Abstract
The S1 mRNA of avian reovirus is functionally tricistronic, encoding three unrelated proteins, p10, p17 and σC, from three sequential, partially overlapping open reading frames (ORFs). The mechanism of translation initiation at the 3′-proximal σC ORF is currently unknown. Transient RNA transfections using Renilla luciferase reporter constructs revealed only a modest reduction in reporter expression upon optimization of either the p10 or p17 start sites. Insertion of multiple upstream AUG (uAUG) codons in a preferred start codon sequence context resulted in a substantial retention of downstream translation initiation on the S1 mRNA, but not on a heterologous mRNA. The S1 mRNA therefore facilitates leaky scanning to promote ribosome access to the σC start codon. Evidence also indicates that σC translation is mediated by a second scanning-independent mechanism capable of bypassing upstream ORFs. This alternate mechanism is cap-dependent and requires a sequence-dependent translation enhancer element that is complementary to 18S rRNA. Downstream translation initiation of the tricistronic S1 mRNA is therefore made possible by two alternate mechanisms, facilitated leaky scanning and an atypical form of ribosome shunting. This dual mechanism of downstream translation initiation ensures sufficient expression of the σC cell attachment protein that is essential for infectious progeny virus production.
INTRODUCTION
Avian reovirus (ARV), a member of the fusogenic orthoreoviruses, is a nonenveloped virus that possesses a segmented, double-stranded RNA genome (1,2). The S1 genome segment encodes a functionally tricistronic mRNA that contains a 5′-terminal m7GpppG cap, a short (24 nt) 5′-untranslated region (UTR) followed by three sequential, partially overlapping open reading frames (ORFs), and no poly(A) tail (Figure 1A) (3). The 5′-proximal ORF encodes p10, the smallest member of the fusion-associated small transmembrane (FAST) protein family, that is responsible for cell–cell fusion and syncytium formation induced by the fusogenic orthoreoviruses (4). The second ORF encodes p17, a nonstructural nucleocytoplasmic shuttling protein of undetermined function (5). The 3′-proximal ORF resides 630 nt downstream from the 5′-cap and encodes the viral cell attachment protein σC, an essential structural protein present as trimers at the 12 vertices of the icosahedral virus particle (6). The mechanism of translation initiation at the σC start site on this unusual tricistronic eukaryotic messenger RNA (mRNA) has yet to be elucidated.
Figure 1.
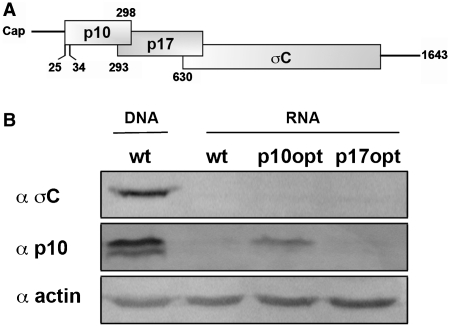
S1 RNA transfection results in inefficient protein expression for detection by western blot. (A) Diagrammatic representation of the tricistronic ARV S1 mRNA gene arrangement. Shaded rectangles indicate the locations of the p10, p17 and σC ORFs. Numbers refer to nucleotide positions of the genome segment, and indicate the first nucleotide position of the 5′-proximal AUG codons. (B) Western blot analysis of QM5 cells transfected with either plasmid pARV-S1 (wt DNA) or in vitro transcribed RNA from the same plasmid (wt) or a plasmid containing optimized p10 (p10opt) or p17 (p17opt) start codons. Blots were probed with the antisera indicated on the left.
The majority of eukaryotic translation initiation occurs in a scanning- and cap-dependent manner at the 5′-proximal AUG on a monocistronic mRNA (7,8). A pre-initiation complex, composed of the 40S ribosomal subunit and various canonical eukaryotic initiation factors (eIFs), binds to the 5′-cap and scans the 5′-UTR, initiating translation at the first AUG codon encountered (9). Context-dependent leaky scanning occurs when upstream AUGs (uAUGs) occur in a sub-optimal context, allowing scanning subunits to bypass these potential start codons and initiate translation at a downstream initiator methionine codon. The preferred context for an initiator methionine codon in vertebrates is ccRccAUGG (10), although more recent studies suggest considerable diversity in this ‘consensus’ sequence among different species (11). The purine in the −3 position (preferably an A) is functionally the most important nucleotide, augmented by a G in the +4 position which can exert a strong effect in the absence of A in the −3 position (7,12). Studies in diverse systems largely confirm the tenets of the leaky scanning hypothesis, with an uAUG in an optimal context serving to dramatically reduce or eliminate translation initiation from a downstream start site (13–16). An optimal context surrounding the AUG start codon is insufficient, however, to eliminate leaky scanning on some mRNAs (17–19), and evidence suggests additional cis and trans factors can influence leaky scanning past upstream ORFs (uORFs) (14,15).
Three additional mechanisms to explain translation initiation at downstream start sites have been described (20). Reinitiation occurs when the 40S subunit resumes scanning the mRNA upon termination of a short uORF (21). This relatively inefficient process operates under a number of constraints, most notably, the translated uORF must be short (<30 codons) to avoid loss of essential eIFs and the intergenic distance must be a minimum length (>70 nt) to allow the scanning 40S subunit time to reacquire the eIF2-GTP-Met-tRNAi cofactor necessary for translation initiation (22). Internal ribosome entry sites (IRESs) are usually found within the 5′-UTR and allow the pre-initiation complex to bind the mRNA at an internal position in a cap-independent manner (23). While viral IRESs are highly structured elements of several hundred nucleotides, some cellular IRESs may be multi-partite, comprised of short nonoverlapping nucleotide fragments (24). Ribosome shunting, best described in cauliflower mosaic virus, is an adapted form of reinitiation (25). Following translation termination of a short uORF, the 40S ribosomal subunit encounters a stable secondary structure and is shunted to an internal position normally located at, or slightly upstream of, an internal AUG codon (26). There are also examples of ribosome shunting that do not require translation of an uORF, but may involve a complementary interaction between the mRNA and 18S ribosomal RNA (rRNA) (27). In view of the large numbers of eukaryotic mRNAs predicted to contain uAUGs or uORFs (28), it is unclear whether the above mechanisms fully define all of the ways that ribosomes can access downstream translation start sites.
We previously demonstrated that expression of the first two ORFs of the ARV S1 mRNA (i.e. p10 and p17) is coordinately regulated via context-dependent leaky scanning (29). Results further indicated that σC expression is only modestly affected by translation initiation events occurring at the upstream p10 and p17 ORFs, leading to the proposal that translation of the σC ORF occurs via a novel scanning-independent mechanism (29). To further define the nature of this scanning-independent mechanism of translation initiation, we set out to identify features within the S1 mRNA that influence translation of the downstream σC ORF using a quantitative Renilla luciferase (RL) reporter system developed for use with RNA-transfected cells. Results indicate that ribosomes can access the σC start site using two alternate cap-dependent mechanisms: (i) facilitated leaky scanning that is capable of efficiently leaking past multiple uAUGs in a preferred context for a start codon; and (ii) a modified version of ribosomal shunting that bypasses uAUGs/uORFs to directly transfer the 40S subunit from the 5′-cap to an internal region downstream of the p17 start codon. This dual mechanism of downstream translation initiation ensures the tricistronic S1 mRNA directs sufficient expression of the σC protein, which is essential for infectious virus production.
MATERIALS AND METHODS
Cells and antibodies
The quail fibroblast cell line, QM5, was maintained as previously described (30), and used for all transfection experiments. Rabbit antisera that recognize σC was previously described (3). Antisera specific for p10 was raised in rabbits against the C-terminal p10 domain using a purified maltose-binding protein-p10 chimeric protein approach, as reported previously (4). Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (KPL, Gaithersburyg, MD, USA) was used as a secondary antibody for western blot analysis that was performed as described previously (29).
Plasmids
Plasmid pARV-S1, containing the full-length complementary DNA (cDNA) of the ARV S1 genome segment cloned into the plasmid pcDNA3 using restriction sites HindIII and NotI, was described previously (4). The AUG start codon of the RL ORF from plasmid phpRLFL (kindly provided by Dr. Richard Lloyd, Baylor College of Medicine) was point substituted to AGG and the RL ORF was inserted into pARV-S1 in-frame with the σC ORF between S1 nucleotides 751 and 752 using the XbaI restriction site present within the S1 sequence at this location, resulting in the production of pσC-RL. A stable hairpin structure (ΔG = –60 kcal/mol) was amplified from plasmid phpRLFL and was inserted into the 5′-end of pARV-S1 between S1 nucleotides 1 and 2 using an NheI restriction site. Plasmid pβ-RL was created by subcloning into the pcDNA3 vector the RL ORF from pσC-RL, with its authentic AUG start site, and the S1 sequence 3′ of the RL ORF using primers that created BamHI and XbaI restriction sites at the 5′- and 3′-ends, respectively. The β-globin 5′-UTR was then inserted upstream of the RL ORF using restriction sites HindIII and BamHI. Site-directed mutagenesis was used to make a single-point substitution within the β-globin UTR to create a start codon with an optimal context (plasmid β-ORF-RL). The same β-globin UTR was also inserted upstream of the S1 segment within plasmid pσC-RL creating pβ-σCRL. The plasmid pσC-RL was used as template for polymerase chain reaction (PCR) to create 5′-terminal truncations using forward primers that deleted 19, 27, 79, 147 or 303 nt from the S1 sequence. Plasmids with internal deletions in the S1 sequence were created by PCR amplification of the S1 5′-terminal region of interest from pARV-S1 using a forward primer with a HindIII restriction site and a reverse primer with a BamHI restriction site. These PCR amplicons and plasmid pRL were digested and ligated together producing plasmids pΔ628–751RL, pΔ593–751RL, pΔ483–751RL, pΔ423–751RL, pΔ393–751RL, pΔ366–751RL and pΔ312–751RL. Plasmid pσC-RL was also modified through the insertion of either 24 (mini-ORF1) or 78 (mini-ORF2) nt between S1 nucleotides 304 and 305 using an ApaI restriction site on both termini of the insert. Plasmid pΔ366–751intRL was created through the insertion of 114 nt of heterologous sequence between S1 nucleotide 365 and the A of the RL start codon. Inserts in all plasmid constructs were confirmed by sequencing.
Where specified, parental plasmids were subjected to site-directed mutagenesis using the QuikChange kit (Stratagene) to optimize the context (ccAccAUGgG) of the p10 or p17 start codons (p10opt and p17opt), to convert the four AUG codons that occur upstream of the σC start site to AUC codons (M1-M4), to convert the two AUG codons present within the insert of miniORF1 to AUC codons, to create additional AUG start sites in a strong context within the p17 region (M5, gcAaaAtGg; M6, ttaacatGg; M7, caacaAtgG; capitalized letters indicate the position of nucleotide point substitutions and the created AUG start codon is underlined), and to remove the three AUG codons that occur in the σC region upstream of the RL ORF in pσC-RL and to introduce a stop codon in the p17 ORF (stp 517) to create plasmid pS1-RL. Point substitutions were also created in plasmids pΔ366–751RL and pΔ423–751RL within regions RCR1 and RCR2 to disrupt 18S complementarity. All plasmids were used as templates for PCR reactions utilizing a forward primer containing the T7 polymerase promoter and an S1-specific reverse primer. These amplicons were used as templates for in vitro transcription. The term construct refers to either the plasmid that was manipulated (e.g. pS1-RL) or the mRNA derived from this plasmid (e.g. S1-RL).
In vitro transcription and RNA purification
PCR amplicons possessing the T7 polymerase promoter at their 5′-ends were used for in vitro transcription utilizing the mMessage mMachine T7 kit (Applied Biosystems) according to the manufacturer’s instructions. Except where specified, these mRNAs contained the authentic 5′-termini of the S1 mRNA with a functional 5′-terminal m7Gppp cap and authentic, non-polyadenylated 3′-termini. The MEGAscript T7 kit (Applied Biosystems) was utilized to in vitro transcribe RNA with the non-functional (ApppA) cap analog (Sigma). The mRNA was purified utilizing the RNeasy Mini kit (Qiagen) on-column purification protocol. The quality and quantity of the mRNA were assessed using denaturing agarose gels and ethidium bromide staining and UV spectrophotometry at 260 nm.
RNA transfection and RL assay
QM5 cells in a 48-well cluster plate were transfected in duplicate with 1 μg of in vitro transcribed RNA and 1.5 μg polyethylenimine (PEI) (Polysciences Inc). Four hours post-transfection, cell lysates were prepared according to the Renilla Luciferase Assay System (Promega). A 20-μl aliquot of the lysate was mixed with 50 μl of RL substrate in a GloMax 20/20 luminometer. Relative light units (RLUs) were captured over a period of 10 s, after a 2-s delay. All constructs tested were in vitro transcribed a minimum of three times and were only compared to their respective parental clone in vitro transcribed on the same day to minimize potential effects of variations in capping efficiency. The RLUs obtained from parental constructs averaged 4 × 106 RLUs for pβ-RL that expressed authentic RL from a moncistronic mRNA, 9 × 105 RLUs for pS1-RL that expressed authentic RL from the downstream ORF in a tricistronic mRNA, and 3 × 105 RLUs for pσC-RL that expressed a less enzymatically active σC-RL chimeric protein from the downstream ORF in a tricistronic mRNA. The mean ± SE from a minimum of three independent experiments were used to determine the fold-change in expression levels and assessed for statistical significance using an unpaired, two-tailed Student’s t-test.
RNA extraction and qRT–PCR
At 4 h post-transfection, the RNA remaining on the surface of QM5 cells in six-well cluster plates was digested with RNase A at a final concentration of 1 μg/μl for 4 min at room temperature with intermittent rocking. Control experiments determined that these treatment conditions were sufficient to digest all of the input mRNA. The QM5 cells were washed extensively with RNase free phosphate-buffered saline (PBS) and total RNA was extracted from cells using Trizol (Invitrogen). The aqueous phase of the phenol–chloroform extraction was further purified using the RNeasy Mini kit (Qiagen) and on-column DNase digestion using the RNase free DNase kit (Qiagen). The RNA concentration was measured with a UV spectrophotometer (Eppendorf Biophotometer) at 260 nm and immediately used for cDNA production using Moloney murine leukemia virus reverse transcriptase and random hexamers. Reverse transcription reactions used 0.25 or 0.5 μg of purified total RNA in a final volume of 10 μl. The cDNA was subjected to real-time PCR using primers specific for the RL ORF (Fwd: 5′-ATCGGACCCAGGATTCTTTT-3′; Rev: 5′-ACTCGCTCAACGAACGATTT-3′), which amplified nucleotides 768–917 (amplicon length 150 nt), and primers specific for Gallus gallus β-actin (Fwd: 5′-AGCAACATGTGTGACGAGGA-3′; Rev: 5′-ACCCCACCCTGCTCACTGA-3′), which amplified nucleotides 7–339 (amplicon length 333 nt). The QuantiFast SYBR Green PCR kit (Qiagen) was used according to manufacturer’s specifications in a final reaction volume of 20 µl. The reaction, with an amplification protocol of a 5-min hot start at 95°C followed by 40 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 30 s, was performed using the MX3000p thermocycler (Stratagene) and analyzed using MxPro software (Stratagene). Melting curves, as well as separation of PCR products on a 1% Tris acetate-EDTA agarose gel, were used to ensure the formation of a single product of the appropriate size. Relative gene expression normalized to β-Actin expression was calculated using the ΔΔCT method (31). In no instance did the RNA concentration vary outside the range of statistical significance.
RESULTS
A cap-dependent, IRES-independent mechanism is responsible for translation initiation of the 3′-proximal ORF from a full-length S1 mRNA
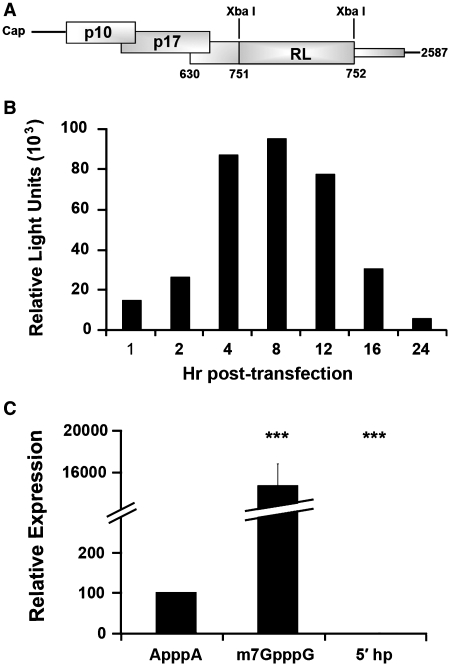
Previous analysis of translation initiation on the ARV tricistronic S1 mRNA utilized western blotting of cell lysates from cells transfected with plasmid DNA containing the full-length S1 cDNA (29). These plasmid-derived S1 transcripts contained vector sequences at the 5′- and 3′-termini and a 3′-poly(A) tail not found on reovirus mRNAs. An RNA transfection approach was therefore developed to generate S1 mRNA with authentic termini. In vitro transcription of the S1 cDNA generated full-length, capped, non-polyadenylated S1 mRNA with authentic 5′- and 3′-termini. This mRNA was transfected into QM5 cells and cell lysates were analyzed by western blotting using polyclonal antisera specific for the p10 or σC proteins. As previously reported (29), the p10 and σC proteins were easily detected in cells transfected with plasmid DNA encoding the S1 mRNA (Figure 1B). This was not the case in cells transfected with the S1 mRNA, although an S1 mRNA containing an optimized p10 translation start site did generate sufficient p10 protein to be barely detected by western blotting (Figure 1B). To address sensitivity issues with the RNA transfection approach, the S1 mRNA was modified by insertion of sequences containing the ORF for RL. This insertion created an ORF encoding a chimeric σC-RL protein containing the N-terminal 41 residues of σC fused to RL (Figure 2A). A time-course analysis of translation of the σC-RL ORF in RNA-transfected cells revealed significant levels of luciferase activity in cell lysates transfected with the σC-RL mRNA, which peaked 4–8 h post-transfection at ∼1–3 × 105 RLUs (Figure 2B). The σC-RL reporter ORF therefore provided a suitable system for analysis of S1 mRNA translation in RNA transfected cells, and all subsequent studies were performed at 4 h post-transfection.
Figure 2.
RNA transfection with a σC-RL chimera functions as an efficient reporter of σC expression. (A) Diagram depicting the mRNA generated by T7 polymerase in vitro transcription of pσC-RL, indicating the location of the modified RL ORF (lacking its own AUG start codon but containing its stop codon) inserted between S1 nucleotides 751 and 752, in-frame with the σC ORF. The thin shaded rectangle depicts the presence of sequences corresponding to the 3′-end of the σC ORF that are not translated due to the presence of the upstream RL stop codon. (B) QM5 cells were transfected with in vitro transcribed capped σC-RL RNA and cell lysates were harvested at the indicated times and analyzed for RL activity. Results are from a single representative experiment conducted in duplicate. (C) QM5 cells were transfected with σC-RL RNA in vitro transcribed either with a functional cap analog (m7GpppG), a nonfunctional cap analog (ApppA), or with a stable hairpin structure inserted between S1 nucleotides 1 and 2 downstream of a functional cap analog (5′ hp). Cell lysates were harvested 4 h post-transfection and analyzed for luciferase activity. Results are reported as mean ± SE (n = 3–6) of the relative level of σC-RL translation, normalized to the parental σC-RL construct with a functional cap analog or a non-functional cap analog. Expression levels significantly different from the parental construct (***P < 0.001) are indicated.
Previous results indicated that σC translation was eIF4G-dependent, suggesting that a cap-dependent mechanism may be responsible for σC translation (29). To directly test this prediction and to validate the σC-RL reporter system, S1 mRNA was in vitro transcribed either with a functional (m7GpppG) or with non-functional (ApppA) cap-analog. The presence of a functional cap-analog at the 5′-terminus of the σC-RL mRNA resulted in a ∼140-fold increase in RL expression compared to the presence of the non-functional cap analog (Figure 2C), indicating that ribosomes access the σC AUG codon in a cap-dependent manner. This result was further confirmed through the addition of a stable hairpin structure (ΔG = –60 kcal/mol) in the 24-nt 5′-UTR of the S1 mRNA. Stable hairpin structures present near the 5′-cap are known to dramatically reduce translation efficiency by interfering with binding of the pre-initiation complex (18,32). As indicated (Figure 2C), insertion of this hairpin eliminated translation of the σC-RL ORF, further confirming that a cap-dependent, IRES-independent mechanism is responsible for translation initiation at the internal σC start codon on the tricistronic S1 mRNA. The cap analysis and hairpin data also confirmed our previous northern blotting results (29), indicating that σC expression occurs from a full-length S1 mRNA.
Optimized upstream start codons have differential effects on translation of downstream ORFs
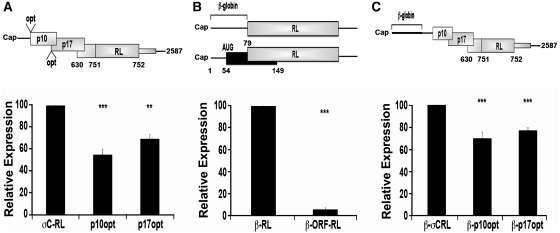
The influence of the upstream p10 and p17 ORFs on translation initiation at the internal σC-RL start site was determined using luciferase assays of cells transfected with the S1, p10opt or p17opt σC-RL mRNAs. As previously reported using plasmid DNA transfections (29), optimization of the upstream start codons adversely affected translation from the σC start codon (Figure 3A). However, there was a substantial retention of internal translation initiation, with optimized p10 or p17 start sites only reducing translation of the σC-RL ORF by ∼46 ± 5% or ∼30 ± 4%, respectively (Figure 3A). The relative mRNA levels, as determined by qRT–PCR, indicated that the variation in luciferase levels were not due to differences in intracellular mRNA concentrations (data not shown). To determine whether the inability of optimized p10 or p17 start codons to exert a pronounced inhibitory effect on downstream translation initiation was a specific reflection of the S1 mRNA, a similar start codon was examined in a heterologous mRNA. A monocistronic mRNA was designed that contained the RL ORF preceded by a 77-nt 5′-UTR from the rabbit β-globin mRNA (Figure3B). The β-globin 5′-UTR is known to efficiently mediate cap-dependent translation and to lack any sequence elements or secondary structure that may significantly affect translation initiation (33). This parental β-globin-RL (β-RL) construct was also modified to introduce an optimized start codon (ccAccAUGG) 54 nt downstream from the 5′-cap (β-ORF-RL construct), creating a 48-codon ORF that overlapped, but was out of frame, with the RL ORF. As shown (Figure 3B), the optimized uORF reduced translation of the downstream RL ORF in the β-globin transcript by >95%. Since the 40S ribosomal subunit occupies approximately the first 30–40 nt upon cap binding (34), the relatively short, 24-nt 5′-UTR present in the S1 mRNA might interfere with ribosome recognition of an optimized p10 start site. To test this possibility, the rabbit β-globin 5′-UTR was inserted at the 5′-termini of the full-length σC-RL, p10opt and p17opt mRNAs (Figure 3C). Extension of the S1 5′-UTR did not affect σC-RL translation from the p17opt construct, and slightly increased, rather than decreased, σC-RL translation from the p10opt construct (67 ± 9% residual translation from the σC start codon). Optimized uAUGs therefore present an effective barrier to leaky scanning, but not in the context of the ARV S1 mRNA.
Figure 3.
Differential effects of optimized upstream start codons on translation of downstream ORFs. (A) Diagram indicating the location of the optimized (opt) (ccAccAUGG) p10 or p17 start codons (top panel). RNA-transfected QM5 cells and cell lysates were harvested and assayed for luciferase activity (bottom panel) as described in Figure 2C. (B) Diagram depicting a capped RL gene (shaded rectangle) with a β-globin 5′-UTR, either with (β-ORF-RL) or without (β-RL) an upstream ORF (black rectangle) (top panel). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity (bottom panel) as described in Figure 2C. (C) The σC-RL constructs represented in (A) were 5′-terminally extended through the addition of the β-globin 5′-UTR, and RNA-transfected cells were harvested and analyzed for luciferase activity (bottom panel) as described in Figure 2C. All results are reported as mean ± SE (n = 4–5) of the relative level of RL (B) or σC-RL (A and C) activity, normalized to the parental σC-RL (A), β-RL (B) or β-σCRL (C) constructs. Expression levels significantly different from the parental constructs (***P < 0.001 and **P < 0.01) are indicated.
The p17 start codon exerts a negative effect on downstream translation initiation
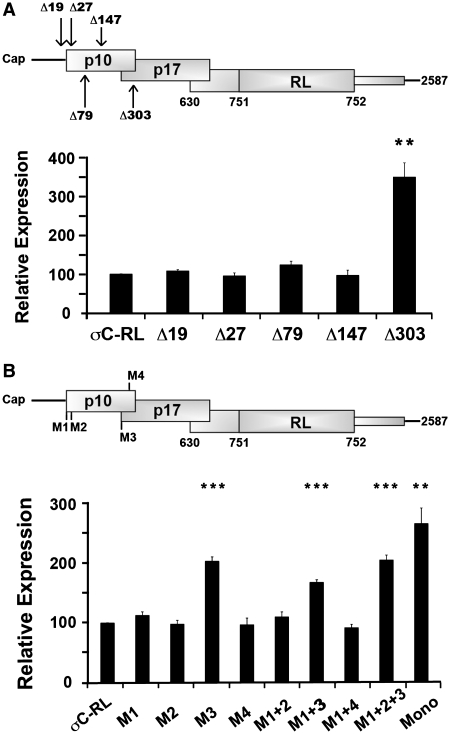
To examine features of the S1 mRNA that contribute to downstream translation initiation, the σC-RL mRNA was progressively truncated from the 5′-terminus. Deletion of the 5′-terminal 147 nt of the σC-RL mRNA, which removes the 5′-UTR and half of the p10 ORF, including the two in-phase p10 start codons at nucleotides 25 and 34, had no significant effect on σC-RL translation (Figure 4A). The two p10 initiator AUG codons are therefore not recognized efficiently as start codons, presumably due to their suboptimal context with pyrimidines in both the −3 and +4 positions. However, plasmid DNA transfections indicated the same 5′-terminal deletion eliminated σC expression (29). While the reason for this discrepancy is unknown, different transcription methods (i.e. transcription in the nucleus versus in vitro transcription) can result in different mRNP complexes and differential folding, ultimately producing contradictory results (35,36). The RNA transfection results are likely a more accurate reflection of the situation in virus-infected cells since the S1 mRNA contains authentic termini and no poly(A) tail. Deletion of the 5′-proximal 303 nt of the S1 mRNA resulted in an approximate 3.5-fold increase in σC expression (Figure 4A), implying that a negative regulatory element resides between nucleotides 148 and 303.
Figure 4.
The p17 translation start site obstructs downstream translation initiation. (A) The parental σC-RL transcript was modified by progressive 5′-truncations (Δ19, Δ27, Δ79, Δ147, Δ303; numbers refer to the number of 5′-terminal nucleotides deleted from the σC-RL mRNA sequence) (top panel). Luciferase activity (bottom panel) of the parental construct (σC-RL) and the 5′-truncations were determined as described in Figure 2C. (B) The σC-RL construct was modified to eliminate the four AUG start codons (M1–M4; AUG to AUC) that reside upstream of the σC start codon, either individually or in various combinations (top panel). Luciferase activity (bottom panel) of the parental (σC-RL) and modified constructs were determined as described in Figure 2C. All results are reported as mean ± SE (n = 3) of the relative level of σC-RL translation, normalized to the parental σC-RL construct. Expression levels significantly different from the parental construct (***P < 0.001 and **P < 0.01) are indicated.
There are only four methionine codons (designated M1–M4) in any of the three reading frames upstream of the σC start site. M3, which initiates the p17 ORF, is in a strong context for translation initiation with an A residue in the −3 position, as is M4 near the end of the p10 ORF with a purine in the −3 position and a G residue at +4. Both of these AUG codons were removed by the Δ303 truncation suggesting either or both might serve as barriers to scanning ribosomes. To examine the influence of these codons on downstream translation initiation at the σC start site, single-nucleotide substitutions (AUG to AUC) were introduced into these methionine codons, individually and in various combinations. Consistent with the 5′-truncation results, removal of the two AUG codons that initiate the p10 ORF (M1 and M2), either singly or in combination, had no effect on σC-RL translation (Figure 4B). The same situation applied to M4, even though it is in a preferred context to serve as a start codon. However, removal of the p17 start codon (M3), either alone or in combination with the other AUG codons, resulted in a 2–3-fold increase in translation of the σC-RL ORF (Figure 4B). The variation in translation efficiency was not attributed to differences in mRNA levels according to qRT–PCR results (data not shown). Together, the 5′-truncation and AUG point substitution results indicated the p17 start codon can serve as a barrier to ribosome access to the σC ORF. Although consistent with a context-dependent leaky scanning model of σC translation initiation, this conclusion is counterbalanced by the 70% residual level of σC-RL translation in the presence of a fully optimized p17 start codon, which effectively inhibits leaky scanning in mRNAs other than the S1 mRNA (Figure 3).
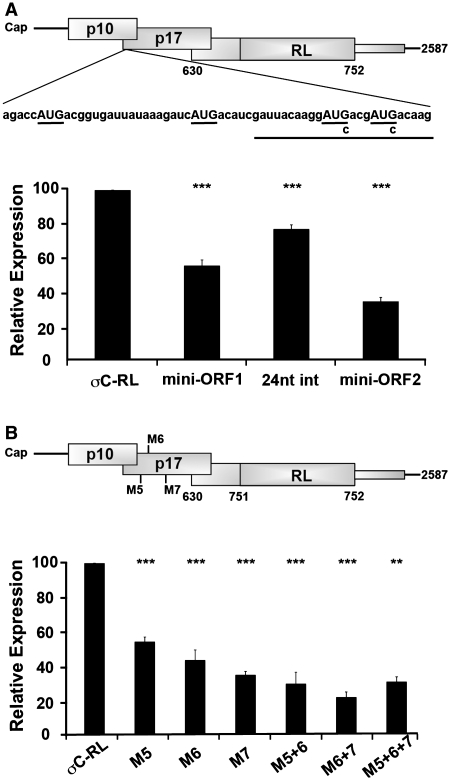
Multiple uAUG codons in a preferred start codon sequence context fail to abolish downstream translation initiation on the S1 mRNA
To examine whether other features of the p17 ORF, beyond its start codon, influence ribosome access to the σC start codon, 24 heterologous nucleotides were inserted between S1 nucleotides 298 and 299, immediately downstream of the p17 start codon at nucleotide 293. This inserted sequence contained two additional AUG codons out of frame with the p17 ORF and in a strong context with A in the −3 position, resulting in the creation of a mini-ORF 31 codons long that terminated 240 nt upstream of the σC start site (mini-ORF1; Figure 5A). The creation of this mini-ORF with two strong start sites reduced the ability of ribosomes to access the σC start site by 45 ± 3% (Figure 5A). To determine whether the inserted sequence alone affected downstream translation initiation, two separate point substitutions within the 24-nt insert (AUG to AUC) were created to eliminate the potential start sites (24 nt int). Analysis of luciferase activity for this insertion construct demonstrated a slight decrease in σC production (23 ± 3%), a possible effect of altering the sequence context of the p17 start codon past the +5 position. If the level of luciferase activity obtained from the insertion alone is considered as the parental level of σC-RL translation (100%), the inhibitory effect of the two additional strong AUG start codons was relatively minor, reducing σC production by only 28 ± 2%. An additional 34 nt were inserted immediately upstream of the previous insertion. This second insertion contained two AUGs in a strong context and in-frame with mini-ORF1, creating mini-ORF2 that contained four potential start codons, all in a strong context to serve as start codons. These additional start codons had an additive effect on inhibiting reporter gene expression, although translation of the downstream ORF was still maintained at ∼35% of maximal levels (Figure 5A). The presence of eight uAUGs (four from the insertions plus the M1–M4 AUG codons naturally present in the S1 mRNA), six of which were in a preferred context to serve as translation initiation codons, therefore still did not abolish σC-RL expression.
Figure 5.
Insertion of additional AUG codons inhibits but does not abolish downstream translation initiation. (A) Diagram depicting the insertion of 24 (underlined) or 78 exogenous nucleotides between S1 nucleotides 298 and 299 containing zero (24 nt int), two (mini-ORF1) or four (mini-ORF2) additional AUG triplets (top panel). Luciferase activity (bottom panel) of the parental (σC-RL) and modified capped mRNAs were determined as described in Figure 2C. (B) Diagram depicting the location of the AUG codons (M5–M7) created within the p17 reading frame (top panel). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity (bottom panel) as described in Figure 2C. Results are reported as the mean ± SE (n = 3) of the relative level of σC-RL translation, normalized to the parental σC-RL construct. Expression levels significantly different from the parental construct (**P < 0.01 and ***P < 0.001) are indicated.
The mini-ORFs in the above constructs terminated 240 nt upstream of the σC start codon, raising the possibility that σC-RL translation might arise via scanning and reinitiation upon termination of these small ORFs. To eliminate this possibility, and to avoid potential complications due to inserted sequences, point substitutions were introduced into the p17 ORF to create additional in-frame AUG codons. Ribosomes initiating translation at these start codons would terminate at the p17 stop codon, downstream of the σC start codon. Point substitutions were used to create three in-frame AUG codons in a preferred context (AxxAUGG) in the p17 ORF, at nucleotides 362 (M5), 419 (M6) or 527 (M7), either singly or in combination. The M5–M7 designations were used to reflect the presence of the four upstream methionine codons naturally present in the S1 mRNA. Each of these additional strong start sites had some inhibitory effect on ribosome access to the σC start site (Figure 5B). However, even the presence of all three additional strong start codons in the p17 ORF resulted in substantial retention of σC-RL translation (31 ± 5%). The S1 mRNA therefore facilitates leaky scanning past six uAUGs in a preferred start codon sequence context, while only one such uAUG can function as an effective barrier to scanning 40S subunits in the context of other mRNAs (Figure 3B).
An internal translation enhancer in the S1 mRNA promotes downstream translation initiation
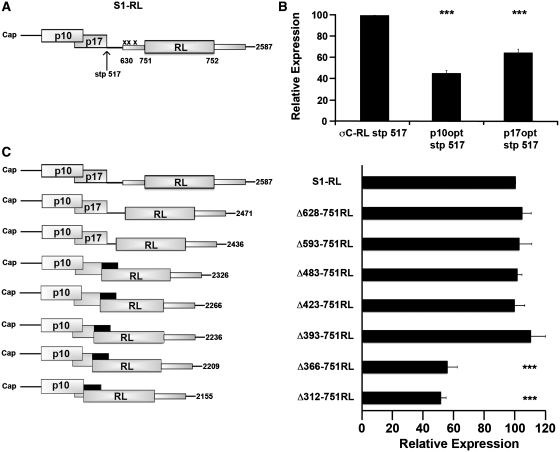
To further define features of the S1 mRNA that promote downstream initiation, progressively larger internal deletions were created within the S1 mRNA, downstream of the p17 start codon. To address whether sequences flanking the σC start codon exert an effect on translation initiation, the internal deletions encompassed the region flanking the σC start codon. Since these constructs would generate an authentic, non-chimeric RL with different enzymatic activity than the chimeric σC-RL form of the protein, a new parental version of the σC-RL reporter construct first had to be created. To this end, the three AUG codons within the 121 nt of the σC region present upstream of the RL ORF were substituted to AUC, thereby largely maintaining the S1 nucleotide sequences flanking the σC start codon while making the RL start codon the first AUG codon downstream of the p17 AUG triplet (Figure 6A). Point substitution of the third σC AUG codon unfortunately also eliminated the p17 stop codon, creating a chimeric p17-RL ORF. To prevent the formation of this chimeric protein, site-directed mutagenesis at nucleotide 517 was used to create a stop codon within the p17 ORF. To ensure that creation of the stop codon at nucleotide 517 would not alter ribosome access to the internal σC start site, the same stop codon was first tested in the existing σC-RL construct, in the absence and presence of optimized p10 or p17 start codons. Transfection of the in vitro transcribed RNA and subsequent luciferase assays demonstrated the same trend in translation from the σC start codon as previously noted, with the optimized p10 and p17 start codons reducing translation of the σC-RL ORF by ∼55% and 30%, respectively (Figure 6B).
Figure 6.
An internal translation enhancer in the S1 mRNA is required for maximal downstream initiation. (A) Diagrammatic representation of a new parental mRNA (S1-RL), where the three AUG codons in the σC ORF between S1 nucleotides 630 and 732 (represented by x) were point substituted to AUC, and a point substitution was made at S1 nucleotide 517, creating a stop codon in the p17 ORF. (B) Constructs σC-RL lacking or containing optimized p10 or p17 start codons were modified at S1 nucleotide 517 to create a stop codon within the p17 ORF. RNA-transfected QM5 cells were harvested and analyzed for luciferase activity as described in Figure 2C. (C) Diagrammatic depiction of the S1-RL cDNA modified by internal deletion. The Δ312–751RL, Δ366–751RL, Δ393–751RL, Δ423–751RL and Δ483–751RL constructs removed the stop insertion at nucleotide 517 such that the p17 ORF (black rectangles) now terminated 71 nt downstream of the RL AUG codon (left panel). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity (right panel) as described in Figure 2C. Results are reported as the mean ± SE (n = 3–5) of the relative level of RL translation, normalized to the parental S1-RL stp 517 (B) or S1-RL construct (C). Expression levels significantly different from the parental construct (***P < 0.001) are indicated.
Using the new S1-RL reporter construct, the region from 628 to 751 was deleted from the S1 mRNA. The 124-nt Δ628–751 internal deletion removed the 3′-end of the p17 ORF, starting immediately upstream and extending 121 nt downstream of the σC start codon at S1 nucleotide 630. This construct displayed the same level of luciferase activity as the full-length parental S1-RL construct (Figure 6C), as did a 159-nt deletion that extended further toward the 5′-terminus (Δ593–751RL construct). The region flanking the σC start site therefore does not contain any cis elements required for ribosome access to a downstream start codon. The internal deletion was further extended toward the 5′-end of the S1 mRNA, deleting a total of 269 (Δ483–751RL), 330 (Δ423–751RL), 359 (Δ393–751RL), 386 (Δ366–751RL) or 440 (Δ312–751RL) nt. These constructs removed the stop insertion site introduced at nucleotide 517 in the S1-RL parental construct, generating a chimeric p17 ORF that terminates 71 nt downstream of the RL AUG codon, a sufficient distance downstream to prevent the terminating ribosomes from interfering with initiation at the RL start codon. While internal deletions up to 359 nt had no effect on translation of the downstream RL ORF (constructs Δ483–751RL, Δ423–751RL and Δ393–751RL), removal of an additional 27 nt (Δ366–751RL construct) decreased expression of the downstream ORF by 45 ± 6% (Figure 6C). Removal of a further 54 nt (Δ312–751RL construct) had no additional inhibitory effect on translation of the downstream RL ORF. As before, the variations in translation of the major downstream ORF were not due to changes in mRNA concentration according to qRT–PCR results (data not shown). Sequences immediately downstream of the p17 start codon, from nucleotides 366 to 392, therefore enhance the ability of ribosomes to bypass the barrier presented by the strong p17 start codon. These sequences, however, are not essential for ribosome access to the downstream start codon since ∼50% of RL translation was retained in the absence of this region.
The S1 translation enhancer functions in a sequence-dependent manner to promote scanning-independent downstream translation initiation
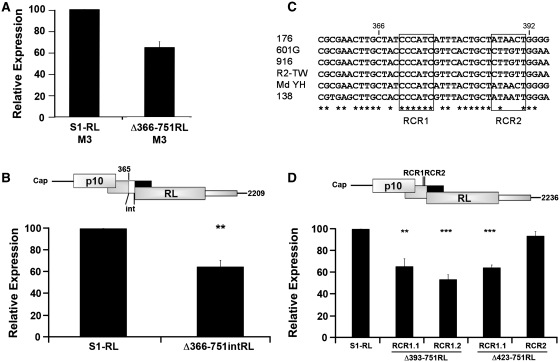
The ability of S1 nucleotides 366–392 to enhance downstream translation initiation was further investigated in different S1 mRNA backgrounds and following point substitutions within this sequence. If the S1 translation enhancer functions to facilitate leaky scanning, the luciferase activity of S1 mRNAs containing or lacking the translational enhancer element should be identical in the absence of the p17 start codon that serves as a barrier to context-dependent leaky scanning. The p17 start codon was therefore point substituted to AUC, both in the S1-RL parental construct and the Δ366–751RL construct. As shown (Figure 7A), translation of the downstream RL ORF in the absence of the translational enhancer region was not restored to maximal levels by removal of the p17 start codon. The Δ366–751RL transcripts containing (Figure 6C) or lacking (Figure 7A) the p17 start codon both showed approximately the same level of luciferase activity relative to their respective parental constructs. To exclude the possibility that the Δ366–751 deletion affected σC expression by merely altering the distance of the downstream start codon from the 5′-terminus, the Δ366–751RL construct was re-extended to the length of the Δ483–751RL construct (that had maximal luciferase activity) by insertion of 114 heterologous nucleotides after nucleotide 365 in the S1 sequence. This Δ366–751intRL construct displayed the same 40% reduction in luciferase activity (Figure 7B) as the Δ366–751RL construct (Figure 6C), indicating that S1 nucleotides 366–392 harbor an element that is required for maximum ribosome access to the 3′-proximal ORF. Taken together, the above results suggest that nucleotides 366–392 in the S1 mRNA function as a translation enhancer by promoting something other than facilitated leaky scanning.
Figure 7.
The S1 translation enhancer functions in a sequence-dependent manner to promote scanning-independent downstream translation initiation. (A) The S1-RL and Δ366–751 cDNAs were modified by site-directed mutagenesis of the p17 start site (AUG to AUC). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity as described in Figure 2C. (B) Diagram indicating the location of the 114 heterologous nucleotides (denoted by white box and labeled int) inserted into the Δ366–751RL construct between S1 nucleotide 365 and the first nucleotide of the RL ORF (top panel). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity (bottom panel) as described in Figure 2C. (C) Alignment of S1 nucleotides 356–395 from six ARV isolates. Consensus nucleotides are indicated (asterisk) and RCR1 and RCR2 regions are boxed. (D) Diagram of the Δ393–751RL mRNA indicating the location of the two 18S rRNA complementarity sites (RCR1 and RCR2) (top panel). The indicated construct was modified by point substitution of S1 nucleotides 370/1/5 (RCR1.1) or nucleotides 372/3 (RCR1.2). Construct Δ423–751RL was also modified by point substitution at S1 nucleotides 370/1/5 (RCR1.1) and 388/9 (RCR2). RNA-transfected QM5 cells were harvested and analyzed for luciferase activity (bottom panel) as described in Figure 2C. All results are reported as the mean ± SE (n = 3–4) of the relative level of RL translation, normalized to the parental S1-RL construct. Expression levels significantly different from the parental construct (**P < 0.01 and ***P < 0.001) are indicated.
Closer examination of the 27-nt S1 translation enhancer region revealed the presence of two 6-nt sequences (CCCAUC and AUAACU) that are complementary to human 18S rRNA nucleotides 1600–1605 and 107–112. Alignment of the S1 cDNA sequences of 27 ARV isolates from chickens and ducks indicated complete conservation of rRNA complementary region 1 (RCR1) but not RCR2 (Figure 7C). Various reports have demonstrated that base-pairing between the mRNA and 18S rRNA can contribute to recruitment of 40S ribosomal subunits and translation efficiency (27,37,38), and that this complementary region can be as small as 7–9 nt (39). Point substitutions were therefore introduced into RCR1 and RCR2 in the S1 translation enhancer region to disrupt potential base-pairing with 18S ribosomal RNA. Replacement of either 2 or 3 nt in RCR1, in either the Δ393–751RL or Δ423–751RL transcripts, decreased luciferase activity to approximately the same level as deletion of the entire 27-nt translation enhancer (Figure 7D). This was not the case for RCR2, where a 2-nt replacement had no adverse effect on translation of the downstream RL ORF (Figure 7D). Specific regions within the S1 mRNA translational enhancer therefore function in a sequence-specific manner to promote scanning-independent ribosome access to a downstream translation start site.
DISCUSSION
A previous study of the tricistronic S1 mRNA of ARV demonstrated that translation initiation at the 3′-proximal σC ORF occurs in an IRES- and reinitiation-independent, eIF4G-dependent manner (29). Based on the inability of optimized p10 or p17 start codons to abrogate translation of the σC ORF, we proposed that a scanning-independent mechanism was responsible for translation initiation at the internal σC start site. The present study necessitates a refinement of this previous model of translation control of the S1 mRNA. The ability of uAUGs to decrease σC translation clearly implicates leaky scanning in translation of the σC ORF. However, ribosomal subunits are capable of leaking past multiple uAUGs in the S1 mRNA, even though the optimal context of these uAUGs exerts a pronounced inhibitory effect on translation of a downstream ORF when present in mRNAs other than the S1 mRNA. The S1 mRNA therefore facilitates leaky scanning, which occurs relatively independent of the sequence context of uAUGs. At the same time, the inhibitory effect exerted by progressive addition of uAUGs in a strong context plateaued at ∼70% inhibition, suggesting effective elimination of facilitated leaky scanning (i.e. additional inhibitory elements have no additional inhibitory effects). The ∼30% residual level of σC translation under these conditions therefore implies a second, scanning-independent mechanism for translation of the downstream ORF. This second mechanism of σC translation initiation is cap-dependent, it is not influenced by the presence of uORFs, and it requires a sequence-dependent translation enhancer element that occurs downstream of the p17 start codon, but several hundred nucleotides upstream of the σC start site. We propose that translation initiation of the 3′-proximal σC ORF is therefore made possible by two alternate mechanisms, facilitated leaky scanning and an atypical form of ribosome shunting.
Several different approaches indicated that features inherent in the S1 mRNA facilitate the ability of scanning ribosomal subunits to access the σC start site. The inhibitory effects of optimized uAUGs and uORFs on σC expression suggests that at least some ribosomes access the downstream σC ORF via leaky scanning. However, a single optimized uAUG in the β-RL mRNA construct (Figure 3B) and in numerous other mRNAs (7,14,16,18) decreases downstream translation initiation by >90%, while significant levels of σC translation are retained even in the presence of multiple uAUGs in a preferred context (Figure 5). Factors other than the immediate sequence context of AUG codons must therefore influence the scanning capability of 40S subunits on the S1 mRNA. While the importance of the sequence context of translation start codons on leaky scanning is well established, there are examples where sequence-context alone cannot explain the efficiency of leaky scanning to a downstream translation start codon (17–19,40,41). The ribosome filter hypothesis of Mauro and Edelman (42,43) accommodates such discrepancies by suggesting that, beyond cap- and context-dependent leaky scanning, diverse mRNA–ribosome interactions may differentially regulate translation of particular mRNAs. The present results add additional weight to the concept that considerable heterogeneity in the interactions of different mRNAs with components of the cellular translation machinery may influence translation start site selection. How ribosomes are able to leak past traditional barriers to scanning on the S1 mRNA remains to be determined. Studies indicate that eIF1 is responsible for preventing premature engagement with a putative start codon, while eIF1A facilitates pausing at correct start codons long enough for initiation to proceed (44,45). It is therefore conceivable that recruitment of a cellular factor by the S1 mRNA or direct interaction of the S1 mRNA with the 40S subunit could alter the conformation of the 40S subunit or function of eIF1 or eIF1A, thus enabling enhanced leaky scanning. Continued examination of the mechanisms that regulate leaky scanning on the ARV S1 mRNA should provide additional insights into the cis and trans factors that influence ribosomal scanning, which may be of considerable interest in view of the growing appreciation that numerous eukaryotic mRNAs are likely to contain AUG codons upstream of the major ORF (19,28,46,47).
In addition to facilitated leaky scanning, the present results confirmed previous predictions that transit of the 40S subunit from the 5′-cap to the internal σC start codon also involves some form of nonlinear subunit migration, and provided additional insights into the mechanism of ribosome handoff. The insertion of just one (M7) or two (M5 + M6) uAUGs in the p17 ORF reduced σC expression up to 70% (Figure 5B), while the insertion of three uAUGs showed no greater reduction in expression (Figure 5B). This result suggests that facilitated leaky scanning from the 5′-cap was maximally inhibited, implying that the residual ∼30% translation of the downstream ORF occurs by a separate cap-dependent mechanism that is capable of bypassing uORFs. Further support for a scanning-independent pathway was provided by results indicating that extension of the 24-nt 5′-UTR on the S1 mRNA in the presence of an optimized p10 start site increased, rather than decreased, translation of the downstream reporter ORF (Figure 3C). 40S subunits bound to the cap are positioned immediately adjacent to the p10 AUG codon. Optimizing the context of the p10 start codon might therefore be expected to recruit ribosomes into the scanning pathway and away from the handoff pathway. This prediction is consistent with the more pronounced inhibitory effect of an optimized p10 versus p17 start codon (Figure 3A), and by the increase in translation of the downstream ORF when the S1 5′-UTR was extended. By lengthening the 5′-UTR, 40S subunits recruited to the 5′-cap would no longer be positioned immediately adjacent to the p10 start site, increasing the likelihood of ribosome handoff downstream of the p17 start codon that serves as a barrier to scanning 40S subunits.
The scanning-independent mechanism of translation initiation on the S1 mRNA requires the presence of a translation enhancer element located within S1 nucleotides 366–392. This 27-nt region located downstream of the p17 start codon promotes maximum ribosome access to a downstream ORF, independent of leaky scanning (Figure 7A). The translation enhancer element functions in a length-independent manner (Figure 7B), and a conserved 6-nt sequence with complementarity to 18S rRNA (RCR1) was sensitive to point substitution (Figure 7D). The region of the 18S rRNA that is complementary to RCR1 corresponds to a loop between helices 46 and 47 (helices 41 and 42 in human 18S rRNA). Fluorescent in situ hybridization using oligonucleotide probes complementary to the 18S rRNA of Saccharomyces cerevisiae also identified the region complementary to RCR1 as highly accessible (48). While point substitutions within RCR1 may perturb RNA secondary structure and thus cause the inhibition of downstream expression, the above results are also consistent with the hypothesis that RCR1 may promote ribosome handoff from the 5′-cap to an internal region on the S1 mRNA via interactions with 18S rRNA. A similar situation occurs in the 5′-leader of the homeodomain protein Gtx mRNA, where a 9-nt sequence is capable of acting as a ribosome recruitment site by base pairing with 18S rRNA (49). Base-pairing between mRNA and rRNA may also promote ribosome shunting from the 5′-cap to a downstream start codon in human adenovirus IVa2 mRNA and adenovirus late mRNAs, and in human heat shock protein 70 (hsp70) mRNA (27). Growing evidence, therefore, suggests that specific interactions between mRNA cis elements comprised of short nucleotide sequences and rRNA or ribosomal proteins may regulate translation initiation at downstream ORFs (42,43).
The present results suggest nonlinear ribosome migration on the S1 mRNA represents an atypical form of ribosomal shunting. As opposed to the classical shunting pathway typified by the plant pararetroviruses (26), the short length of the S1 5′-UTR indicates that 40S subunits would have to be handed off directly from the 5′-cap to an internal location downstream of the p17 start codon without the need to translate a short uORF that precedes a stable stem–loop structure. While shunting on the adenovirus late mRNAs does not require translation of a short uORF, it does involve scanning a short distance to encounter a complex group of stable hairpin structures that are essential for shunting to occur (50). Shunting also generally positions the shunted 40S subunit immediately adjacent, or in close proximity, to the downstream start site. If the S1 translation enhancer functions as a shunt acceptor site, shunted 40S subunits would need to scan >200 nt to reach the σC start codon. There is some evidence to suggest this may be the case. The absence of AUG codons in any of the three reading frames between the p17 and σC start codons is consistent with the possibility that 40S subunits may scan this region following shunting. The internal deletion results also indicated there is no essential cis element flanking the σC start codon and ribosomes can readily access a start codon located >100 nt further downstream (Figure 6). Lastly, the M6 and M7 constructs, which had start codons created in a preferred context 27 or 135 nt downstream of the RCR1 motif, exerted the strongest inhibitory effect on translation of the downstream ORF, particularly when present in combination (Figure 5), again consistent with 40S subunits scanning downstream of the translation enhancer element. We therefore favor the hypothesis that shunting from the 5′-cap positions the 40S subunits in proximity to the internal translation enhancer, which then scan the intervening region to initiate translation at the σC ORF.
As to why the ARV S1 mRNA would evolve this dual mechanism of translation initiation, we are reminded that the S1 mRNA is an exceedingly unusual eukaryotic mRNA that encodes three functionally distinct proteins. The p10 FAST protein is an integral membrane protein responsible for the virus-induced syncytium formation that contributes to viral dissemination and pathogenesis, the p17 protein is a nonstructural, nucleocytoplasmic shuttling protein, and σC is an essential, structural protein that functions in virus-to-cell attachment (5,51,52). In the absence of transcription control to regulate expression of these three genes, this tricistronic mRNA uses translation control to regulate production of both an integral membrane protein and two soluble viral proteins. The σC protein is only present at 36 copies per virion (6) and therefore while essential, does not need to be synthesized at relatively high levels in virus-infected cells. The dual mechanism of downstream translation initiation may be required to ensure that in diverse cell types, and under the changing translation conditions that arise in reovirus-infected cells, that sufficient σC is expressed to ensure infectious virus progeny production.
FUNDING
The Natural Sciences and Engineering Research Council of Canada (NSERC; to R.D.); a Nova Scotia Health Research Foundation (NSHRF) scholarship (to T.R.). Funding for open access charge: The Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Jingyun Shou for expert technical assistance.
REFERENCES
- 1.Day JM. The diversity of the orthoreoviruses: molecular taxonomy and phylogentic divides. Infect. Genet. Evol. 2009;9:390–400. doi: 10.1016/j.meegid.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Benavente J, Martinez-Costas J. Avian reovirus: structure and biology. Virus Res. 2007;123:105–119. doi: 10.1016/j.virusres.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Shmulevitz M, Yameen Z, Dawe S, Shou J, O'Hara D, Holmes I, Duncan R. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 2002;76:609–618. doi: 10.1128/JVI.76.2.609-618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shmulevitz M, Duncan R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000;19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costas C, Martinez-Costas J, Bodelon G, Benavente J. The second open reading frame of the avian reovirus S1 gene encodes a transcription-dependent and CRM1-independent nucleocytoplasmic shuttling protein. J. Virol. 2005;79:2141–2150. doi: 10.1128/JVI.79.4.2141-2150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grande A, Rodriguez E, Costas C, Everitt E, Benavente J. Oligomerization and cell-binding properties of the avian reovirus cell-attachment protein sigmaC. Virology. 2000;274:367–377. doi: 10.1006/viro.2000.0473. [DOI] [PubMed] [Google Scholar]
- 7.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 14.Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WB, Mackenzie SA. Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell. 2009;21:157–167. doi: 10.1105/tpc.108.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBratney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol. Cell. Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futterer J, Rothnie HM, Hohn T, Potrykus I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 1997;71:7984–7989. doi: 10.1128/jvi.71.10.7984-7989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frederiks F, Heynen GJ, van Deventer SJ, Janssen H, van Leeuwen F. Two Dot1 isoforms in Saccharomyces cerevisiae as a result of leaky scanning by the ribosome. Nucleic Acids Res. 2009;37:7047–7058. doi: 10.1093/nar/gkp765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XQ, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 21.Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 24.Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 2003;278:33793–33800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 25.Futterer J, Kiss-Laszlo Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 26.Ryabova LA, Pooggin MM, Hohn T. Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yueh A, Schneider RJ. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- 29.Racine T, Barry C, Roy K, Dawe SJ, Shmulevitz M, Duncan R. Leaky scanning and scanning-independent ribosome migration on the tricistronic S1 mRNA of avian reovirus. J. Biol. Chem. 2007;282:25613–25622. doi: 10.1074/jbc.M703708200. [DOI] [PubMed] [Google Scholar]
- 30.Corcoran JA, Duncan R. Reptilian reovirus utilizes a small type III protein with an external myristylated amino terminus to mediate cell-cell fusion. J. Virol. 2004;78:4342–4351. doi: 10.1128/JVI.78.8.4342-4351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Features in the 5′ non-coding sequences of rabbit alpha and beta-globin mRNAs that affect translational efficiency. J. Mol. Biol. 1994;235:95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- 34.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 35.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 36.Koh DC, Mauro VP. Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems. RNA Biol. 2009;6:54–58. doi: 10.4161/rna.6.1.7567. [DOI] [PubMed] [Google Scholar]
- 37.Chappell SA, Dresios J, Edelman GM, Mauro VP. Ribosomal shunting mediated by a translational enhancer element that base pairs to 18S rRNA. Proc. Natl Acad. Sci. USA. 2006;103:9488–9493. doi: 10.1073/pnas.0603597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panopoulos P, Mauro VP. Antisense masking reveals contributions of mRNA-rRNA base pairing to translation of Gtx and FGF2 mRNAs. J. Biol. Chem. 2008;283:33087–33093. doi: 10.1074/jbc.M804904200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chappell SA, Edelman GM, Mauro VP. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc. Natl Acad. Sci. USA. 2004;101:9590–9594. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castano A, Ruiz L, Hernandez C. Insights into the translational regulation of biologically active open reading frames of Pelargonium line pattern virus. Virology. 2009;386:417–426. doi: 10.1016/j.virol.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 41.Sen N, Cao F, Tavis JE. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol. 2004;78:11751–11757. doi: 10.1128/JVI.78.21.11751-11757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc. Natl Acad. Sci. USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell SF, Lorsch JR. Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition. J. Biol. Chem. 2008;283:27345–27349. doi: 10.1074/jbc.R800031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peri S, Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 48.Behrens S, Ruhland C, Inacio J, Huber H, Fonseca A, Spencer-Martins I, Fuchs BM, Amann R. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 2003;69:1748–1758. doi: 10.1128/AEM.69.3.1748-1758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 51.Brown CW, Stephenson KB, Hanson S, Kucharczyk M, Duncan R, Bell JC, Lichty BD. The p14 FAST protein of reptilian reovirus increases vesicular stomatitis virus neuropathogenesis. J. Virol. 2009;83:552–561. doi: 10.1128/JVI.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salsman J, Top D, Boutilier J, Duncan R. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J. Virol. 2005;79:8090–8100. doi: 10.1128/JVI.79.13.8090-8100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]