Abstract
In this contribution, an electrochemical aptameric sensing scheme for the sensitive detection of small molecules is proposed using adenosine as a target model. A ferrocene (Fc)-functionalized thiolated aptamer probe is adapted and immobilized onto an electrode surface. Introducing a recognition site for EcoRI into the aptamer sequence not only suppresses the peak current corresponding to blank sample but also provides a signal-on response mechanism. In the absence of adenosine, the aptamer can fold into a hairpin structure and form a cleavable double-stranded region. Fc is capable of being removed from electrode surface by treatment with endonuclease, and almost no peak current is observed. The adenosine/aptamer binding induces the conformational transition of designed aptamer, dissociating the cleavable double-stranded segment. Therefore, the integrated aptamer sequence is maintained when exposing to endonuclease, generating a peak current of Fc. Utilizing the present sensing scheme, adenosine even at a low concentration can give a detectable current signal. Thus, a detection limit of 10−10 M and a linear response range from 3.74 × 10−9 to 3.74 × 10−5 M are achieved. The proposed proof-of-principle of a novel electrochemical sensing is expected to extend to establish various aptameric platforms for the analysis of a broad range of target molecules of interest.
INTRODUCTION
Designing sensors has become a focus of research because it can provide on-site, real-time detection and quantification of target analytes of interest for civilian, clinical and military applications (1). Toward this goal, the wide repertoire of recognition elements (e.g. antibodies) with desirable specificities and affinities are generated by natural and artificial immune systems for preparing biorecognition devices. However, those methods to produce the large recognition elements are often not applicable to the development of anti-small analyte ‘antibody’ because small analytes are toxic to the host animal or trigger a minimal immunogenic response (2). On the other hand, detecting and quantifying small target molecules such as drugs, hormones, adenosine and the corresponding derivatives are of fundamental importance in environmental analysis and clinical assay (3). Thus, demonstrating a desirable recognition element and developing a robust, simple and cost-effective screening device are of great value.
Aptamers are single-stranded DNA/RNA oligonucleotides with characteristic 3D structures artificially selected from synthesized random-sequence nucleic acid libraries by in vitro evolution process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment). Aptamers are able to bind their targets with high affinity and specificity (4), and they themselves by and large undergo the conformational transition that can be generally employed for designing analysis systems (5–7). Moreover, aptamers display numerous advantages such as simple synthesis, good stability, easy labeling and wide applicability. Thus, aptamers are becoming promising recognition elements for the sensor fabrication, disease diagnosis, protein analysis, new drug development and molecular switch design, etc. This type of versatile recognition elements brings new opportunities for the development of sensing systems for the small target detection.
Research efforts in the detection of small molecules have been fundamentally focused on the development of aptameric biosensing platforms with several transducing techniques, including fluorescent (8), electrochemical (9) and colorimetric (10) transducers. Occasionally, surface plasmon resonance (11), ion-selective field-effect transistor (12), capillary electrophoresis (13) and microfluidic platform (14) were applied to screen small target molecules. Albeit progress was accomplished, a poor detection limit is often achieved because of the relatively low-association constant of aptamers with their ligands. Therefore, developing target recognition schemes capable of amplifying signal readout is essential. For this purpose, several strategies have been introduced to amplify the aptamer/target interaction, such as autonomous aptamer-based machine (15), catalytic DNAzyme (16), rolling cycle amplification (RCA) (17) and additional oligonucleotide-based target binding amplification (3). Although these small molecule detection methods exhibit an improved performance, they suffer from the related shortcomings to some extent (e.g. the low-catalytic activity of DNA peroxidase, time-consuming and complex detection procedure).
Recently, the electrochemical detection method serving as a powerful detection approach has attracted increasing attention because of its numerous merits, including high sensitivity, low cost, small dimensions, rapid response, low-power requirements and compatibility with microfabrication technology (18). By and large, electrochemical aptameric assays are based on two signal transduction mechanisms: target binding-induced conformational change (aptasensor A) (19,20) and strand displacement originating from competitive binding of target molecules with complementary oligonucleotides for recognition elements (aptasensor B) (21,22). For example, Baker et al. (20) developed an electronic signal-on aptameric sensor for cocaine detection by self-assembling a methylene blue-tagged aptamer on a gold electrode. In the absence of target molecules, the aptamer remains partially unfolded, generating a low-peak current. Adding target onto the sensing interface makes the surface-confined aptamer change its conformation and fold into the cocaine-binding three-way junction, increasing the observed reduction peak. Zhang et al. (21) reported a sandwich electrochemical signal-off aptameric biosensor, capture DNA/adapted aptamer/reporter DNA-modified Au nanoparticles (nanoAu-probe). The redox-active moieties were bound to the modified electrode through electrostatic interactions. In the presence of target molecules, the nanoAu-probes can be displaced from the electrode surface because the aptamer is designed to exhibit a preference for target molecules, decreasing the peak current and creating a detectable electrochemical signal. Unfortunately, a high-blank peak current (peak current corresponding to a blank sample) and residual peak current are often encountered by aptasensor A and aptasensor B, respectively. This leads to a deteriorated assay capability. The improvement of electronic aptasensors for the highly sensitive detection of small target molecules remains a considerable challenge. Suppressing substantially the residual peak current of signal-off architecture or the blank peak current of signal-on biosensor should represent a potential breakthrough.
Adenosine as an endogenous nucleoside exhibits potential of vasodilator and anti-arrhythmic activities. This molecule performs crucial signaling functions in both the peripheral and central nervous system (23–26). As a ‘retaliatory metabolite’, adenosine can also equalize energy intake to metabolic demand (27). Additionally, there is good evidence that it has some important function in the immune system (28). Thus, the efficient detection of adenosine is of importance.
In this study, we developed a blank peak current-suppressed sensitive aptameric assay system for signal-on detection of small molecules. In this biosensor, adenosine and an adapted aptamer were used as the target model and recognition element, respectively. Via designing a restriction cleavage into the signaling process, this adenosine detection platform can afford a sensitive current response. A low-detection limit and wide linear response range were obtained. Additionally, high selectivity toward adenosine was observed. The signaling design and the preparation process of the proposed aptasensor were represented in detail, and analytical performance was evaluated by comparing with the existing screening systems. The measurement reproducibility and the feasibility for an adenosine assay were also verified.
MATERIALS AND METHODS
Chemicals
The designed aptamer, adenosine, cytidine, uridine, guanosine and EcoRI (15 U/μl) accompanied by 10× buffer were all obtained from Takara Biotechnology Co., Ltd. (Dalian China). The aptamer sequence was described as follows:
5′-HS-(CH2)6-GAATT CACAC CTGGG GGAGT ATTGC GGAGG AAGGT GAATT C-NH2-3′.
The underlined portion is the sequence of original anti-adenosine aptamer (9,11,21). The thiol and amine are covalently attached to the 5′- and 3′-ends, respectively. Ferrocene (Fc) monocarboxylic acid and N-hydroxysuccinimide (NHS) were obtained from Acros organics, while N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) was received from Sigma.
Two buffered solutions involved are 0.3 M buffer (50 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM MgCl2) and 0.1 M buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl,10 mM MgCl2 and 1 mM DTT). Unless otherwise stated, 0.1 M buffer was used to prepare the endonuclease solution while other buffered solutions mentioned in this work were 0.3 M buffer.
All other reagents were of analytical grade and used without further purification. Deionized and sterilized water (resistance >18 MΩ·cm) was used throughout the experiments.
Apparatus and electrochemical measurements
Electrochemical experiments were conducted on CHI 760B electrochemical workstation (Shanghai, China). A normal three-electrode configuration was involved, where the modified gold electrode was used as the working electrode while the platinum foil and a saturated calomel electrode (SCE) were as the counter electrode and reference electrode, respectively.
The differential pulse voltammogram (DPV) measurements were performed in 5 ml of 0.1 M NaClO4 solution. DPV parameters were detailed as follows: initial potential 0.6 V, final potential 0.1 V, incr potential 4 mV, pulse amplitude 50 mV, sample width 16.7 ms, pulse period 0.2 s, pulse width 0.05 s, quiet time 2 s, sensitivity 10−7 A/V. The electrochemical signal provided in this work was recorded by drawing a tangent between both sides of current peak. Impedance spectra were collected over a voltage frequency range of 1 to 100 000 Hz at an initial potential of 240 mV with the AC potential amplitude of ±5 mV. The supporting electrolyte was 10 mM PBS containing 0.1 M KCl and 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] redox couple.
Preparation of Fc-functionalized aptamer
The Fc-conjugated aptamer was prepared according to the literature method (9) with a minor modification. Briefly, 1 mg of Fc monocarboxylic acid was added to 1 ml of 0.3 M buffer containing EDC/NHS (0.1 M each) solution followed by being immediately mixed. After 100 μl of 7.8 μM aptamer was injected, the resulting solution was stirred at room temperature for 2 h. Subsequently, the mixture (Fc-aptamer solution) was stored in refrigerator at 4°C until use.
Fabrication of sensing interface
A gold electrode (1.0 mm diameter) received from Chenhua Instruments C. (Shanghai, China) was polished to a mirror smoothness successively with 0.3 and 0.05-µm alumina powder on a clean polishing microcloth and then was thoroughly washed with water. Subsequently, the electrode was cleaned by sonicating successively in water, absolute alcohol and water for 5 min each, followed by being incubated with piranha solution (3:1 of H2SO4:H2O2, warning: ‘piranha solution reacts violently with organic solvents’) for 15 min. After washing with water, the resulting electrode was electrochemically treated by cycling the potential between −0.3 and +1.55 V in 0.1 M H2SO4 until a cyclic voltammogram characteristic of a clean Au electrode was observed. Next, the cleaned gold electrode was dried in a stream of nitrogen followed by being washed with water.
The fabrication of electrochemical sensing interface is accomplished by thiol-gold self-assembly. Briefly, a 20-µl droplet of the Fc-aptamer solution was dropped onto the surface of freshly cleaned gold electrode that was held upside down and then was kept in a water-saturated atmosphere. The self-assembly reaction was kept for 120 min. Subsequently, the aptamer-functionalized modified gold electrode was exposed to a 1 mM mercaptohexanol (MCH) solution for 10 min. After the physically adsorbed molecules were removed from the electrode surface by washing with buffered solution, the sensing interface was ready for the adenosine detection.
Detection of adenosine sample and assessment of analytical characteristics
The sensing interface was immersed in target solution at specific concentration and was incubated at 45°C for 60 min. After the working electrode was allowed to cool to room temperature, 20 μl of endonuclease solution containing 0.75 U/μl EcoRI was placed onto the resulting electrode surface and was incubated at 37°C for 120 min. Subsequently, the treated electrode surface was washed with buffered solution prior to electrochemical measurements. The peak current observed at 0.29 V was used to estimate the amount of adenosine in sample and to assess the analytical characteristics of aptameric screening system.
RESULTS AND DISCUSSION
Design of electrochemical sensing scheme and improved signaling capability
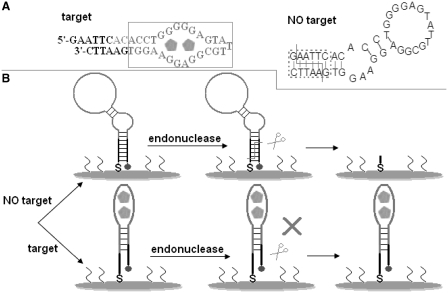
The target binding often causes the conformational change of aptamers. However, transferring the small analyte binding-induced limited conformational transition of recognition probe into a physically detectable signal remains a great challenge. In this contribution, the design of aptamer probe and the detection principle of electrochemical biosensor are shown in Scheme 1. The used aptamer was designed via lengthening the original anti-adenosine aptamer sequence by 8-base segment at the 5′-end and by 6-base segment at the 3′-end, and the secondary structure was predicted by the program ‘mfold’ (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi). As shown in the left panel of Scheme 1A, in the presence of adenosine, the designed aptamer can specifically bind to target molecule and form folded structure (called target recognition folding here) with a double-helix stem (30). However, in the absence of target molecules, the aptamer can fold into a hairpin structure (right panel of Scheme 1A) due to the hybridization of the two complementary arm sequences designed by our lab, forming a cleavable double-stranded segment by the enzyme EcoRI. For the synthesized aptamer-immobilized electrode (Scheme 1B), a high-peak current is observed in DPV because the aptamer is modified with the Fc and this terminal redox-active probe can approach the electrode surface due to the formation of a hairpin structure. When exposing the sensing interface without target to EcoRI, the endonuclease can cleave the double-stranded stem, and cleavage products, including Fc-attached fragments, can be removed from the surface by washing with buffered solution. Consequently, almost no peak current is obtained, suppressing the blank peak current. In contrast, introduction of target molecules induces the conformational change of aptamer probe on the electrode surface, deforming EcoRI recognition site. The endonuclease can not cleave the probe sequence in this case. Moreover, the Fc probe is still located close to the electrode to some extent in spite of the ligand-induced structural rearrangement. In this case, a high-peak current is observed, indicating a signal-on signaling scheme that is preferable to signal-off one (31). Additionally, a striking signal-to-noise ratio is also achieved. According to the classical signaling results (32), an assay system with a high-signal-to-noise ratio is predicted to offer an attractive analytical performance. Utilizing the present signaling scheme, a powerful sensing interface for the highly sensitive detection of adenosine is expected to be accomplished.
Scheme 1.
Design of anti-adenosine aptamer and the working principle of electrochemical biosensor. (A) Predicted secondary structure of the aptamer used in this study with (left panel) and without (right panel) target molecules. The sequence in the solid rectangle and that in the dotted rectangle are the original anti-adenosine aptamer sequence and the cleavable double-stranded region, respectively. The folded line indicates the cuts made by the enzyme EcoRI (29). (B) Schematic representation of the adenosine assay. For the upper half, the endonuclease cleaves the folded aptamer sequence with enzyme recognition site, and the redox-active species (Fc) is removed during washing the resulting electrode, almost completely eliminating the blank peak current. In contrast, for the lower half, the target binding deforms the EcoRI recognition site, conserving the integrity of the aptamer probe when exposing the electrode surface to endonuclease solution. Consequently, a peak current is observed. The details are seen in text.
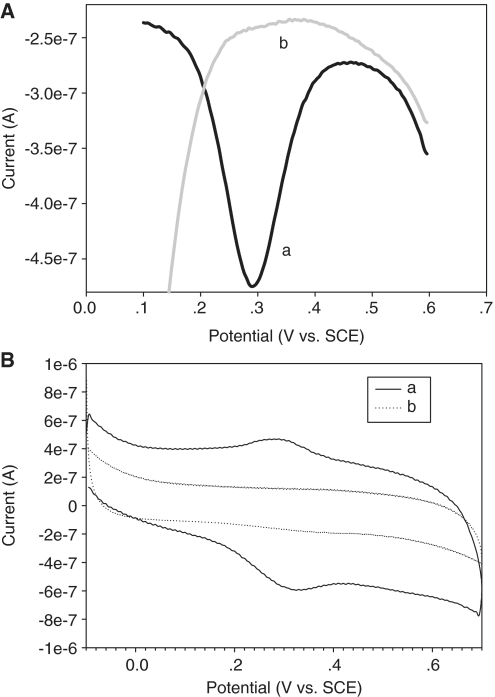
Figure 1 confirms the feasibility of electrochemical aptameric system for screening the target adenosine. As shown in Figure 1A, target sample causes a remarkable peak current (line a) that is almost equal to the peak current value of freshly prepared sensing interface, while Blank sample is incapable of inducing detectable peak current (line b). This observation is consistent with that seen from their corresponding cyclic voltammograms (presented in Figure 1B). The Fc-functionalized aptamer-modified electrode obtained shows a typical current signal in the presence of target molecule in comparison to the previous reports (19,22). Even though the geometric area (closely related to the amount of Fc-functionalized aptamer) of the working electrode involved is much smaller than the literature one (22), the prepared sensing interface can give a higher peak current. To some extent, this phenomenon should be attributed to a fact that the terminal Fc moiety is kept in the close vicinity of electrode surface due to the target binding induced head-to-tail folding of aptamer probe. Importantly, a nearly zero blank peak current and a signal-on signaling mechanism are simultaneously achieved in this aptameric sensor.
Figure 1.
Electrochemical response to target adenosine for the described aptameric sensing system. (A) The lines a and b indicate representative DPVs for the sensing interfaces with and without 374 µM adenosine, respectively. (B) The cyclic voltammograms a and b correspond to lines a and b in (A), respectively. The suppression of blank peak current makes the signal-to-noise ratio a large value, promoting the improvement in analytical characteristics of aptasensor.
Construction and impedance characterization of sensing interface
The immobilization of the aptamer probe onto an electrode surface is of critical importance in developing an effective detection system. There are certain universal requirements for a desirable aptameric sensing interface. First, the recognition aptamer must be strongly anchored onto the electrode surface for characterization and subsequent target detection. Second, a high-target-binding efficiency of the immobilized aptamer probe should be achieved. Third, the nonspecific adsorption process of target molecules can be eliminated. Furthermore, the proper strategy for the preparation of sensing interface should be simple and easy to control. The thiol-gold self-assembly technique has proven to be successful for the fabrication of DNA recognition probe-based biosensor (9,22). In the present work, the sufficient interstitial space between surface-confined aptamer sequences for adenosine binding is expected to be achieved when immobilizing aptamer probe onto a gold electrode via thiol-gold self-assembly technique since the designed aptamer can pre-fold into a hairpin structure under selected conditions while the target is a small molecule in comparison to aptamer probe. Although the nitrogen-containing nucleotide side chains could interact with the gold surface (33) and might result in a disordered self-assembled monolayer, treatment with thiol molecule (for example, MCH) can achieve a well-ordered mixed self-assemblyed monolayer of thiol molecule and aptamer probe, because less strongly adsorbed bases could be displaced by thiol groups possessing the stronger affinity for gold. Consequently, the performance of the aptameric sensing interface is predicted to be improved. In fact, the mixed self-assembly method has been used to prepare promising aptasensors (3,19). To meet the requirements mentioned above, the aptamer-based assay system reported in our work was fabricated by the mixed self-assembly technology (see ‘Materials and Methods’ for details). The surface coverage (Γap) of the Fc-modified aptamer confined on the gold electrode could be estimated from the amount of charge (Q) of the redox process of electroactive Fc during measuring cyclic voltammogram using the following formula: Γap = Q/nFA, in which n is number of electrons per mole of reaction, F is Faraday constant and A is electrode area. The total charge obtained from the anodic peak of Figure 1B line a is ∼l.7 × 10−7 C while he real surface of the gold electrode used was calculated as 62.5 × 10−3 cm2 from the corresponding charge for the reduction of the oxide monolayer. Assuming that all of the Fc labels adsorbed on the electrode surface were electrochemically active, the aptamer sequence surface coverage is about 2.8 × 10−11 mol/cm2 (1.7 × 1013 molecules/cm2) that exceeded the literature value (22). The possible reason for the high-surface density of aptamer probe is that the rigid aptamer with a hairpin-shaped structure easily forms a well-ordered monolayer on gold electrode. Additionally, because the electrochemical signal is induced exclusively by the aptamer/adenosine interaction that determines the DNA cleavage efficiency for this screening scheme, it is not difficult to achieve the high-detection specificity.
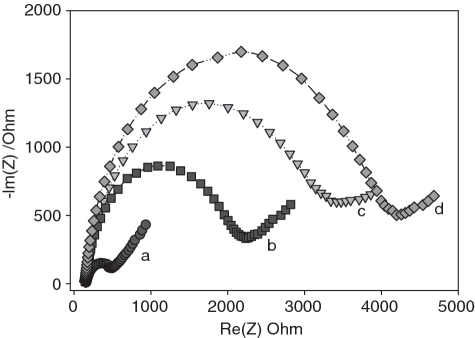
As an effective method to probe the interface properties of modified electrodes, impedance spectroscopy was used to evaluate the interfacial electron transfer efficiency at different stages of biosensor preparation as well as adenosine detection. Figure 2 displays Faradaic impedance spectra (presented as Nyquist plot) of the same electrode with different modified surfaces. The bare gold substrate exhibits a very small impedance (curve a), reflecting excellent electrochemical conductivity of the treated electrode. The self-assembly of aptamer probe onto the electrode surface results in a substantial increase in the electrochemical impedance (curve b), an indicator of the anchoring of thiolated molecules on the electrode surface. MCH treatment induces a further increase in the electron transfer impedance (curve c). This observation is in good agreement with previous reports (34): MCH can take up the residual active sites of gold surface and lift the aptamer molecules from the electrode surface. Due to electrostatic repulsion between the net negative dipole of the alcohol terminus on the gold surface and the negatively charged ferricyanide, the MCH submonolayer formed on the electrode surface acts as the blocking layer in electron transfer, hindering the diffusion of ferricyanide toward the electrode surface (34). After exposure to adenosine solution, the sensor shows a slight increase in impedance value (curve d). This was due to the fact that the target-aptamer binding brought about an additional negative charge on the electrode surface, which slightly prevented the electroactive probes from reaching the electrode interface. In this case, the accurate quantification of adenosine can not be accomplished because there is only a small impedance signal.
Figure 2.
Typical Nyquist diagrams obtained for the working gold electrode prepared at different stages and used for adenosine detection: (a) bared electrode; (b) aptamer-modified electrode; (c) aptamer/MCH-modified electrode; (d) adenosine-exposued electrode.
The measured data indicated that this designed aptamer can function as an active recognition element and the sensing interface for adenosine screening can be prepared via the present protocol. Nevertheless, target binding cannot trigger a substantial change in the electrochemical behavior of surface-confined biomolecule layer because target molecules are small molecules compared with recognition probes. Developing a versatile aptameric sensing scheme is necessary for the practical detection of small molecules.
Analytical performance of signal-on interrogating scheme
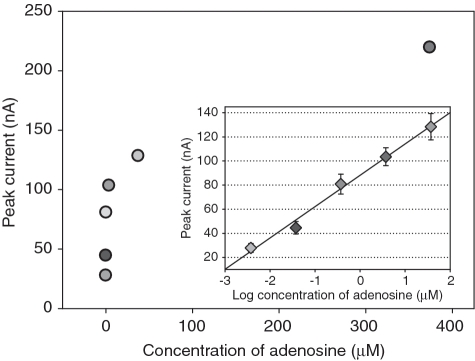
The experimental conditions are optimized as shown in Supplementary Data. To validate the utility of this aptameric sensor, a high concentration of target sample was prepared by dissolving adenosine in 0.3 M buffer and the low concentration of samples were obtained by diluting the high concentration of sample with the same buffer. The target samples at different concentrations gained were detected using the developed aptasensor, and the measured data are given in Figure 3. The peak current recorded for the present sensing interface increases with increasing the concentration of adenosine. Figure 3 Inset shows the representative calibration curve. There is a good linear relationship between the peak current value and logarithm of target concentration ranging from 3.74 × 10−9 to 3.74 × 10−5 M. The peak current ‘point’ corresponding to target adenosine at higher or lower concentration is beyond the linear response range. The average SD of at least triplicate determinations for each concentration of target sequence is 7.1%. The regression equation is Y = 26.00 logC + 88.14 with a correlation coefficient of 0.9905, where Y and C represent the value of peak current (nA) and target concentration (µM), respectively. The detection limit is 10−10 M, at which adenosine can trigger a peak current slightly higher than the Blank. Compared with colorimetric measurement (1), surface-enhanced Raman scattering method (35) and electrochemical technique with different transducing strategies (9,36), an improvement by at least two orders of magnitude in the detection limit was obtained, and the linear dynamic range was widened by >10-fold, demonstrating the excellent detection capability of the designed sensing scheme. Essentially, the intrinsic feature of a signaling scheme determines its analytical performance. Presumably, the detection limit lowered by several orders of magnitude should be attributed to the following two reasons: (i) the advantages, especially a high sensitivity, of electrochemical measurement over other detection techniques (18) that could contribute to the improvement of sensor performance; (ii) a large difference between peak currents in the absence and presence of target adenosine. Compared with electrochemical aptasensors reported in previous studies (9,36), the present interrogating strategy seems to be the only one that not only almost completely eliminates the blank peak current accompanied by a signal-on response mechanism (preferable to signal-off one) (31) but also gives a high-response current because the redox active probes (Fc) are designed to approach the electrode surface in the presence of target molecules. The achievement of unexpected detection ability suggests an opportunity to break through the bottleneck in the development of powerful aptameric assay system for small molecules and the ultrasensitive detection of target adenosine would be accomplished.
Figure 3.
Peak currents recorded in DPVs for the sensing interfaces upon addition of target adenosine at various concentrations. Inset: a calibration curve corresponding to the electrochemical detection of the variable concentrations of target molecules. The peak current is logarithmically related to target molecule concentration ranging form 3.74 nM to 37.4 µM. The error bars denote the SD of three measurements.
Measurement reproducibility
As an indispensable element in developing a promising sensing tool, the measurement reproducibility was further evaluated by intra- and inter-assay precision of peak current recorded in DPV. The intra-assay precision was estimated from four repetitive detections of each sample at the same electrode, while the inter-assay precision was from the measurements of the same sample at four different electrodes. All the sensing interfaces, including ones for the intra-assay measurements, were prepared according to the method described in the section of ‘Fabrication of sensing interface’. The unwanted elements on the used sensing interfaces can be removed during electrode treatment (for example, polishing with alumina powder and incubation in piranha solution). Three target samples at various concentrations (0.374, 3.74 and 37.4 µM) within the dose-response curve were involved. The maximum value of the relative SDs was 10.9% for intra-assay and 13.8% for inter-assay. The results are partly attributed to the change of the electrode positions and/or the difference of the surface areas from electrode to electrode.
Selectivity of sensing system and recovery test
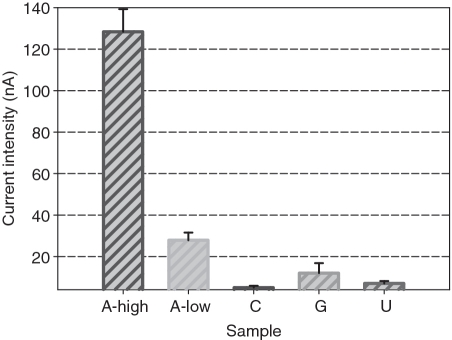
To validate the selectivity of the electrochemical aptameric sensor toward adenosine, the peak currents corresponding to cytidine, uridine and guanosine were evaluated because these molecules belonged to the nucleoside family and possessed a similar structure to adenosine (36). As shown in Figure 4, the intensity of peak currents upon the interferents is not >10% of the current value triggered by 37.4 µM target adenosine. Importantly, the peak current value corresponding to such high concentration of interferents is substantially low compared with that induced by 3.74 nM adenosine. Such high detection selectivity achieved in the present aptameric sensing system should be attributed to the recognition feature of integrated aptamer and high fidelity of EcoRI.
Figure 4.
Selectivity of the descripted electrochemical aptameric ensing system. A, C, U and G represent adenosine, cytidine, uridine and guanosine, respectively. The concentration of C, U and G are 3.5 µM. A-high and A-low indicate the current responses upon 37.4 µM and 3.74 nM adenosine, respectively. The detection experiments were conducted under identical conditions. The error bars represent the SD of three replicate electrochemical analyses.
To evaluate the applicability and reliability of the present assay system for adenosine small molecule, the recovery experiments of adenosine samples at different concentrations within the dynamic range were carried out. A certain amount of adenosine sample at a known concentration was added to a blank sample to make the final concentration of adenosine reach to 0.374, 3.74 or 37.4 µM. The target detections were carried out according to the general procedure, and the ‘Found’ concentration was estimated from corresponding DPV according to the regression equation. All the measurements were conducted four times, and the results are shown in Table 1. The recoveries afforded are in the range of 94–106%, and the average relative SD is ∼7.2%.
Table 1.
Recovery of adenosine assay
| Sample | Added adenosine (µM) | Found adenosine (µM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 0.374 | 0.396 | 106 | 8.1 |
| 2 | 3.74 | 3.50 | 94 | 3.6 |
| 3 | 37.4 | 38.1 | 102 | 9.8 |
CONCLUSIONS
In this contribution, a new highly selective electrochemical aptameric sensing scheme for the highly sensitive detection of small molecule has been proposed, in which a cleavable restriction site for EcoRI is introdued into the anti-adenosine aptamer sequence. The sensing interface is prepared by assembling directly an Fc-tagged aptamer onto a gold electrode via the gold-thiol linkage. This surface-confined aptamer probe plays a dual role: specifically binding target molecule as well as transuding the binding event into a physically detectable signal. The developed aptameric system can not only provide a low-detection limit but also exhibit a wide linear response range. Moreover, the target analyte in samples can be selectively identified and quantified within the whole linear concentration range. This should originate from the designed aptamer probe based on the combination of the recognition properties of aptamer with the high efficiency of EcoRI. Additionally, the straightforward preparation of sensing interface makes this signaling protocol desirable. Because aptamers can essentially target any class of molecules (37,38), success in the present electrochemical signaling scheme is expected to promote the development of aptameric screening probes in a wide range of areas, including biomolecule analysis, clinical diagnostic tests, drug discovery and so on.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (grants No. 90817101, 20775023 and 20905022); ‘973’ National Basic Research Program of China (No. 2007CB310500); Science Commission of Hunan Province. Funding for open access charge: ‘973’ National Basic Research Program of China (No. 2007CB310500).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Liu J, Lu Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal. Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 2.Willner I, Zayats M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Nie Z, Xu XH, Shen Q, Deng C, Chen J, Yao S. A sensitive, label free electrochemical aptasensor for ATP detection. Talanta. 2009;78:954–958. doi: 10.1016/j.talanta.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Ho H-A, Leclerc M. Optical sensors based on hybrid aptamer/conjugated polymer complexes. J. Am. Chem. Soc. 2004;126:1384–1387. doi: 10.1021/ja037289f. [DOI] [PubMed] [Google Scholar]
- 5.Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. DNA aptamer folding on gold nanoparticles: from colloid chemistry to biosensors. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 6.Nutiu R, Li Y. Structure-switching signaling aptamers: Transducing molecular recognition into fluorescence signaling. Chem. Eur. J. 2004;10:1868–1876. doi: 10.1002/chem.200305470. [DOI] [PubMed] [Google Scholar]
- 7.Nutiu R, Li Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 8.Stojanovic MN, de Prada P, Landry DW. Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc. 2000;122:11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 9.Wu ZS, Guo MM, Zhang SB, Chen CR, Jiang JH, Shen GL, Yu RQ. Reusable electrochemical sensing platform for highly sensitive detection of small molecules based on structure-switching signaling aptamers. Anal. Chem. 2007;79:2933–2939. doi: 10.1021/ac0622936. [DOI] [PubMed] [Google Scholar]
- 10.Liu JW, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Zhou HS. Aptamer-based Au nanoparticles-enhanced surface plasmon resonance detection of small molecules. Anal. Chem. 2008;80:7174–7178. doi: 10.1021/ac801281c. [DOI] [PubMed] [Google Scholar]
- 12.Zayats M, Huang Y, Gill R, Ma CA, Willner I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006;128:13666–13667. doi: 10.1021/ja0651456. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Li XF, Le XC. Differentiation and detection of PDGF isomers and their receptors by tunable aptamer capillary electrophoresis. Anal. Chem. 2009;81:7795–7800. doi: 10.1021/ac901471w. [DOI] [PubMed] [Google Scholar]
- 14.Liu B-F, Ozaki M, Hisamoto H, Luo Q, Utsumi Y, Hattori T, Terabe S. Microfluidic chip toward cellular atp and atp-conjugated metabolic analysis with bioluminescence detection. Anal. Chem. 2005;77:573–578. doi: 10.1021/ac0490447. [DOI] [PubMed] [Google Scholar]
- 15.Shlyahovsky B, Li D, Weizmann Y, Nowarski R, Kotler M, Willner I. Spotlighting of cocaine by an autonomous aptamer-based machine. J. Am. Chem. Soc. 2007;129:3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- 16.Shlyahovsky DL, Elbaz J, Willner I. Amplified analysis of low molecular weight substrates or proteins by the self-assembly of DNAzyme-aptamer conjugates. J. Am. Chem. Soc. 2007;129:5804–5805. doi: 10.1021/ja070180d. [DOI] [PubMed] [Google Scholar]
- 17.Cho EJ, Yang L, Levy M, Ellington AD. Using a Deoxyribozyme Ligase and Rolling Circle Amplification To Detect a Non-nucleic Acid Analyte, ATP. J. Am. Chem. Soc. 2005;127:2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]
- 18.Radi AE, Sanches LA, Baldrich E, O’Sullivan CK. Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J. Am. Chem. Soc. 2006;128:117–124. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 19.Ferapontova EE, Olsen EM, Gothelf KV. An RNA aptamer-based electrochemical biosensor for detection of theophylline in serum. J. Am. Chem. Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 20.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An electronic, aptamer-based small molecule sensor for the rapid, reagentless detection of cocaine in adulterated samples and biological fluids. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Xia J, Li X. Electrochemical biosensor for detection of adenosine based on structure-switching aptamer and amplification with reporter probe DNA modified Au nanoparticles. Anal. Chem. 2008;80:8382–8388. doi: 10.1021/ac800857p. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, Li X, Zhang L, Yu P, Su L, Mao L. Readily regenerated aptamer-based electrochemical sensors with electrode-confined redox-labeled complementary DNA as probe. Anal. Chem. 2008;80:1883–1890. doi: 10.1021/ac7018014. [DOI] [PubMed] [Google Scholar]
- 23.Phillis JW. Adenosine in the control of the cerebral circulation. Cerebrovasc. Brain Metab. Rev. 1989;1:26–54. [PubMed] [Google Scholar]
- 24.Portellos M, Riva CE, Cranstoun SD, Petrig BL, Brucker AJ. Perivascular adenosine in the choroid most likely accounts for the increase in blood flow in this tissue. Invest. Ophthalmol. Vis. Sci. 1995;36:1904–1909. [PubMed] [Google Scholar]
- 25.McMillan MR, Burnstock G, Haworth SG., Br Vasodilatation of intrapulmonary arteries to P2 receptor nucleotides in normal and pulmonary hypertensive newborn piglets. J. Pharmacol. 1999;128:543–548. doi: 10.1038/sj.bjp.0702815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunwiddie TV, Masino SA. The role and regulation of adenosine in the central nervous system. Annu. Rev. Neurosci. 2001;24:31–55. doi: 10.1146/annurev.neuro.24.1.31. [DOI] [PubMed] [Google Scholar]
- 27.Newby AC. Adenosine and the concept of retaliatory metabolites. Trends Biol. Sci. 1984;9:42–44. [Google Scholar]
- 28. “Damping the flames: inflammation control mechanism determined by NIH researchers”, to be found under http://www.aarda.org/infocus_article.php?ID=12. [Google Scholar]
- 29.Bath J, Turberfield A. DNA nanomachines. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Zhou C, Fang X. Aptamer-based ATP assay using a luminescent light switching complex. Anal. Chem. 2005;77:3542–3546. doi: 10.1021/ac050165w. [DOI] [PubMed] [Google Scholar]
- 31.March G, Noël V, Piro B, Reisberg S, Pham M-C. Nanometric layers for direct, signal-on, selective, and sensitive electrochemical detection of oligonucleotides hybridization. J. Am. Chem. Soc. 2008;130:15752–15753. doi: 10.1021/ja8047255. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama S, Yan L, Sintim HO. Junction Probes - sequence specific detection of nucleic acids via template enhanced hybridization processes. J. Am. Chem. Soc. 2008;130:12560–12561. doi: 10.1021/ja803146f. [DOI] [PubMed] [Google Scholar]
- 33.Herne TM, Tarlov MJ. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 1997;1119:8916–8920. [Google Scholar]
- 34.Li B, Wang Y, Wei H, Dong S. Amplified electrochemical aptasensor taking AuNPs based sandwich sensing platform as a model. Biosens. Bioelectron. 2008;23:965–970. doi: 10.1016/j.bios.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Chen JW, Liu XP, Feng KJ, Liang Y, Jiang JH, Shen GL, Yu RQ. Detection of adenosine using surface-enhanced Raman scattering based on structure-switching signaling aptamer. Biosens. Bioelectron. 2008;24:66–71. doi: 10.1016/j.bios.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Wang F, Dong S. Methylene blue as an indicator for sensitive electrochemical detection of adenosine based on aptamer switch. J. Electroanal. Chem. 2009;626:1–5. [Google Scholar]
- 37.Liu J, Lee JH, Lu Y. Quantum dot encoding of aptamer-linked nanostructures for one-pot simultaneous detection of multiple analytes. Anal. Chem. 2007;79:4120–4125. doi: 10.1021/ac070055k. [DOI] [PubMed] [Google Scholar]
- 38.Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan WH. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl Acad. Sci. USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.