Abstract
A novel, site-specific, DNA backbone S-modification (phosphorothioation) has been discovered, but its in vivo function(s) have remained obscure. Here, we report that the enteropathogenic Salmonella enterica serovar Cerro 87, which possesses S-modified DNA, restricts DNA isolated from Escherichia coli, while protecting its own DNA by site-specific phosphorothioation. A cloned 15-kb gene cluster from S. enterica conferred both host-specific restriction and DNA S-modification on E. coli. Mutational analysis of the gene cluster proved unambiguously that the S-modification prevented host-specific restriction specified by the same gene cluster. Restriction activity required three genes in addition to at least four contiguous genes necessary for DNA S-modification. This functional overlap ensures that restriction of heterologous DNA occurs only when the host DNA is protected by phosphorothioation. Meanwhile, this novel type of host-specific restriction and modification system was identified in many diverse bacteria. As in the case of methylation-specific restriction systems, targeted inactivation of this gene cluster should facilitate genetic manipulation of these bacteria, as we demonstrate in Salmonella.
INTRODUCTION
The high G+C Gram-positive bacterium Streptomyces lividans 66 contains DNA that is degraded by oxidative cleavage during electrophoresis in Tris-acetate buffer [the so-called DNA degradation (Dnd) phenotype] (1). Double-strand cleavage occurs at specific sites which contain a non-bridging S atom attached to backbone phosphorus at rare but specific sites (2,3). We have cloned and sequenced the S. lividans dnd gene cluster responsible for DNA S-modification. The proposed biochemical pathway leading to DNA S-modification has been shown to involve five putative proteins encoded by the dnd gene cluster (4–6), but the physiological function of DNA backbone S-modification in vivo has remained obscure. Previously, post-replicative host-specific cytosine and adenine modification (7,8) of DNA by methylation, hydroxymethylation and glucosyl-hydroxymethylation has been shown to be involved in DNA replication and repair, control of genomic imprinting, regulation of gene expression, and host-specific DNA restriction–modification (RM) systems in almost all organisms (9–12).
Here, we selected one of at least 10 Salmonella enterica serovar strains among more than 100 strains (13,14) from phylogenetically diverse bacteria which exhibited the Dnd phenotype (15) and contained homologs of the S. lividans dnd gene cluster to investigate whether a corresponding DNA restriction system related to S-modification system could exist in S. enterica, functioning like DNA methylation in prokaryotes for the protection of homologous DNA against host-specific restriction of foreign DNA (12,16).
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are described in Table 1. Salmonella enterica serovar Cerro 87 isolated from an egg-producing farm was a non-pathogenic strain displaying the Dnd phenotype.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Salmonella enterica | ||
| 87 | Serotype Cerro, Dnd+ | (4) |
| XTG101 | 87 derivative, dptC interruption mutant | This work |
| XTG102 | 87 derivative, dptB-E deletion mutant | This work |
| XTG104 | 87 derivative, dptF in-frame deletion mutant | This work |
| XTG105 | 87 derivative, dptG in-frame deletion mutant | This work |
| Escherichia coli | ||
| DH5α | F- recA lacZ ΔM15 | (17) |
| EPI300TM-T1R | used for construction of genomic library | EPICENTER |
| B7A | Dnd+ | |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector, Ampr | (18) |
| pMD18-T | vector for DNA sequencing | TaKaRa |
| pUC18 | cloning vector, Ampr | (19) |
| pKD46 | temperature-sensitive replication, Ampr | (20) |
| pKOV-Kan | temperature-sensitive replication, sacB, Cmr, Kanr | (21) |
| pCC1FOSTM | Cmr oriV ori2 | EPICENTER |
| 8F4 | pCC1FOSTM derived fosmid carrying dpt cluster | This work |
| 6G12 | pCC1FOSTM derived fosmid carrying dpt cluster | This work |
| pJTU1231 | pMD-18T derivative carrying a 0.9-kb PCR product of p10/p11 | This work |
| pJTU1232 | pUC18 derivative carrying a 4.4-kb EcoRI fragment from 8F4 | This work |
| pJTU1233 | pUC18 derivative carrying a 3.4-kb EcoRI fragment from 8F4 | This work |
| pJTU1237 | pBluescript II SK(+) derivative carrying a 2.5-kb EcoRI–ScaI/EcoRV fragment from pJTU1233 | This work |
| pJTU1238 | pJTU1237 derivative carrying 6.7-kb dptB-E region | This work |
| pJTU1239 | pKD46 derivative carrying a 0.7-kb EcoRI–EcoRV fragment of dptC from pJTU1231 | This work |
| pJTU2489 | pUC18 derivative carrying a 5.9-kb PvuII-SmaI fragment from 6G12 | This work |
| pJTU2492 | pJTU2489 derivative carrying a 8.9-kb NsiI fragment from 6G12 | This work |
| pJTU3809 | pJTU2492 derivative with 4.7-kb SalI region deletion | This work |
| pJTU3818 | pCC1FOSTM derivative carrying a 20-kb BglII fragment from 6G12 | This work |
| pJTU2468 | pKOV-Kan derivative carrying a 1.5-kb BamHI/XhoI digested PCR product with deletion in dptB-E | This work |
| pJTU3825 | pUC18 derivative carrying a 1.1-kb PCR product of FLL/FRR for sequencing | This work |
| pJTU3826 | pUC18 derivative carrying a 1.1-kb PCR product of GLL/GRR for sequencing | This work |
| pJTU3828 | pKOV-Kan derivative carrying 1.1-kb BamHI-SalI region from pJTU3825 with in-frame deletion in dptF | This work |
| pJTU3829 | pKOV-Kan derivative carrying 1.1-kb BamHI-SalI region from pJTU3826 with in-frame deletion in dptG | This work |
oriT, origin of transfer of plasmid RK2; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, Kanamycin resistance; sacB, levansucrase gene of Bacillus amyloliquefaciens.
Generation of dpt gene probes from S. enterica serovar Cerro 87
Degenerate oligonucleotide primers p1/p2 (p1: 5′ CTACTCGTTTCCGGCTATHCGNGG 3′; p2: 5′ ATCCTAGTTGCCCAAGNGCNTGCA 3′), p3/p4 (p3: 5′ GCGTNGNCAGCAGATGTTCGCCGA 3′; p4: 5′ GGAAGAATCTTTNCCNCCGCTGTA 3′) and p7/p8 (p7: 5′ TAGTTTCCGTTGGTGCACNGANCG 3′; p8: 5′ GCAATTTTCTCGCCRTCGAARAAG 3′), designed based on the highly conserved putative dnd genes in S. lividans 66, Streptomyces avermitilis NRRL8165, Pseudomonas fluorescens Pf0-1, Nostoc PCC7120, Nostoc punctiforme and Bacillus cereus E33L, were synthesized and used to amplify the corresponding fragments from S. enterica serovar Cerro 87 genomic DNA. The specific primers of p10/p11 (p10: 5′ GATTTGGCCGAATACGAAGA 3′; p11: 5′ TTTCCATCGCTTTATCTTTG 3′), according to the sequences of fragments p3p4 and p7p8, were used as a probe to amplify a 0.9-kb fragment and cloned into pMD-18T, giving pJTU1231 and sequenced (Supplementary Figure S1B). The association of this cloned probe with the Dnd phenotype was confirmed by the construction of mutant XTG101 through homologous recombination, which interrupted the dptC region and abolished the Dnd phenotype (Supplementary Figure S1D).
Localization of dpt gene cluster
The genomic fosmid library of S. enterica serovar Cerro 87 was constructed using CopyControl™ Fosmid Library Production Kit from EPICENTER® Biotechnologies.
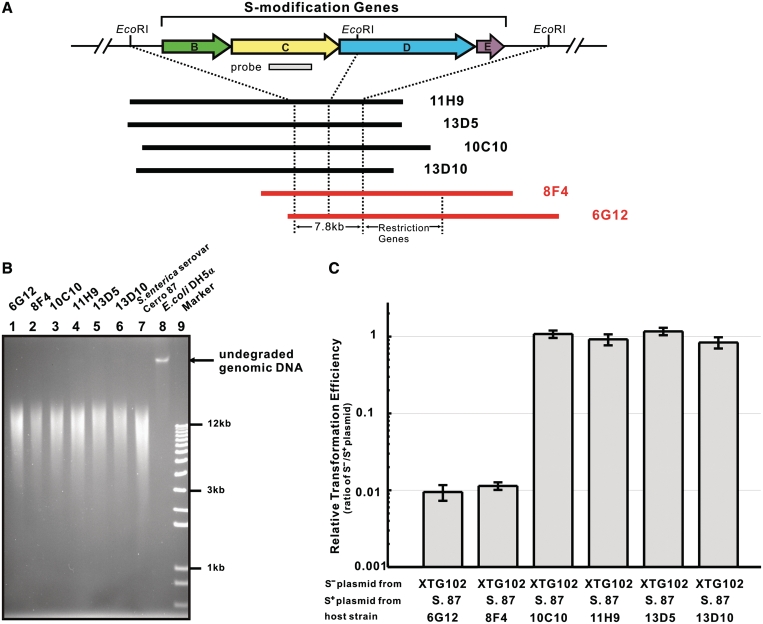
Six fosmids were picked using polymerase chain reaction (PCR) sib-selection with primers p10/p11 (Supplementary Figure S1B). Restriction patterns and sequence analysis localized the dpt gene cluster on a 7.8-kb core region comprised of a 4.4-kb and a 3.4-kb EcoRI fragments, which were digested from fosmid 8F4 and inserted into the EcoRI site of pUC18 for the generation of pJTU1232 and pJTU1233, respectively.
A 2.5-kb EcoRI–ScaI fragment from pJTU1233 was ligated into EcoRI–EcoRV-digested pBluescript II SK (+) to generate pJTU1237. A 4.2-kb BglII–EcoRI fragment from pJTU1232 was inserted into the BamHI–EcoRI sites of pJTU1237, generating pJTU1238, carrying a 6.7-kb functional dptB-E gene cluster.
Fosmid 6G12 was selected to sequence based on the restriction phenotype (Figure 2). A c. 20-kb BglII fragment carrying the modification and restriction gene cluster from 6G12 was ligated into BamHI-digested pCC1FOSTM to generate pJTU3818.
Figure 2.
Fosmids containing inserts of S. enterica serovar Cerro 87 DNA express a restriction phenotype in E. coli DH5α. (A) dptB-E region of S. enterica serovar Cerro 87 with the probe DNA that was used to identify six fosmid clones shown below at a reduced scale. The fosmids were aligned relative to each other according to their EcoRI (and other) restriction patterns. (B) Genomic DNA of E. coli DH5α containing the six S. enterica serovar Cerro 87 fosmids fractionated using PFGE. Extensive degradation occurred in all the DNA samples except in the sulfur-free sample from E. coli DH5α. (C) Restriction of S-free and S-modified pKOV-Kan in different E. coli DH5α containing the six S. enterica serovar Cerro 87 fosmids. The results were presented as relative transformation efficiencies (ratios of S−/S+ plasmid) obtained by parallel transformation of S− and S+ plasmid DNA. Error bars represent the standard deviation from three repeat experiments. S. 87, S. enterica serovar Cerro 87; XTG102, S. enterica serovar Cerro 87 mutant strain with deletion of dptBCDE.
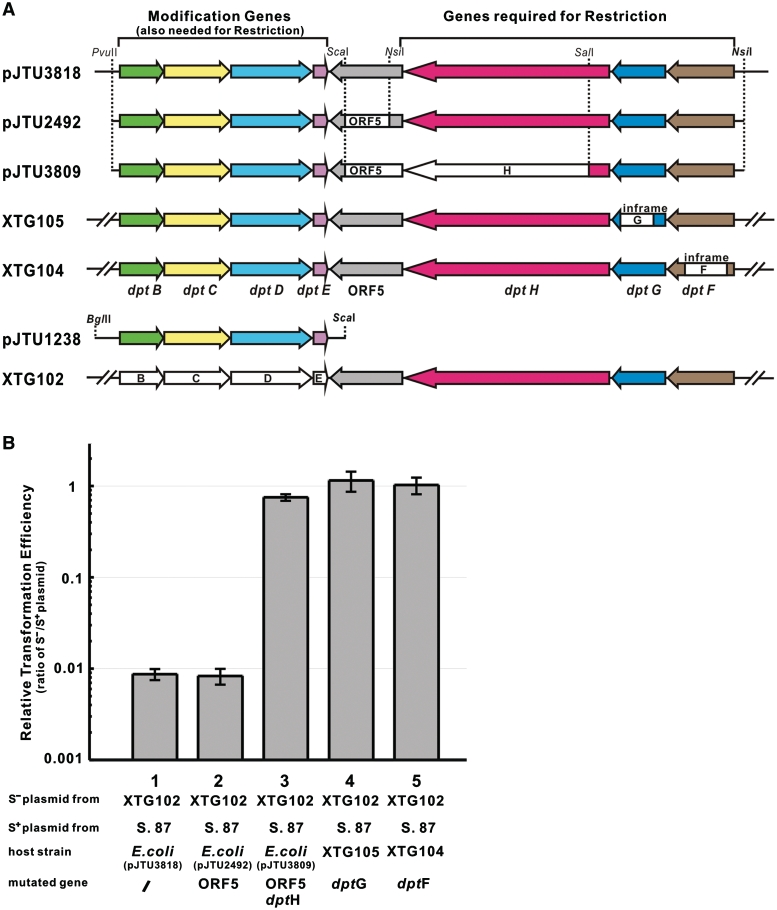
A 5.9-kb PvuII/ScaI fragment (modification genes) from pJTU1238 was cloned into the SmaI site of pUC18, generating pJTU2489. An 8.9-kb NsiI-digested fragment (restriction genes) from 6G12 was then ligated into the PstI site of pJTU2489, giving pJTU2492, carrying the whole cluster except for ORF5. pJTU2492 was digested with SalI and self-ligated to generate pJTU3809 with partial deletion of dptH.
Construction of mutants of S. enterica serovar Cerro 87
Construction of XTG101
A 0.7-kb EcoRI–EcoRV fragment from pJTU1231, carrying a 0.9-kb PCR product originated from using primers p10/p11, was cloned into the corresponding sites of pKD46, resulting in pJTU1239. After introduction by transformation into S. enterica serovar Cerro 87 at 30°C, mutant XTG101 was obtained by single crossover at 42°C selection, which disrupted dptC (Supplementary Figure S1).
Construction of XTG102, XTG104, XTG105
The following were the strategies to construct mutants XTG102, XTG104 and XTG105: first, amplifying left and right arms using total DNA isolated from S. enterica serovar Cerro 87 as template and introducing the corresponding restriction sites; second, amplifying the recombinant fragment using the mixture of left and right arms in the proper ratio as templates, overlapping by 40 bp, and then cloning into the thermo-sensitive plasmid pKOV-Kan digested with BamHI–SalI; third, introducing the generated plasmids by transformation into Escherichia coli DH5α (pJTU1238) to be phosphorothioated; fourth, introducing the phosphorothioated plasmids into S. enterica serovar Cerro 87 at 30°C and obtaining single crossover intermediates at 43°C; finally, obtaining the mutant through double crossing over on an LA plate with 15% sucrose at 43°C.
For XTG102, dptLL/dptLR (dptLL: 5′ GACCTCGAGTTGGTTTTCAATA 3′, XhoI underlined; dptLR: 5′ AAGCAACCGTGTCAAGGTAATTGGCGTTGCTGCGTGGTTA 3′) were used for the 752-bp left arm, dptRL/dptRR (dptRL: 5′ TAACCACGCAGCAACGCCAATTACCTTGACACGGTTGCTT 3′; dptRR: 5′ ATAGGATCCACGACGCCTACAAAT 3′, BamHI underlined) for the 819-bp right arm and dptLL/ dptRR for the 1531-bp entire homologous recombination region with introduced BamHI and XhoI sites, generating pJTU2468 with deletion of the modification genes dptB-E.
For XTG104, FLL/FLR (FLL: 5′ AATGTCGACTATCGTCCGAATAAATA 3′, SalI underlined; FLR: 5′ AGCACCAGCACTTTCCCTGCTGCCGTAGATCCCGCACTTA 3′) were used for the 535-bp left arm, FRL/FRR (FRL: 5′ TAAGTGCGGGATCTACGGCAGCAGGGAAAGTGCTGGTGCT 3′; FRR: 5′ CTTGGATCCTGTTAATCAGGGTATCG 3′, BamHI underlined) for the 580-bp right arm and FLL/FRR for the 1075-bp entire homologous recombination region with introduced BamHI and SalI sites, generating pJTU3828 with a 921-bp deletion in dptF.
For XTG105, GLL/GLR (GLL: 5′ TGGGTCGACTTGCGAAGTCATCTT 3′, SalI underlined; GLR: 5′ GGACGATAGCCATTATTCAACTGGAATTTAGGGTAGTGTT 3′) were used for the 537-bp left arm, GRL/GRR (GRL: 5′ AACACTACCCTAAATTCCAGTTGAATAATGGCTATCGTCC 3′; GRR: 5′ CGCGGATCCAATTGGCAAACAGCGTAC 3′, BamHI underlined) for the 604-bp right arm and GLL/GRR for the 1101-bp entire homologous recombination region with introduced BamHI and SalI sites, generating pJTU3829 with a 762-bp deletion in dptG.
Restriction test
The concentrations of plasmid DNA for restriction test, purified using Qiagen Plasmid Midi Kit, were determined by the absorbance at 260 nm and the purity was estimated with the ratio of A260/A280.
Electroporation-competent cells of S. enterica strains were prepared as described in Sambrook et al. Electroporation was done using a Cell-Porator with a voltage booster and 1-mm gap cuvette according to the manufacturer's instructions (GIBCO/BRL) by using 50 μl of cells and 5 µl plasmid DNA (∼10 ng). Shocked cells were added to 1 ml LB, incubated at 30°C for 30 min, and then spread onto agar containing antibiotic to select transformants.
The CaCl2-heat shock method was used for introducing plasmids into E. coli. The competent cells were prepared according to Sambrook et al. 5 µl (∼10 ng) of plasmid DNA and 95 µl of competent cells were mixed well and kept in ice for 30 min before being heat-shocked for 90 s at 42°C and placed in ice for another 5 min till adding 900 µl of LB. The mixture was incubated at 30°C for 30 min and plated on L-agar containing antibiotic(s).
RESULTS
Salmonella enterica serovar Cerro 87 restricts plasmid DNA from E. coli
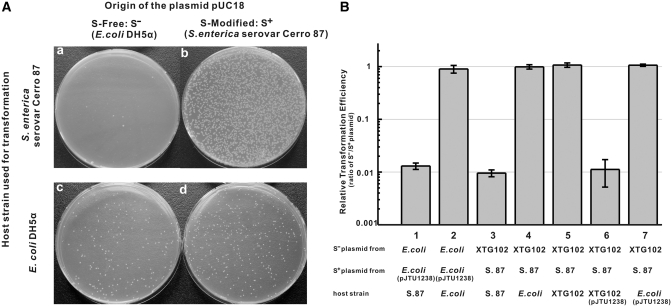
In bacteria, site-specific DNA modification (usually methylation) is often, but not always, associated with a sequence-specific endonuclease. These host-specific restriction systems prevent the introduction of heterologous DNA from phages and plasmids that lack the specific DNA modification. We first tried to test whether pUC18 DNA isolated from E. coli was restricted by S. enterica serovar Cerro 87. Figure 1A shows the results of transformations of S. enterica- or E. coli-competent cells using equal amounts of pUC18 isolated from E. coli (S− DNA) or S. enterica (S+ DNA). Salmonella enterica reproducibly yielded about 100 times fewer colonies with S− DNA from E. coli than with S+ homologous DNA from S. enterica, while about the same number of transformants could be obtained when E. coli DH5α was used as the host.
Figure 1.
Restriction in S. enterica. (A) Restriction of non-phosphorothioated (S-free) pUC18 isolated from S. enterica. Escherichia coli DH5α and S. enterica serovar Cerro 87 were used as hosts for transformation by equal amounts of S-free pUC18 isolated from E. coli (S−) or S-modified pUC18 isolated from S. enterica (S+). (B) Restriction of S-free and S-modified plasmids of pKOV-Kan in different host strains. The results were presented as relative transformation efficiencies (ratios of S−/S+ plasmid) obtained by parallel transformation of S− and S+ plasmid DNA. Error bars represent the standard deviation from three repeat experiments. S. 87, S. enterica serovar Cerro 87; E. coli, E. coli DH5α; XTG102, S. enterica serovar Cerro 87 mutant strain with deletion of dptBCDE.
Plasmids from E. coli that had escaped restriction by S. enterica were no longer restricted by S. enterica. When these plasmids were passaged through E. coli, they were again restricted by S. enterica, indicating that the plasmids were not mutants lacking the S-modification site. These results suggested that S. enterica might specifically restrict DNA lacking the S-modification.
DNA S-modification protects DNA against restriction by S. enterica serovar Cerro 87
To investigate whether this restriction phenotype was related to DNA S-modification, we cloned the dnd homologous gene cluster of the enteropathogenic S. enterica serovar Cerro 87. The S. lividans gene cluster was not suitable as a hybridization probe because it has a higher G+C content (66%) than Salmonella (48%). We designed degenerate oligonucleotide primers according to the most highly conserved region of the S. lividans dnd genes and amplified a fragment of the expected size (0.9 kb) within the S. enterica DptC coding sequence. Using the fragment, an S. enterica mutant strain that had S-free (unmodified) DNA was obtained by single crossover integration of a temperature-sensitive plasmid. This proved that the amplified fragment originated from the dnd homologous dpt (DNA phosphorothioation) gene cluster responsible for the S-modification of S. enterica DNA (Supplementary Figure S1A and B). The dpt gene cluster probe was then used to screen a fosmid library from the genome of S. enterica serovar Cerro 87. PCR sib-selection identified six overlapping clones covering four genes (dptB-E) in the same order as the dndB-E genes of S. lividans, whose encoded proteins were significantly similar.
The S-modification cluster was cloned into pBluescriptII SK(+) to give pJTU1238, which is compatible with pKOV-Kan, a vector with a kanamycin resistance marker. The presence of this plasmid conferred S-modification on pKOV-Kan and prevented its restriction by S. enterica (Figure 1B-1 and B-2). The S. enterica mutant strain XTG102, from which the genomic dptB-E operon was removed by gene replacement, gave S-free (Dnd−) pKOV-Kan DNA that was 100-fold restricted by the wild-type strain (Figure 1B-3 and B-4). These experiments proved that the S-modification and not another, unknown, DNA modification protected heterologous DNA from the as-yet unknown S. enterica serovar Cerro 87 restriction system.
The dptB-E S-modification gene cluster is required for host-specific restriction
It was surprising that S. enterica XTG102, which lacks the entire gene cluster responsible for DNA S-modification, was viable because it was thought to contain restriction genes without the protective S-modification genes, or a mutation might have removed the restriction function when the strain was generated by double-crossover recombination. Lack of restriction was confirmed by transformation (Figure 1B-5), and the cloned dptB-E gene cluster in pJUT1238 did not confer restriction on E. coli (Figure 1B-7). However, XTG102 became normally restricting after the cloned dptB-E gene cluster was introduced using pJUT1238 (Figure 1B-6). This suggested that the S. enterica restriction system may require the presence of the S-modification proteins for activity, and it seemed likely that additional genes required for restriction alone could be found on the fosmid clones near the dptB-E genes.
Identification of restriction genes in S. enterica serovar Cerro 87
The six fosmids mentioned earlier carrying dptB-E were introduced by transformation into E. coli DH5α. Two conferred the restriction phenotype on E. coli DH5α (Figure 2). A 20-kb fragment from fosmid 6G12 was cloned into pUC18 to generate pJTU3818, which retained the ability to confer the restriction phenotype on E. coli DH5α. This suggested that the restriction genes were located on this fragment (Figure 3). The 20-kb region of pJTU3818 was sequenced (Genbank Accession number GQ863484), and eight open reading frames (ORFs), including dptB-E, related to DNA S-modification were identified. Four other genes, ORF5, dptF, dptG and dptH, showed no convincing similarity to any proteins in the databases, nor any pfam matches. In order to precisely identify these genes, a series of chromosomal in-frame (to avoid polar effects) deletion mutants were generated in S. enterica serovar Cerro 87. The strains XTG104 and XTG105, with deletions in dptF and dptG, respectively, lost the restriction phenotype. Surprisingly, pJTU2492, carrying a DNA fragment within which ORF5 was deleted, still restricted foreign plasmids when expressed in E. coli DH5α, suggesting that ORF5 was not essential for restriction. Escherichia coli, DH5α containing pJTU3809 (ORF5 and dptH deleted), did not restrict foreign plasmids. These results unambiguously demonstrated that dptF-H were absolutely essential for restriction in S. enterica serovar Cerro 87 (Figure 3). In summary, this is a complex restriction system that depends for its activity on the presence of S-modification (phosphorothioation) genes.
Figure 3.
Mutational analysis of the dpt gene cluster of S. enterica serovar Cerro 87. (A) Mutational analysis of the dpt gene cluster. The complete, functional dptB-H cluster (including ORF5) was cloned in pJTU3818 and expressed in E. coli DH5α. Mutations were created in plasmids and in the S. enterica serovar Cerro 87 genome (XTG strains). White boxes with gene designations indicate deleted regions. (B) Restriction of S-free and S-modified pKOV-Kan in different mutantion strains. The results were presented as relative transformation efficiencies (ratios of S−/S+ plasmid) obtained by parallel transformation of S− and S+ plasmid DNA. Error bars represent the standard deviation from three repeat experiments. S. 87, S. enterica serovar Cerro 87; XTG102, S. enterica serovar Cerro 87 mutant strain with deletion of dptBCDE; XTG105, S. enterica serovar Cerro 87 mutant strain with in-frame deletion of dptG; XTG104, S. enterica serovar Cerro 87 mutant strain with in-frame deletion of dptF.
Similar host-specific restriction systems exist in many other bacteria
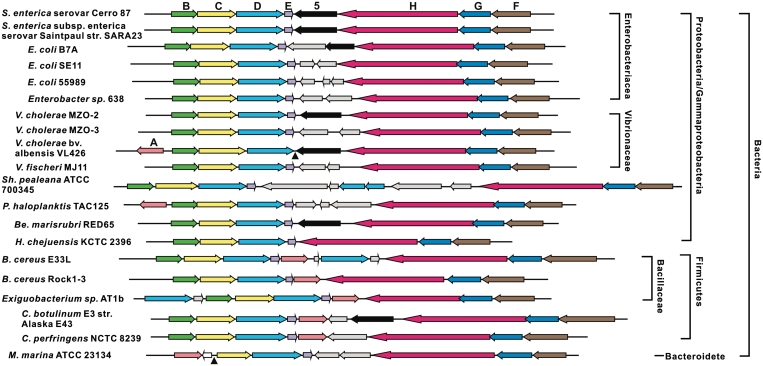
DNA from many bacteria is degraded during normal electrophoresis or pulsed-field gel electrophoresis in the presence of Tris, which is diagnostic for DNA S-modification (15). Nineteen dpt-like gene clusters from diverse strains were found in the DNA databases (Figure 4). Escherichia coli B7A was selected for further tests: it possesses the Dnd phenotype typical of S-modification in its DNA (Supplementary Figure S5A) and, in addition, S-free plasmids from S. enterica serovar Cerro 87 mutant XTG102 transformed E. coli B7A with 100-fold lower efficiency than S-modified plasmid DNA from S. enterica serovar Cerro 87 (Supplementary Figure S5B). This confirmed the prediction from sequence comparisons that a host-specific restriction system associated with DNA S-modification exists in E. coli B7A, and probably in many other bacteria of diverse origin.
Figure 4.
Organization of 20 bacterial dpt homologs. Colored arrows indicate very similar ORFs. Light gray arrows, diverse ORFs without predicted function that are not homologous to each other. Note that ORF5 (black arrows) is missing from most gene clusters, and dptA homologs (pink arrows) were found in only eight of the clusters. Filled triangles position of presumed deletions in the dptB-F homologous gene clusters. Strains and Genbank accession numbers in the order shown on the figure: S. enterica serovar Cerro 87 (GQ863484); Salmonella enterica subsp. enterica serovar Saintpaul str. SARA23 (NZ_ABAM02000001); E. coli B7A (AAJT02000066); E. coli SE11 (AP009240); E. coli 55989 (CU928145); Enterobacter sp. 638 (CP000653); Vibrio cholerae MZO-2 (AAWF01000002); Vibrio cholerae MZO-3 (AAUU01000003); Vibrio cholerae bv. albensis VL426 (ACHV01000001); Vibrio fischeri MJ11 (CP001133); Shewanella pealeana ATCC 700345 (CP000851); Pseudoalteromonas haloplanktis TAC125 (CR954247); Bermanella marisrubri RED65 (AAQH01000003); Hahella chejuensis KCTC 2396 (CP000155); Bacillus cereus E33L (CP000001); Bacillus cereus Rock1-3 (NZ_ACMG01000043); Exiguobacterium sp. AT1b (CP001615); Clostridium botulinum E3 str. Alaska E43 (CP001078); Clostridium perfringens NCTC 8239 (ABDY01000007); Microscilla marina ATCC 23134 (AAWS00000000).
DISCUSSION
Our results have established, at least for S. enterica, a biological role for the previously identified DNA backbone phosphorothioation: in a cluster of eight genes, seven are evidently the determinants for host-specific R–M. The dptB-E genes are essential for phosphorothioation, and the dptF-H genes are all required for restriction of unmodified DNA. The novel host-specific restriction and S-modification system described here is similar to the earlier extensively studied R–M system as prokaryotic immune systems that attack foreign DNA entering the cell. The R–M systems are traditionally classified into four major groups: type I, II, III and IV on the basis of subunit composition, sequence recognition, cleavage position and cofactor requirements. In types I, II and III, foreign DNA is inactivated by endonucleolytic cleavage, the host DNA is normally methylated within specific sequence by the congnate methyltransferase and protected against restriction. In type IV, only modified foreign DNA is cleaved (22,23). Here, experiment results suggested the host-specific R–M system might be close to type I R–M system. The host-specific restriction system is composed of seven genes, dptBCDE are essential for phosphorothioation and the dptFGH genes are all required for restriction of unmodified DNA. Restriction activity required dptFGH genes in addition to at least four contiguous dptBCDE genes necessary for DNA S-modification. This functional overlap ensures that restriction of heterologous DNA occurs only when the host DNA is protected by phosphorothioation. The composition of genes and functional overlap are very similar to the type I R–M system, which are composed of three subunits that typically contain two REase subunits that are required for DNA cleavage, one specificity subunit that specifies the DNA sequence recognized, and two MTase subunits that catalyse the methylation reaction. The genes for DNA S-modification and restriction also seemed to form a complex which was more complicated than type I R–M system. In addition, genes responsible for DNA sequence specificity and DNA cleavage site still remained obscure.
In methylation-specific R–M systems, a restriction gene is often linked to a modification gene, forming a so-called restriction–modification gene complex, but often solitary methyltransferases that do not have a restriction-enzyme counterpart (7) could also be found. This also seems to be true for the phosphorothioation-specific restriction systems. Among 20 taxonomically diverse bacteria, including low G+C Gram-positive Bacillus species (Figure 4), identified as carrying the syntenous dptB-E homologous gene clusters, all possessed dptF-H homologs (required for restriction only) in same order, but many other bacteria contain only dptB-E (for DNA S-modification) without dptF-H homologs. We suspect that, as in methylation-specific restriction systems, DNA S-modification may act not only as a sort of immune system, allowing the bacteria to protect themselves from infection by bacteriophages, but also as an epigenetic signal for new biological function(s).
We do not know which of the dndA counterparts is functional for phosphorothioation in S. enterica serovar Cerro 87, but dndA counterparts, whose function could likely be recruited from shared pathway(s), were found to be separated from eight of the 20 syntenous dptB-H counterpart clusters. The high degree of synteny of the dptB-H counterparts therefore provides strong support for the idea that dptB-H counterparts might be a later addition by horizontal transfer of genomic island(s) (15). This agrees well with the fact that many restriction–modification gene complexes reside on plasmids and prophages, which lends mobility (24), and host-specific restriction systems may be part of a mobile element near to an integrase (15) and therefore able to move freely between genomes. Additionally, methylated DNA is known to be an essential factor in Salmonella virulence, and its absence causes severe attenuation (25), but it was not clear whether S-modified DNA could play a similar role in host–pathogen interactions. We also attempted in vitro restriction enzyme assays to detect the presence of a host-specific restriction system, but these enzymes did not produce any distinctive DNA fragments in the crude extracts.
The discovery described here is, to our knowledge, a first report of a host-specific restriction system associated with S-modification of DNA instead of methylation to protect homologous DNA. The systems might be useful for engineering host resistance against in vivo exo- or endo-nucleolytic degradation, and/or for transfer into heterologous cells to control the detrimental effects of phage contamination in the fermentation industry.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation of China; the Ministry of Science and Technology; China Ocean Mineral Resources R & D Association (DYXM11502203); the Shanghai Municipal Council of Science and Technology (05ZR14069); and Shanghai Leading Academic Discipline Project B203. Funding for open access charge: Ministry of Science and Technology of China (grant 2009ZX09501-008).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Tobias Kieser, Prof. Peter C. Dedon and Prof. Sir David Hopwood, FRS for valuable discussion and critical reading of the manuscript and Prof. Toshiyuki Murase for supplying S. enterica serovar Cerro 87.
REFERENCES
- 1.Zhou X, Deng Z, Firmin JL, Hopwood DA, Kieser T. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 1988;16:4341–4352. doi: 10.1093/nar/16.10.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang J, Wang Z, He X, Li J, Zhou X, Deng Z. DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res. 2007;35:2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. A novel DNA modification by sulphur. Mol. Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 5.You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46:6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
- 6.Yao F, Xu T, Zhou X, Deng Z, You D. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009;583:729–733. doi: 10.1016/j.febslet.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 7.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 10.Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 11.Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr. Biol. 1997;7:R305–307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 12.Song HK, Sohn SH, Suh SW. Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex. EMBO J. 1999;18:1104–1113. doi: 10.1093/emboj/18.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liesegang A, Tschape H. Modified pulsed-field gel electrophoresis method for DNA degradation-sensitive Salmonella enterica and Escherichia coli strains. Int. J. Med. Microbiol. 2002;291:645–648. doi: 10.1078/1438-4221-00180. [DOI] [PubMed] [Google Scholar]
- 14.Murase T, Nagato M, Shirota K, Katoh H, Otsuki K. Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp. enterica serovar Livingstone and serovar Cerro isolates obtained from a chicken layer farm. Vet. Microbiol. 2004;99:139–143. doi: 10.1016/j.vetmic.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 15.He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol. Microbiol. 2007;65:1034–1048. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 16.Wilson GG, Murray NE. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalioti M, Heath J. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:E14. doi: 10.1093/nar/29.3.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome–restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 25.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.