Abstract
Hematopoietic stem cells in the bone marrow give rise to lymphoid progenitors, which subsequently differentiate into B and T lymphocytes. Here we show that the proto-oncogene LRF plays an essential role in the B versus T lymphoid cell fate decision. We demonstrate that LRF is key for instructing early lymphoid progenitors to develop into B lineage cells by repressing T cell-instructive signals produced by the cell fate signal protein, Notch. We propose a new model for lymphoid lineage commitment, in which LRF acts as a master regulator of B versus T lineage fate decision.
All hematopoietic cells are generated from a small subset of pluripotent stem cells (HSCs) via lineage-restricted progenitors. In adult mice, HSCs reside in the bone marrow (BM) and give rise to lymphoid restricted-progenitors (1), which subsequently develop into B and T lymphocytes in the BM and thymus, respectively. This developmental process is coordinated by the differentiation-stage specific expression of distinct sets of genes. Although some of the transcriptional regulators that play key roles in early stages of lymphocyte development are known (2, 3), the precise molecular mechanisms by which lymphoid restricted-progenitors are instructed towards B or T cell fates are still largely undefined.
The proto-oncogene LRF (4), encoded by the Zbtb7a gene, (formerly known as Pokemon (5) and also described as FBI-1 (6) and OCZF (7)) is a transcriptional repressor that belongs to the POK (POZ/BTB and Krüppel) protein family. Promyelocytic Leukemia Zinc Finger (PLZF) and B-Cell Lymphoma 6 (BCL6), two members of the POK family, are involved in chromosomal translocations associated with Acute Promyelocytic Leukemia (APL) and Non-Hodgkin’s Lymphoma (NHL), respectively (8, 9). In a similar manner, we recently reported that LRF plays a pivotal proto-oncogenic role and is highly expressed in human non-Hodgkin’s lymphoma tissues (5). Emerging experimental evidence indicates that the POK family is indispensable for normal hematopoiesis and immune system developments (9–12). BCL6 has been shown to be essential for germinal center (GC) formation and for Th2-type inflammatory responses (9). More recently, two independent groups reported that a close homologue of LRF (Th-POK, also known as cKrox, Zbtb7b) is a master regulator of CD4/8 T cell lineage specification (11, 12).
Given the fact that LRF is broadly expressed in multiple hematopoietic lineages (Fig. S2A), especially in the GC B cells (5), forms complexes with BCL6 (4), and is highly expressed in human Non-Hodgkin lymphoma tissues, we hypothesized that this gene could play a key role in B cell development. We therefore investigated both fetal and adult lymphopoiesis using LRF gene deletion in mice.
B cell development
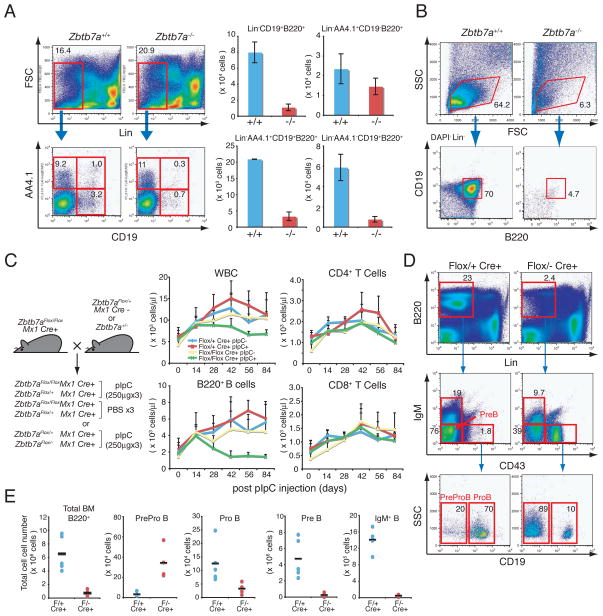
Deletion of the Zbtb7a gene in mouse was carried out with a conventional gene knockout approach (Fig. S1A and S1B). While we didn’t observe a gross defect in the heterozygous mutant, homozygous deletion of the Zbtb7a gene (Zbtb7a−/−) resulted in embryonic lethality around 16.5 d.p.c. due to severe anemia. Examination of fetal B lymphopoiesis in 14.5 d.p.c fetal livers (FLs) from Zbtb7a−/− revealed a reduction in the total number of CD19+B220+ B cells (Fig. 1A). This was mainly due to reduction of B cells after the ProB stage of differentiation. Absolute numbers of the earliest B cell precursors (Lin−AA4.1+CD19−B220+) were comparable to that of wild type (WT) littermate controls, while total numbers of Lin−AA4.1+CD19+B220+ and Lin−AA4.1−CD19+ B220+ B cells were markedly decreased in Zbtb7a−/− FLs (Fig. 1A, right). Hematopoietic stem cell (HSC) and common lymphoid progenitor (CLP) populations were intact in Zbtb7a−/− FLs (Fig. S2B).
Fig. 1.
LRF is indispensable for both fetal and adult B lymphopoiesis. (A) 14.5 d.p.c FL cells were stained with fluorochrome-conjugated anti-B220, CD19, AA4.1 and lineage markers. Representative FACS profiles of the Zbtb7a+/+ and Zbtb7a−/− FL cells are shown (left). Total numbers of FL mononuclear cells were counted and absolute number of B cells in each developmental stage was calculated. Average cell numbers of three independent embryos for each genotype are presented with +/− standard deviations (SD). (B) The 14.5 d.p.c. FL-HSCs (Lin−Sca1+c-Kit+) were cultured on OP9 stromal cell layers in the presence of IL-7 and Flt3 ligand. After 10 days of culture, cells were isolated and analyzed by FACS. (C) Schematic representations of mouse breeding strategy for conditional LRF knockout experiments (left). Follow-up of the PB counts after pIpC (or PBS) injections overtime (right). 4 groups of mice were examined according to genotype and treatment. WBC counts in the PB were measured by a hematology analyzer and total numbers of B and T cells were subsequently calculated based on the percent positivity of B220 and CD4/8 expression, respectively. The average cell count of 5 animals was plotted on each time point with error bars (+/− SD). (D) BM cells were stained with fluorochrome-conjugated anti-B220, CD19, IgM, CD43 and lineage markers (21) one month after the last pIpC injection. Representative FACS profiles for each genotype are demonstrated. (E) Absolute cell number of each population was calculated according to FACS profiles. Black horizontal bars represent mean cell counts among 5 animals.
ProB cells can be propagated in vitro on OP9 stromal cell layers in the presence of IL-7 and Flt3 ligand (13). While substantial numbers of ProB cells could be propagated from Zbtb7a+/+ fetal liver HSCs (FL-HSCs), Zbtb7a−/− ProB cells were barely detectable (Fig. 1B), indicating a cell autonomous defect in early B cell development. In contrast, Zbtb7a−/− FL-HSCs retained their capacity for T cell development with Zbtb7a−/− FL-HSCs successfully giving rise to T cells in vitro, after culture on OP9-DL1 stromal cells overexpressing the Notch ligand Delta-like 1 (14)(Fig. S2C). To further investigate the defect in early B cell development in Zbtb7a−/− FLs, we performed bone marrow competitive repopulation assays (15). Thus, Zbtb7a+/+ and Zbtb7a−/− FL cells were transplanted separately into lethally irradiated recipient mice along with WT BM cells from a congenic strain expressing the CD45.1 antigen (Fig. S3A, left). In these experiments Zbtb7a+/+ FL cells successfully gave rise to peripheral blood (PB) B cells in the recipients, while Zbtb7a−/− FL derived B cells were virtually undetectable (Fig. S3A, right).
The role of LRF in adult lymphopoiesis was next explored using conditional deletion of the Zbtb7a gene (Fig. S1C and D). Mx1-Cre transgenic mice were used, in which Cre recombinase is induced in HSC upon polyinosinic-polycytidylic acid (pIpC) administration (16). A series of double mutant mice were treated with pIpC at 3 weeks of age as previously described (Fig. 1C, left) (16) and peripheral blood was analyzed at two week intervals. Upon pIpC treatment, a significant decline of white blood cell (WBC) counts in the Zbtb7aFlox/Flox Mx1cre+ mice was observed (Fig. 1C) primarily due to a considerable reduction in circulating B220+ B cells, while T cell numbers remained similar to controls (Fig. 1C).
Like the defect seen in Zbtb7a−/− mice (Fig. 1A), B cell development in the BM of Zbtb7aFlox/− Mx1cre+ mice was also severely impaired (Fig. 1D). Thus, ProB, PreB and IgM+B cells were drastically reduced, while absolute numbers of the PreProB cells (Fig. S3B) (17) was increased in the pIpC-treated Zbtb7aFlox/−Mx1cre+ BM (Fig. 1D and 1E). Of note, LRF mRNA expression in the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells was essentially undetectable, as revealed by quantitative real-time RT-PCR assays (q-RT-PCR) (Fig. S3C). Furthermore, proportions of the HSCs and CLPs were not grossly affected in the pIpC-treated Zbtb7aFlox/− Mx1cre+ mice (Fig. S3D), whilst absolute numbers of their HSCs and CLPs were slightly increased as compared to control mice (Fig. S3E).
Extrathymic T cell development
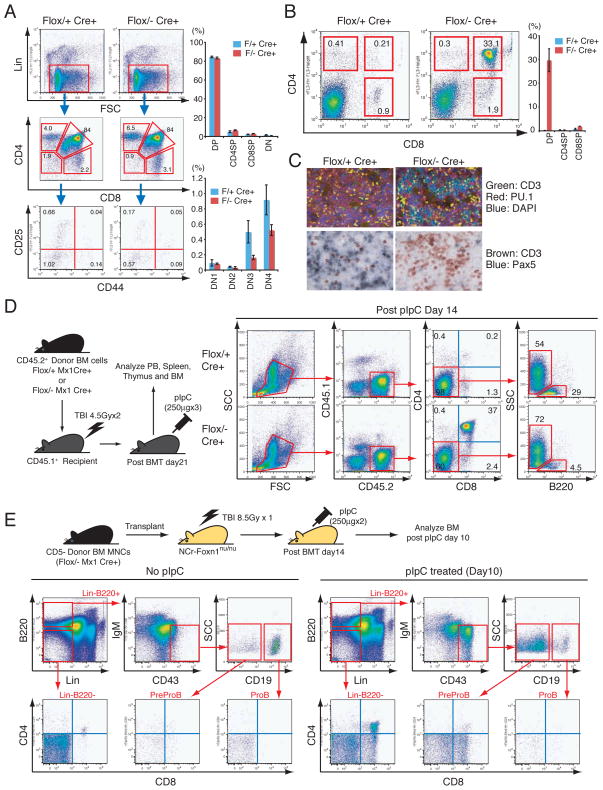
Both T and B cells share their origins with a common lymphoid progenitor (CLP) (1). However, we did not observe a gross defect in the T cell compartment in pIpC-treated Zbtb7aFlox/− Mx1cre+ thymus (Fig. 2A). Although a slight decrease was observed in double negative 3 (DN3) and DN4 thymocyte populations, the proportions of CD4 single positive (CD4-SP), CD8-SP and CD4/8 double positive (DP)-T cells were comparable to those of control mice (Fig. 2A). Unexpectedly however, an accumulation of extrathymic DP-T cells in the BM of pIpC-treated Zbtb7aFlox/− Mx1cre+ mice was detected (Fig. 2B) comprising nearly 30% of the BM mononuclear cells (BMMNCs) at one month after pIpC treatment (Fig. 2B). Immunohistochemical/fluorescent analyses further demonstrated that CD3dim DP-T cells accumulated in the BM of Zbtb7aFlox/− Mx1cre+ mice (Fig. 2C). These extrathymic BM DP-T cells were polyclonal in origin, as revealed by Dβ1-to-Jβ1 rearrangement status of the T cell receptor beta locus (Fig. S4A). Moreover, quantitative measurement of gene dosage indicated that the Zbtb7a gene was at almost undetectable levels in both thymic and BM DP-T cells in the pIpC-treated Zbtb7aFlox/− Mx1cre+ mice (Fig. S4B and S4C). Notably, extrathymic T cell development appeared to be limited to the BM, as these cells were not observed in spleen or Peyer’s patches (Fig. S4D and S4E).
Fig. 2.
Extrathymic DP-T cell development in the BM upon LRF loss. (A) Thymic T cells were analyzed one month after pIpC injection. Representative FACS profiles for each genotype are shown (left). Proportions of CD4/8 double negative (DN), CD4/8 double positive (DP), CD4 single positive (CD4SP) and CD8 single positive (CD8SP) populations were examined and the DN fraction was further stratified according to CD44 and CD25 expression. Three mice were analyzed for each genotype (right). (B) BMMNCs were analyzed for CD4/8 expression one month after pIpC treatment. Representative FACS profiles for each genotype are shown (left). Average percent positivity of three mice for each genotype is demonstrated with +/− SD (right). (C) Immunofluorescent analysis of CD3 and PU.1 expression in BM sections (top). Immunohistochemical analysis of T cell and B cell markers (CD3 and Pax5, respectively) in BM sections one month after pIpC treatment (bottom). (D) Either Zbtb7aFlox/+ Mx1cre+ or Zbtb7aFlox/− Mx1cre+ donor BMMNCs (CD45.2+) were transplanted into lethally irradiated recipient mice (CD45.1+). Upon engraftment, recipient mice were treated with pIpC. Recipients’ BMMNCs were then collected and analyzed two weeks after the last pIpC administration. Representative FACS profiles are presented. (E) Zbtb7aFlox/− Mx1cre+ donor BMMNCs (CD45.2+) were transplanted into lethally irradiated recipient nude mice. Two weeks after transplantation, mice were treated with either pIpC or PBS. Recipients’ BMMNCs were harvested and subsequently analyzed 10 days after the last pIpC administration. Representative BM FACS profiles are presented.
To investigate whether the extrathymic DP-T cell accumulation seen upon loss of the LRF gene was caused by defects in a cell intrinsic mechanism, LRF inactivation was induced in BM-reconstituted recipient mice. Thus, Zbtb7aFlox/+ Mx1cre+ or Zbtb7aFlox/− Mx1cre+ BMMNCs were transplanted into lethally irradiated recipient mice, and upon engraftment recipient mice were treated with pIpC to induce Cre expression (Fig. 2D, left). In Zbtb7aFlox/−Mx1cre+ reconstituted mice, an accumulation of donor-derived (CD45.2+) DP-T cells was seen in the BM with a significant reduction of B cells both in BM and PB in a cell autonomous manner (Fig. 2D and S4F). To determine if extrathymic BM DP-T cell development was thymus-independent, Zbtb7aFlox/− Mx1cre+ BMMNCs were transferred into lethally irradiated athymic nude mice, and the LRF gene subsequently inactivated after engraftment by pIpC administration. DP-T cell accumulation was observed in the BM 10 days after the last pIpC administration (Fig. 2E), indicating that in the absence of LRF, lymphoid progenitors in the BM gave rise to BM DP-T cells in a thymus independent fashion.
Aberrant lymphocyte commitment
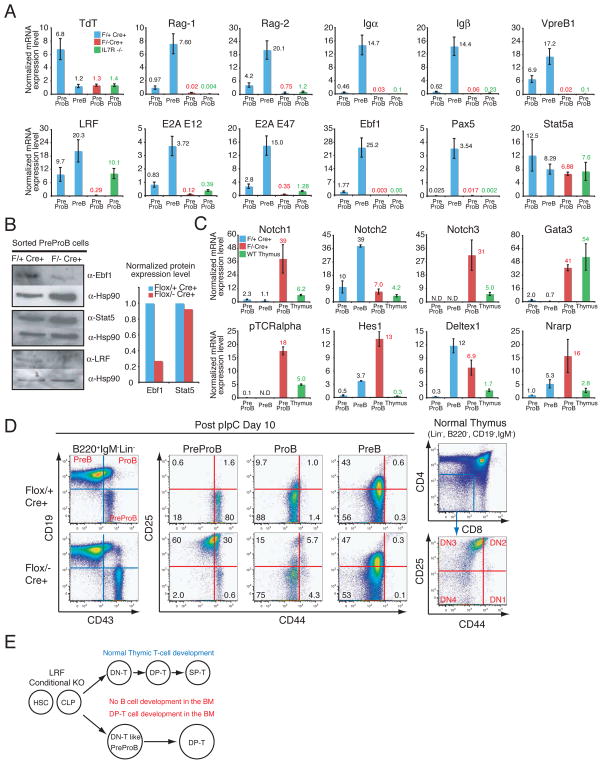
To explore whether the early B cell developmental program takes place correctly in pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells, the expression of the genes encoding pre-BCR components, terminal deoxynucleotidyl transferase (TdT) and Rag recombinases was examined. In these experiments, mRNA levels of pre-BCR components (Igα, Igβ VpreB1), Rag recombinases (Rag-1, Rag-2) and TdT in Zbtb7aFlox/− Mx1cre+ mice were markedly reduced as compared to those of control mice (Fig. 3A).
Fig. 3.
pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells are defective in early B cell development and demonstrate a DN-T cell signature. (A) q-RT-PCR analysis of the genes encoding pre-BCR components, TdT, the Rag recombinases and the critical transcription factors in early B cell development. mRNA expression levels were normalized to Hprt mRNA amount and are represented by bar graphs. Each sample was analyzed in duplicate and error bars indicate +/− SD. BMMNCs were collected and flow-sorted one month after the last pIpC injection. (B) Western blot analysis for Ebf1 and Stat5 protein in the pIpC-treated PreProB cells. Bar graph represents normalized protein expression level over corresponding Hsp90 protein level. (C) q-RT-PCR analysis of Notch and Notch target genes in the PreProB cells. q-RT-PCR was performed as described in (A). (D) CD25 and CD44 expression in PreProB cells was examined 10 days after the last pIpC injection. FACS profiles of normal thymic DN-T cell populations are also presented. (E) Schematic representation of lymphoid lineage development in LRF conditional knockout mutants.
Early B cell development is governed by a small set of cytokines and transcription factors. Both PU.1 and Ikaros are essential for the maintenance of HSCs and CLPs (18, 19), while Flt3 ligand is required for the generation of CLPs but not HSCs (20). Both IL-7 and its receptor (IL-7R) are indispensable for PreProB to ProB transition (21) and transcription factors Bcl11a, E2A, Stat5 and Ebf1 also play critical roles at this developmental stage (3)(Fig. S5A). pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells showed a significant down-regulation of E2A, Ebf1 and Pax5 mRNA (Fig. 3A). Similarly, low amounts of Ebf1 protein were detected in pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells, while Stat5 protein was abundant (Fig. 3B). Since the enforced expression of Ebf1 is able to rescue B cell developmental defects in PU.1−/−, IL-7R−/− and E2A−/− mice (3), we examined whether the overexpression of Ebf1 in Zbtb7a−/− FL-HSCs might have a similar effect. Positive control LRF-transduced Zbtb7a−/− FL cells successfully gave rise to splenic B cells in recipient mice (Fig. S5B). However, neither GFP–vector nor Ebf1-transduced FL cells were able to rescue the Zbtb7a−/− B cell phenotype (Fig. S5B).
Given that pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells were unable to progress further in the B cell developmental program, we speculated that they might have become aberrantly committed to the T cell lineage, thus generating the extrathymic DP-T cells found in the BM. To test this, we examined mRNA expression levels of T cell specific target genes in the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells. In these analyses, mRNA levels of Notch1, Notch3, but not Notch2, and their downstream target genes were profoundly elevated in the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells (Fig. 3C). Despite expressing the cell surface B cell marker B220 (Fig. 1D), pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells appeared aberrantly committed to the T cell rather than B cell lineage. To determine whether the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells could differentiate into more mature stages of T cell development, sorted PreProB cells were cultured on OP9 stromal cell layers. Upon co-culture with control OP9-GFP cells, the pIpC-treated Zbtb7aFlox/+Mx1cre+ PreProB cells, in which one allele of LRF remained intact, efficiently differentiated into ProB cells, while the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells did not give rise to ProB cells (Fig. S6A, left). In the case of cells expressing the Delta notch ligand (OP9-DL1) however, control PreProB cells still differentiated into ProB cells, even though activation of the Notch pathway drives T cell development (Fig. S6A). This is likely due to the fact that normal PreProB cells express very low levels of the Notch1 receptor and thus cannot respond to the DL1 signal (see Fig. 3C for Notch1 mRNA). On the contrary, the pIpC-treated Zbtb7aFlox/− Mx1cre+ PreProB cells immediately lost B220 expression on the cell surface and effectively differentiated to CD4/8 DP-T cells (Fig. S6A, right). In agreement with these findings, Zbtb7aFlox/− Mx1cre+ PreProB cells were mostly positive for CD25 and negative for the CD44 surface markers, which is reminiscent of normal thymic DN3 T cells (Fig. 3D). Taken together, these data indicate that in the absence of LRF lymphoid progenitors give rise to aberrant B220 positive DN-T like cells, which subsequently differentiate to DP-T cells in the BM at the expense of normal B cell development (Fig. 3E).
Notch repression by LRF
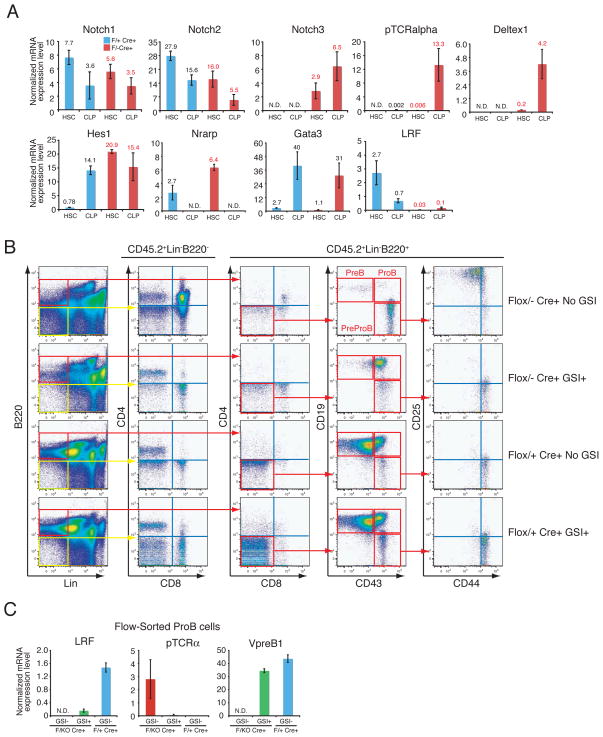
Notch signaling is critical for T cell development and the perturbation of this pathway can result in cellular transformation (22). Furthermore, Notch is obligatory for correct B versus T lineage fate decision in the lymphoid progenitors (2) and was recently reported as the most commonly mutated gene in human T cell acute lymphoblastic leukemia (T-ALL) (23). Notch1 deletion in mouse HSCs results in a marked reduction in thymic T cells and simultaneous B cell development in the thymus (24). Conversely, constitutive activation of Notch1 pathways in HSCs/CLPs results in a block of B cell development and a profound expansion of extrathymic DP-T cells in the BM. While these mice eventually develop T cell leukemia in the BM (22), thymic T cell development is primarily not impaired (22). Given that the phenotype of LRF conditional knockout mice is seen in mice overexpressing the intracellular domain of Notch1 that leads to constitutive Notch pathway activation (22), we hypothesized that LRF might oppose Notch1 function at HSC/CLP stages. To test this directly, we examined expression of Notch genes and their targets in HSCs/CLPs. Upon pIpC treatment LRF mRNA was efficiently eliminated both in HSCs and CLPs (Fig. 4A) and was followed by the up-regulation of all Notch target genes in the pIpC-treated Zbtb7aFlox/− Mx1cre+ HSCs and CLPs (Fig. 4A). Corresponding Notch1 mRNA levels were comparable to control mice (Fig. 4A). This “Notch signature” was evident mainly at the HSC and CLP stages, as relatively low levels of Notch target genes were detected in myeloid/erythroid progenitor compartments (Fig. S6B). To further elucidate whether the aberrant T cell commitment in the absence of LRF was Notch dependent, LRF conditional KO mice were treated with a gamma secretase inhibitor (GSI). GSIs are potent inhibitor of Notch signaling that act by preventing the cleavage and release of the intracellular moiety of the Notch receptor (25). Strikingly, this almost completely rescued abnormal B/T cell commitment seen in LRF conditional KO mice (Fig. 4B). Upon GSI treatment, neither the DP-T cells nor aberrant DN-T-like PreProB cells were observed (Fig. 4B). Furthermore, Zbtb7aFlox/− Mx1cre+ PreProB cells could give rise to ProB cells upon GSI treatment (Fig. 4B). Importantly, in these ProB cells expression of VpreB1, a component of the pre-BCR, resumed upon GSI treatment, while LRF mRNA became barely detectable confirming correct gene targeting (Fig. 4C).
Fig. 4.
LRF opposes Notch pathways at the HSC/CLP stage. (A) BM-HSCs and CLPs were flow-sorted from pIpC-treated animals one month after the last pIpC injection. q-RT-PCR analysis were performed as described in Fig. 3A. (B) In vivo GSI treatment rescued aberrant lymphoid development in LRF conditional KO mice. Either Zbtb7aFlox/+ Mx1cre+ or Zbtb7aFlox/− Mx1cre+ BMMNCs (CD45.2+) were transplanted into lethally irradiated recipient mice (CD45.1) as described in Fig. 2D. Mice were subsequently treated with pIpC. Either GSI or vehicle alone control was orally administered as described in Methods. Recipients’ BMMNCs were collected and analyzed 3 weeks after the last pIpC administration. (C) RNA was extracted from flow-sorted ProB cells and q-RT-PCR analysis was subsequently performed as described in Fig. 3A.
Discussion
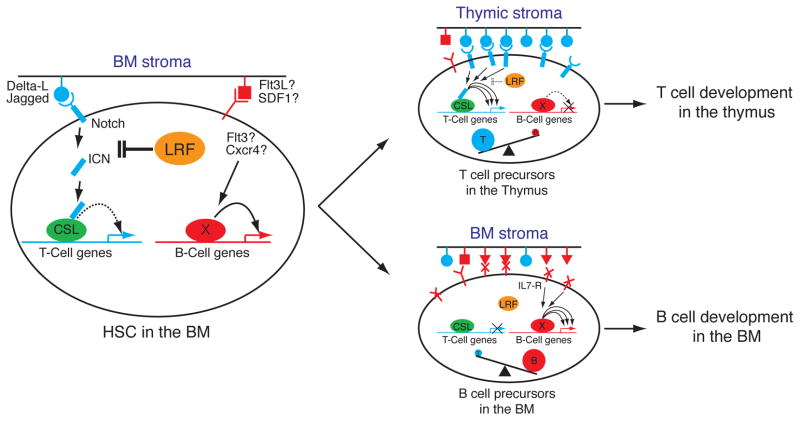
Our findings allow us to reach two conclusions: Firstly, we identify LRF as a master regulator in B versus T lymphoid fate decision with loss of LRF in HSCs/CLPs resulting in an absence of B cell development and spontaneous extrathymic DP-T cell development in the BM. Secondly, we demonstrate that loss of LRF results in aberrant activation of the Notch pathway, with Notch target genes becoming strongly up-regulated in HSCs/CLPs. Taken together, we propose a working model for B versus T cell lineage fate decision, in which LRF plays a pivotal role as negative regulator of T lineage commitment by opposing Notch function (Fig. 5). In normal HSCs/CLPs, LRF opposes Notch function. LRF blocks basal Notch signaling triggered from BM stromal cells, which express moderate level of Notch-ligands (26). BM stromal cells also express molecules that support B cell commitment and development, such as Flt3 and SDF1 (27). Therefore, HSCs/CLPs in the BM are committed to the B cell lineage by default and differentiate into B cells in the presence of signals, such as IL-7 (21). After homing to the developing thymus, in which Notch ligands are abundantly expressed (26), HSCs and/or lymphoid progenitors efficiently give rise to thymic T cells, since at this point Notch signaling would overrule the repressive role of LRF on Notch function. However, in the absence of LRF the low levels of Notch ligands expressed by the BM stroma would now be sufficient to activate Notch target genes normally repressed by LRF in HSCs/CLPs, thus aberrantly specifying T cell fate (Fig. 5). In support of this working model, exogenous expression of LRF in HSCs was seen to result in inefficient DN T cell production, as compared to mock-infected HSCs in an OP9-DL1 culture system (Fig. S6C).
Fig. 5.
Proposed model for the role of LRF in B versus T lineage fate decision. In BM, where stromal cells express moderate levels of Notch ligands, LRF expression in HSCs and lymphoid progenitors functions to repress T cell-instructive signals produced by Notch (left). However, upon homing of progenitors to the thymus where Notch ligands are more abundantly expressed, this repressive role of LRF on Notch function is overruled hence allowing efficient production of T cell precursors (top right).
Our data provide strong evidence that LRF can oppose the Notch signaling pathway. Given that GSI treatment, which blocks the Notch pathway upstream, was sufficient to resume normal B versus T cell commitment in mutant HSC/CLPs, LRF likely targets upstream components of the pathway rather than repressing downstream Notch target genes. As we observed high LRF expression in non-Hodgkin’s lymphoma patients (5) and Notch signaling is known to play a tumor suppressive role in human B cell malignancies (28), it is tempting to speculate that LRF can also exert its oncogenic activity by opposing Notch function in the B cell compartment. In addition, given that LRF is widely expressed in the organism and Notch has described roles in regulating the development and differentiation of multiple tissues, future studies will seek to address the extent of LRF’s repressive role on Notch-dependent processes.
Supplementary Material
Acknowledgments
We would like to thank Giulio Draetta, Peter Strack and Victoria Richon for providing us with GSI, MRK-003. We are grateful to Zuniga-Pflucker for OP9-GFP and OP9-DL1 cells. We thank Jia-Hui Dong and other MSKCC Transgenic core facility members for help and advice on the generation of Zbtb7a mutant mice; Yuri Igarashi-Alexander for assistance with transplantation experiment; Jan Hendrikx and other MSKCC Flow Cytometry core facility members for assistance with FACS analysis and cell sorting; Davide Robbiani and Anna Gazumyan for advice and helpful discussion; Carmela Gurrieri, Jose Costoya, Francesco Piazza, Luipa Khandker, Ilhem Guernah, Linda DiSantis and other P.P.P. lab members for assistance and helpful discussion. This work is supported in part by the NCI grant CA-102142 to P.P.P.
Footnotes
References
- 1.Kondo M, Weissman IL, Akashi K. Cell. 1997;91:661. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Maillard I, Fang T, Pear WS. Annu Rev Immunol. 2005;23:945. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 3.Medina KL, Singh H. Curr Opin Hematol. 2005 May;12:203. doi: 10.1097/01.moh.0000160735.67596.a0. [DOI] [PubMed] [Google Scholar]
- 4.Davies JM, et al. Oncogene. 1999 Jan 14;18:365. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 5.Maeda T, et al. Nature. 2005 Jan 20;433:278. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 6.Pessler F, Pendergrast PS, Hernandez N. Mol Cell Biol. 1997;17:3786. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kukita A, et al. Blood. 1999 Sep 15;94:1987. [PubMed] [Google Scholar]
- 8.Chen Z, et al. Embo J. 1993;12:1161. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye BH, et al. Nat Genet. 1997;16:161. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 10.Piazza F, Costoya JA, Merghoub T, Hobbs RM, Pandolfi PP. Mol Cell Biol. 2004 Dec;24:10456. doi: 10.1128/MCB.24.23.10456-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun G, et al. Nat Immunol. 2005 Apr;6:373. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 12.He X, et al. Nature. 2005 Feb 24;433:826. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 13.Vieira P, Cumano A. Methods Mol Biol. 2004;271:67. doi: 10.1385/1-59259-796-3:067. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt TM, Zuniga-Pflucker JC. Immunity. 2002 Dec;17:749. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 15.See materials and methods, available as supporting material on Science Online.
- 16.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 17.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. J Exp Med. 1991 May 1;173:1213. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki H, et al. Blood. 2005 Sep 1;106:1590. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allman D, et al. Nat Immunol. 2003 Feb;4:168. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 20.Sitnicka E, et al. Immunity. 2002 Oct;17:463. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi K, Lai AY, Hsu CL, Kondo M. J Exp Med. 2005 Apr 18;201:1197. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pui JC, et al. Immunity. 1999 Sep;11:299. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 23.Weng AP, et al. Science. 2004 Oct 8;306:269. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 24.Radtke F, et al. Immunity. 1999 May;10:547. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 25.Lewis HD, et al. Chem Biol. 2007 Feb;14:209. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Harman BC, Jenkinson EJ, Anderson G. Semin Immunol. 2003 Apr;15:91. doi: 10.1016/s1044-5323(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 27.Nagasawa T. Nat Rev Immunol. 2006 Feb;6:107. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 28.Zweidler-McKay PA, et al. Blood. 2005 Dec 1;106:3898. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.