Abstract
In prokaryotes, regulation of gene expression is predominantly controlled at the level of transcription. Transcription in turn is mediated by a set of DNA-binding factors called transcription factors (TFs). In this study, we map the complete repertoire of ∼300 TFs of the bacterial model, Escherichia coli, onto gene expression data for a number of nonredundant experimental conditions and show that TFs are generally expressed at a lower level than other gene classes. We also demonstrate that different conditions harbor varying number of active TFs, with an average of about 15% of the total repertoire, with certain stress and drug-induced conditions exhibiting as high as one-third of the collection of TFs. Our results also show that activators are more frequently expressed than repressors, indicating that activation of promoters might be a more common phenomenon than repression in bacteria. Finally, to understand the association of TFs with different conditions and to elucidate their dynamic interplay with other TFs, we develop a network-based framework to identify TFs which act as markers, defined as those which are responsible for condition-specific transcriptional rewiring. This approach allowed us to pinpoint several marker TFs as being central in various specialized conditions such as drug induction or growth condition variations, which we discuss in light of previously reported experimental findings. Further analysis showed that a majority of identified markers effectively control the expression of their regulons and, in general, transcriptional programs of most conditions can be effectively rewired by a very small number of TFs. It was also found that closeness is a key centrality measure which can aid in the successful identification of marker TFs in regulatory networks. Our results suggest the utility of the network-based approaches developed in this study to be applicable for understanding other interactomic data sets.
INTRODUCTION
Organisms respond to continuous variations in internal and external conditions by orchestrating their transcriptional responses depending on the environmental challenges they are faced with. This involves the usage of a subset of a complex network of transcriptional interactions, which undergo rewiring from condition to condition and is commonly called the transcriptional regulatory network (TRN) of an organism (1,2). In bacteria, where regulation of gene expression is primarily believed to occur at the level of transcription, the protein complement that can sense these variations in internal and external cellular status is termed as the collection of transcription factors (TFs) (3–5). It is through the activity of TFs which can respond to specific signals resulting in allosteric modifications, that their affinities to specific DNA-binding sites (operators) or with the rest of the transcriptional machinery change (3).
Although several recent studies have successfully employed the extent of cross-species conservation of regulatory elements or regulatory network structure to show that there is extensive rewiring of transcriptional machinery even in closely related organisms (6–11), our understanding of species specific aspects of transcriptional regulation and their dynamics across conditions is rather limited. Therefore, to gain further insights into bacterial TRNs and to quantify properties of TFs which govern their function under different experimental conditions, we exploited the publicly available expression data for the best characterized bacterial model, Escherichia coli (12,13).
Recent advancements in deciphering the expression patterns of genes across an entire genome using microarray technologies have allowed us to characterize the transcriptomes of several model organisms. Indeed, previous efforts have shown that microarray expression patterns can be successfully used to study the transcriptional network in E. coli (14). However, our understanding of the expression patterns of TFs, which are themselves responsible for the dynamics of gene expression in an entire genome, is limited. Therefore, in order to have a comprehensive overview and comparative perspective of the properties of TFs, which are expressed under different experimental conditions, we analyze in this study the repertoire of TFs active under different conditions and show that a relatively small fraction of the complete repertoire of TFs are active in any given condition. We then use the set of active TFs in each condition to study their mode of regulation, ability to sense intracellular or extracellular status and connectivity in the TRN. We further show that most conditions can be associated with a small set of marker TFs using a dynamic network of TF–TF interactions generated in respective conditions. Our results provide a first comprehensive overview of the transcriptional landscape of TFs in a bacterial model system demonstrating the dynamic nature of sequence-specific DNA-binding factors across conditions.
MATERIALS AND METHODS
Transcriptional regulatory network of E. coli
The currently known network of transcriptional regulatory interactions in the complete genome of E. coli was obtained from RegulonDB (15). The network contained 1420 nodes and 3461 edges after removing sigma-mediated interactions. We found that the network comprised of 165 TFs regulating a set of 1255 target genes. Since some TFs act as dimers, for the sake of calculating the number of genes regulated by a TF in such cases we included targets of dimers as the target of each of the monomeric subunit involved. In addition, monomers of these dimeric TFs are often expressed in different transcription units and might be subject to distinct regulation.
Data set of TFs and their classification based on the number of regulated genes, functional role and sensing environment
The complete set of E. coli TFs analyzed in this study was obtained from RegulonDB (15), which is a manually curated database containing information on transcriptional regulation in E. coli. However, since several TFs in E. coli are uncharacterized, we also included predictions of TFs (16) made available through this database. Our final data set comprised of 296 known and predicted TFs in the whole genome which was used for all the subsequent analysis. This data set is available as Supplementary Data along with literature evidence confirming the DNA-binding activity of the TF where available.
To characterize a TF based on the number of genes it regulates, we have first calculated the degree of all the TFs in the complete TRN and grouped them into high (H)-, medium (M)- and low (L)-degree TFs. H-degree TFs were defined as those which regulate more than mean(degree) + 2 standard deviation(degree), while the set of L-degree TFs comprised those with degrees less than mean(degree). M-degree TFs corresponded to those with degrees in between these two groups. This classification resulted in 24 and 7 TFs to belong to the M- and H-degree groups and the rest to the L-degree. TFs can modulate the expression of a gene either positively or negatively and this often depends on the site of action on the DNA with respect to the transcription start site (3,17). In order to understand whether activators or repressors or dual regulators are abundant in each experimental condition examined, we classified TFs into activators (positive mode of regulation), repressors (negative mode of regulation) and dual regulators (TFs which exhibit both modes of regulation on their promoters without preference for one or the other). TFs were classified as activators or repressors if at least 60% of all the promoters it controls are known to be positively or negatively regulated, otherwise it was considered as a dual regulator which does not have a preference for either mode of regulation. Such a classification resulted in identifying 79 and 78 TFs as repressors and activators, respectively, with the remaining belonging to the dual class of regulators. The basic unit of transcriptional sensing system is composed of a TF and its corresponding effector genes; the former encodes for a TF sensing the effector signal produced or obtained by the product of the second gene (4,5,18). The main characteristics of the subclasses of the genetic sensing machinery in E. coli are shown in Supplementary Data and a more complete discussion is presented elsewhere (4,8). We mapped experimental or annotated information for 96 TFs, which were previously classified into one of the five different classes namely internal sensing metabolites (ISMs), internal DNA-bending (IDB) or nucleoid-associated proteins (NAPs), hybrid (H; sensing transported and synthesized metabolites), external sensing two-components (ETCs) and external sensing transported metabolites (ETMs).
Microarray data and processing for analyzing TF activity
To compare the expression levels of TFs across different experimental conditions, we obtained a large compendium composing of 445 microarray data sets available as a public resource for E. coli in the form of M3D database (Build 4 of E. coli expression data) (13). These data were available in the form of Robust Multi Array (RMA) normalized profiles (19), thus enabling us to directly calculate the average expression value of protein coding genes across all experimental conditions tested. Therefore, averaged gene expression values were used to compare the levels of expression of TFs and other protein coding genes. Expression data could be obtained and mapped for 4125 genes in the complete genome of E. coli K12 (NCBI reference genome sequence NC_000913.2), while all TFs could be mapped onto the expression compendium. Since a number of conditions available as part of this compendium are redundant or minor variations of the standard conditions, we have calculated the correlation of expression for all genes between all arrays using Pearson’s correlation as the similarity metric between arrays and performing a hierarchical linkage clustering in the cluster package (20). This enabled us to identify conditions which are highly correlated to each other and to include only one of the repeated conditions as a representative. We found that at a correlation threshold of 0.95 a total of 62 conditions could be considered as nonredundant representatives of the compendium which we use for the entire analysis. This threshold allows sequential snapshots in time-course experiments to be considered as different, while a stricter threshold of 0.90, which yields only 25 conditions, filters out these experiments. A list of these conditions is available in Supplementary Table 5.
Identifying TFs which are significantly expressed in each nonredundant condition
It has been recently found by a number of studies that there is a relationship between the number of genes regulated by a TF and its concentration (21–23), suggesting that the number of active TFs in a condition cannot be determined purely based on a comparison of their messenger RNA (mRNA) concentrations in a given condition. Therefore, we first launched a detailed analysis on whether the expression profiles of TFs across conditions vary and found that most TFs show a variation in their expression profile. These expression values were sorted, plotted and finally inspected for all the nonredundant conditions, and it was observed that a variety of expression patterns emerged from the data, suggesting that each TF should be handled separately. In particular, we found that the dominant trend comprised of a truncated normal distribution with varying ranges of expression. Accordingly, an expression vector corresponding to each TF was used to calculate the mean (M) and standard deviation (SD), which were subsequently employed to define the significant expression threshold (SET) in the form of SET = M + SD. In other words, TFs were labeled as significantly expressed in a given condition if their measured expression value surpassed SET. Dot plots showing the expression profiles for the experimentally verified TFs in RegulonDB (15) are available as Supplementary Data with SET values indicated. Such a cross-condition comparative approach to identify active TFs not only takes into account the differences in the levels of expression of global versus local TFs but also sensitive to variations across conditions.
Estimating the significance for the enrichment of different properties of TFs across conditions
To estimate the significance for the enrichment of TFs in each condition, we calculated the hypergeometric probability using the dhyper function in R. This was done by identifying the total number of genes present on the microarray chip and the number of protein coding genes which are detected to be expressed at the same thresholds used for TFs in a given condition. The total pool of TFs (297) was also used as a parameter for estimating this probability. The same approach was used for estimating significance of different sub-populations /classes of TFs in various sections of the manuscript. For instance, to understand whether there is enrichment for activators, repressors or dual factors in each condition, we computed the P-values using the reference distribution of these classes from the static network. P-values estimated using this approach are shown in the figures and a more complete list for different sections is available as Supplementary Data.
Defining active TF–TF subnetworks in each condition
A recent study mapped the static network of interactions between different TFs in E. coli providing a compendium of information for studying the dynamic nature of regulatory cross-talk between TFs (21). Therefore, to understand whether TFs can be associated to different conditions based on their interplay with other TFs in a given condition, we first constructed active sub-networks of TFs for each nonredundant condition using this static network, which comprised of 171 regulatory interactions between TFs after excluding autoregulatory interactions. The procedure to create active subnetworks essentially involved two steps, first of which is to identify TFs which are active in a given condition as described in a previous section and then mapping them onto the static TF–TF network. The second step involved finding all interactions where in at least one of the TFs participating in a static regulatory interaction was found to be active in the condition of interest. Such an approach yielded an active subnetwork for each of the nonredundant conditions. The number of interactions in subnetworks varied from 14 observed in the condition where the predicted biofilm formation regulatory protein (yceP) is knocked-out, to 95 interactions in one of the mid-log growth aerobic conditions of the E. coli wild-type strain BW25113. Supplementary Data shows the set of active subnetworks identified as a result of this procedure for different conditions.
Calculating network properties and identifying statistically significant TFs in the active TF–TF network in each condition
To study the properties of each active subnetwork and the variation of the network properties of different TFs across conditions, we used igraph, a publicly available R package for analyzing graphs (http://cneurocvs.rmki.kfki.hu/igraph/ and http://www.r-project.org). In particular, we used the igraph functions degree, transitivity, betweenness and closeness for calculating the degree, clustering coefficient, betweenness and closeness centralities of a node, respectively. The clustering coefficient of a node (within a directed graph) of interest was calculated locally, as the proportion of links between its neighbors divided by the maximum number of links that could theoretically exist between them. Betweenness centrality, which is the number of shortest paths going through a node, was calculated using the Brandes algorithm (24) implemented in R. Similarly, closeness, measured as the inverse of the average length of the shortest paths to all other vertices in the graph, was obtained using the implementation in R. Since the centrality measures betweenness and closeness use the shortest path lengths between all pairs of nodes in a graph, for cases where no path exists between a particular pair of nodes, shortest path length was taken as one less than the maximum number of nodes in the graph. Note that this is also the default assumption for calculating centrality measures in igraph. Since different subnetworks have different sizes, degree and betweenness need to be normalized before they can be compared across conditions, we employed the following normalization formulas:
 |
(1) |
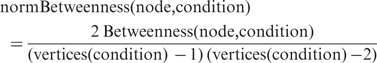 |
(2) |
To find associations between TFs and conditions we compared the network properties of a given TF across different conditions and identified conditions which showed significant variation of the network property with respect to what is expected in an average profile. In particular, for each TF we calculated the degree, clustering coefficient, betweenness and closeness values in the active subnetworks representing the different conditions and identified conditions where a TF exhibited a significant centrality threshold (SCT) ≥ mean (M) + standard deviation (SD) of the particular network property. TF-condition associations were considered significant only if two or more of these network descriptors were found to cross the significant threshold. This network significance parameter had a considerable effect on the number of predicted markers, with a stringent SCT cutoff of 2 yielding 179 potential markers, as explained in the ‘Results’ section, while a relaxed cutoff of 1 resulted in 728 potential markers. By contrast, a very stringent threshold value of 3 uncovered only 27 markers.
Using regulon expression to validate identified marker TFs
The previous section described a network-based procedure that produces a list of markers for any given condition. In this section, a protocol is presented to further check these marker TFs, by testing whether they have a detectable effect on the expression of their target genes. More specifically, this benchmark consists of estimating the expression footprint of markers in comparison with randomly chosen transcription factors. It takes several steps to calculate the expression footprint of a marker m in condition C :
Take the static regulatory network of E. coli.
Identify all target genes T1…r known to have direct regulatory interactions with m.
-
For each Ti check expression state in C :
Check expression level, which we term as expression(Ti,C).
Calculate the mean and standard deviation of the expression values of Ti across all nonredundant conditions C1.c.
When expression(Ti,C) < Mean(Ti) – SD(Ti) or expression(Ti,C) > Mean(Ti) + SD(Ti) we say that Ti has significantly changed its expression in condition C; otherwise, we say that this target gene has not significantly changed its expression in C.
Calculate the fraction of the m regulon (of size r) that significantly changes its expression in C.
In the case of transcription factors with both positively and negatively regulated target genes, the protocol is applied separately to the activated and repressed regulons, excluding genes with dual regulation. Furthermore, in order to get reliable expression measurements, only regulons of at least five genes were considered, which is equivalent to sample the effect of a marker gene 5 or more times per condition.
The same protocol is repeated with 100 randomly sampled TFs in order to estimate the mean (background) regulon state in C, so that we can now calculate: (i) the percentage change (expression ratio) between the regulon state of m and the background regulon state and (ii) the associated normal distribution P-value for each marker m (Supplementary Table S3). In order to classify predicted markers that effectively rewire the transcriptional network, a cutoff value of expression ratio was enforced. In our tests, the preferred minimum expression change value was 15%, which selects a total of 107 effective markers. A cutoff of 25% was also tested, which still reported 97 markers.
RESULTS AND DISCUSSION
Expression of TFs across conditions
Organisms react with numerous transcriptional responses depending on the fluctuations in their internal and external conditions by controlling the expression of their genes. The cellular components that sense these variations are linked to the transcriptional machinery through the activity of TFs. TFs can respond to specific signals resulting in allosteric modifications that change their affinities to specific DNA-binding sites upstream of genes, thereby controlling their expression. These effector signals can be classified as exogenous or endogenous depending on their origin in the cellular context—i.e. whether the cell can take them from the milieu or produce them in the cytoplasm (4,25). The network of interactions between TFs and the set of genes they regulate have been studied in great detail in several model organisms at varying levels (17,26,27). In particular, TRNs have been shown to possess a multilayer hierarchical modular structure using either a top-down or a bottom-up approach for determining hierarchy (28,29) at the global level, encompassed with motifs, which are formed of patterns constituting one or more TFs modulating the activity of a set of target genes, at the local level (17,27,30). Indeed, each of the different types of network motifs was found to exhibit distinct dynamical functions (27). However, our understanding on whether TFs are more expressed than other functional classes or how TFs belonging to different layers of this hierarchical network and different sensing abilities are expressed across conditions is not clear. In what follows, we first compare the expression of TFs as a class compared to other functional groups and then address a series of questions on whether the set of TFs identified to be active in each experimental condition show distinct trends depending on the condition.
TFs are frequently less expressed than other classes of genes
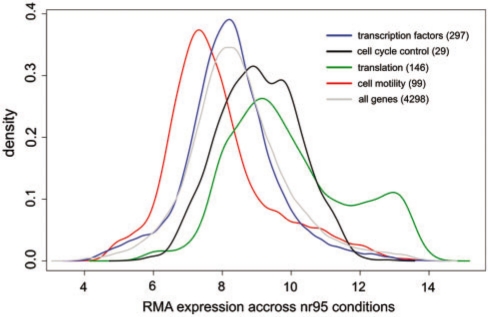
It is now a known notion that not all genes are expressed to the same extent in a cell. Some functional classes such as ribosomal genes or genes involved in core metabolic processes are known to be expressed in higher levels than others because of their frequent use. In general, TFs are thought to be expressed in lower levels based on anecdotal observations from well-studied lac system where it was shown that the number of protein copies of LacI (a dedicated TF for lactose utilization) rises from around five to a maximum of 20 molecules upon induction of lactose (31). However, NAPs and other global regulators such as crp, lrp and fur in E. coli reach protein concentrations of more than 1000 units per cell (32,33), suggesting that some TFs can be expressed in higher concentrations. Therefore, to learn whether TFs as a class are expressed differently to other functional groups, we compared their mRNA expression levels using two alternate functional schemas available for E. coli, namely COGs (34) and the Multifun classification of genes by Riley and co-workers (35). Figure 1 highlights some COG functional classes which exhibited the largest differences in expression with respect to TFs (see Supplementary Data for a comparison with all classes, including Multifun). Among these, we found that ‘translation’ and ‘cell cycle control’ classes clearly showed enrichment for highly expressed genes (mean RMA expression values are 9.95 and 9.06, respectively). We also identified some classes such as cell motility, which need to be sporadically expressed under specific conditions, to be less expressed in general than TFs (mean RMA expression values are 7.89 and 8.23, respectively). Figure 1 also includes the combined expression profile of all E. coli genes, plotted in gray, with a mean expression value of 8.45, showing that TFs are weakly expressed even when compared to the average expression profile of all protein coding genes. While the TF expression density appears to be only slightly shifted toward smaller values, a Wilcoxon test confirms that both classes indeed have significantly different medians (P-value = 7.589E-81), and therefore different distributions. Overall, these results suggest that most TFs are poorly expressed across conditions by triggering their activity only when needed, although the absolute difference in expression is small. In contrast, global transcription factors are known to achieve relatively high expression levels (21,22,36) but nevertheless have short transcript half-lives (37).
Figure 1.
Kernel density estimates of RMA normalized microarray expression for various gene subsets in E. coli. Subsets are defined according to COG (34) (cell cycle control, translation, cell motility) and RegulonDB (transcription factors) annotations and their size is indicated within parenthesis. Median expression values across 62 nonredundant conditions are significantly different among gene subsets, as evaluated by a Kruskal–Wallis nonparametric test (P < 2.2E-16). Only some classes which exhibited the largest differences in expression with respect to TFs are shown, together with the combined expression distribution of all E. coli genes. This figure was generated using the R language for statistical computing.
Conditions exhibit varying number of active TFs
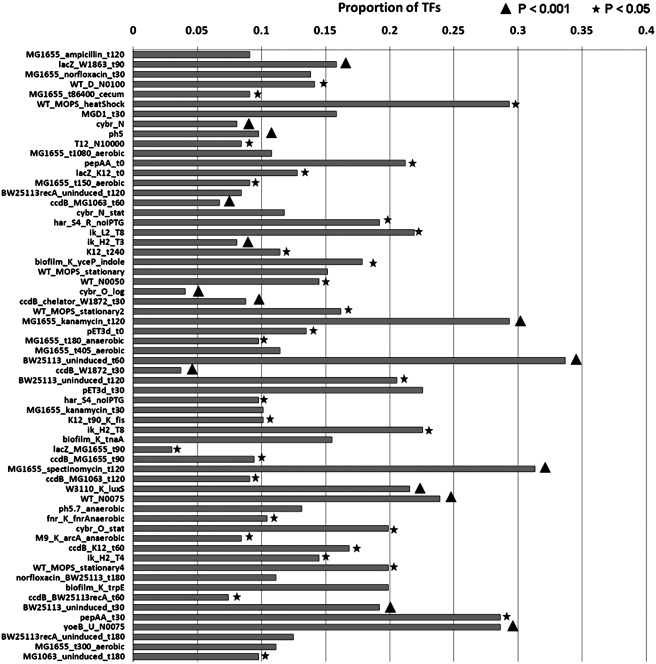
TFs are known to be highly dynamic in their expression, thereby providing timely response to external perturbations using a range of network sub-structures from motifs to signal processing units (27,38–40). Therefore, to assess the number of TFs, which are active in each condition, and to analyze whether different conditions exhibit varying proportions, we identified the set of active TFs in each of the 62 nonredundant conditions (see ‘Materials and Methods’ section). Above the SET of each TF, we found that different conditions harbored varying proportions with the lowest observed in lacZ upregulated condition 90 min after mid-log growth induction of the riboregulated CcdB plasmid (Figure 2, condition lacZ_MG1655_t90). We also found six conditions where the proportion of active TFs exceeded 25% of the total TF repertoire. These conditions are: aerobic growth of wild-type cells in log phase using MOPS media with 10 min heat shock at 50° (WT_MOPS_heatShock) (41); yoeB upregulated condition under high concentrations of norfloxacin in LB (yoeB_U_N0075) (12); an experiment in which the synthetic peptide pepAA, containing least abundant E. coli amino acids, was overexpressed and expression was measured 30 min postinduction (pepAA_t30) (42); E. coli MG1655 wild-type 120 min after treatment with 5 ug/ml kanamycin (MG1655_kanamycin_t120); and 400 ug/ml spectinomycin (MG1655_spectinomycin_t120) (43). Despite the variations in the proportion of TFs expressed across conditions, we found that the maximum number of active TFs was limited to 100, accounting for about 33% of the total TFs, observed in the uninduced condition of the wild-type strain BW25113 post 60 min (BW25113_uninduced_t60), suggesting that much less than one-third of the total collection of TFs in an organism might be employed for transcriptional responses specific to a condition. Indeed, an analysis of the average number of TFs expressed across conditions suggests that about 15% of the total TFs might be active, indicating that most conditions might be exploiting no more than 50 TFs, with stress induced conditions like heat shock or translational burden (42) and drug resistance-associated conditions exhibiting an increase in the number of expressed TFs. These observations suggest that under stress and drug-induced conditions, organisms might undergo a significant change in their transcriptional circuitry. In order to understand whether the number of expressed TFs in a given condition is significant when compared to the total number of protein coding genes detected to be expressed, we computed its significance using a background hypergeometric distribution (see ‘Materials and Methods’ section). As shown in Figure 2 (also see Supplementary Data for all conditions), we found that 46 conditions (75%) showed higher than expected number of TFs at a P-value threshold of 0.05, suggesting that although the proportion of TFs identified across conditions is small, they form a significant component of the expressed pool of genes.
Figure 2.
Proportion of the total repertoire of TFs expressed in each nonredundant condition. Conditions were found to harbor varying number of TFs, with percentages ranging from 3% to as high as 33%, suggesting enormous variation in the number of active TFs across conditions. P-values reflecting the enrichment for the occurrence of TFs in a particular condition, computed using a hypergeometric distribution (‘Materials and Methods’ section), are shown in two different ranges. P < 0.001 are indicated with a triangle while those <0.05 were marked with a star.
Activators are more abundant across conditions than repressors
Transcription initiation in bacteria requires that RNA polymerase (RNAP) recognizes and binds specific DNA sequences upstream of transcription units called promoters. The recognition of promoter sequences by RNAP occurs when it associates with sigma (σ) factor. The primary or housekeeping sigma factor in E. coli is encoded by the rpoD gene and is known as σ70 (44). A bacterial promoter is defined as the segment of DNA that enables a gene or set of genes to be transcribed and is located immediately proximal (6–8 bp) to the transcription start site. However, in addition to sigma factors, TFs also bind to these regions to mediate the process of transcription and hence play a central role in governing the activity of a gene. In particular, TFs recognize their target genes (TGs), whose transcription they control, due to the presence of the binding sites in the promoter regions. Typically, a TF, upon binding to the promoter regions of its target genes or transcription units, can control the expression of the genes positively or negatively. While repressor sites which can inhibit the transcription of genes are known to occur downstream of transcription start site, activators generally attach to DNA upstream of the start site (45–47). In E. coli and several other bacteria it has been predicted, based on the location of the helix–turn–helix DNA-binding protein motif in the protein sequence of the TF, that there is an enrichment for factors which act as transcriptional repressors and hence postulated that significant fraction of the genes in the transcriptional network might be negatively regulated (46,48,16). However, it is not known how the proportion of TFs based on their mode of regulation varies across different experimental conditions.
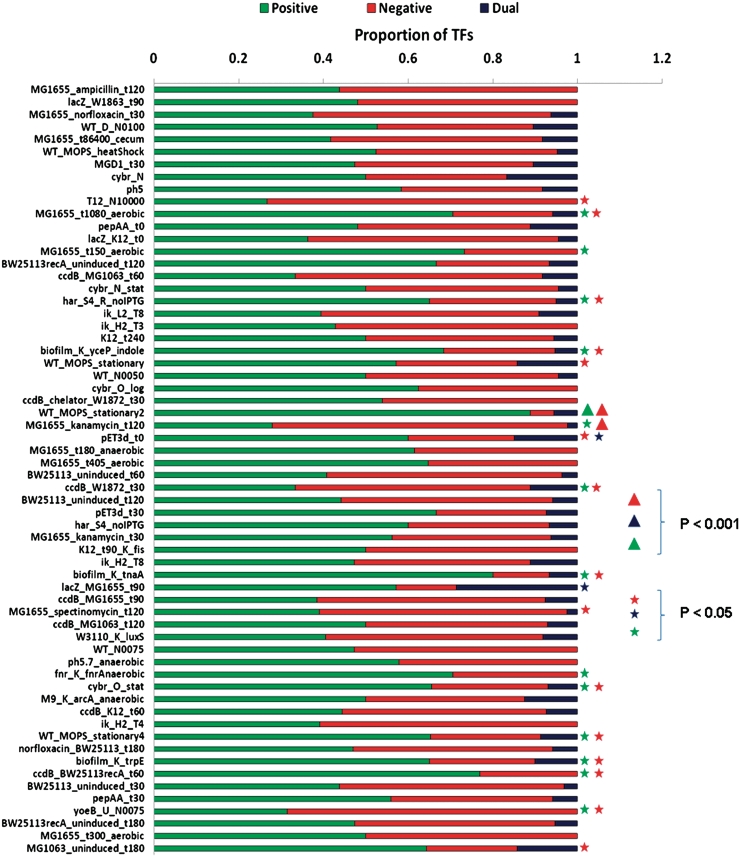
Therefore, we sought to address this by grouping experimentally characterized TFs for which transcriptional regulatory interactions are well documented into activators, repressors and dual regulators (see ‘Materials and Methods’ seciton). Figure 3 shows the proportion of TFs belonging to different modes of regulation in each condition of growth. Although most conditions show a similar distribution of activators and repressors, it is easy to note that there are some conditions which exhibit marked enrichment for either class. For instance, contrary to the expectation that most conditions might be overrepresented for repressors due to their genomic abundance and high conservation in closely related species (16,49), we found that only four conditions showed more than 60% of the TFs working as repressors, while 17 conditions had more than 60% of the TFs represented as activators, indicating that activation is the most common mode of regulation for TFs in most conditions. Indeed, nearly 50% of the conditions exhibited more than 50% of the TFs working as activators, while only 30% of the conditions showed the same frequency of TFs acting as repressors. A closer look at the conditions suggests that most of these conditions associated with high number of activators correspond to E. coli cells in the later phases (mid-log to late-log) of growth representing: aerobic (MG1655_t1080_aerobic, MG1655_t150_aerobic, MG1655_t405_aerobic); anaerobic (MG1655_t180_anaerobic, fnr_K_fnrAnaerobic); recombinant protein expression cultures (har_S4_R_noIPTG) in the absence of isopropyl-1-thio-β-d-galactopyranoside (IPTG) (50); recombinant protein production of E. coli abundant amino acid encoded peptides (pET3d_t30) (42); or biofilm-associated conditions (biofilm_K_yceP_indole, biofilm_K_tnaA, biofilm_K_trpE), suggesting that most of the activators are upregulated in the later phases of growth or in conditions where there is a metabolic burden on the cell. Similarly, we found that repressors are abundant in E. coli cells at 12 min posttreatment with norfloxacin (T12_N10000), at 120 min posttreatment with kanamycin (MG1655_kanamycin_t120), upregulation of yoeB under norfloxacin-induced conditions (yoeB_U_N0075) or in LB with high concentrations of glucose 4 h post-incubation (ik_H2_T4). These observations indicate that while metabolic repressors might be expressed in order to turn off the corresponding metabolic operons (21), stress and antibiotic response regulators might be upregulated in the former conditions. Again we used a background hypergeometric distribution to estimate the significance of these populations of activators, repressors and dual TFs when compared to their abundance in the static network. As indicated in Figure 3 (see also Supplementary Data), we found 14, 17 and two conditions which exhibited significant numbers of activators, repressors and dual regulators respectively at a P-value threshold of 0.05, further supporting the protocol for identification of active TFs.
Figure 3.
Proportion of activators, repressors and dual regulators in different conditions. TFs predominantly regulating positively and negatively the expression of their promoters were defined as activators and repressors, respectively, while those which do not show this tendency were considered as dual regulators (‘Materials and Methods’ section). Although a number of conditions exhibited roughly equal proportions of activators and repressors, some conditions were clearly found to exhibit biases in their repertoire as discussed in the text. P-values reflecting the enrichment for the occurrence of different types of TFs in a particular condition, computed using a background hypergeometric distribution, are shown in two different ranges. P < 0.001 are indicated with a triangle, while those <0.05 were marked with a star.
These observations suggest that most of the normal conditions of growth invoke activators, while stress or metabolic response to particular carbon sources might induce a number of repressors. Overall, our results based on expression of TFs across conditions suggest that activators are more abundant and hence promoters might be predominantly activated in majority of the conditions contrary to the holistic notion that promoters are mostly repressed (47,51).
Nucleoid-associated TFs are consistently expressed across conditions
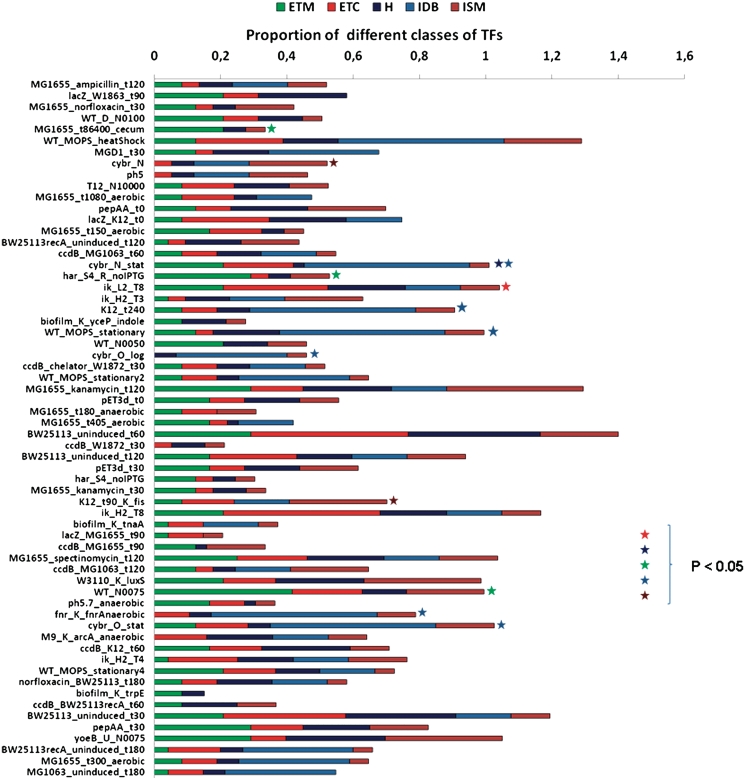
In bacterial cells, the dynamics of TFs is controlled by signals which can have origin both within the cell or exterior to the cell (4,5,18). The basic unit of this sensing machinery at the genomic level is constituted by TF and effector genes; the former encode for a TF sensing the effector signal produced or obtained by the product of the second gene (4,25). The main characteristics of the subclasses of the genetic sensing machinery in E. coli are described elsewhere (4,8). Using a literature-curated data set of 96 TFs and their effectors (see ‘Materials and Methods’ section), we asked whether different conditions show distinct patterns of preference for different classes of sensing. As a result of this analysis, we found that except IDB TFs, which seem to be consistently expressed in most conditions to remodel the bacterial nucleoid, all other classes were represented with <30% of the total TFs in most conditions (Figure 4).
Figure 4.
Distribution of different sensing classes (4,5) of transcription factors across conditions, showing proportions of each class which are found to be active. Different conditions were found to exhibit different combinations of sensing classes depending on their growth environment, but the associated hypergeometric distribution P-values show that there is very little enrichment of sensing classes across conditions beyond internal DNA-bending (IDB) TFs. The remaining classes are: internal sensing metabolites (ISMs), hybrid (H; sensing transported and synthesized metabolites), external sensing two-components (ETCs) and external sensing transported metabolites (ETMs).
In order to further validate these observations, we computed their significance using a background hypergeometric distribution. As shown in Figure 4 (also see Supplementary Data for all conditions), we found few conditions which exhibited significant enrichment for any class of sensing at a P-value threshold of 0.05, possibly due to the small number of TFs which could be associated to sensing classes; however, as expected, the most frequent enriched class was found to be IDB.
Dynamics of TFs across conditions
There is convincing evidence that, similar to eukaryotic transcriptional regulators, bacterial TFs work in a combinatorial fashion to control their promoters by integrating external and internal signals (5,30). However, our ability to unravel the interplay between TFs and the association of TFs with specific conditions has been limited to specific conditions. Therefore, to understand the association between physiological states and the subset of active regulatory proteins, we developed a network-based framework to link each condition with a specific set of TFs which were found to be central to the condition under investigation.
To assess the association of a TF with a particular condition, we first mapped the known static network of TF–TF interactions (21) onto each of the nonredundant microarray conditions, and as a result condition-specific TF networks were obtained as explained in ‘Materials and Methods’ section. These subnetworks were then employed to study the centrality and clustering coefficient of each of the nodes across conditions. Briefly, three centrality measures have been described in the literature: (i) degree or connectivity, which is interactions a protein has in the TRN—the higher the connectivity (i.e. hub nodes) the more targets it has; (ii) betweenness centrality, which measures the number of shortest path lengths between all pairs of TFs in the network that pass through a TF of interest—the higher the number of paths that pass through a TF, the more important it is; and (iii) closeness centrality, which provides the inverse of the average length of all the shortest paths from a TF of interest to all other TFs in the network. Likewise, the clustering coefficient of a TF gives an idea of the proportion of immediate neighbors to that theoretically expected. As explained in more detail in ‘Materials and Methods’ section, a TF was classified as associated to a microarray condition if any two of these network descriptors achieved values that were significantly higher than their average values across conditions. Such TFs were called marker TFs.
Different conditions have distinct subsets of marker TFs
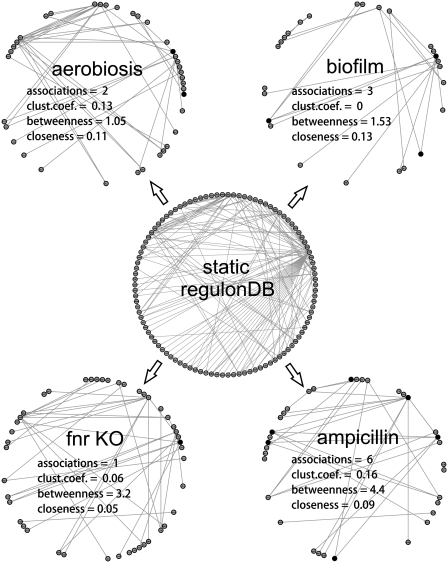
In total, we found 179 TF-condition associations across 52 experimental conditions (listed in Supplementary Tables 1 and 2). On average, each condition has nearly three marker TFs, of which one is a global hierarchical regulator. A few representative conditions, displaying one to six TF associations, are discussed below in more detail and are also shown in Figure 5.
Figure 5.
Active subnetworks in four representative conditions: aerobiosis, biofilm, anaerobiosis (fnr knockout) and ampicillin induction. The static network of TF–TF associations from RegulonDB, which is the basis for the generation of dynamic networks in respective conditions, is shown at the center. Different network properties are displayed for each active sub-network to illustrate the variation in the parameters among conditions, and the predicted markers are shown as black nodes.
First we analyze two examples from standard experimental conditions:
-
(i)
Experiment MG1655_t300_aerobic corresponds to growth in rich medium under anaerobiosis for 300 min. In such conditions, our protocol identifies a couple of upregulated genes encoding TFs (gadX and arcA) that have been previously reported to be associated to aerobiosis/anaerobiosis transitions (52,53).
-
(ii)
Experiment biofilm_K_trpE monitors the formation of biofilms after knocking out the gene trpE. Our approach could detect three marker TF-encoding genes: biofilm repressor, rcsA, found to be downregulated (54); gadE, a regulator of acid resistance (55), upregulated; and global regulator lrp, found to be downregulated, which regulates the development of type I fimbriae (56).
Next, we present two more examples with the purpose of illustrating the value of this approach when the goal is to understand mutant phenotypes:
-
(iii)
Experiment fnr_K_fnrAnaerobic measures the anaerobic growth of an fnr knockout strain of E. coli, and the upregulated csgD gene was found to be the marker, in good agreement with a literature report showing that this gene is repressed by the fnr product (57) in wild-type strains.
-
(iv)
The same marker gene is identified in a related experiment, M9_K_arcA_anaerobic, performed with an arcA knockout strain. Similarly, the observation that the expression levels of arcA and csgD are linked has already been reported (58).
Finally, we describe an example of drug inhibition, a condition in which a culture of E. coli is exposed to a drug which results in a subsequent rewiring of the regulatory network:
-
(v)
Experiment MG1655_ampicillin_t120 reports the growth of a wild-type E. coli strain in a rich medium when ampicillin is added. In this case, two global regulators (crp and fis) are highlighted as markers, but the upregulated marA and marR genes, known to be activated by fnr and fis, possibly have greater diagnostic value, as they are key components in the antibiotic resistance mechanism (59). Likewise, oxyR is assumed to be involved in responses to oxidative stress (60). Finally, metJ is also predicted to be a marker in this condition.
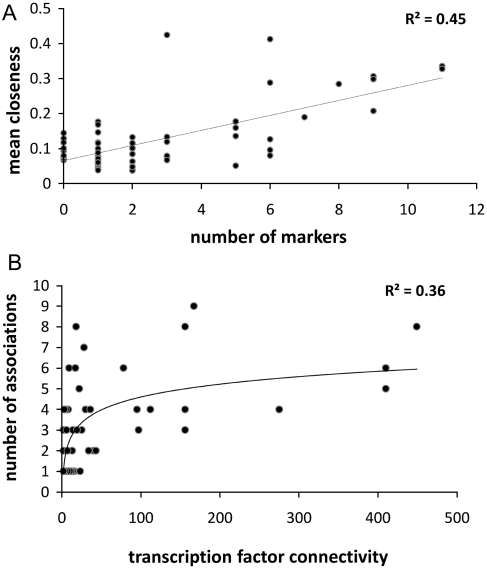
Inspection of these examples suggests that a network-based approach, as the one presented in this work, is able to identify biologically meaningful associations between TFs and environmental conditions. Nevertheless, this approach could not find significant associations in 10 conditions. A possible explanation is that some conditions might capture an equilibrium (or amorphous) state of the regulatory network in which no single active TF can be identified as a marker. However, it is also plausible that some conditions exhibit much smaller numbers of active TFs, which would result in smaller regulatory sub-networks. In order to further investigate this, we examined several network descriptors for all 62 regulatory subnetworks (maximum diameter, average path length, mean degree, mean closeness, mean clustering coefficient and mean betweenness; see Supplementary Table S2) to observe to what extent the condition-specific network topology imposes restrictions on the number of markers found. The only property that correlates significantly with the number of associated marker TFs is mean closeness (R2 = 0.45, P-value = 1.47E-09; see also Figure 6A), suggesting that this variable can be a predictive estimate for conditions to have significant number of associated markers. This means that conditions in which the active subnetwork has a larger fraction of TFs with short paths to all other nodes (a higher closeness centrality) are more likely to produce markers TFs that are responsible for re-shaping regulatory networks.
Figure 6.
(A) Number of marker TFs identified in microarray conditions as a function of the mean closeness of the active transcriptional network. The linear regression P-value is 1.47E-09. (B) Relationship between transcription factor connectivity and the number of condition associations found with centrality descriptors, which can be fitted to a logarithmic regression with a P-value of 6.13E-07.
Overall, this analysis shows that the conditions studied in this work clearly exhibit distinct network structure and properties, indicating that a distinct subset of the transcriptional network might be employed by bacteria depending on the environment. Furthermore, while this work does not attempt to estimate the number of possibly different regulatory states in the cell, the observation that diverse experimental conditions show redundant transcriptional footprints (383 out of 445, see ‘Materials and Methods’ section) suggests that the regulatory state repertoire is somewhat limited when compared to the theoretical state space.
Highly connected TFs are frequently found associated to conditions
Transcriptional networks are scale-free in their structure with a small set of TFs regulating most of the genes and this results in the identification of a set of TFs which can be identified as hubs or global regulators (61,62). Although a number of approaches and criteria have been developed for identifying global regulators (63), here we have classified TFs into three different classes, namely H-, M- and L-out-degree, depending on the number of genes controlled by them, as described in ‘Materials and Methods’ section. In terms of active TFs, we note that most microarray conditions capture <30% of the H, M or L classes, which is obviously in agreement with the previously presented global pattern of expression and again insinuates that only a small subset of the TFs from each class might be exerting regulatory roles in any one physiological state (Supplementary Figure S1). However, as there are only a small number of highly connected TFs to sample, they are found to be active in more conditions, in contrast with a majority of L-degree TFs which seem to be just sporadically expressed. Therefore, after applying the network methodology described above, we observe that regulatory proteins are more frequently found to be central (associated) as their connectivity increases, as plotted in Figure 6B. In summary, it seems that the diagnostic value of TFs increases with their connectivity, presumably as they integrate a larger fraction of the physiological signals.
Most marker TFs effectively re-wire the regulatory network
In order to further evaluate the relevance of the representative marker TFs presented in the previous section, which were derived from the analysis of the network of TF–TF interactions, we set to measure their effect over the transcriptional network. A way of doing this for any experimental condition is by monitoring the expression levels of target genes that are part of a marker’s regulon, provided the regulon contains a minimum number of genes. It must be stressed that this experiment uses an independent data set, i.e. the microarray expression values of target genes, which was not used to define the markers that are going to be validated. As explained in ‘Materials and Methods’ section, the expression level of randomly sampled regulons can be taken as a reference and those markers with regulon changes deviating from background expression will confirm their role as condition landmarks.
We are aware that this approach oversimplifies the regulatory network of E. coli, since combinatorial regulatory interactions, in which several TFs effectively control a single promoter, are frequent. In these cases, the regulatory effect of a given marker TF, which we are measuring, might be shadowed by the regulatory action of its regulatory partners. Indeed we find that regulatory proteins with large regulons, i.e. highly connected TFs from the H class defined above, which tend to have more regulatory partners, induce relatively smaller expression changes across their regulons than local TFs (see Supplementary Figure S2, R2 = 0.37, P-value = 0.0014).
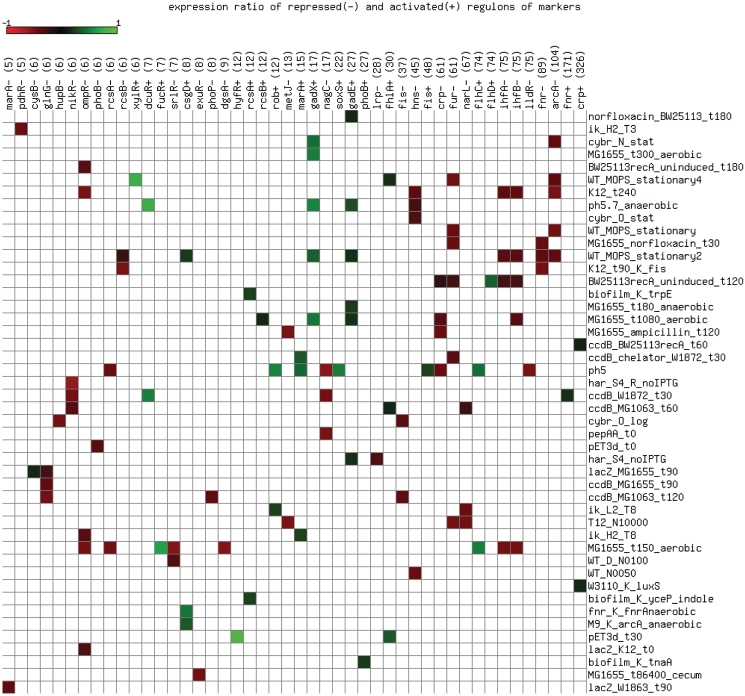
Despite these methodological drawbacks, out of 179 potential markers identified by means of centrality properties, as explained in the previous section, 141 have regulons with at least five target genes and could therefore be further evaluated by checking their regulon expression (Supplementary Table S3). In 107 cases (76%), significant regulon changes were observed, with a mean observed expression change of 37.4% and an SD of 10%. Figure 7 shows a heat map of these confirmed markers, dissecting the activated and repressed fraction of each regulon, which were considered independently.
Figure 7.
Heat map of regulon expression ratios for 141 marker transcription factors. Rows represent conditions, while columns describe activated (+) and repressed (–) regulons of markers, which are handled separately. Green cells highlight regulons over-expressed with respect to the background (0 to 1 scale), while red cells denote under-expressed target genes (0 to −1 scale). The size of each regulon is shown in parenthesis. Note that some markers are monomers of transcription factors which are active as protein dimers (RcsAB, IHF, FlhDC).
If we filter out markers with small regulon changes, on average there are two markers per condition, of which one is expected to be highly connected. These numbers illustrate that the network-based approach put to the test in this study might single out TFs (24%), which display significant changes in terms of network centrality but show little regulon expression changes. This might be caused by limitations of the approach or by the inherent noise in gene expression measurements, but the complexity of the transcriptional network, in which frequently several TFs co-regulate the same promoter, must also be included in the equation. Nevertheless, we found that the network methodology was successful in robustly identifying marker transcription factors in 46 experimental conditions and we find it remarkable that the expression state of a bacterium such as E. coli can be summarized by looking at, on average, just two or three transcription factors. This mean number of two markers per condition must be handled with caution, as some condition-specific regulatory networks were found to be effectively rewired by up to nine TFs (see, for instance, condition ph5 in Supplementary Table S3). Moreover, as explained in ‘Materials and Methods’ section, if we relax the significance level of the network parameters then it is possible to predict a larger set of markers in 61 conditions (Supplementary Table S4). If we apply the same validation protocol to this larger set of markers, on average six will be rewiring the network in each condition. While these analyses show that the thresholds on the parameters of the network-based approach have an effect on its performance, they also support that the number of TFs responsible for adapting the regulatory network to each condition is rather small.
CONCLUSIONS
In this study, we have used the publicly available gene expression data for the model bacterium, E. coli, to understand the dynamic properties of its regulatory network. This detailed analysis involved the identification of active set of TFs in each of the representative set of growth conditions and enabled us to address for the first time on a genomic scale the dynamic properties of TFs across a number of different environmental conditions. In particular, our analysis indicated that TFs are generally less expressed than other functional classes. The previous report that TFs regulating different number of genes are expressed at different levels (21,22) guided us to develop a TF-centric approach to identify the set of active TFs in each condition. We note that different conditions exhibit different number of TFs with an average of about 15% of the TFs per condition and a maximum of about one-third of the total TF repertoire identified in certain stress-induced conditions such as heatshock or translational burden and drug-induced conditions. These observations suggest that under stress and drug-induced conditions, organisms might express a higher proportion of TFs compared to their normal growth conditions to counter the challenges they are faced with. Our analysis also suggests that activators are generally more abundant than repressors across conditions, contrary to the expectations that bacterial promoters are mostly repressed and the observed higher number of repressors in the genome. It is possible to interpret from our analysis that only in certain stress and drug-induced conditions the proportion of repressors is higher than activators, indicating that in most conditions activators play a dominant role in controlling the expression of genes in bacteria.
To understand the dynamic nature of TFs and their association with different conditions, we studied the experimentally characterized set of regulatory interactions between TFs in the transcriptional network of E. coli by mapping it onto different experimental conditions. The network-based methodologies employed here unveiled a landscape in which the adaptation of bacterial populations to their environment could be monitored at the transcriptional level. The repertoire of experimental conditions tested could be mirrored by a repertoire of transcriptional subnetworks which, we suspect, reflected the ability of E. coli to survive in changing niches. The results presented suggest that the response to these changes can be mapped by using a rather small number of marker TFs that usually have a clear biological interpretation supported in the literature. For instance, our analysis could clearly predict the association of antibiotic resistance regulators such as marR and marA with drug-induced conditions or arcA and gadX with anaerobiosis associated conditions, suggesting the utility of the proposed method for identifying regulatory markers specific to different perturbations. Nevertheless, analysis of some conditions did not produce any markers, as it was found that a minimum value of subnetwork closeness is required for marker identification. Therefore, it is possible to suggest that closeness is a centrality measure of high interest for finding markers in transcriptional networks.
This study not only provides a venue for improving our understanding of the gene expression dynamics of TFs in bacteria but also allows us to apply the network-based approaches developed in this study to be used for studying other well-characterized systems for which there is abundant transcriptomic and network topology data available. In particular, we believe that the application of network parameters employed in this study to identify marker TFs can be a more general approach to study other kinds of cellular dynamic networks like those of protein–protein interactions or metabolic pathways, or even cell-type specific networks in higher eukaryotes, and hence has the potential for improving our ability to exploit the noisy expression and interactomic data for their meaningful interpretation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cambridge Commonwealth Trust (to S.C.J) Gobierno de Aragón to the research group of José María Lasa in 2010 (to B.C.M.). Funding for open access charge: MRC Laboratory of Molecular Biology (to S.C.J.)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rosa María Gutiérrez-Ríos, Cristhian Ávila-Sánchez, Miguel Ángel Ramírez, Heladia Salgado, Gabriel Moreno-Hagelsieb and Julio Collado-Vides for their feedback and fruitful discussions in the early stages of this work. We would also like to thank Guilhem Chalancon, Joseph Marsh, Nitish Mittal, Subhajyoti De, Tina Perica and Vladimir Espinosa-Angarica for critically reading the manuscript and providing helpful comments.
REFERENCES
- 1.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 2.Balazsi G, Oltvai ZN. Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals. Sci. STKE. 2005;2005:pe20. doi: 10.1126/stke.2822005pe20. [DOI] [PubMed] [Google Scholar]
- 3.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Janga SC, Salgado H, Martinez-Antonio A, Collado-Vides J. Coordination logic of the sensing machinery in the transcriptional regulatory network of Escherichia coli. Nucleic Acids Res. 2007;35:6963–6972. doi: 10.1093/nar/gkm743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgado H, Martinez-Antonio A, Janga SC. Conservation of transcriptional sensing systems in prokaryotes: a perspective from Escherichia coli. FEBS Lett. 2007;581:3499–3506. doi: 10.1016/j.febslet.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- 10.Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 11.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, Schneider SJ, Gardner TS. Many Microbe Microarrays Database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 2008;36:D866–D870. doi: 10.1093/nar/gkm815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez-Rios RM, Rosenblueth DA, Loza JA, Huerta AM, Glasner JD, Blattner FR, Collado-Vides J. Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janga SC, Collado-Vides J. Structure and evolution of gene regulatory networks in microbial genomes. Res. Microbiol. 2007;158:787–794. doi: 10.1016/j.resmic.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wall ME, Hlavacek WS, Savageau MA. Design of gene circuits: lessons from bacteria. Nat. Rev. Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 20.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. J. Mol. Biol. 2008;381:238–247. doi: 10.1016/j.jmb.2008.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J. Mol. Biol. 2008;379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seshasayee AS, Fraser GM, Babu MM, Luscombe NM. Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res. 2009;19:79–91. doi: 10.1101/gr.079715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandes U. A faster algorithm for betweenness centrality. J. Math. Sociol. 2001;25:163–177. [Google Scholar]
- 25.Janga SC, Salgado H, Collado-Vides J, Martinez-Antonio A. Internal versus external effector and transcription factor gene pairs differ in their relative chromosomal position in Escherichia coli. J. Mol. Biol. 2007;368:263–272. doi: 10.1016/j.jmb.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Gelfand MS. Evolution of transcriptional regulatory networks in microbial genomes. Curr. Opin. Struct. Biol. 2006;16:420–429. doi: 10.1016/j.sbi.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 28.Ma HW, Buer J, Zeng AP. Hierarchical structure and modules in the Escherichia coli transcriptional regulatory network revealed by a new top-down approach. BMC Bioinformatics. 2004;5:199. doi: 10.1186/1471-2105-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl Acad. Sci. USA. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 31.Droge P, Muller-Hill B. High local protein concentrations at promoters: strategies in prokaryotic and eukaryotic cells. Bioessays. 2001;23:179–183. doi: 10.1002/1521-1878(200102)23:2<179::AID-BIES1025>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Hao Z, Bieniek E, Calvo JM. Modulation of Lrp action in Escherichia coli by leucine: effects on non-specific binding of Lrp to DNA. J. Mol. Biol. 2001;314:1067–1075. doi: 10.1006/jmbi.2000.5209. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serres MH, Goswami S, Riley M. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 2004;32:D300–D302. doi: 10.1093/nar/gkh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janga SC, Salgado H, Martinez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009;37:3680–3688. doi: 10.1093/nar/gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang E, Purisima E. Network motifs are enriched with transcription factors whose transcripts have short half-lives. Trends Genet. 2005;21:492–495. doi: 10.1016/j.tig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nat. Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 39.Balazsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl Acad. Sci. USA. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez-Rios RM, Freyre-Gonzalez JA, Resendis O, Collado-Vides J, Saier M, Gosset G. Identification of regulatory network topological units coordinating the genome-wide transcriptional response to glucose in Escherichia coli. BMC Microbiol. 2007;7:53. doi: 10.1186/1471-2180-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen TE, Herrgard MJ, Liu M, Qiu Y, Glasner JD, Blattner FR, Palsson BO. Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets. J. Bacteriol. 2003;185:6392–6399. doi: 10.1128/JB.185.21.6392-6399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonomo J, Gill RT. Amino acid content of recombinant proteins influences the metabolic burden response. Biotechnol. Bioeng. 2005;90:116–126. doi: 10.1002/bit.20436. [DOI] [PubMed] [Google Scholar]
- 43.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 44.deHaseth PL, Nilsen TW. Molecular biology. When a part is as good as the whole. Science. 2004;303:1307–1308. doi: 10.1126/science.1095483. [DOI] [PubMed] [Google Scholar]
- 45.Madan Babu M, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
- 46.Moreno-Campuzano S, Janga SC, Perez-Rueda E. Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes–a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gralla JD, Collado-Vides J. In: Cellular and Molecular Biology: Escherichia coli and Salmonella, Chap. 79. 2nd edn. Neidhardt FC, Curtiss R III, Ingraham J, Lin ECC, Low KB, Magasanik B, Reznikoff W, Schaechter M, Umbarger HE, Riley M, editors. Washington, DC: American Society for Microbiology; 1996. pp. 1232–1245. [Google Scholar]
- 48.Perez-Rueda E, Collado-Vides J. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J. Mol. Evol. 2001;53:172–179. doi: 10.1007/s002390010207. [DOI] [PubMed] [Google Scholar]
- 49.Hershberg R, Margalit H. Co-evolution of transcription factors and their targets depends on mode of regulation. Genome Biol. 2006;7:R62. doi: 10.1186/gb-2006-7-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haddadin FT, Harcum SW. Transcriptome profiles for high-cell-density recombinant and wild-type Escherichia coli. Biotechnol. Bioeng. 2005;90:127–153. doi: 10.1002/bit.20340. [DOI] [PubMed] [Google Scholar]
- 51.Collado-Vides J, Magasanik B, Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 2005;187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J. Biol. Chem. 2005;280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 54.Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, Latour-Lambert P, Ghigo JM. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl Acad. Sci. USA. 2006;103:12558–12563. doi: 10.1073/pnas.0605399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sayed AK, Odom C, Foster JW. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology. 2007;153:2584–2592. doi: 10.1099/mic.0.2007/007005-0. [DOI] [PubMed] [Google Scholar]
- 56.Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009;5:e1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 59.Gambino L, Gracheck SJ, Miller PF. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 1993;175:2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tartaglia LA, Storz G, Ames BN. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 61.Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. BioEssays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Freyre-Gonzalez JA, Alonso-Pavon JA, Trevino-Quintanilla LG, Collado-Vides J. Functional architecture of Escherichia coli: new insights provided by a natural decomposition approach. Genome Biol. 2008;9:R154. doi: 10.1186/gb-2008-9-10-r154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.