Abstract
Structural information on RNA, emerging more and more as a major regulator in gene expression, dramatically lags behind compared with information on proteins. Although NMR spectroscopy has proven to be an excellent tool to solve RNA structures, it is hampered by the severe spectral resonances overlap found in RNA, limiting its use for large RNA molecules. Segmental isotope labeling of RNA or ligation of a chemically synthesized RNA containing modified nucleotides with an unmodified RNA fragment have proven to have high potential in overcoming current limitations in obtaining structural information on RNA. However, low yields, cumbersome preparations and sequence requirements have limited its broader application in structural biology. Here we present a fast and efficient approach to generate multiple segmentally labeled RNAs with virtually no sequence requirements with very high yields (up to 10-fold higher than previously reported). We expect this approach to open new avenues in structural biology of RNA.
INTRODUCTION
RNA has become widely recognized not only as protein coding information carrier but also as an important regulator of gene expression such as riboswitches, miRNAs or large non-coding RNAs (1). Although only 1.5% of the human genome encodes for proteins while 60–70% of it is transcribed into RNA, only 2% of the structures deposited in the Protein Data Bank account for RNA compared to 95% for proteins. NMR has proven to be a method of choice to solve RNA structures; however, large RNA structures are difficult to tackle due to the important spectral overlap observed in NMR spectra of RNA (2). Segmental isotopic labeling of RNA is therefore essential to study RNAs of moderate to large size by NMR spectroscopy, especially in combination with measurements of residual dipolar couplings (RDC) and paramagnetic relaxation enhancement (PRE) (3).
RNA segmental labeling can be also very useful for other biophysical techniques as site-specific incorporation of heavy atoms at internal positions within longer RNAs showed to have high potential for solving phases in X-ray crystallography (4,5). Similarly, single molecule experiments with RNA often require the incorporation of modified nucleotides at specific positions within a long RNA to study its structure or folding (6). Therefore, enzymatic ligation between a short synthetic RNA containing the modified nucleotides and longer fragments produced by in vitro transcription is expected to become the method of choice for studying biologically important RNAs (7). However, methods for incorporating modified nucleotides into internal positions of longer RNAs (>100 nt) for structural studies or segmental isotope labeling for NMR structural studies remain very time-consuming, costly as the yield they provide is low and not always applicable because of the sequence-dependence of most protocols (8,9) (see Supplementary Table S1).
Therefore, we have developed an alternative approach for segmental labeling of RNA. With this method, we can obtain very rapidly (5–7 days) high amounts (up to 10-fold higher than previously reported) of segmentally labeled RNAs without sequence requirement. The power of this method is demonstrated with a 72-nt non-coding RNA containing four stem-loops, for which we could obtain four distinct NMR samples in which only one of the four stem-loops is isotopically labeled. This could not have been achieved with current segmental labeling methods (Supplementary Table S1). For generating these four samples, only one labeled transcription reaction is required. We anticipate this method to become widely used in the emerging fields of structure determination of large biologically relevant RNAs by NMR and X-ray crystallography or of single molecule spectroscopy.
MATERIALS AND METHODS
Vector construction and plasmid amplification
The construct used for co-transcriptional cleavage of the RsmZ RNA is composed of a T7 promoter, an optimal transcription starting sequence (GGGAUC), an MS2 stem-loop structure (10), a HH ribozyme (11), the coding sequence for a truncated version (72 nt) of the RsmZ RNA of Pseudomonas fluorescence CHA0, a minimal sequence for recognizing the VS RNA in trans (12) and finally a BamHI restriction site for linearization (see Supplementary Figure S6). The MS2 stem-loop structure was included to extend the length of the fragment containing the HH ribozyme in order to obtain optimal separation from the RsmZ RNA during anion-exchange HPLC. To check that both HH and VS ribozyme are predicted to fold properly M-fold (13) was used to check the secondary structure of the whole transcribed RNA. The full construct was obtained by ligating an insert obtained by three consecutive PCR reactions into a pUC19 vector. To obtain the insert of around 300 nt we performed a first PCR reaction using only one overlapping primer pair and no template. After purification, this first PCR product was used as template for a second PCR reaction using primers extending on both ends. After a second purification, this extended primer PCR was repeated once by again using the product of the previous reaction as template for the next PCR to finally obtain the insert sequence of ∼300 nt. After sequencing, the plasmid was amplified by performing a large plasmid purification to obtain typically 5–8 mg of DNA. The plasmid was linearized using 0.2 U BamHI/μg of DNA for overnight digestion.
The DNA sequence coding for the VS ribozyme RNA (12) was obtained with the same extended primer PCR as described above and was also cloned into a pUC19 vector. After its amplification, the plasmid was linearized with HindIII restriction enzyme.
RNA purification
All RNAs were purified by anion-exchange chromatography on a preparative Dionex DNAPac PA-100 column (22 × 250 mm) at 85°C. Flow rate: 20 ml/min; eluent A: 12.5 mM Tris–HCl (pH = 8.0), 6 M urea; eluent B: 12.5 mM Tris–HCl (pH = 8.0), 0.5 M NaClO4, 6 M urea; detection at 260 nm; 30–75% B gradient within 18 min. Fractions containing the purified RNA were determined by 16% urea acrylamide gels and were liberated from urea and desalted by dialysis against water or by n-butanol extraction of the aqueous phase until RNA precipitation (14). The precipitate was resuspended in few hundred microliters of water and freeze-dried overnight. The lyophilized RNA was dissolved into an appropriate buffer.
RNA transcription and co-transcriptional ribozyme cleavage
Transcription yields and ribozyme cleavage efficiencies were optimized on 40 μl small scale reactions with changing concentrations of MgCl2, plasmid DNA, NTPs and T7 polymerase and testing the influence of the addition of pyrophosphatase and/or GMP. The best condition has been scaled-up to a large-scale reaction of 20 ml, which contained 42.5 mM MgCl2, 4.5 mM of each NTP, 33 ng/μl linearized HH ribozyme plasmid, 10 μM separately transcribed VS RNA and 1.7 μM in-house produced T7 Polymerase in a transcription buffer containing 40 mM Tris–HCl pH = 8.0, 1 mM spermidine, 0.01% Triton X-100 and 5 mM DTT. After 4–6 h of transcription it was sufficient to heat the reaction mix to 65°C for 15 min to complete the ribozyme cleavage. The reaction was stopped with the addition of 100 mM EDTA pH = 8.0. After 0.22 μm filtration the RNA was purified by anion-exchange HPLC followed by n-butanol extraction and lyophilization.
The labeled NTPs were either purchased from Spectra Stable Isotopes or prepared according to the protocol from Batey et al. (15). The NTPs were prepared by first extracting the nucleic acids with phenol/chloroform from 13C,15N-labeled Escherichia coli cultures. After precipitation with sodium acetate and isopropanol, they were hydrolyzed with S1 nuclease. The separation of NMPs and dNMPs was accomplished on a boronate affinity gel column. The NMPs were converted to NTPs by an enzymatic phosphorylation. The NTPs were not separated. Finally, they were desalted on an additional boronate affinity column.
Sequence-specific RNase H cleavage
Sequence-specific RNase H cleavage was performed by annealing a 2′-O-methyl-RNA/DNA chimera to the site of ligation (16). All chimeras used in this study are presented in Supplementary Figure S1. The chimeras were designed such that the DNA segment is flanked by two 2′-O-methyl-RNA sequences of 3–9 nt. Except for the cleavage in SL2 only one chimera had to be designed to get only specific cleavage. For the cleavage in SL2 a chimera flanked by only three 2′-O-methyl-RNA nt 5′ of the DNA segment yielded 30–40% unspecific cleavage occurring in SL4. Increasing the length of the 5′ 2′-O-methyl-RNA segment to 11 nt reduced the unspecific cleavage in SL4 to around 15% (see Supplementary Figures S1 and S3). The best conditions for cleavage were determined by small scale reactions (typically 500 pmol RNA in 15 μl reaction volume) mainly optimizing the RNase H enzyme concentration [NEB, or in-house produced (17)] and the ratio between the RNA and 2′-O-methyl-RNA/DNA chimera. The reaction temperature and the reaction time did not have a big influence on the cleavage efficiencies except for the cleavage in SL2, which has been run for 1 h at 4°C. All other reactions have been conducted for 1 h at 37°C. Most RNase H reactions could be performed using only 5% stoichiometric amount of 2′-O-methyl-RNA/DNA chimera. However, some reactions were less sensitive to unspecific cleavage when using stoichiometric amounts of chimera. For the large-scale reactions (20–200 nmol) the RNase H enzyme concentration had to be down-scaled 10 times compared to the small scale reaction to prevent potential unspecific cleavage. A typical reaction to cleave 200 nmol of RNA was performed in 6 ml volume containing 33 μM RNA, 1.65 μM chimera, 80 nM in-house produced RNase H in 50 mM Tris–HCl pH = 7.5, 100 mM NaCl and 10 mM MgCl2. The reactions were directly loaded onto the anion-exchange HPLC followed by n-butanol extraction and lyophilization.
RNA ligation using T4 RNA and DNA ligase
Non-splinted T4 RNA based ligations were first performed on small scale reactions (typically 400 pmol RNA fragments in 10 μl reaction volume) mainly by optimizing the T4 RNA enzyme concentration (NEB) and testing the addition of BSA, whereas splinted T4 DNA based small scale ligation reactions (typically 200 pmol RNA fragments in 20 μl reaction volume) were performed by optimizing the T4 DNA enzyme concentration (NEB, fermentas or in-house), the reaction time, the reaction temperature and testing the influence of PEG-4000. The DNA splints, which were added in a 1.5-fold excess compared to the RNA fragments, were annealed to the RNA fragments prior to ligation. An overview of all DNA splints used in this study is presented in Supplementary Figure S1. The best reaction conditions were scaled up for the large-scale reactions. A typical large-scale ligation reaction using T4 RNA ligase was 40 μM in both RNA fragments in 1× NEB ligation buffer (50 mM Tris–HCl pH = 7.8, 1 mM ATP, 10 mM MgCl2, 10 mM DTT), 1x in BSA using 5 U T4 RNA ligase per nmol of RNA to be ligated. The reaction was performed for 2 h at 37°C. A typical large-scale ligation reaction using T4 DNA ligase was 10 μM in RNA fragments, 15 μM in DNA splint oligo, 10% PEG-4000 in 40 mM Tris–HCl pH = 7.8, 0.5 mM ATP, 10 mM MgCl2, 10 mM DTT using 50 U T4 DNA ligase (fermentas) per nmol of RNA to be ligated or 2 μM final concentration of in-house produced T4 DNA ligase. The reaction was performed for 2 h at 37°C. The reactions were subjected to HPLC purification followed by n-butanol extraction and lyophilization.
NMR spectroscopy
The lyophilized RNAs were dissolved into 250 μl Buffer containing 10 mM sodium phosphate at pH = 6.0 containing 10% 2H2O with RNA concentrations of 0.2 mM. 1H-15N-hetero-nuclear single quantum correlation spectra (1H-15N-HSQC) were recorded at 10°C on a 600 or 700 MHz Bruker Avance spectrometer equipped with a cryoprobe.
RESULTS
Principle of the method
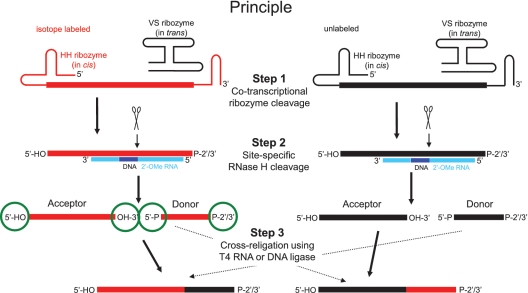
Our approach is based on the transcription of two full-length RNAs with identical sequence, one isotopically labeled and one unlabeled (Figure 1). The transcribed RNAs are flanked at the 5′-end by a hammerhead (HH) ribozyme in cis and at the 3′-end by the minimal sequence required by the Neurospora Varkud satellite (VS) ribozyme for cleavage in trans (11) (Step 1 in Figure 1). Both ribozymes (in cis or in trans) cleave co-transcriptionally leading to two homogenous termini, a 5′-hydroxyl and 2′/3′-cyclic phosphate for the full-length RNA. After purification, the two transcribed RNAs are subjected to site-specific RNase H cleavage with a guide 2′-O-methyl-RNA/ DNA splint yielding an acceptor fragment (5′-fragment) with two hydroxyl termini and a donor fragment (3′-fragment) with a phosphate at its 5′-end and a cyclic 2′/3′-phosphate at its 3′-end (18) (Step 2 in Figure 1). After separation of the two fragments of each cleavage reaction, the subsequent two cross-religations between the labeled fragment and the unlabeled fragment using T4 RNA or T4 DNA ligase results in two segmentally labeled RNAs, in which either the 5′- or the 3′-fragment is isotopically labeled (19) (Step 3 in Figure 1). Each reaction step is followed by a fast and efficient denaturing anion-exchange HPLC purification followed by a n-butanol extraction or a dialysis to get rid of urea and salts. For a two-piece ligation, 5–7 days are required in total, whereas 2–3 days are needed for Step 1 (1 day transcription optimization, 1–2 days large-scale transcription and purification), 1.5–2 days for Step 2 (0.5–1 day RNase H cleavage optimization, 1 day large-scale cleavage and purification) and 1.5–2 days for Step 3 (0.5–1 day ligation optimization, 1 day large-scale ligation and purification).
Figure 1.
Principle of the method comprising three reaction steps. The isotope labeled material is highlighted in red, the unlabeled material in black. In the 2′-O-methyl RNA/DNA chimera, the DNA is in dark blue and the 2′-O-methyl RNA in light blue. The termini of both acceptor and donor fragments are encircled in green. Scissors indicate RNase H cleavage sites. P-2′/3′ stands for a 2′/3′-cyclic phosphate.
One main advantage of this method compared to others is that we can obtain from only one labeled transcription reaction two homogenous fragments, which are correctly engineered at all four termini (Figure 1 after Step 2). This is essential to obtain the highest possible yields during the ligation step (Figure 1, Step 3) since no self-ligation or ligation in the wrong sequential order is possible and only the correct product can be obtained. Furthermore, there are no sequence restrictions on the identity of the fragments. A comparison of the principle of our method with the different published methods for segmental isotope labeling of RNA is shown in Supplementary Table S1. We will now demonstrate the practical feasibility of this method for a two-piece ligation by T4 RNA ligase and for a multiple ligation by T4 DNA ligase of a 72-nt ncRNA.
In vitro transcription, co-transcriptional ribozyme cleavage and purification by denaturing anion-exchange HPLC (Step 1)
Below, we demonstrate the applicability of the method using a truncated version of the ncRNA RsmZ from Pseudomonas fluorescens CHA0 (72 nt). In vitro transcription by T7 RNA polymerase was performed from a linearized plasmid coding for the ncRNA RsmZ flanked at its 5′-end by a HH ribozyme in cis (which has no sequence requirements at the terminus of the RNA of interest, when placed at the 5′-end) and at its 3′-end by an RNA sequence that can be cleaved by a VS ribozyme added in trans (11) (Figure 2a). The VS ribozyme has a very minimal sequence requirement since it will cut efficiently after any nucleotide other than a cytosine. If a cytosine would be required at the 3′-end of the RNA of interest, the hepatitis delta virus RNA ribozyme could be used as it has no sequence requirements (20). Both ribozymes cleave co-transcriptionally to a high extend and almost to completion after one thermal cycle (heating to 65°C for 10 min and subsequent cooling to 37°C for 1 h) without substantial degradation (Figure 2a). Thermal cycling ensures proper folding of the ribozyme, which is necessary for an efficient cleavage. The VS ribozyme in trans has been added either as a separate linearized plasmid (which serves as template for the VS RNA transcription) or as a separately transcribed and purified RNA. The latter approach has the advantage to save isotopically labeled NTPs that otherwise would be used to transcribe the VS ribozyme from the plasmid. The yield of the RsmZ ncRNA was about 2-fold higher if the VS ribozyme was added as a purified RNA rather than cotranscribed.
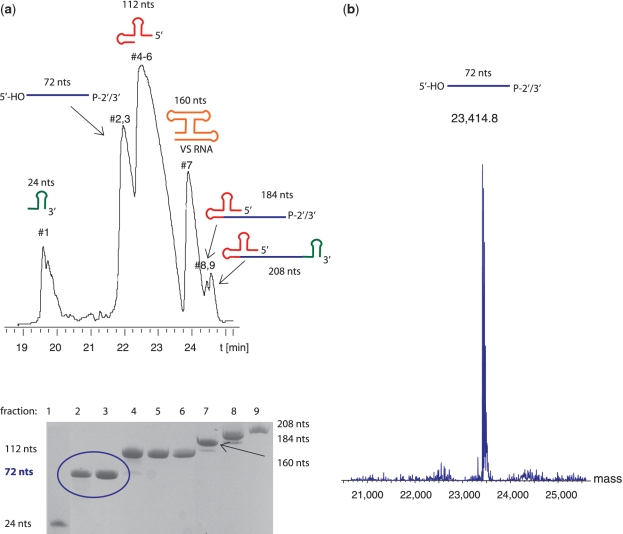
Figure 2.
Co-transcriptional ribozyme cleavage (Step 1) and its purification. (a) Denaturing anion-exchange HPLC profile of a 10 ml transcription mix (which corresponds to 200 nmol of the 72-nt RsmZ RNA product after purification) (top) and analytical 16% denaturing PAGE gel of the corresponding elution fractions (bottom). The different fragments obtained by co-transcriptional ribozyme cleavage are shown on the top of their corresponding peak (blue: target RsmZ RNA, red: hammerhead ribozyme, green: 24 nt VS stem-loop sequence required for VS ribozyme cleavage in trans, orange: VS ribozyme). The retention time of the HPLC profile is indicated on the x-axis. The purification conditions used are presented in the Materials and methods section. (b) ESI–MS spectrum of the homogenous unlabeled 72-nt RsmZ RNA with correct 5′- and 3′-termini. The measured mass is 23 414.8 daltons, whereas the calculated mass of the RsmZ RNA, which has a 5′-OH and 2′/3′-cyclic phosphate at its termini, is 23 415 daltons.
The three-step approach of the entire protocol (Figure 1) requires after each step (transcription, cleavage and religation) to separate and purify the different RNA fragments. The most commonly used purification method for large quantities of RNA needed for NMR spectroscopy is still polyacrylamide gel electrophoresis (PAGE) under urea denaturing conditions, which typically achieves single nucleotide resolution for RNAs up to 30 nt on a preparative scale (8,9). This procedure is laborious, suffers from low recovery yields and would be impractical for our three step segmental labeling procedure. We therefore use a fast purification method based on anion-exchange HPLC under denaturing conditions followed by n-butanol extraction or dialysis of the RNA to remove urea and salts. This approach has the advantage to allow the purification of large quantities of RNA within a few hours and to provide recovery yields >90%. To obtain optimal separation between the target RNA (72 nt) and the HH ribozyme (63 nt), we included a MS-2 stem-loop structure preceding the HH ribozyme to obtain a longer construct of 112 nt (see ‘Materials and methods’ section). We can easily separate the 72 nt target RNA from the different RNA products of the co-transcriptional ribozyme cleavage and of the VS ribozyme added in trans on a preparative scale (>200 nmol) (Figure 2a). We expect to be able to separate fragments differing in length by <5–10 nt (for RNAs up to 150 nt), when smaller reaction scales are purified. We confirmed that the purified RNA has correctly engineered ends and is properly folded using electrospray ionization mass spectrometry (ESI–MS) (Figure 2b) and NMR spectroscopy (Figure 4c), respectively.
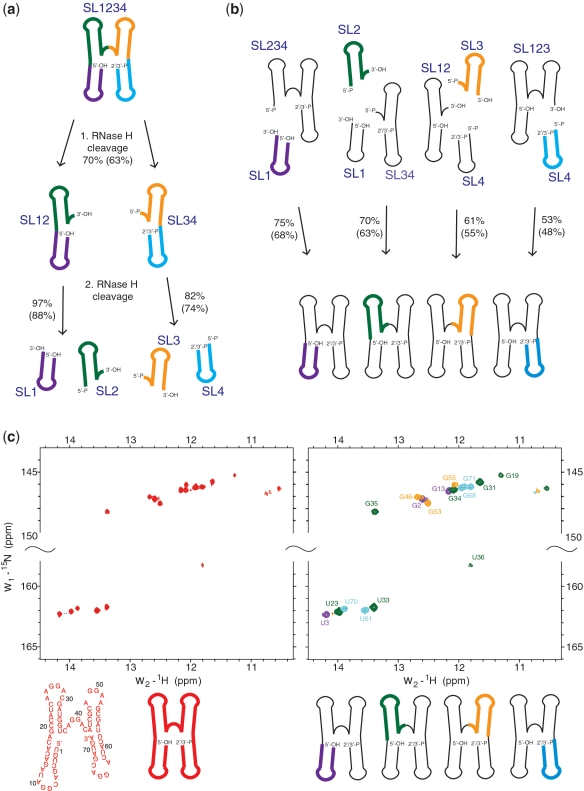
Figure 4.
Principle, reaction efficiencies and NMR evidence for isotope labeling of each stem-loop of the RsmZ RNA separately. (a) Sequence-specific RNase H cleavages to obtain all four isotopically labeled stem-loop fragments. The yields of the cleavage reactions before HPLC purification are indicated, the values in brackets are expressing the yield after purification. The different stem-loops are colored (SL1: magenta, SL2: green, SL3: orange, SL4: cyan). (b) Splinted T4 DNA ligase mediated ligations of isotope labeled (in color) and unlabeled (in black) fragments. The unlabeled fragments were obtained in a similar way as the labeled fragments. (c) NMR evidence for the successful segmental isotope labeling of each stem-loop separately. 1H-15N-HSQC NMR spectrum of the uniformly 15N-labeled RsmZ RNA (left) and overlay of the 1H-15N-HSQC NMR spectra of the four segmentally labeled RsmZ RNAs with each stem-loop labeled separately (right). The spectra were recorded on a Bruker 600 MHz spectrometer at 10°C.
With this procedure, we obtained for the first step of our protocol 400 nmol of unlabeled RsmZ ncRNA from a 20 ml transcription reaction (at a concentration of 4.5 mM for each NTP). When using in-house produced labeled NTPs, we obtained around 230 nmol of RNA from a 20 ml transcription reaction. The lower yield for the labeled RNA is due to the non-stochiometric amount of the four different NTPs. However, we obtained >100 nmol of labeled RNA from a 5 ml transcription reaction with commercially available 13C,15N-labeled NTPs.
Sequence specific RNase H cleavage (Step 2) and cross-religation using T4 RNA ligase (Step 3)
Step 2 of the protocol involves the cleavage of both the unlabeled and the labeled RNAs by RNase H. Here we will first demonstrate Step 2 and Step 3 of the method on a non-splinted two-piece ligation with T4 RNA ligase using only unlabeled material. In order to achieve sequence-specific cleavage, the RNA is hybridized with a 2′-O-methyl-RNA/DNA chimera composed mostly of 2′-O-methyl nucleotides interrupted by four deoxyribonucleotides (18,21) (Supplementary Figure S1). The cleavage site on the RNA target is the phosphodiester linkage opposite to the 5′-end of the DNA segment of the chimera (Supplementary Figure S1). In contrast to ribozyme and DNA enzyme mediated RNA cleavage, RNase H has the advantage to produce 5′-monophosphates and 3′-hydroxyls as termini (Figure 3b). After sequence-specific RNase H cleavage of an unlabeled and of a labeled RNA followed by a purification (Figure 3a), direct cross religation of the labeled and unlabeled cleaved products can therefore be done using T4 RNA or T4 DNA ligase (19) (Figure 3d). The fact that both donor and acceptor fragments have correctly engineered ends prevents either a self-ligation or a ligation between two fragments in the wrong sequential order which improves the yield.
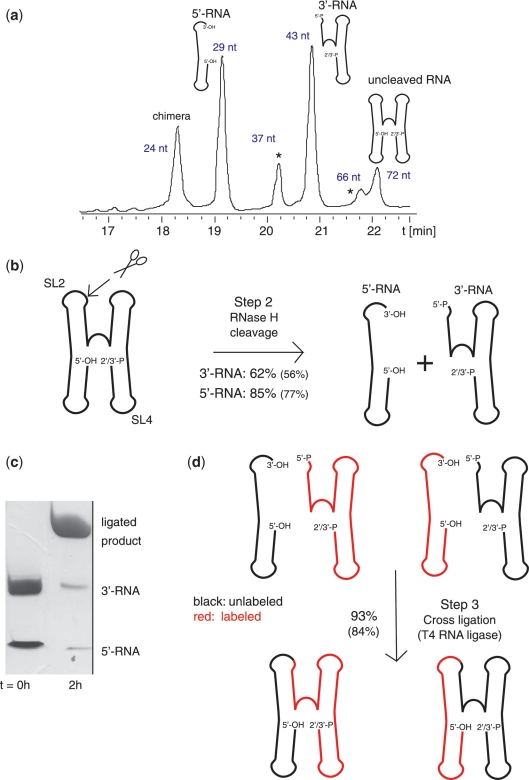
Figure 3.
RNase H cleavage (Step 2) and direct non-splinted cross-religation using T4 RNA ligase (Step 3). (a) Denaturing anion-exchange HPLC profile of fragments obtained by site-specific RNase H cleavage in SL2 between A29 and C30 of the RsmZ RNA (60 nmol reaction). The RNase H cleavage was performed with a chimera/RNA ratio of 0.75:1. The different fragments obtained by RNase H cleavage are shown on the top of their corresponding peak. Side-products occurring because of ‘unspecific’ cleavage in SL4 are marked by asterisks (see Supplementary Figure S3). The retention time of the HPLC profile is indicated on the y-axis. The purification conditions used are presented in the methods section. (b) Scheme of RNase H cleavage reaction and corresponding reaction yields. The yield of the cleavage reaction before HPLC purification is indicated, the values in brackets are expressing the yield after purification. The site of cleavage is shown by scissors. (c) Analytical 16% denaturing PAGE gel of the ligation reaction. Left lane: 400 pmol of each 5′-RNA (29 nt) and 3′-RNA (43 nt) before ligation, right lane: after ligation. (d) Reaction scheme and corresponding reaction yields for T4 RNA ligase mediated non-splinted cross-ligation of both a labeled (in red) and an unlabeled (in black) fragment, respectively. The ligation yield determined with a reaction using only unlabeled fragments is indicated.
We first chose to cleave and religate within the loop of SL2 of the ncRNA RsmZ between A29 and C30 (Figure 3b and d). After sequence-specific RNase H cleavage at this site, we obtained two homogenous fragments of 29 nt (5′-RNA) and 43 nt (3′-RNA) (Figure 3a) containing correctly engineered ends as confirmed by ESI–MS (Supplementary Figure S2). Because the sequence surrounding the cleavage site in SL2 is very similar to SL4, we observed also partial unspecific cleavage in SL4 leading to lower yields for the 3′-RNA (62%) compared to the 5′-RNA (85%) (Figure 3a, b and Supplementary Figure S3a, d). However, sequence-specific RNase H cleavage in SL2 was close to 100%, when a construct missing SL4 was used (Supplementary Figure S3b and c). We performed RNase H cleavage reactions at several sites on the full-length RNA and obtained no or only minor unspecific cleavage or degradation (Supplementary Figure S4). The cleavage efficiencies were between 70 and 97% (Figure 4a). Compared to other methods our approach requires 2′-O-methyl-RNA/DNA chimeras that might be costly if obtained commercially. However, it must be mentioned that for most RNase H cleavage reactions small amounts of chimera compared to the RNA one to be cleaved are sufficient. For example, in order to cleave between SL1 and SL2 or between SL3 and SL4, only 5% of chimera compared to the RNA substrate was needed to obtain complete cleavage.
After purification of the two RNA fragments cleaved by RNase H, the 3′-RNA fragment was ligated with the 5′-RNA fragment using T4 RNA ligase. The RNA ligation efficiency was close to 100% using only 500 Units T4 RNA ligase for ligating 100 nmol of RNA for 2 h at 37°C (Figure 3c). With such high yields for cleavage and ligation, we could potentially obtain by scaling up the transcription reactions to 20 ml up to 190 and 260 nmol for two segmentally labeled 72 nt RsmZ RNAs in only 5–7 days.
Successive RNase H cleavage followed by two- or three-piece ligations with T4 DNA ligase allows multiple segmental isotope labeling
We aim at understanding how the global regulatory protein RsmE is binding the ncRNA RsmZ, which contains several conserved binding consensus sequences ANGGA in loops of several RNA hairpins (22,23). However, the NMR study of the 72 nt at the 5′-end of this RNA in complex with RsmE has been hampered by the fact that all four stem-loops of this RNA contain very similar sequences leading to severe spectral overlap. We therefore require a method, which allows segmental isotope labeling of each SL separately and also allows to obtain yields high enough for NMR studies. In contrast to segmental isotope labeling methods of RNA published to date (Supplementary Table S1), our approach offers this possibility.
To this end, we therefore started with two transcriptions, one of 20 ml 15N-labeled and one of 60 ml unlabeled RsmZ RNA (Figure 2). Subsequently, we subjected the full-length 15N-labeled RNA to three sequence specific RNase H cleavages to separate the four labeled stem-loops (SL) (Figure 4a). First, we cleaved between SL2 and SL3 to obtain SL12 (40 nt) and SL34 (32 nt). After denaturing anion-exchange HPLC purification, the two fragments were cleaved separately by RNase H to obtain SL1 (16 nt), SL2 (24 nt), SL3 (18 nt) and SL4 (14 nt). Using our HPLC system, we could separate easily SL1 (16 nt) from SL2 (24 nt) (Supplementary Figure S4a) or SL3 (18 nt) from SL4 (14 nt) (Supplementary Figure S4b) generated during RNase H cleavage of SL12 (40 nt) or SL34 (32 nt) in preparative scales of >100 nmol, respectively. We expect to be able to separate also small RNA fragments (up to 30 nt) that differ in length by only 1 or 2 nt in a >100 nmol preparative scale. Although the chimera for RNase H cleavage must break down the secondary structures of both SL1 and SL2, we found that RNase H cleavage between SL1 and SL2 is close to 100% (88% yield after purification) (Figure 4a and Supplementary Figure S4). An overview of all cleavage efficiencies is shown in Figure 4a and all the chimera sequences used for sequence-specific RNase H cleavage is presented in Supplementary Figure S1. We could confirm by ESI–MS that all fragments are homogenous and contain the correctly engineered ends (Supplementary Figure S2). Similarly, we cleaved the unlabeled RNA into the necessary fragments required for the different cross-religations (Figure 4b).
Although ligation by T4 RNA ligase can be highly efficient like in SL2 (Figure 3c and d), T4 RNA ligation is only efficient when both donor and acceptor fragments can be brought into close proximity via base-pairing. Furthermore, T4 RNA ligation is highly sequence-dependent with preference for a purine at the acceptor 3′-end and a pyrimidine at the donor 5′-end (24). Therefore, ligation with T4 RNA ligase revealed to be impractical for our purpose. To our aim, ligation with T4 DNA ligase with the help of a splint oligo at the ligation site appeared to be the method of choice since no preformed secondary structure bringing the acceptor and donor fragments into proximity is required (24) and since this is sequence independent with only marginal structure-dependence on the ligation efficiency. Such ligations by T4 DNA ligase in presence of a splint oligo have been shown previously to work efficiently also for highly structured RNAs such as tRNA (25). Two two-piece ligations by T4 DNA ligase and splint DNAs were necessary to obtain RsmZ with SL1 or SL4 15N-labeled with 75 and 53% ligation efficiency, respectively. To obtain RsmZ molecules with the internal SL2 and SL3 15N-labeled, two three-piece ligations were necessary with 70 and 61% ligation efficiency, respectively. Note that for the three-piece ligation, only one reaction is required using the three pieces of RNA and the two DNA splints. With such high ligation efficiencies, yields between 90 and 150 nmol for each of the four segmentally labeled RNAs could be obtained from one 20 ml labeled and one 60 ml unlabeled transcription reactions within 7–9 days (Figure 4a and b).
NMR evidences of this successful segmental labeling strategy are shown in Figure 4c. The overlay of the four 1H-15N-HSQC spectra recorded with the segmentally labeled RNAs is identical to the spectrum of the uniformly 15N-labeled ncRNA RsmZ. This shows that the four segmentally labeled RNAs adopt the same fold as the uniformly labeled RNA (Figure 4c). As expected, the spectra show that with segmental labeling, we can drastically reduce the number of resonances observed in one spectrum of this 72 nt RNA. With this approach, NMR structural information (NOE, RDCs) from each individual stem-loop of this RNA can now be measured unambiguously, which will be essential for studying this RNA in complex with several RsmE protein dimers.
DISCUSSION
Although several methods for segmental isotope labeling of RNA have already been reported in the literature, the reported low yields (<25 nmol of segmentally labeled RNA were obtained from 20 ml transcription with the method of Lukavsky and coworkers (26,27) (see Supplementary Table S1 for a comparison of the different methods), or the sequence requirements at the sites of ligation [such as the need of a guanosine 3′ to the ligation site (27)], or the requirement that the different fragments to be ligated need to fold into a structure for efficient ligation (28), or the tedious purification steps by PAGE or the need to use additional enzymes such as T4 polynucleotide kinase for engineering the 3′- and 5′-termini (8) resulted in a so far very limited use of segmental isotope labeling in NMR studies of RNA (29).
We present here the principle of a fast, efficient and sequence-independent method for segmental labeling of RNA and its application on a 72 nt RNA that was isotopically labeled segmentally for NMR investigations. This method is based on a combination of co-transcriptional ribozyme cleavage, sequence-specific RNase H cleavage, cross-religation using either T4 RNA or T4 DNA ligase (Figure 1). Although our approach requires one more step (RNase H cleavage) compared to other methods (27,28), it allows to purify the RNA after each reaction step by fast denaturing anion-exchange HPLC, which makes this new approach much less time consuming (at least two to three times faster) and also provides much higher yields (up to 10-fold) compared with the most recently published methods (Supplementary Table S1). Using RNA ligation by T4 RNA or T4 DNA ligase, we can obtain, starting from only two 20-ml transcription reactions (one labeled and one unlabeled), between 190 and 260 nmol for two segmentally labeled RNAs, in which only one of the two fragments is isotopically labeled within just one week. It has to be mentioned that this high yield is barely sequence dependent compared to all methods published so far, in which the sequence of the different fragments has a large influence on the final yield (Supplementary Table S1). Most importantly, this method is flexible enough to allow not only to ligate two differentially labeled fragments but also to introduce any labeled fragment within an unlabeled RNA via three- or more-piece ligations. Within <10 days, we can produce four segmentally labeled RsmZ RNAs, in which only one of the four stem-loops embedded in this RNA is 15N-labeled at a time. Between 90 and 150 nmol can be obtained for these four RNAs using T4 DNA ligase for the ligation. Another important advantage of our method is that there are virtually no sequence requirements at the sites of ligation and that only two transcription reactions are needed, one with labeled NTPs and one with unlabeled NTPs. The method is flexible as any new ligation site can be introduced by just performing a new splint directed site-specific RNase H cleavage followed by cross-ligation, which prevents the need for recloning or even doing a new transcription reaction. Producing a new segmental labeled RNA sample with a different ligation site can therefore be performed within 2–4 days. Considering that RNase H cleavage and splinted T4 DNA ligation have been shown to be practical and efficient also for highly structured RNAs such as tRNA (16,25), our method is expected to be efficient for virtually any RNA.
We therefore expect our method to be applicable outside NMR spectroscopy. In the context of RNA structure determination by X-ray crystallography or of single molecule studies by fluorescence spectroscopy, our method could be used to incorporate short synthetic RNA fragments containing modifications for heavy atom binding at specific sites to help phasing (4,5) or for fluorescence labeling at defined positions (6), respectively (Supplementary Figure S5). Similarly, site-specific incorporation of modified nucleotides for attaching a paramagnetic tag to measure PRE by NMR (30) or for introducing a paramagnetic center for distance and dynamics measurements using EPR spectroscopy (31) could also be done using this method.
In summary, we present a fast and efficient method allowing segmental labeling of RNA sequence-independently at multiple sites with up to 10-fold higher yields than previously reported. The possibility to introduce via two- or more-piece ligations any labeled fragment within an otherwise unlabeled RNA in only 5–9 days makes us confident that this new approach will be widely used and could have a high impact in the field of structural biology of RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
SNF grant number (3100A0-118118); the SNF-NCCR Structural Biology. The Open Access publication charges for this article has been waived by Oxford University Press–NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Scott Stroebel (Department of Chemistry, Yale University, New Haven) and Kiyoshi Nagai (MRC-LMB, Cambridge, UK) for providing the plasmid coding for T4 DNA ligase and Luc Ponchon and Frédéric Dardel (Faculty of Pharmacy and Medicine, University Paris Descartes, Paris) for the plasmid encoding RNase H. The authors thank Peter Lukavsky (MRC-LMB, Cambridge, UK) for providing the protocol to make labeled NTPs. The authors also thank Serge Chesnov and Yolanda Auchli from the Functional Genomics Center Zurich for measuring mass spectra. The authors appreciate the help of Bouthaina Alila and Nana Diarra dit Konté in cloning and producing RNA. The authors thank Stefan Pitsch (EPFL Lausanne) for giving as access to the PhD thesis of Frédéric Meylan about splinted T4 DNA ligase meditated ligation. Finally, the authors acknowledge Pierre Barraud, Erich Michel and the whole Allain lab for helpful discussions.
REFERENCES
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 3.Tzakos AG, Grace CRR, Lukavsky PJ, Riek R. NMR techniques for very large proteins and RNAs in solution. Annu. Rev. Biophys. Biomol. Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 4.Hobartner C, Rieder R, Kreutz C, Puffer B, Lang K, Polonskaia A, Serganov A, Micura R. Syntheses of RNAs with up to 100 nucleotides containing site-specific 2'-methylseleno labels for use in X-ray crystallography. J. Am. Chem. Soc. 2005;127:12035–12045. doi: 10.1021/ja051694k. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Hobartner C, Polonskaia A, Phan AT, Wombacher R, Micura R, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama BM, Stone MD. Assembly of complex RNAs by splinted ligation. Methods Enzymol. 2009;469:27–46. doi: 10.1016/S0076-6879(09)69002-9. [DOI] [PubMed] [Google Scholar]
- 8.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2009;46:113–25. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dayie KT. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008;9:1214–1240. doi: 10.3390/ijms9071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batey RT, Kieft JS. Improved native affinity purification of RNA. RNA. 2007;13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferre-D'Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo HC, Collins RA. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cathala G, Brunel C. Use of n-butanol for efficient recovery of minute amounts of small RNA fragments and branched nucleotides from dilute solutions. Nucleic Acids Res. 1990;18:201. doi: 10.1093/nar/18.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- 16.Hayase Y, Inoue H, Ohtsuka E. Secondary structure in formylmethionine tRNA influences the site-directed cleavage of ribonuclease H using chimeric 2'-O-methyl oligodeoxyribonucleotides. Biochemistry. 1990;29:8793–8797. doi: 10.1021/bi00489a041. [DOI] [PubMed] [Google Scholar]
- 17.Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat. Protoc. 2009;4:947–959. doi: 10.1038/nprot.2009.67. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Hayase Y, Iwai S, Ohtsuka E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987;215:327–330. doi: 10.1016/0014-5793(87)80171-0. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Lapham J, Crothers DM. Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc. Natl Acad. Sci. USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapham J, Crothers DM. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA. 1996;2:289–296. [PMC free article] [PubMed] [Google Scholar]
- 22.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 24.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 25.Kurschat WC, Muller J, Wombacher R, Helm M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzakos AG, Easton LE, Lukavsky PJ. Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J. Am. Chem. Soc. 2006;128:13344–13345. doi: 10.1021/ja064807o. [DOI] [PubMed] [Google Scholar]
- 27.Tzakos AG, Easton LE, Lukavsky PJ. Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat. Protoc. 2007;2:2139–2147. doi: 10.1038/nprot.2007.306. [DOI] [PubMed] [Google Scholar]
- 28.Nelissen FH, van Gammeren AJ, Tessari M, Girard FC, Heus HA, Wijmenga SS. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008;36:e89. doi: 10.1093/nar/gkn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 30.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sowa GZ, Qin PZ. Site-directed Spin Labeling Studies on Nucleic Acid Structure and Dynamics. Prog. Nucleic Acid Res. Mol. Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.