Abstract
The advances of next-generation sequencing technology have facilitated metagenomics research that attempts to determine directly the whole collection of genetic material within an environmental sample (i.e. the metagenome). Identification of genes directly from short reads has become an important yet challenging problem in annotating metagenomes, since the assembly of metagenomes is often not available. Gene predictors developed for whole genomes (e.g. Glimmer) and recently developed for metagenomic sequences (e.g. MetaGene) show a significant decrease in performance as the sequencing error rates increase, or as reads get shorter. We have developed a novel gene prediction method FragGeneScan, which combines sequencing error models and codon usages in a hidden Markov model to improve the prediction of protein-coding region in short reads. The performance of FragGeneScan was comparable to Glimmer and MetaGene for complete genomes. But for short reads, FragGeneScan consistently outperformed MetaGene (accuracy improved ∼62% for reads of 400 bases with 1% sequencing errors, and ∼18% for short reads of 100 bases that are error free). When applied to metagenomes, FragGeneScan recovered substantially more genes than MetaGene predicted (>90% of the genes identified by homology search), and many novel genes with no homologs in current protein sequence database.
INTRODUCTION
Microbes are ubiquitous in nature and co-exist with other organisms including humans, and play a critical role in sustaining biological and environmental cycles (1–5). As such, the analysis of microbial genomes is necessary for a better understanding of the functionality of the microbes and their interactions within the microbe community as well as with the environment or the host (6). Most microbes, however, are difficult to culture—previous studies have indicated that <1% of microbes in many environments can be cultivated (4,7,8). Environmental sequencing reads can be assigned into specific genomes and then assembled for further analysis. High-complexity communities that contain diverse species in a metagenomic sequencing project (thus low coverage reads for each composite genome), however, make this problem extremely challenging. In addition, the sequencing reads generated by next-generation sequencing (NGS) techniques have sequencing error rates of up to 3%, some of which can cause frameshifts and thus make the prediction of protein-coding regions even more difficult (9,10).
Identification of genes is one of the most important and challenging problems in whole-microbial genome-sequencing projects (11–13). In metagenomics, gene finding can provide the opportunity to elucidate the activities and interactions of genes within an environmental sample, from which the metabolic and signaling pathways specific to the environment can be reconstructed and identified (14). To date, only a few methods have been developed for gene prediction in metagenomic sequences (15–19). Most commonly, genes encoded by metagenomes have been identified by using homology-based methods such as BLASTX (20,21). Homology searches against known protein databases, however, cannot be used to predict novel genes, although discovering new genes is one of the most important aspects in metagenomics research. Alternatively, sequence conservation information can be utilized for prediction of novel protein-coding genes (17,22); for example, a Ka/Ks value of ∼1 for a group of similar sequences indicates that these sequences are under no selective pressure and hence unlikely to code for proteins. This way, novel families that have multiple members in a metagenomic dataset can be identified (22). The other straightforward solution to novel gene prediction in metagenomics is to use feature-based approaches such as probabilistic models to evaluate the probabilities of open reading frames (ORFs) being protein-coding regions (16,18,23), in a manner similar to conventional gene finding methods such as Glimmer and GeneMark (24–26).
Short read length and sequencing errors are two major issues that pose significant challenges to gene prediction: incomplete genes (gene fragments) are difficult to predict, and sequencing errors may cause frameshifts that further complicate gene prediction. The average length of genes in microorganisms is about 950 bp (16), which is much longer than the sequencing reads generated by NGS (27,28). Different NGS methods now produce sequencing reads of various length ranging from 35 bp (from Illumina/Solexa Genome Analyzers) to 400 bp (from Roche/454 sequencers) and have different error profiles (27). Sanger sequencers produce reads with an error rate of up to 1%, whereas 454 sequencers produce reads with an error rate of up to 3% (9,10). Illumina Genome Analyzer may produce reads that have high mismatch rates, especially when relatively long reads are acquired (e.g. G is mistaken as T, and in later cycles A, C and G are mistaken as T) (29). In 454 sequencing reads, sequencing errors tend to occur in the homopolymer regions, resulting in frequent insertions and deletions. It has been shown that ORF-based gene prediction methods are more substantially affected by sequencing errors that cause frameshifts (9). As a consequence, programs that are currently available for gene prediction from short reads show a significant decrease in their performance as the sequencing error rate increases. For example, a low sensitivity of 26–43% was observed with sequencing error rate of 2.8% (9).
We propose a probabilistic model combining sequencing error models and codon usages (Figure 1) to improve the accuracy in predicting protein-coding regions from environmental sequences. In a study of the effect of sequencing errors on metagenomic gene prediction (9), all tested gene predictors showed poor performance on short reads that have sequencing errors. The gene predictors tested include GeneMark (26), MetaGene (16), MetaGeneAnnotator (extended from MetaGene aiming to improve gene prediction for whole genomes; 23) and Orphelia (which uses machine-learning technique with features for codon usage, di-codon usage and translation initiation sites, and shows performance comparable with MetaGene for gene prediction in short reads; 30). Taking advantage of this benchmark study, here we focus on the comparison of our program FragGeneScan with Glimmer (24,25) and MetaGene, since they are commonly used in genomics and metagenomics studies, respectively (31–35).
Figure 1.
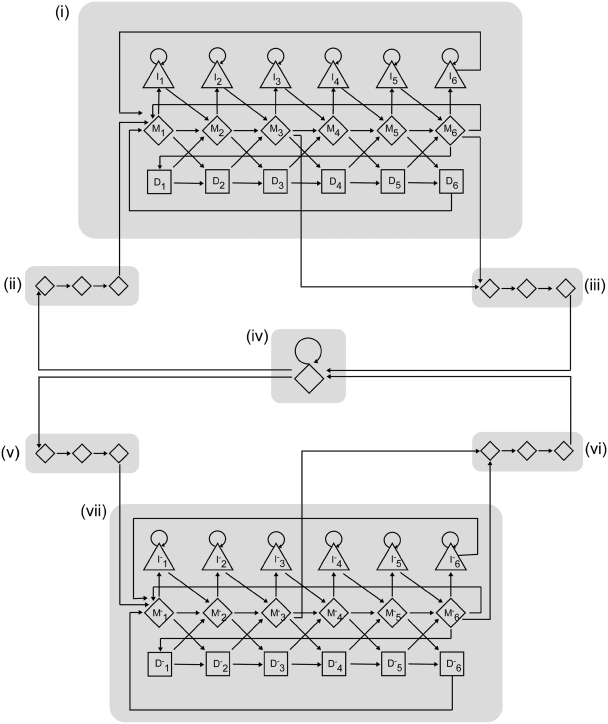
The HMM of FragGeneScan with seven super-states. The super-states are denoted as seven shaded boxes representing gene regions: (i) start codons (ii) and stop codons (iii) for both the forward (i–iii) and backward (v–vii) strands, and non-coding regions (iv). The states for gene regions (i and vii) consist of six consecutive match states represented by diamonds, insertion states by triangles and deletion states by squares, which collectively correspond to a six-periodic inhomogeneous HMM.
MATERIALS AND METHODS
Compared with classical microbial gene finding methods, FragGeneScan has two unique features. The first feature is finding genes fragmented by the boundary of given input sequences such as sequencing reads. The second feature, which is more important for gene prediction from reads generated by the current NGS methods, is correcting frameshifts caused by indel errors in reads. Even though a few recently developed methods (MetaGene and Orphelia) were designed to predict genes from short reads (thus can predict gene fragments), they do not provide a solution for predicting genes with frameshifts and genes in very short reads. We thus developed FragGeneScan to predict fragmented genes and genes with frameshifts in addition to the complete genes.
FragGeneScan algorithm
FragGeneScan is built on a hidden Markov model (HMM) (36), which incorporates codon usage bias, sequencing error models and start/stop codon patterns in a unified model. Given a short read (or a complete genome), the gene prediction problem is to find the best path of hidden states (see below) that is most likely to generate the observed nucleotide sequence, which can be solved by the Viterbi algorithm. FragGeneScan reports genes if they meet the following three conditions: (i) the length of the genes is longer than 60 bp, (ii) the genes start in a start state (start codon) or in a match state (internal region of genes) and (iii) the genes end in a stop state (stop codon) or in a match state (internal region of genes). Therefore, FragGeneScan can predict complete genes as well as partial (fragmented) genes without start and/or stop codons.
FragGeneScan HMM
FragGeneScan HMM consists of two-level representations based on data abstraction (Figure 1). In order to predict genes simultaneously from both strands, FragGeneScan considers separate states representing the gene regions in the forward strand and the reverse strand of a nucleotide sequence. The model has seven super-states (denoted as shaded boxes in Figure 1) representing gene regions (i), start codons (ii) and stop codons (iii) for the forward (i–iii) and backward (v–vii) strands, and non-coding regions (iv), respectively. The states for gene regions consist of six consecutive sets of a match state, an insertion state and a deletion state, which collectively correspond to a six-periodic inhomogeneous HMM. This representation allows using different parameters for each position in a di-codon (i.e. six nucleotides). Notably, by allowing transitions between the insertion/deletion states and the match states, this model effectively detects frameshifts that are caused by indel errors in sequencing. Considering that complete genomic sequences are unlikely to contain indel errors, the transition probabilities to insertion and deletion states are set to 0 when applying FragGeneScan to gene prediction in complete genomic sequences. Each match state in the gene regions [(i) and (v) in Figure 1] uses a second-order Markov chain to model the codon usage. The state for non-coding regions is based on a first-order Markov chain. Since the probability of gene regions and non-coding regions are calculated solely based on the composition of sequences (which is consistent regardless of the read length and gene length), our method is more robust when input sequences are of different lengths (see ‘Results’ section).
FragGeneScan also incorporates the sequence patterns for each start codon (ATG, GTG and TTG) and stop codon (TAA, TAG and TGA) in the start and stop state, respectively. The stop state is modeled by a probability distribution of the stop codons estimated from the training set: P(TAG|stop) = 0.54, P(TAA|stop) = 0.30 and P(TGA|stop) = 0.16. The model of the start states, on the other hand, takes into consideration the sequence pattern within a window surrounding the start codons. Notably, although the start codon model does not necessarily help the prediction of genes in short reads without a start codon, it can improve the gene prediction for complete genomes, as well as the short reads that contain a start codon.
Start codons in bacterial genomes are relatively difficult to predict because several putative start codons are often present around each of the real ones. In order to achieve accurate prediction of start codons, the probability of a start codon in the start states [(ii) and (v) in Figure 1] is modeled by using a positional weight matrix (PWM) over 63 nucleotides centered on a putative start codon ATG, GTG or TTG. In accordance with previous findings [A/T-rich region, Shine–Dalgarno sequence (AGGAG) (37), and triple-A downstream box] (38–40), the sequence patterns around the real start codons are different from those around false start codons that are present upstream or downstream of the real ones. We thus compute the following score for each putative start codon based on its 63 nt window (i.e. 61 overlapping trinucleotides).
 |
(1) |
where trinucleotidei represents the triplet at position i, and P(trinucleotidei|PWM) is the probability of observing the trinucleotide at position i, given the PWM of triplet frequencies, which was trained by using the same complete genomic sequences for HMM parameter estimation (see below). Two Gaussian distributions, one for real start codons and the other for false start codons, were fitted to the scores computed for a collection of annotated start codons (Supplementary Figure S1). For a putative start codon in a read, the probabilities (likelihood) of observing its 63-bp window given the condition that it is a real or false start codon, denoted as P(score|real) and P(score|false), respectively, can then be estimated from the two fitted Gaussian distributions. We calculate the posterior probability of a start codon being a real one given its surrounding 63-bp window, from P(score|real) and P(score|false) by using a naïve Bayesian classifier (41).
Parameter estimation for HMM
A total of 139 complete genomes (collected from the NCBI website; Supplementary Table S1) were used to estimate parameters of second-order Markov chains for all 12 match states in the forward strand [M1–M6 in Figure 1(i)] and in the reverse strand [M’1–M’6 in Figure 1(vii)] (see Supplementary Table S2 for a summary of the training data for different states). The parameters show linear correlation with GC contents, and therefore a linear regression (Supplementary Figure S2) was applied to give estimations of parameters for various GC contents. Note that FragGeneScan does not need training for gene prediction in individual genomes or datasets of short reads. Given a dataset of short reads, FragGeneScan estimates GC contents independently for each read and uses the corresponding set of pre-computed parameters based on the GC content for gene prediction in that read.
The parameters of emission and transition probabilities for insertion and deletion states were estimated for different sequencing methods with different error rates. The current version of FragGeneScan contains different sets of parameters for Sanger, 454 pyrosequencing and Illumina sequencing. Since the error rates directly affect the transition probabilities from match states to insertion/deletion states, we estimated parameters for four sequencing error rates: 0.5% and 1% for Sanger and Illumina sequencing, and 1% and 3% for 454 pyrosequencing, respectively. The sequencing reads used in the estimation were generated using MetaSim (10). If emission and transition probabilities of HMM are needed for error rates different from what we provided, they can be easily obtained and combined into existing models, which are separate from the gene prediction procedure.
Running time of FragGeneScan
The computational complexity of FragGeneScan is O(n), where n is the total length of the input genomic sequences. FragGeneScan is sufficiently fast for predicting genes for genome-wide annotation and metagenomics studies, achieving gene predictions for ∼2 Mb/min on an Intel Xeon CPU 2 GHz. The running time for all the tests shown in the paper ranges from 2 min (simulated reads from the Escherichia coli genome) to 58 minutes (the TS50 dataset).
Benchmark data sets
A total of nine complete genomes (with various GC contents) and their annotations were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/) (Table 1). (This set of genomes does not overlap with the genomes we used for training.) To systematically test FragGeneScan, reads of various lengths (100, 200, 400 and 700 bp) and with various sequencing error rates (0–3%) were simulated from these genomes using MetaSim (10). For each genome, up to 1-fold coverage of reads was sampled for each read length and sequencing error rate. Based on the current estimation of sequencing error rates (10), Sanger sequencing reads of 700 bp were simulated with the error rates ranging from 0% to 1%, and 454 sequencing reads were simulated with the error rates ranging from 0% to 3%.
Table 1.
Genomes of microbial species that were used to evaluate the performance of FragGeneScan
| Species | Gene Bank Acc. | CG (%) | Genome size (Mb) | No. of genes |
|---|---|---|---|---|
| Buchnera aphidicola str. APS | NC_002528 | 26 | 0.6 | 564 |
| Burkholderia pseudomallei K96243 chr1 | NC_006350 | 67 | 4.1 | 3399 |
| Bacillus subtilis subsp. subtilis str. 168 | NC_000964 | 43 | 4.2 | 4105 |
| Corynebacterium jeikeium K411 | NC_007164 | 61 | 2.5 | 2104 |
| Chlorobium tepidum TLS | NC_002932 | 56 | 2.2 | 2252 |
| Escherichia coli str. K-12 substr. MG1655 | NC_000913 | 50 | 4.6 | 4132 |
| Helicobacter pylori J99 | NC_000921 | 39 | 1.6 | 1489 |
| Prochlorococcus marinus str. MIT 9312 | NC_007577 | 31 | 1.7 | 1810 |
| Wolbachia endosymbiont str. TRS | NC_006833 | 34 | 1.1 | 805 |
Three real metagenomes were used for gene prediction in metagenomic sequences (Supplementary Table S3). Two real metagenomes (TS28 and TS50) from the twin obese and lean study (14) were downloaded from the MG-RAST website (http://metagenomics.nmpdr.org). The other real metagenome (SRX007415) from the rumen microbiota response study was downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov). These three metagenomes were BLASTXed against 98% non-redundant protein sequences from prokaryotic genomes, plasmids and phages collected from IMG 3.0 (http://img.jgi.doe.gov) using an E-value cutoff of 1.0e-3 for TS28 and TS50, and 1.0e-1 for SRX007415 (which has shorter reads), respectively. FragGeneScan gene prediction in these metagenomes was compared to the similarity search results.
Performance evaluation and comparison
The performance was measured in terms of sensitivity (the ratio of true positives to all annotated genes) and specificity (the ratio of true positives to all predicted genes). The accuracy was calculated by averaging the sensitivity and specificity. The performance of FragGeneScan was compared to that of Glimmer3 and MetaGene, which were downloaded from http://www.cbcb.umd.edu/software/glimmer/ and http://metagene.cb.k.u-tokyo.ac.jp/metagene/download.html, respectively. For training of Glimmer and MetaGene, we followed the standard procedures provided by the programs. Given a new genome, Glimmer uses its internal long-ORF scanner to predict long genes, which will be used for parameter estimation for the genome. MetaGene has its model parameters pre-trained.
Implementation and availability
FragGeneScan is implemented in C and Perl. In the training phase, we built Markov chains for the intergenic region and match states, and calculated emission and transition probabilities for the HMM. In the main module, the program predicts genes after identifying the best path of the hidden states by using the Viterbi algorithm implemented in C. The package of FragGeneScan includes all the programs, and does not require any other third party programs. FragGeneScan is available as open source at the FragGeneScan website, http://omics.informatics.indiana.edu/FragGeneScan/. All of our predictions are also available for download at the FragGeneScan website.
RESULTS
We tested and compared FragGeneScan with Glimmer and MetaGene by using the nine complete genomes (Table 1). We also tested FragGeneScan on short reads simulated from the same set of genomes, which allowed us to systematically evaluate the performance of FragGeneScan on the reads with various length and error rates. Finally, we tested and compared the performance of FragGeneScan with those of MetaGene and homology-search approach (BLASTX) on three real metagenomic datasets. The performances were measured in terms of sensitivity (the ratio of true positives to all annotated genes), specificity (the ratio of true positives to all predicted genes) and accuracy (the average of sensitivity and specificity). A gene fragment prediction is considered to be a true positive if it is of at least 60 bases (i.e. encoding 20 amino acids), and overlaps with ≥80% of the true protein-coding region in the read. We also compared the performances using different overlap criteria.
Evaluation on complete genomic sequences
The nine complete genomic sequences of various GC contents (Table 1) we used are also widely used for testing gene predictors in previous studies (16,18). Overall, the accuracy of FragGeneScan is comparable with MetaGene, and slightly higher than Glimmer (Table 2). In particular, FragGeneScan and Glimmer showed higher sensitivity, whereas MetaGene showed higher specificity on average. (The conclusion remains the same when a different overlap threshold of 50% was applied, or a perfect match was required; see Supplementary Table S4.)
Table 2.
Comparison of the gene prediction performances of different methods in complete genomic sequences
| Organisms | FragGeneScan |
Glimmer |
MetaGene |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sna | Spb | Accuracy | Sn | Sp | Accuracy | Sn | Sp | Accuracy | |
| B. aphidicola | 97.16 | 94.48 | 95.82 | 98.05 | 92.47 | 95.26 | 98.23 | 93.42 | 95.83 |
| B. pseudomallei | 94.26 | 88.21 | 91.24 | 93.06 | 77.48 | 85.27 | 95.73 | 92.23 | 93.98 |
| B. subtilis | 96.93 | 88.64 | 92.78 | 94.84 | 87.92 | 91.38 | 91.04 | 93.92 | 92.48 |
| C. jeikeium | 93.20 | 90.87 | 92.04 | 94.39 | 92.24 | 93.32 | 92.54 | 93.11 | 92.83 |
| C. tepidum | 82.55 | 90.11 | 86.33 | 83.61 | 89.02 | 86.32 | 80.37 | 92.73 | 86.55 |
| E. coli | 96.13 | 89.56 | 92.84 | 94.58 | 88.71 | 91.65 | 93.64 | 93.50 | 93.57 |
| H. pylori | 96.57 | 92.42 | 94.50 | 97.11 | 88.66 | 92.89 | 93.89 | 94.65 | 94.27 |
| P. marinus | 90.50 | 90.55 | 90.52 | 96.35 | 88.98 | 92.67 | 94.03 | 93.06 | 93.54 |
| W. endosymbiont | 94.53 | 69.18 | 81.86 | 91.30 | 48.58 | 69.94 | 91.06 | 73.67 | 82.36 |
| Average | 93.54 | 88.22 | 90.88 | 93.70 | 83.78 | 88.74 | 92.28 | 91.14 | 91.71 |
aSensitivity.
bSpecificity.
All three methods consistently showed the highest accuracy for Buchnera aphidicola and the lowest accuracy for Wolbachia endosymbiont. The low accuracy obtained for W. endosymbiont is due to very low specificities. This might be caused by the fewer number of genes in W. endosymbiont. However, Glimmer showed significantly lower specificity compared with the other two methods. We note that both FragGeneScan and MetaGene use generalized model parameters obtained by regression across genomes of different GC contents. But Glimmer uses specific model parameters trained from the genes in the testing genome. We thus suggest that more generalized parameter estimation may improve the performance in gene prediction for the cases when only insufficient data is available for training a specific model.
Evaluation on simulated sequencing reads
Table 3 shows the sensitivity, specificity and accuracy of gene prediction in simulated reads of 100 (Figure 2), 200 and 400 bp (Figure 3) with 1% sequencing error rate and 700 bp with 0.5% sequencing error rate. Prediction in longer reads shows higher accuracies with few exceptions (predictions in the 200 bp reads from B. aphidicola and Perkinsus marinus show higher accuracies than those in the 400 bp reads). Overall, FragGeneScan achieved 21–68% higher accuracies as compared to MetaGene. We note that FragGeneScan shows consistently high accuracy ranging from 63% to 89% for all the lengths we tested (100–700 bp); MetaGene, on the other hand, shows highly varied and lower accuracy ranging from 16% to 52%. Additional results on simulated reads with different sequencing error rates, and using different overlap criteria for defining true positive gene prediction, are listed in Supplementary Tables S5 and S6 (Supplementary Table S7 lists the proportion of simulated reads that contain annotated genes for each simulated dataset.)
Table 3.
Gene prediction performance in short reads simulated from complete genomic sequences
| Organisms | Read length (bp)a | FragGeneScan |
MetaGene |
||||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy | ||
| B. aphidicola | 100 | 79.16 | 80.12 | 79.64 | 49.59 | 55.24 | 52.41 |
| 200 | 83.56 | 84.20 | 83.88 | 31.32 | 28.92 | 30.12 | |
| 400 | 84.75 | 81.58 | 83.16 | 17.63 | 13.73 | 15.68 | |
| 700 | 89.92 | 74.64 | 82.28 | 45.89 | 32.42 | 39.16 | |
| B. pseudomallei | 100 | 75.79 | 64.78 | 70.28 | 18.64 | 49.63 | 34.14 |
| 200 | 86.56 | 78.01 | 82.29 | 46.97 | 43.86 | 45.41 | |
| 400 | 90.40 | 82.57 | 86.48 | 31.03 | 25.91 | 28.47 | |
| 700 | 91.57 | 82.50 | 87.04 | 54.42 | 42.10 | 48.26 | |
| B. subtilis | 100 | 72.36 | 65.96 | 69.16 | 31.21 | 55.81 | 43.51 |
| 200 | 83.39 | 79.06 | 81.22 | 34.03 | 36.18 | 35.10 | |
| 400 | 88.24 | 83.51 | 85.88 | 19.83 | 19.25 | 19.54 | |
| 700 | 92.17 | 84.37 | 88.27 | 47.93 | 39.67 | 43.80 | |
| C. jeikeium | 100 | 75.46 | 71.04 | 73.25 | 33.30 | 60.11 | 46.71 |
| 200 | 83.75 | 80.93 | 82.34 | 39.65 | 39.27 | 39.46 | |
| 400 | 86.94 | 84.44 | 85.69 | 24.65 | 22.06 | 23.35 | |
| 700 | 90.21 | 85.72 | 87.97 | 49.81 | 39.14 | 44.47 | |
| C. tepidum | 100 | 73.45 | 65.20 | 69.33 | 28.90 | 58.64 | 43.77 |
| 200 | 81.54 | 77.22 | 79.38 | 40.41 | 40.71 | 40.56 | |
| 400 | 84.37 | 83.02 | 83.70 | 24.42 | 22.73 | 23.58 | |
| 700 | 86.51 | 85.86 | 86.19 | 49.33 | 42.55 | 45.94 | |
| E. coli | 100 | 75.24 | 65.99 | 70.62 | 31.33 | 57.64 | 44.48 |
| 200 | 85.78 | 78.52 | 82.15 | 39.78 | 37.85 | 38.81 | |
| 400 | 89.19 | 82.76 | 85.98 | 23.54 | 19.57 | 21.56 | |
| 700 | 92.86 | 84.19 | 88.53 | 50.97 | 38.26 | 44.62 | |
| H. pylori | 100 | 72.69 | 71.69 | 72.19 | 41.94 | 54.58 | 48.26 |
| 200 | 82.81 | 81.39 | 82.10 | 30.28 | 29.83 | 30.05 | |
| 400 | 84.34 | 78.25 | 81.29 | 17.68 | 15.64 | 16.66 | |
| 700 | 88.63 | 81.79 | 85.21 | 45.79 | 34.87 | 40.33 | |
| P. marinus | 100 | 73.30 | 75.05 | 74.16 | 45.45 | 57.01 | 51.23 |
| 200 | 80.00 | 81.39 | 80.69 | 32.04 | 31.01 | 31.52 | |
| 400 | 80.02 | 77.85 | 78.94 | 18.89 | 16.63 | 17.76 | |
| 700 | 86.63 | 82.35 | 84.49 | 47.27 | 36.51 | 41.89 | |
| W. endosymbiont | 100 | 70.71 | 55.90 | 63.30 | 38.83 | 45.39 | 42.11 |
| 200 | 77.56 | 60.10 | 68.83 | 33.23 | 26.81 | 30.02 | |
| 400 | 80.43 | 61.78 | 71.10 | 18.05 | 13.57 | 15.81 | |
| 700 | 86.66 | 61.16 | 73.91 | 47.90 | 31.11 | 39.51 | |
Figure 2.
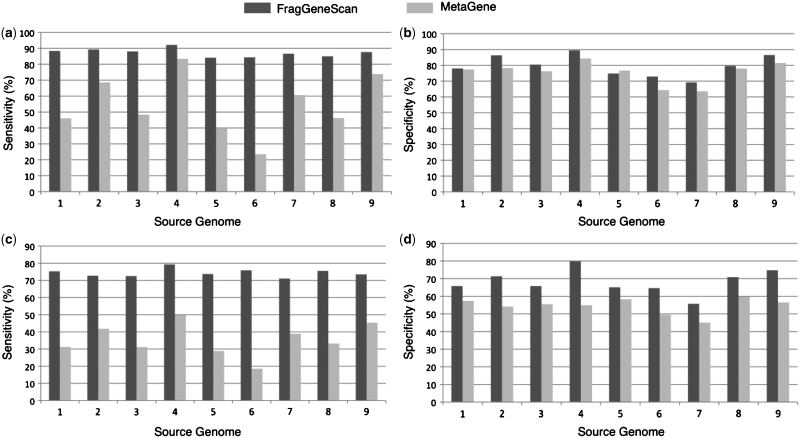
Gene prediction performance in simulated reads of 100 bases without sequencing error (a) and (b), and with 1% sequencing error (c) and (d). The x-axis denotes the source genomes from which the short reads were simulated: 1. E. coli; 2. H. pylori; 3. B. subtilis; 4. B. aphidicola; 5. C. tepidum; 6. B. pseudomallei; 7. W. endosymbiont; 8. C. jeikeium; 9. P. marinus. The y-axis denotes sensitivity in (a) and (c), and specificity in (b) and (d).
Figure 3.
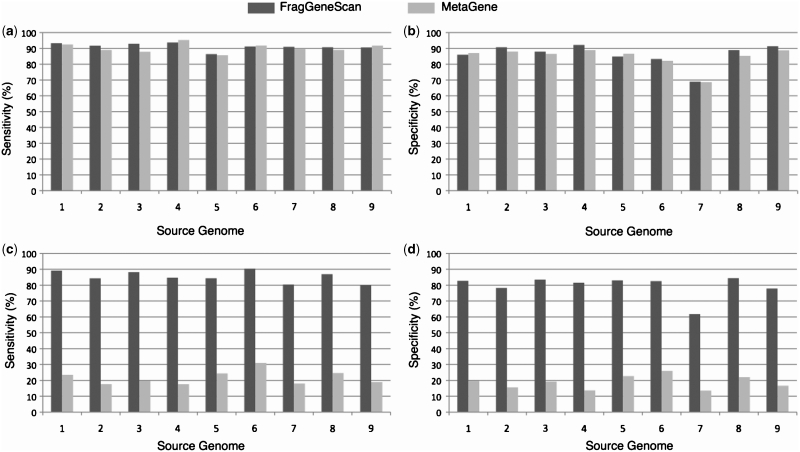
Gene prediction performance in simulated reads of 400 bases without sequencing error (a) and (b) and with 1% sequencing error rate (c) and (d). The x-axis denotes the source genomes from which the short reads were simulated: 1. E. coli; 2. H. pylori; 3. B. subtilis; 4. B. aphidicola; 5. C. tepidum; 6. B. pseudomallei; 7. W. endosymbiont; 8. C. jeikeium; 9. P. marinus. The y-axis denotes sensitivity in (a) and (c), and specificity in (b) and (d).
The performance of FragGeneScan as a function of read lengths and sequencing error rates is summarized in Table 4. For the 400 and 700 bp reads without sequencing error, FragGeneScan and MetaGene show comparable performance with <1% difference. For the shorter reads of 200 and 100 bps that have no sequencing errors, however, FragGeneScan outperforms MetaGene significantly. In particular, the accuracy of FragGeneScan in predicting genes in 100 bp reads without sequencing error is only 5% lower than those in longer reads (200 and 400 bp); in contrast, MetaGene shows 22% decrease in accuracy under the same condition. For all the cases studied with reads of various lengths and sequencing errors, a consistently better performance (up to 65%) was observed for FragGeneScan over MetaGene.
Table 4.
Average gene prediction performance in simulated Sanger and 454 reads
| Read length (bp) | Sequencing error rate | FragGeneScan |
MetaGene |
||||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy | ||
| 100 | 0.00 | 86.97 | 80.17 | 83.57 | 54.60 | 76.45 | 65.53 |
| 0.01 | 74.24 | 68.41 | 71.33 | 35.47 | 54.89 | 45.18 | |
| 0.03 | 57.75 | 56.63 | 57.19 | 19.04 | 34.57 | 26.81 | |
| 200 | 0.00 | 91.38 | 86.84 | 89.11 | 88.29 | 82.10 | 85.20 |
| 0.01 | 82.77 | 77.87 | 80.32 | 36.41 | 34.94 | 35.67 | |
| 0.03 | 69.19 | 67.86 | 68.53 | 13.17 | 15.78 | 14.48 | |
| 400 | 0.00 | 91.31 | 86.05 | 88.68 | 90.34 | 84.69 | 87.51 |
| 0.01 | 85.41 | 79.53 | 82.47 | 21.75 | 18.79 | 20.27 | |
| 0.03 | 72.88 | 69.87 | 71.38 | 5.89 | 7.01 | 6.45 | |
| 700 | 0.00 | 91.38 | 85.49 | 88.44 | 91.04 | 86.30 | 88.67 |
| 0.005 | 89.46 | 81.39 | 85.42 | 48.81 | 37.40 | 43.11 | |
| 0.01 | 86.72 | 78.24 | 82.48 | 31.18 | 23.30 | 27.24 | |
Evaluation on real metagenomes
We also tested FragGeneScan on real metagenomes generated by different sequencing platforms (Supplementary Table S3): two datasets TS28 and TS50 from the twin obese and lean study (sequenced by 454 sequencers) (14), and one dataset SRX007415 from the rumen microbiota response study (sequenced by Illumina sequencers). For these real metagenomes, there are no standard annotations available for comparison. So we compared genes predicted by FragGeneScan and MetaGene with those predicted by a homology-based approach (i.e. a read is considered to contain a protein-coding region if BLASTX finds its homologs in a protein database). Here, we consider a predicted gene as a true positive if the predicted gene overlaps with the entire length of the annotated gene from the BLASTX search (which, however, may not give the precise boundaries of the real gene).
For the TS28 dataset, FragGeneScan successfully predicted 92% of the genes identified by BLASTX search, whereas MetaGene predicted 47% of the genes (Table 5). For the TS50 dataset, FragGeneScan predicted 92% of the genes identified by BLASTX, whereas MetaGene predicted 69% of the genes. Note that MetaGene predicted significantly fewer genes, proportionally, in the TS28 dataset than in the TS50 dataset (with respect to BLASTX). In contrast, these ratios are roughly the same for both datasets for FragGeneScan (∼92%). (The comparison based on 50% gene overlap, i.e. the gene predicted by FragGeneScan or MetaGene overlaps with at least half of the BLAST annotated gene, is also shown in Table 5.)
Table 5.
Gene prediction results in metagenomes of TS28 and TS50 from the twin study and SRX007415 from the rumen microbiota study
| Metagenomes | Total reads | Average read length | Overlap of BLAST results (%) | FragGeneScan |
MetaGene | |
|---|---|---|---|---|---|---|
| With indel model | Without indel modela | |||||
| TS28 | 312 665 | 329 | 50 | 94.54 | 82.26 | 76.10 |
| 100 | 92.08 | 54.98 | 46.47 | |||
| TS50 | 622 554 | 200 | 50 | 93.31 | 88.38 | 81.20 |
| 100 | 92.25 | 81.35 | 68.69 | |||
| SRX007415 | 1 164 805 | 72 | 50 | 85.84 | 68.14 | –b |
| 100 | 85.80 | 68.10 | – | |||
aThis experiment was carried out to demonstrate how much improvement gene prediction gained by considering indels.
bMetaGene only works with reads that are of at least 100 bases.
FragGeneScan also predicted potentially novel genes that were missed by homology searches, including 28% (89 340 out of 317 440) of the putative genes predicted from the TS50 dataset, and 25% (142 007 out of 579 362) from the TS28 dataset. We note that BLASTX searches discovered protein-coding genes in ∼74% of the reads in both datasets (462 815 out of 622 554 reads in the TS50 dataset, and 231 946 out of 312 665 reads in TS28). Considering that on average 90% of the 200 bp simulated reads (which is similar to the read lengths of TS28 and TS50 data sets) contain annotated protein-coding genes (see Supplementary Table S7), and that ∼90% of bacterial genomes encode for proteins (42), the fraction of reads annotated as protein-coding regions by BLASTX searches on these two metagenomes (74%) is rather low. Our observation indicates the potential application of ab initio gene predictors such as FragGeneScan in the discovery of novel genes, which may constitute a significant proportion of protein-coding genes from an environmental sample.
The reads in SRX007415 are much shorter (of 72 bp) than those in TS28 and TS50. Considering that a BLASTX search of the original dataset (which has 4.2 Gb nucleotides) would require a drastic amount of CPU hours, we only carried out gene prediction for a small subset (2%), which has 1 164 805 reads, and compared the results with BLASTX results (MetaGene works only with input sequences of at least 100 bp, thus cannot be used for comparison). BLASTX search predicted protein-coding genes in 8% of the reads (87 431 out of 1 164 805) with E-values <1.0e-3. When a less stringent E-value cutoff was applied (1.0e-1), the ratio increased to 19%. Both ratios are extremely low, which may not be that surprising—it has been shown that the sensitivity of similarity search drops significantly when the reads become shorter (43). But it also indicates that for short reads, gene prediction based on homology search may severely underestimate the gene content in an environmental sample. FragGeneScan predicted 1 099 193 gene fragments in total, among which 189 875 gene fragments were also predicted by BLASTX (i.e. FragGeneScan predicted 86% of the potential genes obtained by BLASTX using E-value cutoff 1.0e-1). It is slightly lower than TS28 and TS50 datasets, which might be caused by the shorter length of the reads (72 bp).
Examples of genes that contain frameshift sequencing errors
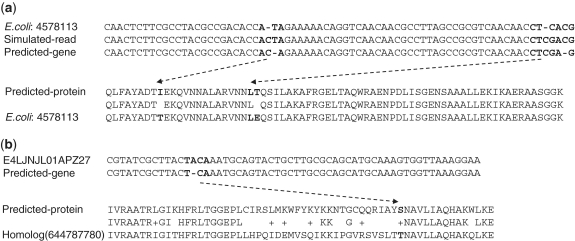
FragGeneScan integrates sequencing error models in its HMM so that it can predict genes broken by frameshift sequencing errors. Here, we show two examples of predicted genes that contain such sequencing errors (Figure 4) to demonstrate the importance of incorporating sequencing error models in gene prediction. Figure 4a shows a gene predicted by FragGeneScan from a simulated read, in which two frameshifts caused by sequencing errors were fixed. The read (r19) was simulated with two insertions of Cs from the E. coli genome (sequence from 4 578 113 to 4 578 339 bp) (Figure 4a). By adding insertion states near the positions of original sequencing errors—for example, FragGeneScan predicted the nucleotide sequence ACTA in the simulated read as ACA, which encodes Threonine, by annotating the T as an insertion—FragGeneScan predicted a gene (without fragmentation) that is almost identical to the annotated gene (see ‘Discussion’ section). Figure 4b shows another gene predicted in a real metagenomic read by FragGeneScan. From the read (E4LJNJL01APZ27) in the TS28 dataset, FragGeneScan predicted an incomplete gene with an insertion state (of nucleotide A, highlighted in Figure 4b). (MetaGene did not predict any gene from this read.) BLAST search of this predicted gene against the IMG 3.0 database resulted in a significant match (YP_002939026 from Eubacterium rectal ATCC 33 656 with an E-value of 2e-28), and the alignment of the predicted protein and the homolog is shown in Figure 4b.
Figure 4.
Examples of fragmented genes that contain frameshift sequencing errors: a gene predicted from a read simulated from the E. coli genome starting at position 4 578 113 (a), and a gene predicted from a metagenomic read from the TS28 dataset (b). The alignments of the nucleotide sequences are partially shown for clarity. The dotted lines connect the regions of nucleotides (with sequencing errors fixed) and the amino acid(s) that they encode. The alignment between the predicted protein from the metagenomic read and its homolog identified in IMG protein database is also shown.
DISCUSSION
Although FragGeneScan was intentionally developed for gene prediction in short and error-prone reads, it provides a versatile method to predict genes in complete microbial genomes, as well as in short reads with or without sequencing errors. The read lengths and sequencing error rates profoundly affect the performance of gene prediction methods. Sequencing errors that cause frameshifts are difficult to be detected by the ORF-based gene finding approaches such as MetaGene, resulting in cases where only fragments of true genes, if anything, are identified; and it may be difficult to interpret the gene fragments. FragGeneScan was developed to overcome the limitations of existing methods in addressing these two major issues, by incorporating sequencing error models into six-periodic inhomogeneous Markov models. FragGeneScan is robust with consistently high performance of predicting genes in reads with widely ranged sequencing error rates.
FragGeneScan is also less affected by the length of sequencing reads. FragGeneScan achieved consistently high gene prediction accuracies in reads of length 100–700 bp, whereas MetaGene showed significant variation in its performance. The robustness of FragGeneScan comes from its design principle: FragGeneScan uses only the probability of emitting a nucleotide at each position throughout the entire sequence (in contrast, existing gene prediction methods use statistical parameters such as the length distribution of genes). This robustness is essential for predicting genes in the reads generated by different NGS methods since each sequencing technique generates reads with different lengths (27).
We used rather stringent criteria that a predicted gene fragment is of at least 60 bp (i.e. 20 amino acids) and has 50–100% overlap with the true gene in the read to be considered as a true positive. In contrast, Hoff (9) used a rather loose definition of true positives: predicted genes that have a BLAT (44) alignment of at least 20 amino acids with the annotated gene and at least 80% sequence identity were called true positives. As a result, truncated gene fragments (due to frameshift-causing sequencing errors) may not be considered as true positives according to our criteria (because they are too short), but still may be considered as true positives according to Hoff’s criteria. So the accuracy reported in our article for MetaGene may be lower than the accuracy reported in Hoff’s paper (9). We want to emphasize that it is important to use stringent criteria for measuring the performance of gene predictors in error-prone short reads, as truncated gene fragments are more difficult to interpret and are less informative.
For gene prediction in Illumina reads (e.g. the SRX007415 dataset with reads of 72 bp), we used the same parameters as for Sanger reads, considering that Illumina sequencers do not show high sequencing errors in the homopolymer regions as 454 sequencers, and produce reads with overall low sequencing error rate. We can learn emission and transition probabilities of HMM for Illumina sequencing when more, longer Illumina reads (e.g. of 125 bp) become available.
The exact amino acids encoded by nucleotide sequences containing frameshift sequencing errors may be difficult to predict (for examples, see Figure 4). But these subtle mistakes in the predicted gene sequences (as long as the overall genes are predicted correctly) will not considerably affect many downstream analyses, such as the similarity search of the predicted genes. We will further explore the possibility of improving the prediction by incorporating the quality score of the reads.
FUNDING
Funding for open access charge: National Institutes of Health (1R01HG004908-02); National Science Foundation (CAREER award DBI-0845685).
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Chad Burrus for reading the article. We are grateful to the authors of MetaGene and Glimmer for providing their programs for comparison.
REFERENCES
- 1.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 2.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:reviews0003. doi: 10.1186/gb-2002-3-2-reviews0003. 0001–0003.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 5.Hattori M, Taylor TD. The human intestinal microbiome: a new frontier of human biology. DNA Res. 2009;16:1–12. doi: 10.1093/dnares/dsn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torsvik V, Øvreås L. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 8.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoff K. The effect of sequencing errors on metagenomic gene prediction. BMC Genomics. 2009;10:520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim – a sequencing simulator for genomics and metagenomics. PLoS ONE. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart AC, Osborne B, Read TD. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinformatics. 2009;25:962–963. doi: 10.1093/bioinformatics/btp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, et al. The comprehensive microbial resource. Nucleic Acids Res. 2001;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause L, Diaz NN, Bartels D, Edwards RA, Pühler A, Rohwer F, Meyer F, Stoye J. Finding novel genes in bacterial communities isolated from the environment. Bioinformatics. 2006;22:e281–e289. doi: 10.1093/bioinformatics/btl247. [DOI] [PubMed] [Google Scholar]
- 18.Hoff KJ, Lingner T, Meinicke P, Tech M. Orphelia: predicting genes in metagenomic sequencing reads. Nucleic Acids Res. 2009;37:W101–W105. doi: 10.1093/nar/gkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yooseph S, Li W, Sutton G. Gene identification and protein classification in microbial metagenomic sequence data via incremental clustering. BMC Bioinformatics. 2008;9:182. doi: 10.1186/1471-2105-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morozova O, Hirst M, Marra M. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genomics Hum. Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 28.Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 29.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoff KJ, Tech M, Lingner T, Daniel R, Morgenstern B, Meinicke P. Gene prediction in metagenomic fragments: a large scale machine learning approach. BMC Bioinformatics. 2008;9:217. doi: 10.1186/1471-2105-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legault B, Lopez-Lopez A, Alba-Casado J, Doolittle WF, Bolhuis H, Rodriguez-Valera F, Papke RT. Environmental genomics of “Haloquadratum walsbyi” in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics. 2006;7:171. doi: 10.1186/1471-2164-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma VK, Kumar N, Prakash T, Taylor TD. MetaBioME: a database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 2010;38:D468–D472. doi: 10.1093/nar/gkp1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl Acad. Sci. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klasson L, Westberg J, Sapountzis P, Näslund K, Lutnaes Y, Darby AC, Veneti Z, Chen L, Braig HR, Garrett R, et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl Acad. Sci. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 37.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starmer J, Stomp A, Vouk M, Bitzer D. Predicting Shine–Dalgarno sequence locations exposes genome annotation errors. PLoS Comput. Biol. 2006;2:e57. doi: 10.1371/journal.pcbi.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tech M, Morgenstern B, Meinicke P. TICO: a tool for postprocessing the predictions of prokaryotic translation initiation sites. Nucleic Acids Res. 2006;34:W588–W590. doi: 10.1093/nar/gkl313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu G-Q, Zheng X, Ju L-N, Zhu H, She Z-S. Computational evaluation of TIS annotation for prokaryotic genomes. BMC Bioinformatics. 2008;9:160. doi: 10.1186/1471-2105-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach. Learn. 1997;29:103–137. [Google Scholar]
- 42.Rogozin IB, Makarova KS, Natale DA, Spiridonov AN, Tatusov RL, Wolf YI, Yin J, Koonin EV. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 2002;30:4264–4271. doi: 10.1093/nar/gkf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl. Environ. Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.