlnRS proximal β-barrel domain (E. coli GlnRS residues 473–494) is circled. (C) Closeup view of the CP domain from M. thermautrophicus ND-GluRS (left) and E. coli GlnRS (right), with the marked structural difference circled. (D) Closeup view of the dβ domain from M. thermautrophicus ND-GluRS (left) and E. coli GlnRS (right), with the marked structural difference circled.
The overall structure closely resembles that of E. coli GlnRS (21) (Figure 1A and B). Superposition of the M. thermautotrophicus ND-GluRS structure onto the E. coli GlnRS structure reveals two insertions in GlnRS at critical points of interaction with the tRNA that are absent in the ND-GluRS structure. There is an extended loop in the GlnRS CP domain (E. coli residues 134–139) that may facilitate discrimination of tRNAGln from tRNAGlu at the first base pair in the acceptor stem (Figure 1C). Also, an anticodon recognition loop in E. coli GlnRS (residues 392–407), which specifically recognizes the G36 base of the anticodon, is absent in ND-GluRS (Figure 1D). In addition, ND-GluRS lacks the protruding loop in the GlnRS proximal β-barrel domain (E. coli GlnRS residues 473–494). In GlnRS, this large loop makes several non-covalent interactions that physically connect the anticodon-binding domain to the catalytic core domain. Upon binding of the cognate tRNA molecule, the loop is thought to be involved in transmitting an allosteric signal to the catalytic center of GlnRS (20,35). Such a signal would explain how the interaction with non-cognate tRNAs lead to significantly lower enzymatic turnovers (kcat) compared to glutaminylation of the cognate tRNA substrate (36) and how mutations to the anticodon bases significantly lower binding affinity of glutamine and the chemical rate constant of aminoacylation (37).
The ND-GluRS structure includes two coordinated ions. A zinc ion is tetrahedrally coordinated by a zinc finger-like structure, Cys191-X-Cys193-X15-Cys208-X-Cys210 (Supplementary Figure S2A), which is observed in close proximity to the binding region for the 3′ CCA end of the tRNA. Zinc finger-like structures are frequently observed in aaRSs, and are thought to stabilize the local protein structure that recognizes the tRNA CCA terminus (38). A calcium ion is coordinated by the main-chain carbonyl group of Gly129 and six water molecules (Supplementary Figure S2B).
Amino acid recognition
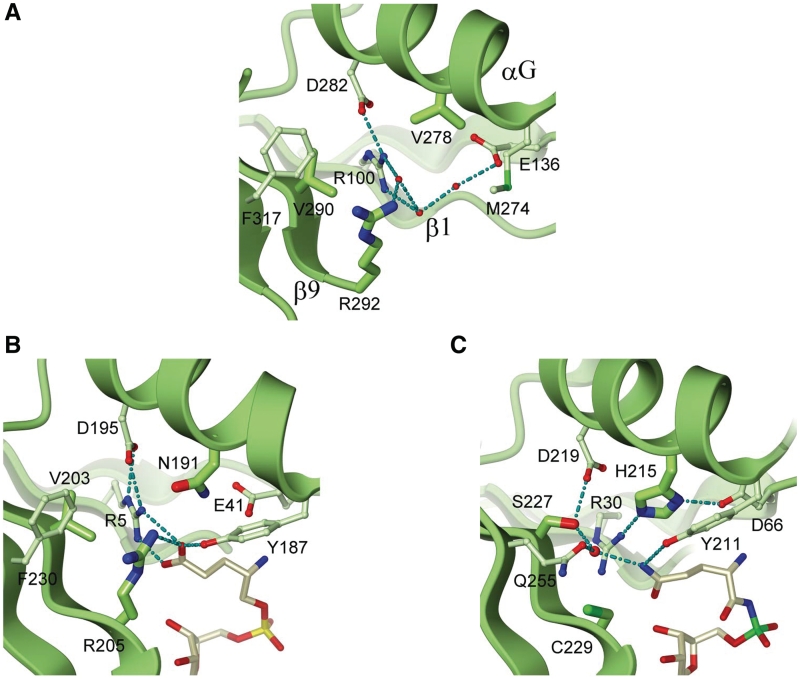
Based on the sequence alignments with Thermus thermophilus D-GluRS (15) and E. coli GlnRS (21), whose structures were solved in the complex with their cognate amino acids, the amino acid-binding pocket of the ND-GluRS resides at β1 (residues 97–103), αG (residues 273–285) and β9 (residues 289–292).
Overall the ND-GluRS amino acid-binding pocket is similar in architecture to the D-GluRS structure, and shows characteristic differences with the GlnRS active site. In GluRSs a strictly conserved hydrogen bonding network, which is generated by residues Arg100, Arg292 and Asp282 in M. thermautotrophicus ND-GluRS and residues Arg5, Arg205 and Asp195 in T. thermophilus D-GluRS, is principally responsible for establishing specific interactions with the substrate glutamate (Figure 2A and B). In the D-GluRS, Arg5 on β1 forms a bi-partite hydrogen bond with the side-chain carboxyl group of substrate glutamate (Figure 2B), Arg205 also forms a hydrogen bond to the carboxyl group of glutamate, and Asp195 interacts with Arg5, presumably stabilizing its productive interaction with the substrate. The GlnRSs show strict conservation of two of these residues, but all GlnRSs replace the residue equivalent to Arg5 (Arg 100 in ND-GluRS) with a cysteine (Cys229 in E. coli GlnRS). In E. coli GlnRS, His215 protrudes from αG and hydrogen-bonds to Arg30 which prevents its interaction with the amino acid substrate by changing the Arg rotamer state (Figure 2C). This His residue is 99% conserved in GlnRSs. In GluRSs, the corresponding residue is highly variable (Asn191 in T. thermophilus D-GluRS, Val 278 in M. thermautotrophicus ND-GluRS), but it is typically a small amino acid that would not interfere with the amino acid recognition by the Arg residue equivalent to Arg5 in T. thermophilus GluRS (Figure 2B).
Figure 2.
Amino acid recognition. (A) amino acid-binding pocket of M. thermautrophicus ND-GluRS. Water molecules involved in the hydrogen-bond network are colored red. (B) The Glu-binding pocket of T. thermophilus discriminating GluRS (15). The bound Glu-AMP is represented by ivory ball-and-stick. (C) The Gln-binding pocket of E. coli GlnRS (21). The bound Gln-AMP analogue is represented by ivory ball-and-stick.
At two other positions, a glutamine and serine residue (Gln255 and Ser277 in E. coli GlnRSs) are strictly conserved in GlnRS sequences, but these positions are highly variable in GluRSs. The Ser and Gln residues in GlnRS recognize the side-chain amide group of the substrate Gln via a water mediated hydrogen bond (Figure 2C), while the corresponding residues are hydrophobic in T. thermophilus D-GluRS (Val203 and Phe230) and M. thermautotrophicus ND-GluRS (Val290 and Phe317) (Figure 2A and B). Finally, in most GluRSs and GlnRSs a conserved Tyr residue makes contact to either the carboxyl group of the substrate Glu (Tyr187 in T. thermophilus GluRS) or the amine moiety of the substrate Gln (Tyr211 in E. coli GlnRS). In contrast, a Met (Met274 in M. thermautotrophicus GluRS) or Leu residue replaces this Tyr in many archaeal GluRSs. The ND-GluRS structure indicates that the Tyr residue is not critical for substrate recognition. The amino acid residues constituting the l-glutamate-specific pocket in GluRSs are more variable compared to the l-Gln-specific pocket in GlnRS. This may be due to the fact that GlnRSs have evolved over a shorter period of time than GluRSs, or possibly because the complicated water-mediated hydrogen bonding network involved in Gln recognition requires a more conserved active site pocket.
Since there are only a small number of residues that differ characteristically between the GluRS and GlnRS active sites, it would seem reasonable that a small number of mutations should suffice to convert a GlnRS to an enzyme specific for glutamylation or convert a GluRS to an enzyme capable of forming Gln-tRNA. Mutagenesis has been applied to the E. coli GlnRS system (39,40), and the experiments showed that replacement of only the residues in first shell of contact with the substrate was insufficient to convert GlnRS to GluRS. Only when residues in the second interaction shell, i.e. those in contact with the active site residues but not in direct contact to the substrate, were also mutated (amounting to 22 amino acid changes and one amino acid deletion) to the equivalent residues found in GluRS, was an enzyme produced that no longer functioned as a GlnRS and did show Glu-tRNAGln formation activity, albeit with 800-fold less aminoacylation efficiency compared to typical aaRSs (40).
tRNA discrimination module
ND-GluRS recognizes both tRNAGlu and tRNAGln without significantly discriminating between them, while D-GluRS and GlnRS are highly specific only for their cognate tRNA. Correspondingly, while the overall structure of the ND-GluRS is highly similar to GlnRS, the ND-GluRS lacks two structural modules that may participate in discrimination between tRNAGlu and tRNAGln.
Superposition of the E. coli GlnRS-tRNAGln complex structure onto the M. thermautotrophicus ND-GluRS structure reveals that the amino acid residues recognizing the first and second anticodon bases are conserved between GlnRS and ND-GluRS. In the GlnRS complex, the C34 nucleotide is recognized through hydrogen bonds by the side chains of Arg410 to the N4 atom and by Arg412 to the O2 atom of C34 (Figure 3B). Arg410 is conserved in ND-GluRS as Arg459 and it likely recognizes C34 in the same manner. Arg412 in GlnRS is not conserved and replaced by Ile461 in ND-GluRS, but the side chain of Arg426 in ND-GluRS occupies a similar spatial position as Arg412 in GlnRS. Also, an additional residue (Asp462 in ND-GluRS) may recognize C34 via a hydrogen-bond to the N3 atom (Figure 3A). In E. coli GlnRS, the U35 nucleobase is recognized through hydrogen bonds with the side chains of Arg341 to the O4 atom and Gln517 to the N3 atom of U35 (Figure 3B). The main-chain amide group of Arg520 interacts with the O2 atom of U35. These three GlnRS residues are conserved in the ND-GluRS as Arg403, Gln530 and Arg533, respectively (Figure 3A).
Figure 3.
tRNA anticodon recognition. (A) The docking model of M. thermautrophicus ND-GluRS and tRNAGln. The residues which may recognize the anticodon bases are represented by ball-and-stick. The same color code as in Figure 1 is used. (B) The structure of E. coli GlnRS·tRNAGln complex.
The first point of significant distinction between the GlnRS and ND-GluRS tRNA recognition involves residues contacting the G36 nucleobase. As noted, the anticodons of tRNAGlu (34YUC36) and tRNAGln (34YUG36) differ at this position. In the GlnRS structure, the side chain of Arg402 (hydrogen bond to the O4 atom), the main-chain carbonyl group of Gln399 (hydrogen bond to the N2 atom), and the side chain of Lys401 (via a π-cation interaction with the base moiety) specifically recognize the G36 nucleobase (Figure 3B). In the ND-GluRS structure, the G36-recognition loop (E. coli GlnRS residues 392–407) is absent (Figure 3A), which may allow the ND-GluRS to recognize both of tRNAGlu and tRNAGln.
The crystal structure of the ND-GluRS reveals a second distinction with the E. coli GlnRS-tRNAGln complex structure that involves a β-hairpin module (unpairing loop; residues 132–141 in E. coli GlnRS) in the CP domain. The unpairing loop disrupts the U1:A72 base pair in the tRNAGln acceptor stem by means of hydrophobic interaction with Leu136 (Figure 1C). In bacteria, this may contribute to the discrimination of tRNAGln from tRNAGlu, since bacterial tRNAGlu has a more stable G1:C72 base pair. Both the U1:A72 in tRNAGln and the unpairing loop in GlnRS are conserved among bacterial representatives. In eukaryotic GlnRS sequences, the unpairing loop is not conserved and the tRNAGln species typically have a G1:C72 base pair. These data indicate that the unpairing loop may be a secondary contribution to discrimination of tRNAGln or that the mode of discrimination is somewhat different for eukaryotic GlnRSs. In M. thermautotrophicus ND-GluRS, this unpairing loop module is absent and replaced by a short turn connecting two α-helices in the CP domain (Figure 1C). Both M. thermautotrophicus tRNAGln isoacceptors have A1:U72 and tRNAGlu has G1:C72, so insertion of the unpairing loop could be detrimental to Glu-tRNAGln formation by ND-GluRS.
Evolution in the GlxRS family
The fact the bacterial GluRS anticodon-binding domain (α-helical cage) is unrelated to that (β-barrel) found in other members of the GlxRS (GluRS/GlnRS) family of enzymes suggested the GluRS in the last universal common ancestor (LUCA) possessed only a catalytic domain (12). To test this hypothesis we used a comparative phylogenetic approach to examine the evolution of the catalytic and anticodon-binding domains in the GlxRS family and related aaRSs (Figures 4 and 5). The phylogenetic analyses were facilitated by our structure of the archaeal ND-GluRS along with the previously solved structures of bacterial GluRS (15,19), GlnRS (20,41,42), CysRS (43) and class I LysRS (18). These data enabled more accurate structure-based sequence alignments and provide an atomic-detailed view of the evolution of this group of aaRSs.
Figure 4.
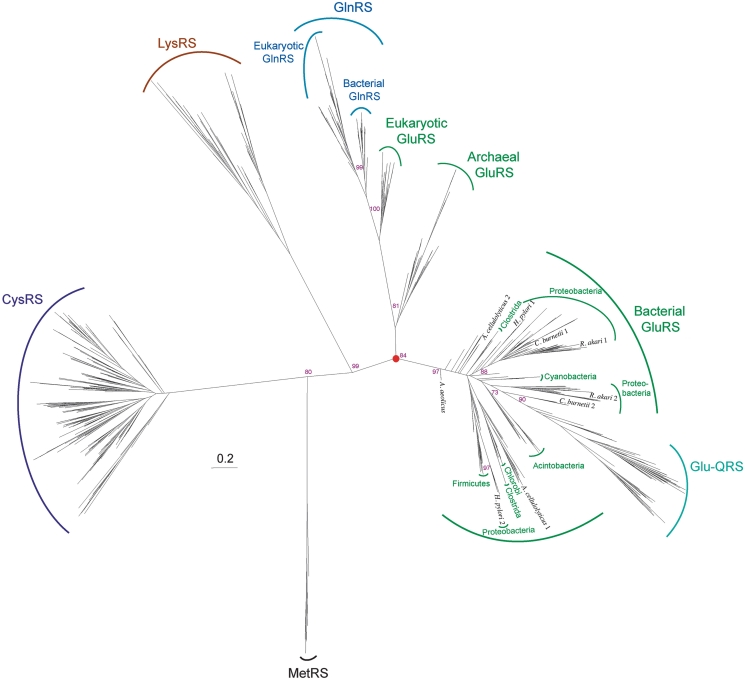
Phylogeny of the catalytic domains of CysRS (purple), LysRS (gold), GluRS (green), GlnRS (blue) and Glu-QRS (turquoise) rooted with representative MetRS (black) catalytic domains. Scale bar represents 0.2 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. Red circle represents LUCA.
Figure 5.
Phylogeny of the anticodon-binding domains in the GlxRS family of enzymes. (A) Unrooted phylogeny of the α-helix anticodon-binding domains of class I LysRS (gold) and bacterial GluRS (green). (B) Unrooted phylogeny of the β-barrel anticodon-binding domains of archaeal and eukaryotic GluRS (green), and GlnRS (blue). Scale bars represent 0.2 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity.
We examined the evolutionary relationship of the GlxRS catalytic domain with those from related aaRSs (CysRS and LysRS) (Figure 4). The phylogeny indicates that LysRS is more closely related to the GlxRS family than CysRS (Figure 4). As previously noted (11–14), GluRSs display a three-domain phylogeny, with GlnRS evolving from a duplication of the eukaryotic GluRS. GlnRS subdivides into two types, eukaryotic and bacterial, suggesting a single transfer of GlnRS from eukaryotes to bacteria followed by subsequent transfer amongst different bacterial lineages.
The phylogeny shows the emergence of glutamyl-queosine tRNAAsp synthetase (Glu-QRS). Glu-QRS is a paralog of GluRS that glutamylates queosine 34 of tRNAAsp in many proteobacteria, cyanobacteria and acintobacteria. Glu-QRS lost the anticodon-binding domain present in its bacterial GluRS predecessor (Figure 4) (44–48). The duplication event that gave rise to Glu-QRS also resulted in the maintenance of a second full length GluRS in some species (e.g. Rickettsia akari and Coxiella burnetii).
Other proteobacteria (e.g. H. pylori) appear to have acquired a second GluRS via horizontal gene transfer from firmicutes. In H. pylori, this enabled the organism to evolve one GluRS for Glu-tRNAGlu formation and the other to attach Glu to tRNAGln as the first step in Gln-tRNAGln synthesis (49,50). In Acidothiobacilli, one GluRS retains the ability to glutamylate tRNAGlu isoacceptors and a tRNAGln species, while the second GluRS favors a different tRNAGln isoacceptor (49,51). This situation may help the organism meet demands from heme biosynthesis and translation that both require Glu-tRNA formation (52).
The phylogeny of the α-helix anticodon-binding domains of the class I LysRS (18) and bacterial GluRS is generally congruent with that observed for the catalytic domains (Figure 5A). The major discrepancy between the phylogenies occurs with regards to the second GluRS encoded in α and γ proteobacteria, whose anticodon-binding domains group with the firmicutes. This may be an artifact of long-branch attraction and the lack of Glu-QRSs in the anticodon domain phylogenetic calculation. The LysRS anticodon-binding domains are separated from the bacterial GluRS anticodon-binding domains by a long-branch (Figure 5A), which is similar to the catalytic domain phylogeny (Figure 4). These data suggest the common ancestor of LysRS and GluRS also possessed an α-helix anticodon-binding domain (Supplementary Figure S1). Furthermore, the tree implies that the ancestor of all GlxRS family members did indeed have an antiocodon-binding domain of the α-helical cage type.
Taken together the phylogenies support the following evolutionary scenario for the GlxRS family (Figure 6). In LUCA, GluRS was a non-discriminating enzyme that had an α-helix anticodon-binding domain and glutamylated both tRNAGlu and tRNAGln. An ancestral Glu-AdT amidated the latter to form the Gln-tRNAGln used in protein synthesis (10). Following the split between the main lines of descent from LUCA, bacteria retained the ancestral type GluRS with an α-helical cage anticodon-binding domain. A common ancestor of eukaryotes and archaea then replaced the α-helix anticodon module with a β-barrel domain giving rise to modern eukaryotic and archaeal GluRS. This was also a non-discriminating enzyme, and the modern archaeal ND-GluRS is most similar to this ancient form of GluRS. Consistent with the scenario, the phylogeny of the β-barrel module is generally congruent with the phylogeny of the catalytic domains of eukaryotic and archaeal GluRS, and GlnRS (Figure 5B). This shows that the archaeal/eukaryotic GluRS catalytic and the β-barrel anticodon-binding domain co-evolved since a time before the divergence of Archaea and eukaryotes. The only significant difference between the phylogenies is in the apicomplexian GlnRS. Their GlnRS anticodon-binding domains grouped outside of the other eukaryotic sequences, which may be an artifact of long-branch attraction due to the accelerated rate of evolution in these parasitic organisms.
Figure 6.
Evolutionary pedigree in the GlxRS family. In LUCA (denoted by the red circle), the ancestral GluRS catalytic domain (green) was fused with an α-helix anticodon-binding domain (blue). In a common ancestor of eukaryotes and archaea, the α-helix anticodon-binding domain was replaced with a β-barrel domain (pink). GlnRS arose from a duplication of the eukaryotic GluRS and was acquired in bacteria via horizontal gene transfer (dashed arrow). In bacteria GluRS with an α-helix anticodon-binding domain was retained. Glu-QRS (grey–green) arose from a duplication of the bacterial GluRS and subsequent loss of the anticodon-binding domain.
Duplication of GluRS in early eukaryotes led to two aaRSs with different tRNA substrate specificities. The second GluRS (ancestral GlnRS) in early eukaryotes not only evolved to recognize tRNAGln over tRNAGlu but also to become specific for Gln over Glu (21) allowing an aaRS to directly attach Gln to its cognate tRNA for the first time in the history of life on earth. This GlnRS in turn enabled eukaryotic GluRS to evolve as a discriminating GluRS as the two-step pathway for Gln-tRNAGln formation was replaced with the direct one (1).
In Saccharomyces cerevisiae, the eukaryotic GluRS, which is a D-GluRS in the cytoplasm, can be imported into the mitochondria where it functions as a ND-GluRS in concert with a Glu-AdT for mitochondrial Gln-tRNAGln formation (8), implying the tRNA discrimination of modern eukaryotic GluRS is context dependent. GlnRS was also acquired in certain bacterial lineages by horizontal gene transfer; first transferred from eukaryotes into bacteria and then among bacteria (Figures 4 and 5B). In eukaryotes as well as in a handful of bacteria that acquired GlnRS, the GluRS evolved to be specific for tRNAGlu (15), and the two-step pathway for Gln-tRNAGln synthesis was replaced with the direct route.
To date, no archaea have acquired GlnRS, and all archaeal GluRS enzymes are non-discriminating in nature (1). Archaeal tRNAGln is a poor substrate for known GlnRSs (3), and it is speculated that this unique archaeal tRNAGln is the barrier preventing the acquisition of the direct route for Gln-tRNAGln synthesis (10).
CONCLUSION
The structure of the archaeal ND-GluRS presented here reveals structural distinctions between itself and the GlnRS structure that underlie tRNA discrimination by GlnRS on the one hand and tRNA promiscuity by the ND-GluRS on the other hand. These data and comparative analyses provide a basis for understanding and potentially manipulating tRNA discrimination in these aaRSs, which can be used to alter or expand the genetic code of organisms with tractable genetic systems.
It is thought that the genetic code evolved before the modern aaRSs because the existence of encoded proteins as complicated as the aaRSs presupposes a genetic code accurate enough to produce such proteins (53). This notion is supported by phylogenetic evidence indicating that tRNA identity elements are older than the aaRSs that use these key nucleotides for tRNA recognition (54,55). In vitro selection experiments have produced ribozymes capable of specific aminoacylation of tRNAs [reviewed in (56) and (57)]. These experiments show that the genetic code could have evolved in an RNA world. While much of the record of the evolution of the genetic code and of the molecules responsible for the coding process has likely been lost, the evolution of the aaRSs has left a record of the final stages in the evolution of the genetic coding process. Archaea retained the ND-GluRS that is the predecessor of the GlnRS and the eukaryotic D-GluRS. The archaeal ND-GluRS structure, therefore, represents an intermediary form or molecular missing link in the evolution of glutamine encoding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japanese Science and Technology Strategic International Cooperative Program, the Ministry of Education, Culture, Sports, Science and Technology National Project on Protein Structural and Functional Analyses (to O.N.); Uehara Memorial Foundation as well as Ministry of Education, Culture, Sports, Science and Technology (to R.I. and O.N.); Institute of General Medical Sciences and the United States Department of Energy (to D.S.). Funding for open access charge: National Institute of General Medical Sciences American Recovery and Reinvestment Act (grant GM022854-34S1).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to beam-line staffs at BL41XU of SPring-8 and NW12A of KEK PF-AR for assistance in data collection.
REFERENCES
- 1.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su D, Hohn MJ, Palioura S, Sherrer RL, Yuan J, Söll D, O'Donoghue P. How an obscure archaeal gene inspired the discovery of selenocysteine biosynthesis in humans. IUBMB Life. 2009;61:35–39. doi: 10.1002/iub.136. [DOI] [PubMed] [Google Scholar]
- 3.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia colitRNAGln1in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 7.Pujol C, Bailly M, Kern D, Maréchal-Drouard L, Becker H, Duchêne AM. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc. Natl Acad. Sci. USA. 2008;105:6481–6485. doi: 10.1073/pnas.0712299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frechin M, Senger B, Brayé M, Kern D, Martin RP, Becker HD. Yeast mitochondrial Gln-tRNAGln is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009;23:1119–1130. doi: 10.1101/gad.518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagao A, Suzuki T, Katoh T, Sakaguchi Y, Suzuki T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc. Natl Acad. Sci. USA. 2009;106:16209–16214. doi: 10.1073/pnas.0907602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J. Mol. Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamour V, Quevillon S, Diriong S, N'Guyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family–rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur. J. Biochem. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown JR, Doolittle WF. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 14.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Hendrickson TL. Divergent anticodon recognition in contrasting glutamyl-tRNA synthetases. J. Mol. Biol. 2004;344:1167–1174. doi: 10.1016/j.jmb.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 18.Terada T, Nureki O, Ishitani R, Ambrogelly A, Ibba M, Söll D, Yokoyama S. Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nat. Struct. Biol. 2002;9:257–262. doi: 10.1038/nsb777. [DOI] [PubMed] [Google Scholar]
- 19.Schulze JO, Masoumi A, Nickel D, Jahn M, Jahn D, Schubert WD, Heinz DW. Crystal structure of a non-discriminating glutamyl-tRNA synthetase. J. Mol. Biol. 2006;361:888–897. doi: 10.1016/j.jmb.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 20.Rould MA, Perona JJ, Söll D, Steitz TA. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 21.Bullock TL, Uter N, Nissan TA, Perona JJ. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. Sect. D. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Usón I, Schmidt B, von Bülow R, Grimme S, von Figura K, Dauter M, Rajashankar KR, Dauter Z, Sheldrick G. M Locating the anomalous scatterer substructures in halide and sulfur phasing. Acta Crystallogr. Sect. D. 2003;59:57–66. doi: 10.1107/s090744490201884x. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou YY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Söll D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc. Natl Acad. Sci. USA. 2009;106:21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 35.Rould MA, Perona JJ, Steitz TA. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 36.Jahn M, Rogers MJ, Söll D. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- 37.Uter NT, Perona JJ. Long-range intramolecular signaling in a tRNA synthetase complex revealed by pre-steady-state kinetics. Proc. Natl Acad. Sci. USA. 2004;101:14396–14401. doi: 10.1073/pnas.0404017101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nureki O, Kohno T, Sakamoto K, Miyazawa T, Yokoyama S. Chemical modification and mutagenesis studies on zinc binding of aminoacyl-tRNA synthetases. J. Biol. Chem. 1993;268:15368–15373. [PubMed] [Google Scholar]
- 39.Agou F, Quevillon S, Kerjan P, Mirande M. Switching the amino acid specificity of an aminoacyl-tRNA synthetase. Biochemistry. 1998;37:11309–11314. doi: 10.1021/bi9805590. [DOI] [PubMed] [Google Scholar]
- 40.Bullock TL, Rodriguez-Hernández A, Corigliano EM, Perona JJ. A rationally engineered misacylating aminoacyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2008;105:7428–7433. doi: 10.1073/pnas.0711812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 42.Deniziak M, Sauter C, Becker HD, Paulus CA, Giegé R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauenstein S, Zhang CM, Hou YM, Perona JJ. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004;11:1134–1141. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- 44.Campanacci V, Dubois DY, Becker HD, Kern D, Spinelli S, Valencia C, Pagot F, Salomoni A, Grisel S, Vincentelli R, et al. The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity. J. Mol. Biol. 2004;337:273–283. doi: 10.1016/j.jmb.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Dubois DY, Blaise M, Becker HD, Campanacci V, Keith G, Giegé R, Cambillau C, Lapointe J, Kern D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc. Natl Acad. Sci. USA. 2004;101:7530–7535. doi: 10.1073/pnas.0401634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salazar JC, Ambrogelly A, Crain PF, McCloskey JA, Söll D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc. Natl Acad. Sci. USA. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaise M, Becker HD, Keith G, Cambillau C, Lapointe J, Giegé R, Kern D. A minimalist glutamyl-tRNA synthetase dedicated to aminoacylation of the tRNAAsp QUC anticodon. Nucleic Acids Res. 2004;32:2768–2775. doi: 10.1093/nar/gkh608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaise M, Olieric V, Sauter C, Lorber B, Roy B, Karmakar S, Banerjee R, Becker HD, Kern D. Crystal structure of glutamyl-queuosine tRNAAsp synthetase complexed with L-glutamate: structural elements mediating tRNA-independent activation of glutamate and glutamylation of tRNAAsp anticodon. J. Mol. Biol. 2008;381:1224–1237. doi: 10.1016/j.jmb.2008.06.053. [DOI] [PubMed] [Google Scholar]
- 49.Salazar JC, Zúñig a R, Raczniak G, Becker H, Söll D, Orellana O. A dual-specific Glu-tRNAGln and Asp-tRNAAsn amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500:129–131. doi: 10.1016/s0014-5793(01)02600-x. [DOI] [PubMed] [Google Scholar]
- 50.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levicán G, Katz A, Valenzuela P, Söll D, Orellana O. A tRNAGlu that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett. 2005;579:6383–6387. doi: 10.1016/j.febslet.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 52.Levicán G, Katz A, de Armas M, Núñez H, Orellana O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc. Natl Acad. Sci. USA. 2007;104:3135–3140. doi: 10.1073/pnas.0611611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woese C. New York: Harper; 1967. The genetic code: the molecular basis for genetic expression. [Google Scholar]
- 54.Ribas de Pouplana L, Turner RJ, Steer BA, Schimmel P. Genetic code origins: tRNAs older than their synthetases? Proc. Natl Acad. Sci. USA. 1998;95:11295–11300. doi: 10.1073/pnas.95.19.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hohn MJ, Park HS, O'Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl Acad. Sci. USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yarus M. On translation by RNAs alone. Cold Spring Harb. Symp. Quant. Biol. 2001;66:207–215. doi: 10.1101/sqb.2001.66.207. [DOI] [PubMed] [Google Scholar]
- 57.Ohuchi M, Murakami H, Suga H. The flexizyme system: a highly flexible tRNA aminoacylation tool for the translation apparatus. Curr. Opin. Chem. Biol. 2007;11:537–542. doi: 10.1016/j.cbpa.2007.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.