Abstract
We had previously exploited a method for targeted DNA methylation in budding yeast to succeed in one-hybrid detection of methylation-dependent DNA–protein interactions. Based on this finding, we developed a yeast one-hybrid system to screen cDNA libraries for clones encoding methylated DNA-binding proteins. Concurrent use of two independent bait sequences in the same cell, or dual-bait system, effectively reduced false positive clones, which were derived from methylation-insensitive sequence-specific DNA-binding proteins. We applied the dual-bait system to screen cDNA libraries and demonstrated efficient isolation of clones for methylated DNA-binding proteins. This system would serve as a unique research tool for epigenetics.
INTRODUCTION
Methylation occurs at the C5-position of cytosine in genomic DNA of various eukaryotes, including plants, fungi and animals, each of which displays a characteristic pattern in genomic distribution of 5-methylcytosine (5mC) (1–3). While some organisms such as budding yeast Saccharomyces cerevisiae and nematode Caenorhabditis elegans are devoid of DNA methylation, it is well established that 5mC functions as a critical epigenetic mark in a variety of organisms, and its distribution pattern along with the genome, or the DNA methylome, constitutes a critical layer of epigenomic information. To understand the encoding mechanisms of this layer, it is essential to reveal how the cell targets DNA methylase/demethylase activities to particular genomic loci (4). To understand the decoding mechanisms, proteins recognizing methylated DNA have to be identified and analyzed (5).
Mammals have at least three distinct classes of methylated DNA-binding proteins, namely methy-CpG binding domain (MBD) proteins, Kaiso-related zinc finger proteins and SET and RING finger-associated (SRA) domain proteins (5). The MBD protein family includes MBD1, MBD2, MBD4 and MeCP2. Intriguingly, MBD4 has been implicated in demethylation, and defects of MeCP2 cause Rett syndrome, both indicating their critical roles. The Kaiso-related family comprises Kaiso, ZBTB4 and ZBTB38, each of which displays a sequence-specific interaction with methylated DNA. The SRA domain protein family includes UHRF1, which binds hemi-methylated DNA and recruits DNMT1 responsible for maintenance methylation. While MBD and SRA domain were found in plants, methylated DNA-binding proteins have not been rigorously explored in organisms other than mammals. It is conceivable that totally different classes of methylated DNA-binding proteins remain to be identified in any eukaryotic species. An efficient method for selective cloning of methylated DNA-binding proteins would accelerate their identification.
A powerful method to clone DNA-binding proteins is the yeast one-hybrid system (6). Taking advantage of the lack of endogenous DNA methylation in S. cerevisiae, we modified the system and successfully detected methylated DNA–protein interactions mediated by MBD1, MeCP2 and Kaiso (7). In this system, we integrated LexA operator (LexAop) at the 5′-flanking region of the bait sequence and expressed LexA-fused SssI CpG methylase (M.SssI), a bacterial de novo DNA methylase. Consequently, LexA-mediated recruitment of M.SssI led to targeted methylation of the bait sequence, to which methylated DNA-binding proteins fused with Gal4 or VP16 activation domain bound and activated the expression of lacZ reporter gene. These results suggested a possible use of the one-hybrid system in screening for methylated DNA-binding proteins. In this study, we pursued the possibility and developed a one-hybrid system to screen cDNA libraries for clones encoding proteins that recognize methylated DNA.
MATERIALS AND METHODS
Construction of strains for one-hybrid screening
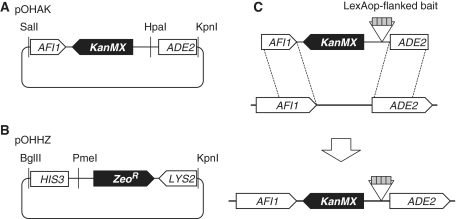
We constructed two integration vectors pOHAK and pOHHZ to generate ADE2 and HIS3 reporter genes, respectively (Figure 1). We amplified and cloned a DNA fragment spanning AFI1-GAL2UAS-GAL2TATA-ADE2 from PJ69-2A (Clontech), which contains the 3′-most 350 nt of AFI1 and the 5′-most 455 nt of ADE2, to pT7-Blue (Novagen) and replaced the GAL2UAS with a fragment containing KanMX and an HpaI site to obtain pOHAK. Similarly, we cloned a fragment spanning LYS2-GAL1UAS-GAL1TATA-HIS3 containing the 3′-most 254 nt of LYS2 and the 5′-most 426 nt of HIS3 and replaced the GAL1UAS with a fragment containing ZeoR and a PmeI site to obtain pOHHZ.
Figure 1.
Integration vectors to construct one-hybrid strains with ADE2 and HIS3 reporter genes. (A) Integration vector pOHAK for ADE2 reporter gene. We cloned a fragment spanning AFI1-GAL2UAS-GAL2TATA-ADE2 from PJ69-2A and replaced the GAL2UAS with a fragment containing KanMX and an HpaI site to obtain pOHAK. (B) Integration vector pOHHZ for HIS3 reporter gene. We cloned a fragment spanning LYS2-GAL1UAS-GAL1TATA-HIS3 from PJ69-2A and replaced the GAL1UAS with a fragment containing ZeoR and a PmeI site to obtain pOHHZ. (C) Schematic representation of reporter gene construction in the case of ADE2 as an example. A methylatable bait sequence is cloned at the HpaI site of pOHAK, and the plasmid linearized by SalI-KpnI digestion is used for transformation of PJ69-2A. Homologous recombination occurs within ORFs for AFI1 and ADE2.
Next, we prepared a DNA fragment LexAop8-Sm4 comprised of eight tandemly iterated copies of LexAop followed by four tandemly iterated copies of Sm (CAG CAG CCG CGC CCA ACG CTG GGA; methylatable CpG residues are underlined) (8). We cloned the LexAop8-Sm4 fragment into the HpaI site of pOHAK, digested the obtained plasmid by SalI and KpnI, used the linearized plasmid DNA to transform PJ69-2A cells (Clonetech) and selected G418-resistant transformants. Following a PCR-based examination of correct integration of the bait sequence between AFI1 and ADE2, we confirmed that the strain fails to grow in the absence of adenine. We then transformed the obtained strain with BglII/KpnI-linearized pOHHZ plasmid bearing the LexAop8-Sm4 fragment at its PmeI site, and isolated Zeocin-resistant clones. We confirmed the correct integration of the second bait sequence between LYS2 and HIS3 as well as the histidine-dependent growth of the strain.
Finally, we transformed the strain with pALMS, a TRP1-marked 2 μ plasmid that expresses LexA-fused M.SssI under the control of ADH1 promoter, and selected the transformants on synthetic complete media (SC) lacking tryptophan (SC–Trp) (9). Consequently, we obtained a single-bait/dual-reporter system. For the construction of a dual-bait/dual-reporter system, we used GAM12 or (GAC)12 (10) as the second bait. To conduct a dual-bait reverse one-hybrid selection, we replaced the ura3-52 allele in the single-bait/dual-reporter strain with a URA3 allele whose promoter bears Sm4 but not LexAop8.
One-hybrid screening of cDNA libraries
Mouse brain and human ovary MatchmakerTM cDNA libraries were purchased from Clontech. The single- and dual-bait strains constructed as above were transformed with the library plasmids using the standard lithium acetate method (9). The transformants were selected on SC–Trp–Leu–His–Ade. On the other hand, an aliquot of the transformants was serially diluted and spread on SC–Trp–Leu agar plates to estimate the size of screening or the number of transformants screened. The library plasmid was isolated from each positive colony and subjected to DNA sequencing.
RESULTS
Construction of a yeast one-hybrid system with auxotrophic reporter genes regulated by a methylatable bait sequence
It is essential for a large-scale yeast one/two-hybrid screening to use auxotrophic markers (e.g. ADE2, HIS3) as reporter genes, since transformants with auxotrophic reporters can be spread on agar plates at much higher density than those with colorimetric reporters (e.g. lacZ). Thus, we developed a host strain in which a promoter carrying a methylatable bait sequence regulates the expression of ADE2 and HIS3 (Figure 2A). Note that these reporter genes were integrated into the host chromosomes, in contrast with the episomal lacZ reporter used in the previous work (7). In this strain, we used Sm4 or four tandemly iterated copies of Sm as the bait (8). We placed eight tandemly iterated copies of LexAop (LexAop8) at the 5′-flanking region of the Sm4 bait sequence to tether LexA-fused M.SssI CpG methyltransferase (LexA-M.SssI) for targeted methylation (7). If a methylated DNA-binding protein fused with Gal4 activation domain (GAD) binds the methylated Sm4 bait, it activates the expression of ADE2 and HIS3. Indeed, transformation with a plasmid that expresses GAD-fused MBD1 made the stain capable of growing in the absence of adenine and histidine, provided that LexA-M.SssI was co-expressed (data not shown).
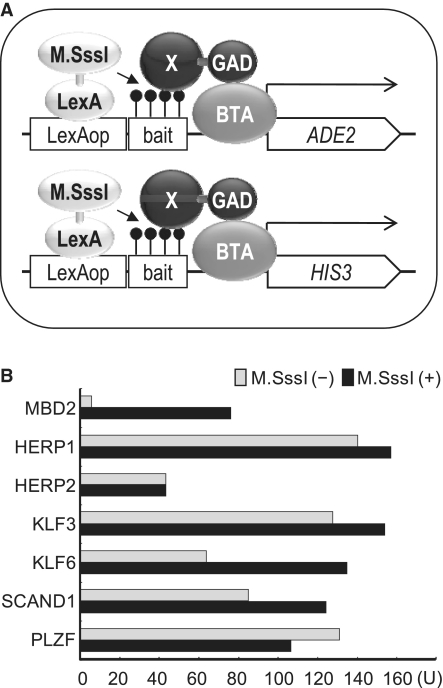
Figure 2.
Methylation dependence of proteins identified by a single-bait one-hybrid screening. (A) Schematic representation of the single-bait/dual-reporter system. The results of the screening using this system are summarized in Table 1. X, methylated DNA-binding protein; BTA, basic transcription apparatus. Filled circle shown on the bait, methylated CpG. (B) Methylation dependence of DNA-binding proteins identified by the single-bait one-hybrid screening. One-hybrid interaction with the Sm4 bait was examined for each protein using a lacZ reporter plasmid pOp8Sm4 (7) in the absence and presence of pALMS that expresses LexA-M.SssI. β-galactosidase activity in the extract of each transformant was measured using a standard method (9).
Identification of methylation-dependent and -insensitive DNA-binding proteins by the single-bait one-hybrid system
Confirming that the single-bait/dual-reporter strain described above behaved as intended, we exploited it to screen a GAD-fusion cDNA library constructed from mouse brain mRNA. We screened ∼3.22 × 106 clones to identify 77 positives capable of growing in the absence of adenine and histidine. We sequenced the plasmids in positive clones to reveal that all of them encode DNA-binding proteins (Table 1). Note that we regarded six PLZF clones as a noise inherent to the system, because PLZF was shown to bind LexAop (11). Of the 71 clones remained, 26 (37%) were derived from MBD2 and judged as true positives. We also screened 2.76 × 106 clones from a human ovary cDNA library and identified 158 positive clones, 49 of which were turned out be PLZF (Table 1). Of the 109 clones remained, 20 (18%) were judged as true positives, including 19 and 1 clones for MBD2 and MBD4, respectively. The other 89 clones were derived from various transcription factors.
Table 1.
Single-bait screening of mouse brain and human ovary cDNA libraries
| Gene | Description | Number of clones |
|---|---|---|
| Mouse brain cDNA library (3.22 × 106 clones) | ||
| MBD2 | Methyl-CpG binding domain protein 2 | 26 |
| KLF6 | Krüppel-like factor 6 | 21 |
| KLF3 | Krüppel-like factor 3 | 7 |
| KLF4 | Krüppel-like factor 4 | 7 |
| HERP1 | HES-related repressor protein 1 | 2 |
| HERP2 | HES-related repressor protein 2 | 2 |
| KLF2 | Krüppel-like factor 2 | 2 |
| KLF5 | Krüppel-like factor 5 | 1 |
| KLF7 | Krüppel-like factor 7 | 1 |
| SCAND1 | SCAN domain protein 1 | 1 |
| ZIC1 | Zinc finger protein of the cerebellum 1 | 1 |
| PLZFa | Promyelocytic leukemia zinc finger protein | 6 |
| Human ovary cDNA library (2.78 × 106 clones) | ||
| KLF2 | Krüppel-like factor 2 | 33 |
| KLF4 | Krüppel-like factor 4 | 24 |
| MBD2 | Methyl-CpG binding domain protein 2 | 19 |
| KLF6 | Krüppel-like factor 6 | 17 |
| KLF7 | Krüppel-like factor 7 | 7 |
| KLF3 | Krüppel-like factor 3 | 5 |
| KLF5 | Krüppel-like factor 5 | 2 |
| HERP2 | HES-related repressor protein 2 | 1 |
| MBD4 | Methyl-CpG binding domain protein 4 | 1 |
| PLZFa | Promyelocytic leukemia zinc finger protein | 49 |
aPLZF was shown to bind LexAop (11). Known methylated DNA-binding proteins are indicated in bold.
To examine whether or not the identified proteins other than MBD2 and MBD4 bind the bait in a methylation-dependent manner, we examined their one-hybrid interaction with the Sm4 sequence using a lacZ reporter plasmid in the absence and presence of LexA-M.SssI (7). As shown in Figure 2B, these proteins bound the bait regardless of its methylation status, thereby failing to show a methylation-dependent interaction mode.
These results demonstrated that the simple single-bait screening can identify methylated DNA-binding proteins but suffers from background clones derived from methylatioin-insensitive DNA-binding proteins.
Discrimination between methylation-dependent and -insensitive DNA-binding proteins by a dual-bait reverse one-hybrid system
Inspired by a dual-bait reverse two-hybrid system to select separation-of-function alleles (12), we intended to examine whether or not the reverse one-hybrid selection (13) can eliminate the backgrounds. We modified the single-bait/dual-reporter strain described above to bear an additional Sm4 bait sequence that is notably preceded by no LexAop to escape methylation and followed by URA3 to enable reverse one-hybrid selection (Figure 3A). If a GAD-fused protein binds the Sm4 bait in a methylation-insensitive manner, it would induce the expression of both ADE2/HIS3 and URA3. Such cells can be eradicated in the presence of 5-fluoro-orotic acid (5-FOA), because URA3 encodes orotidine-5′-phosphate decarboxylase that converts 5-FOA to 5-fluorouracil, a potent cytotoxin to kill the yeast (9,13). In contrast, if a GAD-fused protein binds the Sm4 bait in a methylation-dependent manner, it would induce the expression of ADE2/HIS3 but not URA3, making the cell viable in the presence of 5-FOA and uracil. In other words, 5-FOA-resistant Ade+/His+ cells should bear plasmids encoding methylation-dependent DNA-binding proteins.
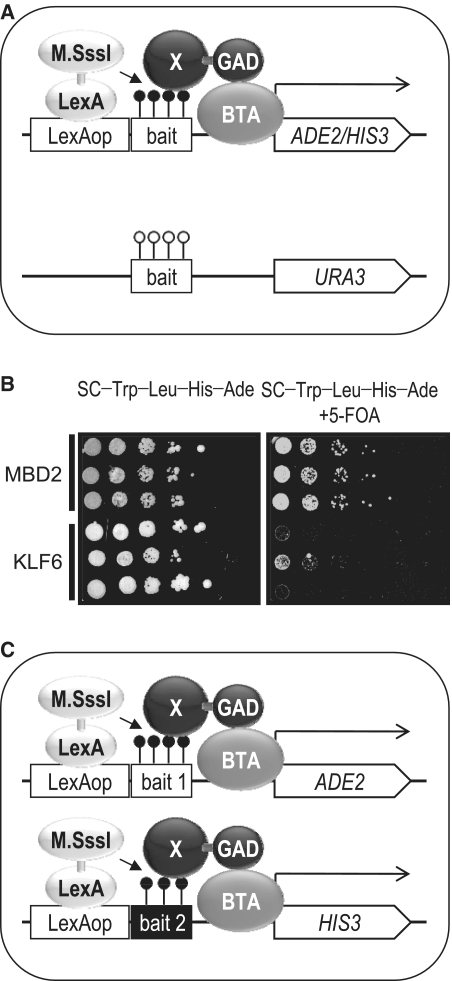
Figure 3.
Dual-bait one-hybrid systems to eliminate background clones derived from methylation-insensitive DNA-binding proteins. (A) Schematic representation of a dual-bait reverse one-hybrid system. In this system, methylatable and unmethylatable versions of the same bait sequence precede ADE2/HIS3 and URA3, respectively. X, methylated DNA-binding protein; BTA, basic transcription apparatus. Filled circle shown on the bait, methylated CpG. Open circle shown on the bait, unmethylated CpG. (B) Discrimination between methylation-dependent and -insensitive DNA-binding by the dual-bait reverse one-hybrid system. In the absence of 5-FOA, both the methylation-dependent DNA-binding protein MBD2 and the methylation-insensitive protein KLF6 made the cell capable of growing in the absence of adenine and histidine (left panel). In the presence of 5-FOA, MBD2, but not KLF6, made the cell capable of growing in the absence of adenine and histidine (right panel). (C) Schematic representation of a dual-bait one-hybrid system using two unrelated methylatable bait sequences. The results of the screening using this system are summarized in Table 2. X, methylated DNA-binding protein; BTA, basic transcription apparatus. Filled circle shown on the bait, methylated CpG.
To test the performance of the dual-bait reverse one-hybrid selection, we used MBD2 and KLF6 as representatives of methylation-dependent and -insensitive DNA-binding proteins, respectively. The strain bearing MBD2, but not KLF6, grew in the medium devoid of adenine and histidine but supplemented with uracil and 5-FOA (Figure 3B). These results provide a proof-of-concept for the dual-bait reverse one-hybrid selection of methylation-dependent DNA-binding proteins. Unfortunately, the selection was not perfect, as we observed leaky growth of a KLF6 clone, which might lead to backgrounds in library screening. The system would be useful rather for examining the methylation dependence of DNA-binding proteins identified by one-hybrid screening.
High-specificity screening for methylated DNA-binding proteins by a dual-bait one-hybrid system
We developed a third system that concurrently uses two independent bait sequences, based on the following simple rationale. If a protein leading to background clones binds the Sm4 bait in a methylation-insensitive, but sequence-dependent manner, it would fail to bind a second bait sequence that lacks any similarity to Sm4, because it is highly unlikely that a single sequence-specific DNA-binding protein can interact with two totally unrelated sequences. To test this idea, we integrated Sm4 and GAM12 upstream of ADE2 and HIS3, respectively, to develop a third system (Figure 3C) and confirmed that the expression of MBD2, but not KLF6, made the cell viable in the absence of adenine and histidine (data not shown).
We used this system to screen the same cDNA libraries as those that we screened using the single-bait system. Screening of 0.32 × 106 clones from the mouse brain library led us to isolate six non-PLZF positives, five (83%) of which were clones for MBD2 or true positives (Table 2). We also conducted a screening of 0.86 × 106 clones from the human ovary library to identify eight non-PLZF-positive clones, eight (100%) of which were true positives, containing 7 and 1 clones for MBD2 and MBD4, respectively (Table 2). When normalized to the number of clones screened, the rate of true positives identified was comparable between the single- and dual-bait systems, indicating that the latter did not compromise the sensitivity of screening. On the other hand, the dual-bait system almost completely eliminated methylation-insensitive DNA-binding proteins but PLZF, an inevitable noise inherent to LexA-based system, thereby achieving much higher specificity than the single-bait system.
Table 2.
Dual-bait screening of mouse brain and human ovary cDNA libraries
| Gene | Description | Number of clones |
|---|---|---|
| Mouse brain cDNA library (0.32 × 106 clones) | ||
| MBD2 | Methyl-CpG binding domain protein 2 | 5 |
| HERP2 | HES-related repressor protein 2 | 1 |
| PLZFa | Promyelocytic leukemia zinc finger protein | 1 |
| Human ovary cDNA library (0.86 × 106 clones) | ||
| MBD2 | Methyl-CpG binding domain protein 2 | 7 |
| MBD4 | Methyl-CpG binding domain protein 4 | 1 |
| PLZFa | Promyelocytic leukemia zinc finger protein | 16 |
aPLZF was shown to bind LexAop (11). Known methylated DNA-binding proteins are indicated in bold.
While the screening described above detected only MBD proteins, we used the same system to successfully isolate a clone for ZBTB38 from another cDNA library, demonstrating its capability to identify Kaiso-type methylated DNA-binding proteins (data not shown).
DISCUSSION
We developed a yeast one-hybrid system for selective cloning of methylated DNA-binding proteins. Although our previous system detected methylated DNA–protein interactions via lacZ reporter activity, it could not screen libraries (7). A single-bait dual-reporter system, which was basically constructed by simply replacing lacZ with ADE2 and HIS3, could screen libraries but suffered high backgrounds (Figure 2 and Table 1). We found that the dual-bait approach was critical to suppress background clones, which were almost always derived from methylation-insensitive sequence-specific DNA-binding proteins (Figures 2, 3 and Table 2). While it is formally possible that proteins showing two-hybrid interactions with LexA or M.SssI lead to backgrounds even in the dual-bait system, we have so far encountered no such clones and, even if we do any, we will be able to discriminate them from true positives through appropriate control experiments.
We recommend the dual-bait system with two independent bait sequences for initial screening (Figure 3C). Each positive clone should be retransformed to the same strain in the absence and presence of LexA-M.SssI as a second screening to confirm methylation dependence of the interaction. Alternatively, one may use the dual-bait reverse one-hybrid system for this purpose (Figure 3A). This two-step approach is critical to select candidates for methylated DNA-binding proteins, especially when screening libraries from organisms with poorly annotated genomes. Finally, the interaction has to be biochemically verified, typically, by an electrophoretic mobility shift assay using methylated and unmethylated DNA.
A matter of concern with this system would be low sensitivity leading to false negatives. For instance, our screening of a brain cDNA library identified MBD2 but not MeCP2 abundantly expressed in neurons (14). Of note, these two proteins show different preferences for methylated DNA; MeCP2 requires an A/T-rich sequence adjacent to methyl-CpG for efficient DNA binding, whereas MBD2 does not (15). Since the bait sequences used for the screening lack any A/T-rich sequence, MeCP2 likely disfavored them. Indeed, MeCP2 required the potent activation domain of VP16 to show a one-hybrid interaction with the Sm4 bait (7). Such sequence preference was recently demonstrated for MBD1 (16) and Kaiso-type proteins (17). In addition, it should be noted that any methylated DNA-binding protein has some affinity to unmethylated DNA. Accordingly, if such unmethylated targets occur frequently in the yeast genome, they may compete with the methylated bait for the protein, especially when its expression level is low. Thus, the sensitivity of screening depends on bait sequences and differs from one protein to another. To increase the general sensitivity, the bait has to be fully methylated. Since nucleosome-wrapped DNA is protected from methylation by M.SssI in yeast (18), the bait should be kept as nucleosome-free as possible. It is, however, conceivable that some methylated DNA-binding proteins rather prefer to cohabitate with nucleosomes for stable DNA binding, as they are typically found in heterochromatic regions. In anyway, we expect that future screening efforts using various bait sequences and cDNA libraries different in terms of expression level and activation domain would provide hints for improvement.
We also foresee several variations of the system. For instance, it is intriguing to use de novo non-CpG methylases instead of M.SssI, because a wide occurrence of non-CpG methylation was found not only in plants and fungi but also in human ES cells (19) and it remains elusive how the cell recognizes it. It would be also possible to use a sequence-specific DNA hemimethylase (20) to screen for hemimethylated DNA-binding protein such as UHRF1. A further challenging modification would be co-targeting of M.SssI with TET1, which converts 5mC to 5-hydroxymethylcytosine (5hmC) (21). While 5hmC attracts increasing attention, it escapes recognition by MBD proteins (22) and how it is recognized remains totally unknown. Identification of proteins that recognize 5hmC would be of great importance in deepening our understanding of this novel epigenetic mark.
Taken together, the one-hybrid system described here and its variants would provide valuable research tools for epigenetics.
FUNDING
Genome Network Project and Cell Innovation Project from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (to T.I.). Funding for open access charge: MEXT.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Kazuyuki Mizushima for his contribution to vector construction.
REFERENCES
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasai N, Defossez PA. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int. J. Dev. Biol. 2009;53:323–334. doi: 10.1387/ijdb.082652ns. [DOI] [PubMed] [Google Scholar]
- 6.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 7.Feng SY, Ota K, Yamada Y, Sawabu N, Ito T. A yeast one-hybrid system to detect methylation-dependent DNA–protein interactions. Biochem. Biophys. Res. Commun. 2004;313:922–925. doi: 10.1016/j.bbrc.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Prokhortchouk A, Hendrich B, Jørgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams A, Gottschling DE, Kaiser CA, Steams T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 10.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sitterlin D, Tiollais P, Transy C. The RARα-PLZF chimera associated with Acute Promyelocytic Leukemia has retained a sequence-specific DNA-binding domain. Oncogene. 1997;14:1067–1074. doi: 10.1038/sj.onc.1200916. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi Y, Ota K, Ito T. A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1: implications for modulation of mating pheromone signaling. J. Biol. Chem. 2007;282:29–38. doi: 10.1074/jbc.M609308200. [DOI] [PubMed] [Google Scholar]
- 13.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein–protein and DNA–protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Clouaire T, de Las Heras JI, Merusi C, Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res. 2010;38:4620–4634. doi: 10.1093/nar/gkq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq280. doi:10.1093/nar/gkq280; April 19, 2010 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kladde MP, Xu M, Simpson RT. Direct study of DNA–protein interactions in repressed and active chromatin in living cells. EMBO J. 1996;15:6290–6300. [PMC free article] [PubMed] [Google Scholar]
- 19.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerasimaite R, Vilkaitis G, Klimasauskas S. A directed evolution design of a GCG-specific DNA hemimethylase. Nucleic Acids Res. 2009;37:7332–7341. doi: 10.1093/nar/gkp772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]