Abstract
Genetic and biochemical data support Kinase Suppressor of Ras 1 (KSR1) as a positive regulator of the Ras-Raf-MAPK pathway, functioning as a kinase and/or scaffold to regulate c-Raf-1 activation. Membrane translocation mediated by the KSR1 CA3 domain, which is homologous to the atypical PKC C1 lipid-binding domain, is a critical step of KSR1-mediated c-Raf-1 activation. In this study, we used an ELISA to characterize the KSR1 CA3 domain as a lipid-binding moiety. Purified GST-KSR1-CA3 protein effectively binds ceramide but not other lipids including 1,2-diacylglyceol, dihydroceramide, ganglioside GM1, sphingomyelin and phosphatidylcholine. Upon epidermal growth factor stimulation of COS-7 cells, KSR1 translocates into and is activated within glycosphingolipid-enriched plasma membrane platforms. Pharmacologic inhibition of ceramide generation attenuates KSR1 translocation and KSR1 kinase activation in COS-7 cells. Disruption of two cysteines, which are indispensable for maintaining ternary structure of all C1 domains and their lipid binding capability, mitigates ceramide-binding capacity of purified GST-KSR1-CA3 protein, and inhibits full length KSR1 membrane translocation and kinase activation. These studies provide evidence for a mechanism by which the second messenger ceramide can target proteins to subcellular compartments in the process of transmembrane signal transduction.
Key Words: KSR1, C1 Domain, Ceramide
Introduction
The cysteine-rich C1 domain is a conserved structure of about 50 amino acids that has the characteristic composition HX12CX2CXnCX2CX4 HX2CX7C, where H is histidine, C is cysteine, X is any other amino acid, and n is 13 or 14 [1]. Two repeat C1 domains located within the same protein are designated C1A and C1B, respectively. C1 domains were initially identified as the phorbol ester and 1,2-dialyglycerol binding moieties of the protein kinase C (PKC) family of serine/ threonine kinases [2, 3]. Elucidation of the solution structure of the C1B domain of PKCα by NMR [4], and co-crystallization of PKCδ C1B domain with bound phorbol 13-acetate [5] revealed a globular structure comprised of two β sheets and a small α helix, with three cysteines and one histidine coordinating a single Zn+ ion at the end of each β sheet. At the top of the domain, there is a ligand-binding cleft composed of positively-charged residues, surrounded by hydrophobic residues. Lipid binding displaces water molecules within the cleft, generating a continuous hydrophobic surface that facilitates tight association with membrane. C1 domains have been classified into two types based on structure and function, the typical C1 domain that binds phorbol ester/1,2-diacylglycerol (DAG) and the atypical C1 domain that does not [6]. Conventional PKCs contain two typical C1 domains in tandem. Alternatively atypical PKCs contain only one C1 domain. The atypical C1 domain has been implicated in interaction with the second messenger ceramide [7, 8], and with other proteins [9, 10]. Further, recent data indicate that C1 domains exist in a variety of non-PKC proteins, including RasGRP, Raf family members and Kinase Suppressor of Ras1 (KSR1), suggesting they may play roles in diverse signaling processes [11]. For example, in T cells DAG activates the Ras signaling cascade by a novel mechanism via interaction with the RasGRP C1 domain [12], and a DAG analogue that preferentially binds RasGRP has been developed [13].
KSR1 is an evolutionarily conserved component of the Ras signaling pathway, originally discovered in genetic screens for novel downstream effectors of Ras in Caenorhabditis elegans and Drosophila melanogaster. KSR1 appears to function as a positive regulator of the MAPK pathway either upstream of or parallel to Raf [14, 15, 16, 17]. While two different mechanisms have been proposed for KSR1 function, that of a scaffold protein coordinating assembly of the MAPK signaling module [18] or a proline-directed Ser/Thr kinase that phosphorylates and activates c-Raf-1 [19], our group routinely detects a kinase activity towards c-Raf-1 using a two-stage in vitro kinase assay in which KSR1 first phosphorylates c-Raf-1, and then activated c-Raf-1 is separated from KSR1 and used in a conventional in vitro MAPK assay to sequentially signal through MEK1, ERK1, and Elk1.
KSR1 contains five conserved domains, including a KSR-unique amino-terminal CA1 domain, a proline-rich CA2 domain, a cysteine-rich CA3 domain that belongs to the atypical C1 domain family, a serine/threonine-rich CA4 domain and a carboxyl-terminal CA5 kinase domain. The overall sequence of KSR1 bears high similarity to c-Raf-1 [16]. The CA3 domain appears critical for KSR1 function, as KSR1 with two cysteine mutations within this domain cannot translocate into plasma membrane in response to PDGF stimulation, and is unable to facilitate Ras signaling [20, 21, 22]. Ceramide, which has been implicated in the process of c-Raf-1 activation [23, 24, 25], has also been found to induce KSR1 activation [19]. While studies from Gulbins and co-workers indicate direct interaction between ceramide and the KSR1 CA3 domain [26], other studies dispute this finding [22].
In the present study, we standardize a ceramide-binding, an enzyme-linked immunosorbent assay (ELISA) using an anti-ceramide antibody, and then use this assay to characterize the KSR1 C1 domain as a lipid-binding moiety that prefers ceramide over 1,2-diacylglycerol. Mutation of two cysteines (C359S/C362S) within this domain, which disrupts its ternary structure, attenuates ceramide binding, and abrogates epidermal growth factor (EGF) EGF-induced KSR1 membrane translocation into sphingolipid-rich membrane platforms, and KSR1 kinase activation. These experiments demonstrate the atypical C1 domain of KSR1 as a bone fide ceramide-binding domain that serves to target proteins into sphingolipid-rich macrodomains for transmembrane signaling.
Materials and Methods
Lipids
Ceramides (C2:0, C8:0, C14:0, C16:0, C18:0, C18:1, C20:0, C24:0 and C24:1 species); 1,2-dioleoyl-sn-glycerol (DAG); (2S,3R,4E)-2-acylaminooctadec-4-ene-3-hydroxy-1-phosphocholine (SM); L-a-phosphatidylcholine (PC); and GalBeta1-3GalNAcBeta1-4(NeuAcAlpha2-3)GalBeta1-4GlcBeta1-1′-Cer (GM1) were purchased from Avanti Polar Lipids. BSA-conjugated C16-ceramide and BSA-conjugated C16-dihydroceramide were from Biomol.
ELISA ceramide binding assay
The CA3 domain (amino acids 320-388) of wild type and C359S/C362S mutant human KSR1 were fused to the glutathione-S-transferase (GST) gene in the pGEX-3T vector (GE Healthcare) using EcoRI and SalI. GST-fusion proteins were purified from E. coli BL21 using glutathione-conjugated agarose according to manufacturer's instructions, eluted in PBS (0.2 g/l KCl, 0.2 g/l KH2PO4, 8 g/l NaCl, 1.15 g/l Na2HPO4) with 20 mM glutathione, and dialyzed in PBS. Purified proteins were separated with 10% SDS-PAGE and purity was assessed by Coomassie blue staining.
ELISA was developed to examine binding of the CA3 domain to ceramide [27]. For these studies, Nunc MaxiSorp 96 well plates were coated with 8 nM lipid in methanol at 4°C overnight, and thereafter the methanol was evaporated for 2 h at room temperature. Plates were washed twice with 0.05% Tween 20 in PBS, and non-specific sites were blocked with 3% lipid-free BSA in PBS for 4 h. Plate-bound lipid was incubated with purified recombinant CA3 protein (0-1600 nM) in 1% lipid-free BSA in PBS in triplicate for 2h, followed by three washes with 0.05% Tween 20 in PBS, incubation with HRP-conjugated anti-GST Ab (1:10,000, GE Healthcare) for 2 h in 1% lipid-free BSA in PBS, and three final washes with 0.05% Tween 20 in PBS. Binding was detected by addition of a mixture of 50 ml hydrogen peroxide (0.02%) and 50 ml tetramethylbenzidine (TMB) (0.4 g/l), and stopped using 50 ml sulfuric acid (2 N). Resulting absorbancies were quantitated at 450 nm with wavelength correction at 570 nm on an ELISA reader (Molecular Devices).
For detection of ceramide binding using anti-ceramide Ab, the procedures are as described above except BSA-conjugated lipid was utilized, and anti-ceramide IgM MID 15B4 (1:10, Alexis Biochemicals) and HRP-conjugated anti-mouse IgM secondary Ab (1:2000, Sigma-Aldrich) were used successively in 2 h incubation.
Expression of KSR1 in COS-7 Cells
Mouse pCMV-Myc-KSR1 was generated as described [28]. Myc-KSR1-C359S/C362S, with a substitution of serine residues for two conserved cysteines, was generated using QuikChange® Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions, and sequenced. For EGF studies, COS-7 cells were plated at a density of 1.5×106 cells in 150-mm plates and grown overnight to ∼70% confluence. Culture medium was replenished with fresh medium 1 h before transfection, and cells were transfected with 8 µg of DNA/ plate using FuGeneTM 6 transfection reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. At 48 h post-transfection, cells were placed in serum-free medium for 12 h prior to treatment with EGF. Cells were then harvested in Nonidet P-40 lysis buffer [25 mM Tris (pH 7.5), 137 mM NaCl, 10% glycerol, 1% Nonidet P-40, 2 mM EDTA, 5 mM NaVO4 and protease inhibitor cocktail (Roche Diagnostics GmbH)]. The homogenate was incubated on ice for 15 min before centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was thereafter collected and protein concentration measured using Protein Assay Reagent (Bio-Rad Laboratories). Lysates were divided into aliquots and stored at −80°C for subsequent use. KSR1 expression was determined by Western blot analysis.
Immunofluorescence Microscopy
KSR1 co-localization with raft markers was detected by indirect immunofluorescence staining. COS-7 cells were seeded onto 12 mm glass slips and transfected with Myc-KSR1 cDNAs. After stimulation with EGF, cells were fixed immediately with freshly prepared 4% paraformaldehyde/PBS for 15 min, washed twice with PBS, and permeabilized by incubation in 0.2% Triton X-100/PBS for 5 min. After blocking in 4% goat serum/PBS for 1 h, cells were stained for KSR1 using anti-Myc Ab (1:1000, Cell Signaling Technology) and detected with Alexa Fluor 488 anti-mouse secondary Ab (1:1000, Molecular Probes), protected from light. After three more washes with PBS, coverslips were mounted onto glass slides with antifade mounting medium containing DAPI (Vector Laboratories) and visualized under a Lecia TCS SP2 AOBS laser confocal microscope. The percentage of cells displaying KSR1 membrane translocation was determined by counting 150-250 cells per point.
For co-staining studies, fixed cells were incubated with rabbit anti-caveolin-1 Ab (1:3000, BD Bioscience) for co-staining of KSR1 with caveolin-1, or with mouse anti-ceramide Ab MID 15B4 (1:50) for co-staining of KSR1 with ceramide. For these studies, staining was detected with anti-mouse Alexa Fluor 488 or anti-rabbit Alexa Fluor 594 secondary Ab (1:1000), respectively. Alternatively, for co-localization of KSR1 with another raft marker, ganglioside GM1, cells were fixed with 2% paraformaldehyde/0.2% glutaraldehyde/PBS, incubated with FITC-conjugated cholera toxin β-subunit (1:250, Sigma-Aldrich) without permeabilization and then permeabilized before subsequent staining with rabbit anti-Myc Ab and Alexa Fluor 594 anti-rabbit Ab as above.
Two-stage KSR1 Activity
Assay-KSR1 kinase activity was measured by the two-stage assay as previously described [29]. Briefly, Myc-KSR1 or Myc-C359S/C362S-KSR1, immunoprecipitated from 500 µg of total cell lysates with 40 µ1 of agarose-conjugated anti-c-Myc antibody (Sigma-Aldrich), were washed with 1 M NaCl containing Nonidet P-40 buffer five times followed by one wash with 20 mM Tris-HCl (pH 7.4). In the first stage of the assay, immunopurified KSR1 was incubated at 30°C in 20 µ1 of reaction buffer containing 60 µM ATP, 7.5 mM MgCl2, and 50 ng of human FLAG-c-Raf-1 purified to homogeneity with agitation. After 30 min, KSR1-containing beads were pelleted (10,000 x g for 1 min at 4°C), and 10 µ1 of supernatant containing activated c-Raf-1 was added in the second stage of the assay to 20 µ1 of reaction buffer containing 60 µM ATP, 7.5 mM MgCl2, 0.1 µg of unactivated murine GST-MEK1 (Upstate Biotechnology), 1.0 µg of unactivated murine GST-ERK-2/MAPK (Upstate Biotechnology), and 2 µg of human GST-Elk-1 fusion protein (Cell Signaling). After 30 min, the reaction was stopped by addition of 10 µ1 of 4x Laemmli buffer. Phosphorylated Elk-1 was resolved by 7.5% SDS-PAGE and visualized by Western blot analysis using rabbit anti-phospho-Elk-1 (Ser383) antibody (Cell Signaling). For the kinase assay using sucrose gradient fractionation, 6 ml light fraction (fractions 4-9 pooled together), 2 ml heavy fraction (fraction 11 and 12 pooled together) or the 50 µ1 pellet fraction were adjusted to 6 ml with MES lysis buffer (25 mM MES, pH 6.5, 0.15 M NaCl, 0.1% Triton X-100 and protease inhibitor cocktail (Roche Diagnostics GmbH). KSR1 was immunoprecipitated from each pooled fraction, and its kinase activity was evaluated using the two-stage assay.
Isolation of Sphingolipid-rich Membrane Platforms
Sphingolipid-rich membrane domains were isolated based on their buoyant density using isopycnic equilibrium sucrose density gradient centrifugation. Low density sphingolipid-rich domains were purified from COS-7 and MCF-7 cells essentially as described [30]. Briefly, cells (two 70% confluent 150-mm dishes) were washed twice with ice-cold PBS, scraped into 1 ml of ice-cold MES lysis buffer and incubated on ice for 30 min. After homogenization by 20 strokes of a tight fitting Dounce homogenizer at 4°C, cells were sonicated at 4°C three times (20 s each at setting 2 with Branson Sonifier 250). Whole-cell lysates were adjusted to 45% sucrose by addition of 1 ml of lysis buffer (minus Triton X-100) containing 90% sucrose and placed at the bottom of 12 ml ultracentrifuge tube. A discontinuous gradient was formed above the lysate by adding 3 ml 35%, 2 ml 25%, 2 ml 15% and 3 ml 5% sucrose solutions, and the tubes were centrifuged at 38,000 g for 17 h in an SW-41 rotor at 4°C. Twelve fractions (1 ml each) were collected beginning at the top of the gradient. The pellet at the bottom of the tube was resuspended in 1 ml of lysis buffer (minus Triton X-100). Sucrose density of each fraction was measured by refractometer and protein content was determined using Protein Assay Reagent (Bio-Rad Laboratories).
For COS-7 cells, proteins in each fraction (except the pellet fraction) were concentrated using trichloroacetic acid precipitation: 10 µ1 sodium deoxycholate (20mg/ml) was added into each fraction, and after 30 min on ice, 250 µ1 trichloroacetic acid was added. After an overnight incubation at 4°C, proteins were pelleted (14,000 x g for 15 min at 4°C) and washed twice with 80% acetone. Pellets were resuspended in 100 µ1 Laemmli sample buffer. A standard immunoblot was established empirically to show the relative concentration of Myc-KSR1 in different fractions. 50 µ1 from concentrated fraction 1 to 10, 10 µ1 from concentrated fraction 11 and 12, and 10 µ1 direct from pellet fraction were separated with 7.5% SDS-PAGE and visualized by Western blot analysis using anti-Flotillin-1 Ab (1:15000, BD Bioscience) or anti-Myc Ab (1:5000).
For MCF-7 cells, 30 µ1 from each 1 ml gradient fraction were separated by 10% SDS-PAGE and visualized by Western blot using anti-Flotillin-2 Ab (1:5000, BD Bioscience) or anti-c-Raf-1 Ab (1:7500, Upstate Biotechnology).
SDS-PAGE and Western blot analysis
Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and probed using appropriate antibodies. Western blots were developed using VisualizerTM Western blot detection kit (Upstate Biotechnology) and bands were quantified by densitometry using ImageJ (NIH).
KSR1 AS-ODN treatment of MCF-7 cells
Human KSR1 antisense ODN (AS-ODN) (5’-CTT TGC CTC TAG GGT CCG-3’) and control sense-ODN (5’-CGG ACC CTA GAG GCA AAG-3’) were generated as phosphorothioate derivatives (Genelink Inc.). MCF-7 cells were transfected with 6 µM AS-ODN or sense-ODN using Oligofectamine (Invitrogen) as described [31].
Cell Proliferation Assay
To generate stable cell lines that overexpress KSR1, KSR1-C359S/C362S or PCMV vector, COS-7 cells were transfected with respective plasmids using FuGeneTM 6 transfection reagent and selected by treatment with G418 (400 µg/ml) for 14 days. Resistant clones were separately propagated and expression of KSR1 or KSR1-C359S/C362S in stable transfectants was analyzed by Western blot using anti-Myc Ab. Stable cell lines were plated at 1000 cells per well using 96-well culture plates. Proliferation was monitored using WST-1 ((4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) (Roche Applied Science) according to manufacturer's instructions. Absorbance of converted dye was measured at a wavelength of 450nm with background subtraction at 650nm.
Statistics
Statistical analysis was performed by student's t test.
Results
C359S/C362S Mutation in KSR1 CA3 Domain Decreases its Ceramide Binding Affinity
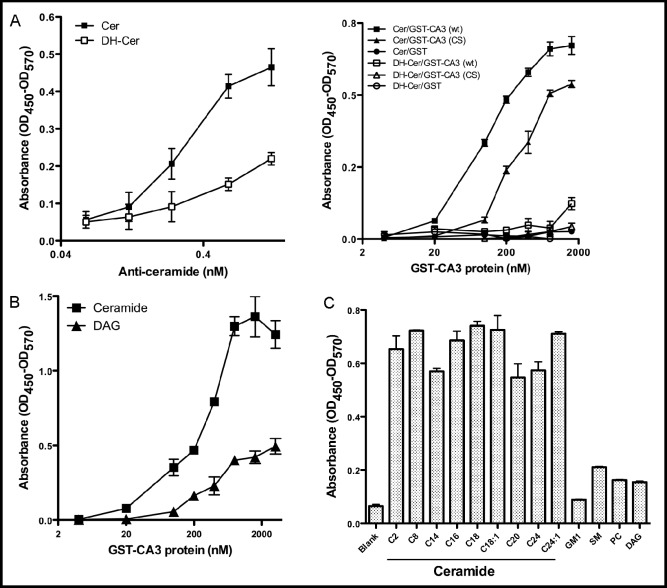
As C1 domains are more and more often being identified as lipid binding moieties that regulate protein localization and activation status, we examined the lipid binding capabilities of the KSR1 CA3 domain, which shares sequence similarity with atypical C1 domains [7]. For these studies, we generated a recombinant human GST-KSR1 CA3 domain construct and an ELISA assay that employs plate-bound lipid [27]. In this assay, commercial anti-ceramide MID15B4 antibody recognized plate-bound C16-ceramide in a dose-dependent manner, while reacting weakly with C16-dihydroceramide, the ceramide precursor that lacks a trans double bond at positions 4-5 of the sphingoid base backbone (Fig. 1A, left panel). Similarly, recombinant GST-CA3 recognized C16-ceramide in a dose-dependent manner, with absorbance detection increasing 100-fold from a baseline of 0.007±0.004 artificial units to 0.67±0.03 over the range of 0-1600 nM GST-CA3 (Fig. 1A, right panel). In contrast, recombinant GST-CA3 bound C16 -dihydro-ceramide minimally, indicating a stringent ligand recognition profile. Next, the role of the two conserved cysteine residues at KSR1 positions 359 and 362 within the CA3 domain were examined for ceramide binding. These two cysteines are conserved in all functional C1 domains and are critical for maintaining three-dimensional structure of the domain, and domain function [22]. Cysteine to serine mutation of those residues (C359S/ C362S) significantly impaired GST-KSR1 CA3 binding to C16-ceramide, right-shifting the dose-response curve and decreasing the maximal binding capacity (Fig. 1A, right panel). Taken together, these data demonstrate that the KSR1 CA3 domain specifically binds C16-ceramide in a conformationally-sensitive manner.
Fig. 1.
CA3 domain mutant CA3 (C359S/C362S) is defective in ceramide binding. An ELISA was used to measure the binding of the KSR1 CA3 domain to ceramide. (A) The wells of the Nunc-MaxiSorp plate were coated with 8 nmol/well BSA-conjugated C16-ceramide (CER) or BSA-conjugated C16-dihydro-ceramide (DH-CER). The binding of anti-ceramide Ab with ceramide (left panel) was detected by sequential incubation with HRP-conjugated anti-mouse IgM secondary Ab and TMB substrate solution as in Materials and Methods; the binding of recombinant GST-CA3, GST-CA3 (CS) and GST proteins with ceramide (right panel) was detected by sequential incubation with HRP-conjugated anti-GST Ab and TMB substrate solution. In (B), microtiter wells were coated with 8 nmol/well non-BSA-conjugated C16-ceramide and DAG or, in (C) with different ceramide species, GM1, SM, PC and DAG. The binding of recombinant GST-CA3, GST-CA3 (CS) and GST proteins with ceramide and other lipids was detected as in (A, right panel). Data are representative of one of three independent experiments in (A-C).
As the atypical C1 domain family manifests minimal DAG binding affinity [32], binding of ceramide and DAG were compared directly using our ELISA assay. For these studies a synthetic DAG, 1,2-dioleoyl-sn-glycerol, biochemically-comparable to C16-ceramide as both contain C16:0 fatty acids, was employed. Fig. 1B shows that GST-CA3 bound C16-ceramide with much greater affinity than DAG. At 200 nM GST-CA3 the isoeffect for DAG binding was 1600 nM GST-CA3, an 8-fold preference for ceramide over DAG. Similar data were derived with the biologically-active DAG, 1-stearoyl-2-arachidonoyl-sn-glycerol (data not shown).
Additional studies examined the ceramide-binding preferences for the KSR1 CA3 domain utilizing a set of ceramides that represent the physiologic profile of ceramide in most mammalian systems. As shown in Fig. 1C, 400 nM recombinant GST-CA3 bound the different ceramide species without preference for fatty acyl chain length. Further, the KSR1 CA3 domain showed little binding of other lipids including the ganglioside GM1, sphingomyelin (SM), phosphatidylcholine (PC) and free sphingosine (not shown). These data suggest the mimimal ligand binding requirements for the KSR1 CA3 domain are a sphingosine backbone in amide linkage to a saturated or monounsaturated fatty acid.
KSR1 CA3 Domain C359S/C362S Mutation Prevents KSR1 Plasma Membrane Translocation and Activation
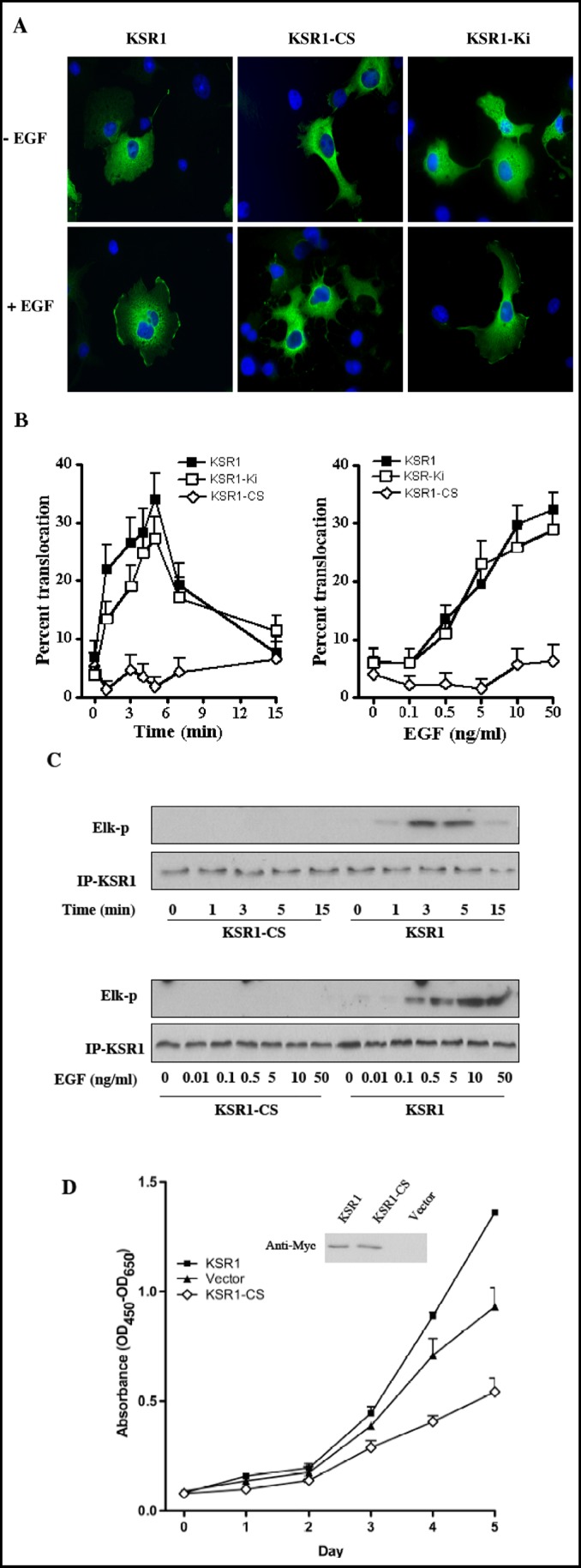
The KSR1 CA3 domain was reported as critical for KSR1 translocation to the plasma membrane and facilitation of Ras-mediated Xenopus oocyte maturation [21]. To gain insight into the mechanism by which the CA3 domain regulates KSR1 localization and kinase activity, we generated a full length KSR1 mutant with the two conserved C1 cysteine residues mutated to serine (KSR1-C359S/C362S), and assessed membrane translocation and kinase activity upon EGF stimulation. For these studies, COS-7 cells, transiently transfected with pCMV-Myc-KSR1 or pCMV-Myc-KSR1-C359S/ C362S (KSR1-CS), were treated with 10 ng/ml EGF for various times (0-15 min), fixed, and transfected proteins localized by confocal fluorescence microscopy. KSR1 and KSR1-CS predominantly exhibited cytoplasmic staining in quiescent cells (Fig. 2A, upper panels). In response to EGF treatment, wild type KSR1 redistributed from cytosol to plasma membrane, demonstrating discrete signal in large domains across the cell surface as shown in the lower panels of Fig. 2A. These results are highly similar to PDGF-induced KSR1 translocation to the plasma membrane of NIH/3T3 cells as reported by Zhou et al [22]. This event was time dependent, with translocation observed as early as 1 min after treatment (Fig. 2B) and a maximal effect at 5 min, decreasing towards baseline at 15 min.
Fig. 2.
An intact CA3 domain of KSR1 is required for its translocation to plasma membrane and kinase activation upon EGF stimulation, and for its enhancement of COS-7 cell proliferation. COS-7 cells transiently transfected with Myc-KSR1, Myc-Ki-KSR1 or Myc-KSR1-CS were serum starved for 12 h before treatment with 10 ng/ml EGF for the indicated times or increasing EGF doses for 3 min. Membrane translocation of KSR1 was determined by indirect immunofluorescence staining with anti-Myc Ab and quantified using fluorescence microscopy. Representative images are shown in (A). Data (mean±SD) are collated from three independent experiments in which 200 cells were analyzed per point (B). In (C), kinase activity of Myc-KSR1 or Myc-KSR1-CS mutant, treated 12 h post serum-starvation with 10 ng/ml EGF for the indicated times or with increasing dose of EGF for 3 min, was measured using the two-stage in vitro reconstitution assay as described in Materials and Methods. Results represent one of three independent experiments. In (D), proliferation of COS-7 cells stably-expressing wild type Myc-KSR1 or mutant Myc-KSR1-CS was monitored using WST-1 reagent. The insert shows the expression level of Myc-KSR1 and Myc-KSR1-CS in the stably-transfected cell lines. Data (mean±SD) are compiled from three independent experiments.
To establish whether translocation of the KSR1-CS mutant to plasma membrane was abrogated or attenuated upon EGF stimulation, KSR1 and KSR1-CS transfected COS-7 cells were treated with varying doses of EGF (0.1-50 ng/ml for 3 min). EGF conferred dose-dependent membrane KSR1 translocation, with significant redistribution observed with as little as 0.5 ng/ml EGF addition, whereas up to 50 ng/ml EGF treatment induced no significant redistribution of KSR1-CS (Fig. 2A and 2B). In contrast, a kinase inactive KSR1 mutant previously reported by our group, KSR1-Ki (D683A/ D700A), generated by substitution of two aspartic acid residues in the kinase domain with alanines, displayed time- and dose-dependent membrane translocation comparable to wild-type KSR1 (Fig. 2A and 2B). These results demonstrate that in COS-7 cells, EGF regulates translocation of KSR1 from cytosol to plasma membrane in a time- and dose-dependent manner, requiring structural integrity of the CA3 domain, but not active KSR kinase, for this process.
Our previous investigation showed that KSR1 kinase activity was activated after EGF treatment [29]. To determine whether an intact CA3 domain is required for KSR1 activation, we studied KSR1 and KSR1-CS kinase activity in response to EGF stimulation. The kinase activity of KSR1 was determined by a two-stage in vitro reconstitution assay [29]. In the first stage, equal amounts of immunoprecipitated KSR1 are used to phosphorylate recombinant c-Raf-1, which is then separated from KSR1 and used in a “classic” c-Raf-1 kinase reaction in the second stage employing recombinant MEK1, ERK2 and Elk-1. Elk-1 phosphorylation serves as readout for activation of the MAPK cascade, visualized by Western blot. In COS-7 cells transiently transfected with pCMV-Myc-KSR1 or pCMV-Myc-KSR1-CS, EGF induced time-and dose- dependent kinase activation of wild type KSR1 but not KSR1-CS (Fig. 2C). While immunoprecipitated KSR1 and KSR1-CS isolated from untreated cells did not display reconstituted kinase activity, EGF (10 ng/ml) induced a detectable increase in KSR1 kinase activity by 1 min of stimulation, with activity peaking by 3-5 min and dissipating rapidly thereafter (Fig. 2C, upper panels). This time course for KSR1 kinase activity correlates closely with that of KSR1 membrane translocation detected by immunostaining (compare Figs. 2B and C). Activation of KSR1 kinase activity was dose-dependent, detectable at 0.5 ng/ml and maximal at 10 ng/ml EGF (Fig 2C, lower panels). In contrast, immunoprecipated KSR1-CS was incapable of reconstituting the MAPK signaling cascade even with 50 ng/ml EGF addition. These studies indicate that in response to EGF stimulation membrane translocation of KSR1 correlates closely with the activation of its kinase activity, with an intact CA3 domain critical for both events.
KSR1 Requires Intact CA3 Domain to Promote Cell Proliferation
KSR1 positively controls duration and intensity of the mitogenic signaling pathway [33, 34]. While murine embryonic fibroblasts cells and T cells from KSR1 knockout mice show decreased proliferation rates [35, 36], KSR1 overexpression in cultured cells significant increases proliferation rates, an effect for which kinase activity is indispensable [31]. To examine the significance of the CA3 domain to the mitogenic function of KSR1, COS-7 cells transfected with pCMV-Myc-KSR1 or pCMV-Myc-KSR1-CS were selected for neomycin resistance to obtain a stably-transfected population of cells. Overexpression of wild type and mutant KSR1 was confirmed by Western blot (Fig. 2D, inset). Compared with vector-transfected controls, a moderate increase in growth rate in KSR1 overexpressing cells was observed (Fig. 2D), consistent with the role of KSR1 in the MAPK mitogenic pathway. In contrast, cells overexpressing KSR1-CS showed a 40% reduction in proliferation, similar to data previously reported using kinase inactive KSR1 in A431 cells [31]. These results demonstrate that an intact CA3 domain, which is critical for KSR1 binding to ceramide, membrane translocation and kinase activity, is also important for KSR1 to deliver a proliferative signal.
KSR1 Co-localizes with Raft Markers in COS-7 Cell Plasma Membranes
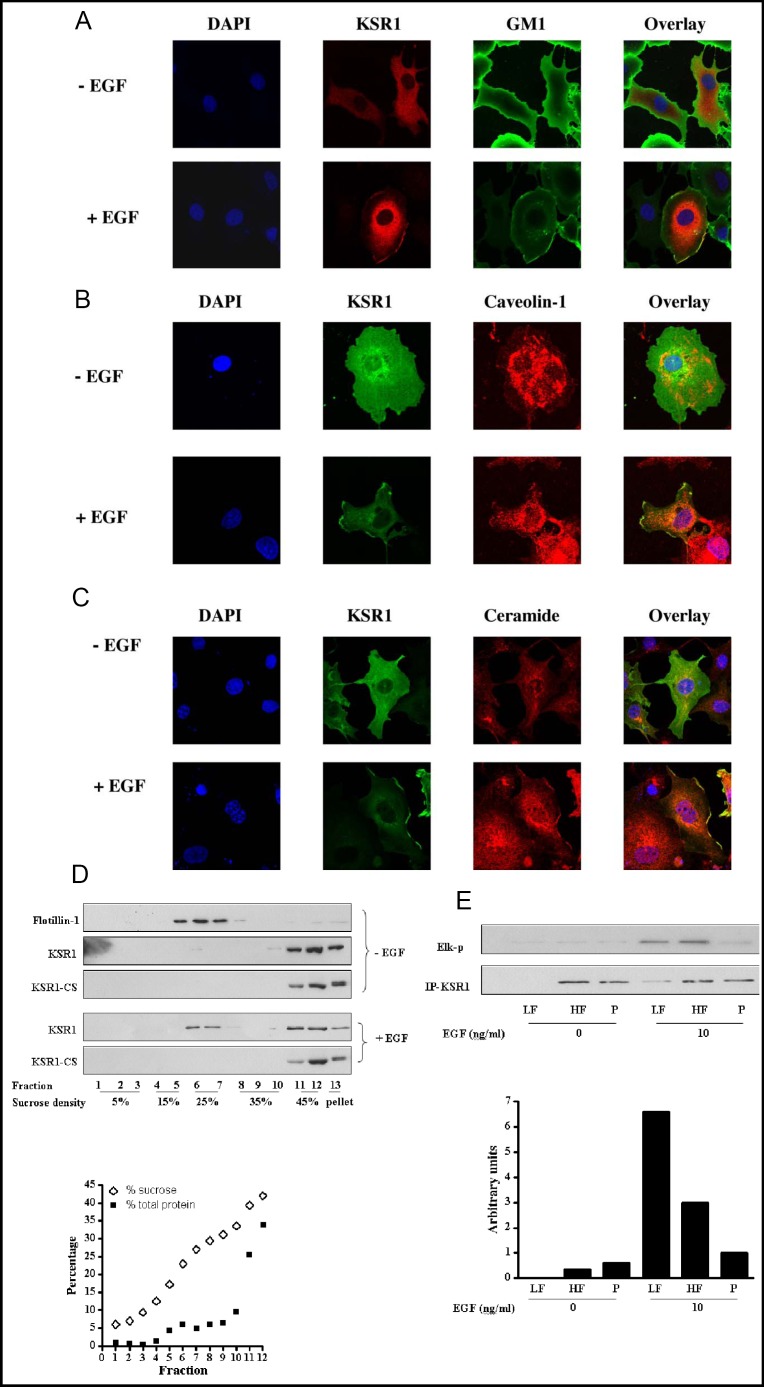
Previous studies indicated macrodomains derived from plasma membrane glycosphingolipid-enriched microdomains (also known as rafts) are involved in Rasmediated MAPK pathway activation providing a specialized environment that facilitates formation of the signaling complex [37, 38, 39]. To investigate whether the surface platforms that contain KSR1 upon EGF stimulation represent sphingolipid-enriched macrodomains, KSR1 localization was compared with that of raft marker GM1, caveolin-1 and ceramide by laser scanning confocal microscopy. For these studies, COS-7 cells transiently transfected with pCMV-Myc-KSR1 were double stained with anti-Myc Ab and FITC-conjugated cholera toxin subunit B (CTXB), which specifically binds ganglioside GM1 (Fig. 3A), anti-caveolin-1 Ab (Fig. 3B) or anti-ceramide Ab (Fig. 3C). In untreated cells, little colocalization of KSR1 and GM1, caveolin-1 or ceramide was observed (Fig. 3 A, B and C, upper panels). However, at 3 min of stimulation with 10 ng/ml EGF, KSR1 was detected in cell surface macrodomains coincident with GM1, caveolin-1 and ceramide (merged yellow signal in Figs. 3 A, B and C, lower panels, respectively). Coloca-lization of KSR1 with another lipid raft marker flotillin-1 was also detected upon EGF treatment (data not shown). These studies indicate that KSR1 translocates into raft-derived plasma membrane platforms upon EGF stimulation.
Fig. 3.
KSR1 is recruited into a ceramide-rich surface macro-domain and activated upon EGF stimulation. COS-7 cells, transiently transfected with Myc-KSR1, either left untreated or stimulated with 10 ng/ml EGF for 3 min, were double-stained for KSR1 and GM1 with anti-Myc Ab and FITC-conjugated cholera toxin subunit B (CTXB), respectively (A); for KSR1 and caveolin-1 with anti-Myc Ab and anti-caveolin-1 Ab, respectively (B); or for KSR1 and ceramide with anti-Myc Ab and anti-ceramide Ab, respectively (C). Immunofluorescent staining was analyzed by confocal microscopy. KSR1 co-localization with GM1, caveolin-1 or ceramide at the plasma membrane after EGF stimulation is identified as cell surface yellow staining in overlay images. Representative images from one of three similar experiments are shown. In (D), cell lysates from Myc-KSR1 or Myc-KSR1-CS transfected COS-7 cells, treated with or without 10 ng/ml EGF, were separated using discontinuous sucrose gradient fractionation. Protein in each fraction was TCA precipitated, separated by SDS-PAGE and immuoblotted with antibodies against Flotillin-1 or Myc. The sucrose and total protein profiles after centrifugation are shown below. These results represent one of three independent experiments. In (E), Myc-KSR1 was immunopre-cipitated from Light Fraction (LF, combined fractions 4-9), Heavy Fraction (HF, combined fractions 11 and 12) and Pellet (P), prepared as described above, and kinase activity was measured using the two-stage in vitro reconstitution assay. Arbitrary KSR1 kinase activity was quantified by densitometry and normalized to levels of immunopre-cipitated KSR1. These results represent one of three independent experiments.
Activation of KSR1 in sphingolipid-enriched platforms
Proteins located in glycosphingolipid-enriched microdomains or platforms derived thereof can be isolated based on their buoyant density using sucrose density gradient centrifugation. To confirm the confocal findings that KSR1 translocates into sphingolipid-enriched platforms upon EGF treatment, a light membrane fraction was isolated from control and EGF-treated COS-7 cells transfected with pCMV-Myc-KSR1 or pCMV-Myc-KSR1-CS by discontinuous sucrose gradient fractionation. After equilibrium centrifugation, the density of each fraction and corresponding protein concentration were measured as shown in Fig. 3D, lower panel. Flotillin-1, a well-defined raft-associated protein, confirmed that fractions 4-9 with sucrose density from 15-30% represent a light fraction enriched for glycosphingolipid-rich domains. Consistent with published data [40], flotillin-1 was nearly absent from heavy membranes found in fractions 11 and 12 with sucrose density between 40-45% representing bulk membrane, and from the pellet fraction 13. The percentage of KSR1 that localized to light, heavy or pellet fractions in the Western blots was quantified using ImageJ densitometry analysis and directly compared after correction for loading. In quiescent COS-7 cells, KSR1 and KSR1-CS were absent from the light membrane fraction, with about 70% in the heavy fraction and 30% in the pellet fraction (Fig. 3D, upper panel). The presence of KSR1 in the pellet fraction, which contains cytoskeleton and nucleus, is consistent with the finding that KSR1 is targeted to the actin filament through leukocyte-specific protein 1 (LSP1) [41]. Upon treatment with 10 ng/ml EGF for 3 min, 3-4% of KSR1 (based on loading dilutions) redistributed into the light membrane fraction. In contrast, no KSR1-CS redistribution was detected.
As substantive evidence indicates that activation of Ras and c-Raf-1 occurs in cell surface raft-like structures [25, 39, 42, 43], we investigated whether the KSR1 that translocated into the light membrane fraction was activated compared to the KSR1 in heavy membrane and pellet fractions. For these studies, Myc-tagged KSR1 was immunoprecipitated from pooled light, heavy and resuspended pellet fractions, before and after EGF stimulation of COS-7 cells. Specific KSR1 kinase activity was determined employing the two-stage KSR1 kinase assay with Elk-1 phosphorylation as readout (Fig. 3E, upper panel). The kinase activity in each fraction normalized for KSR1 protein level is shown in Fig. 3E, lower panel. In the absence of EGF, there is almost no detectable kinase activity associated with KSR1 immune complexes. Upon EGF stimulation, KSR1 was activated in both light and heavy fractions, but not in the pellet fraction. The specific KSR1 kinase activity in the light fraction was more than two fold higher than that in the heavy fraction. The active KSR1 in sphingolipid-enriched macrodomains represents 55% of total cellular KSR1 activity. These results indicate that KSR1 recruited into sphingolipid-rich light membranes after EGF stimulation is highly active.
KSR1 downregulation impairs c-Raf-1 translocation into sphingolipid-rich platforms and activation
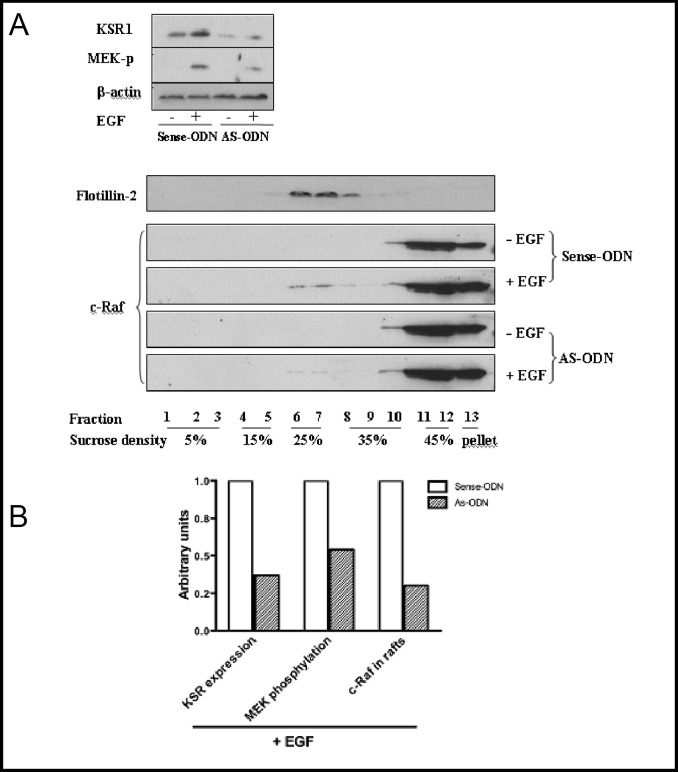
To investigate whether KSR1 regulates c-Raf-1 translocation into sphingolipid-rich surface macrodomains we transfected MCF-7 cells, which manifest high levels of endogenous KSR1 (so that down regulation can be easily quantified), with 6 µM of antisense ODN targeting KSR1, or a corresponding control sense ODN, and evaluated impact on EGF-stimulated c-Raf-1 redistribution into light membranes. As shown in Fig. 4A and quantified in Fig. 4B, treatment of MCF-7 cells with AS-ODN resulted in 60% down-regulation of endogenous KSR1 by Western Blot analysis compared to sense ODN, which was without effect. As in PANC-1 cells [31], KSR1 down-regulation with AS-ODN blocked EGF signaling through the MAPK pathway in MCF-7 cells, with a 50% reduction in EGF-induced MEK1/2 phosphorylation (Fig. 4A and B). While in the absence of EGF, c-Raf-1 was excluded from light membranes, EGF induced redistribution of about 5% of endogenous c-Raf-1 into the light membrane fraction (Fig. 4A, lower panel), similar to published data [44]. Consistent with KSR1 being critical for c-Raf-1 translocation, KSR1 down-regulation resulted in a ∼70% reduction in c-Raf-1 redistribution (Fig. 4A, lower panel, and Fig. 4B).
Fig. 4.
KSR1 antisense ODN impairs EGF-induced c-Raf-1 translocation into sphingolipid-rich light membranes. MCF-7 cells, transfected with 6 µM KSR1 AS-ODN or sense-ODN, were either left untreated or treated with 10 ng/ml EGF for 3 min. The expression level of endogenous KSR1 and the phosphorylation of MEK1/2 were determined by Western blot analysis using anti-KSR1 and anti-phospho-MEK1/2 Abs as described in Materials and Methods. Equal loading of samples were confirmed using β-actin (A, upper panel). Proteins were also subjected to sucrose gradient fractionation as above, and the presence of c-Raf-1 in the light membrane fraction was visualized by Western blot using anti-c-Raf Ab. Flotillin-2 was used as a raft marker (A, lower panel). In EGF-stimulated MCF-7 cells, down-regulation of KSR1 expression and the attenuated MEK1/2 phosphorylation and c-Raf-1 recruitment into the raft fraction after AS-ODN treatment were quantified by densitometry and presented as arbitrary optical density units (B). These results represent one of three similar experiments.
Ceramide down regulation impairs KSR1 translocation into sphingolipid-rich platforms, and KSR1 kinase activity
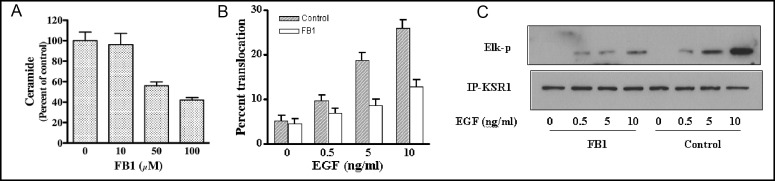
To our surprise, EGF treatment did not increase ceramide levels at any time up to 10 min in COS-7 cells (data not shown). Nonetheless, pre-treatment of these cells with the natural ceramide synthase inhibitor Fumonisin B1 (FB1), a sphingoid base analog that serves as a competitive inhibitor of ceramide synthase activity with an IC50 two logs left shifted compared to the natural sphingoid base substrate [45], prevented KSR1 activation. As shown in Fig. 5A, FB1 treatment resulted in dose-dependent reduction in COS-7 ceramide levels, with 100 µM FB1 reducing ceramide to 40% of baseline at 16 h of incubation. Maximal reduction was observed at 16 h of FB1 treatment, with ceramide levels returning towards baseline by 24 h (data not shown). With 100 µM FB1 pre-treatment, EGF (10 ng/ml)-induced ceramide-rich platforms were similarly reduced by 50-60% (not shown). Consistent with this latter observation, FB1 reduced KSR translocation into platforms (Fig. 5B) measured by the number of KSR1-containing platforms, and markedly reduced EGF-stimulated KSR1 kinase activation using total cellular immunoprecipitated KSR1 in the two-stage kinase assay (Fig. 5C).
Fig. 5.
FB1 inhibits KSR1 membrane translocation and kinase activation. COS-7 cells were treated with increasing doses of FB1 for 16 h and ceramide levels were measured using the diacylglycerol kinase assay as described in Materials and Methods (A). In (B), COS-7 cells transfected with Myc-KSR1 were pretreated with or without 100 µM FB1 for 16 h and then stimulated with increasing doses of EGF. Myc-KSR1 membrane translocation was determined by indirect immunofluorescence staining and quantified using fluorescence microscopy. In (C), Myc-KSR1 kinase activity was analyzed the using two-stage in vitro reconstitution assay. Results represent one of three similar experiments in (A-C).
Discussion
The current studies provide unequivocal evidence for the existence of a C1 domain that prefers ceramide to 1,2-diacylglycerol. This is evidenced by ELISA, and has functional consequence for protein targeting to ceramide-rich macrodomains on the cell surface. Disruption of the integrity of the KSR1 C1 domain by a standard approach, that is mutation of the cysteine bridges that hold it together, attenuates ceramide binding by ELISA, and EGF-induced KSR1 translocation and activation. In contrast, mutation of conserved aspartates within the kinase domain required for ATP catalysis, does not affect KSR1 translocation into sphingolipid-rich platforms. The most parsimonious molecular ordering of these events is that membrane translocation of KSR1 precedes kinase activation, required for c-Raf-1 transactivation. Collectively, these studies identify a previously-unreported mechanism for targeting proteins to sphingolipid-rich signaling compartments, with functional consequence.
These are not the first studies to show that ceramide can activate proteins that contain atypical C1 domains. In this regard, in addition to KSR1, evidence has been presented indicating that ceramide can stimulate PKC? [46], c-Raf-1 [23], and Vav [47] activity. While ceramide is capable of activating PKC? in vitro, and translocating it into glycosphingolipid-rich domains [48, 49, 50], the role of the C1 domain in this process was not examined. Alternately, while ceramide activated c-Raf-1 in intact NIH/3T3 cells, an event that required the c-Raf-1 CR2 domain, the authors emphasized that this did not indicate direct interaction of ceramide with the CR2 domain. In fact, membrane targeting appeared primarily related to the Ras binding domain of c-Raf-1. The only evidence that C1 domains might detect ceramide comes from studies of Gulbins and co-workers that show that a GST-KSR1 CA3 domain construct binds radiolabeled ceramide in an in vitro binding assay, and detects ceramide-rich platforms on the surface of JY B lymphocyte cells [26]. Using an almost identical binding assay Morrison and co-workers found no [3H]ceramide binding [22]. While reasons for these differences between groups is not clear, it should be recognized that the assay used by both groups is complex requiring a ceramide-binding protein such as bovine serum albumin to solvate ceramide, which is extremely hydrophobic. Hence the assay actually represents a competition between the two potential binding proteins. In the past a similar assay also failed to detect direct 1,2-diacylglycerol binding to typical C1 domains [51]. The current studies, which apply ELISA technology and solid phase ceramide bypass the solubility issue, and now provide definitive data to confirm that the KSR1 C1 domain is ceramide binding.
These studies further elucidate the mechanism of c-Raf-1 activation by tyrosine kinase receptors. While previous data indicated that a very small portion of the c-Raf-1 pool in Rat-1 fibroblasts cells translocated into sphingolipid-enriched domains, an event confirmed in the current studies as involved in c-Raf-1 activation, no mechanism for such translocation has been elucidated. The current studies would indicate that KSR1, likely by serving as a scaffold, targets c-Raf-1 into sphingolipid-rich platforms upon EGF stimulation, where additional post-translational modifications are engaged. KSR1 translocation in part or in whole requires ceramide recognition, although not necessarily ceramide elevation as EGF failed to induce cellular ceramide increases within the time frame of KSR1 translocation. A note of caution in this regard however, as macrodomain formation might still be due to a local ceramide elevation or a re-balancing of ceramide species favoring pro-macrodomain species, possibilities that will require substantive additional investigation. While c-Raf-1 translocation into sphingolipid domains is crucial for its activation, our studies are not consistent with the model suggested by Hancock et al. [38] regarding the activation state of the sphingolipid domain-associated c-Raf-1. These investigators concluded that sphingolipid domain-associated c-Raf-1 must traffic out of such domains and into the bulk membrane to achieve maximal activation status. For the studies of Hancock and co-workers, constitutively active H-rasG12V which traffics into sphingolipid domains and then into bulk plasma membrane with associated c-Raf-1 was used and compared to H'ΔhvrG12V, a mutated Ras isoform that cannot egress from sphingolipid domains. Whether this latter mutant also somehow suppresses c-Raf-1 activity is currently unknown, and will require additional study. The current observations are however consistent with the report that the sphingolipid domain-associated c-Raf-1 binding protein prohibitin is a positive regulator of c-Raf-1 activity required for EGF-induced c-Raf-1 activation in HeLa cells [39].
Collectively these studies identify the atypical C1 domain of KSR1 as a bone fide ceramide binding motif involved in the selective translocation of KSR1-bound c-Raf-1 into sphingolipid-rich macrodomains, sites of c-Raf-1 activation. As such these studies provide evidence for a mechanism by which the second messenger ceramide can target proteins into subcellular compartments in the process of transmembrane signal transduction.
Acknowledgements
This work was supported by NIH RO1 #CA105125 to A. H-F., NIH ROI #CA42385 to R.K., and a gift from the Virginia and D.K. Ludwig Fund for Cancer Research to Z. F.
References
- 1.Quest AF, Bloomenthal J, Bardes ES, Bell RM. The regulatory domain of protein kinase c coordinates four atoms of zinc. J Biol Chem. 1992;267:10193–10197. [PubMed] [Google Scholar]
- 2.Driedger PE, Blumberg PM. Specific binding of phorbol ester tumor promoters. Proc Natl Acad Sci U S A. 1980;77:567–571. doi: 10.1073/pnas.77.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase c requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hommel U, Zurini M, Luyten M. Solution structure of a cysteine rich domain of rat protein kinase c. Nat Struct Biol. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- 5.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase c delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of c1 protein kinase c homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Blitterswijk WJ. Hypothesis: Ceramide conditionally activates atypical protein kinases c, raf-1 and ksr through binding to their cysteine-rich domains. Biochem J. 1998;331(Pt 2):679–680. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Silva J, Krishnamurthy K, Tran E, Condie BG, Bieberich E. Direct binding to ceramide activates protein kinase czeta before the formation of a pro-apoptotic complex with par-4 in differentiating stem cells. J Biol Chem. 2005;280:26415–26424. doi: 10.1074/jbc.M501492200. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Meco MT, Municio MM, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase c. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Meco MT, Municio MM, Sanchez P, Lozano J, Moscat J. Lambda-interacting protein, a novel protein that specifically interacts with the zinc finger domain of the atypical protein kinase c isotype lambda/iota and stimulates its kinase activity in vitro and in vivo. Mol Cell Biol. 1996;16:105–114. doi: 10.1128/mcb.16.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazanietz MG. Novel “Nonkinase” Phorbol ester receptors: The c1 domain connection. Mol Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759. [DOI] [PubMed] [Google Scholar]
- 12.Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. Rasgrp is essential for mouse thymocyte differentiation and tcr signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 13.Pu Y, Perry NA, Yang D, Lewin NE, Kedei N, Braun DC, Choi SH, Blumberg PM, Garfield SH, Stone JC, Duan D, Marquez VE. A novel diacylglycerol-lactone shows marked selectivity in vitro among c1 domains of protein kinase c (pkc) isoforms alpha and delta as well as selectivity for rasgrp compared with pkcalpha. J Biol Chem. 2005;280:27329–27338. doi: 10.1074/jbc.M414132200. [DOI] [PubMed] [Google Scholar]
- 14.Downward J. Ksr: A novel player in the ras pathway. Cell. 1995;83:831–834. doi: 10.1016/0092-8674(95)90198-1. [DOI] [PubMed] [Google Scholar]
- 15.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in ras-mediated signaling in c. Elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 16.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. Ksr, a novel protein kinase required for ras signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 17.Sundaram M, Yochem J, Han M. A rasmediated signal transduction pathway is involved in the control of sex myoblast migration in caenorhabditis elegans. Development. 1996;122:2823–2833. doi: 10.1242/dev.122.9.2823. [DOI] [PubMed] [Google Scholar]
- 18.Morrison DK. Ksr: A mapk scaffold of the ras pathway? J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 20.Therrien M, Michaud NR, Rubin GM, Morrison DK. Ksr modulates signal propagation within the mapk cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 21.Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. Ksr stimulates raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci U S A. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich c1 domain of kinase suppressor of ras (ksr) J Mol Biol. 2002;315:435–446. doi: 10.1006/jmbi.2001.5263. [DOI] [PubMed] [Google Scholar]
- 23.Huwiler A, Brunner J, Hummel R, Vervoordeldonk M, Stabel S, van den Bosch H, Pfeilschifter J. Ceramide-binding and activation defines protein kinase c-raf as a ceramide-activated protein kinase. Proc Natl Acad Sci U S A. 1996;93:6959–6963. doi: 10.1073/pnas.93.14.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller G, Storz P, Bourteele S, Doppler H, Pfizenmaier K, Mischak H, Philipp A, Kaiser C, Kolch W. Regulation of raf- 1 kinase by tnf via its second messenger ceramide and cross-talk with mitogenic signalling. Embo J. 1998;17:732–742. doi: 10.1093/emboj/17.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hekman M, Hamm H, Villar AV, Bader B, Kuhlmann J, Nickel J, Rapp UR. Associations of b- and c-raf with cholesterol, phosphatidylserine, and lipid second messengers: Preferential binding of raf to artificial lipid rafts. J Biol Chem. 2002;277:24090–24102. doi: 10.1074/jbc.M200576200. [DOI] [PubMed] [Google Scholar]
- 26.Grassme H, Schwarz H, Gulbins E. Molecular mechanisms of ceramide-mediated cd95 clustering. Biochem Biophys Res Commun. 2001;284:1016–1030. doi: 10.1006/bbrc.2001.5045. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM. Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. Phosphatidic acid regulates the translocation of raf-1 in 12-o-tetradecanoylphorbol-13-acetate-stimulated madin-darby canine kidney cells. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 28.Xing HR, Kolesnick R. Kinase suppressor of ras signals through thr269 of c-raf-1. J Biol Chem. 2001;276:9733–9741. doi: 10.1074/jbc.M008096200. [DOI] [PubMed] [Google Scholar]
- 29.Xing HR, Lozano J, Kolesnick R. Epidermal growth factor treatment enhances the kinase activity of kinase suppressor of ras. J Biol Chem. 2000;275:17276–17280. doi: 10.1074/jbc.C900989199. [DOI] [PubMed] [Google Scholar]
- 30.Lisanti MP, Tang Z, Scherer PE, Sargiacomo M. Caveolae purification and glycosylphosphatidylinositol-linked protein sorting in polarized epithelia. Methods Enzymol. 1995;250:655–668. doi: 10.1016/0076-6879(95)50103-7. [DOI] [PubMed] [Google Scholar]
- 31.Xing HR, Cordon-Cardo C, Deng X, Tong W, Campodonico L, Fuks Z, Kolesnick R. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates rasmediated pancreatic cancer. Nat Med. 2003;9:1266–1268. doi: 10.1038/nm927. [DOI] [PubMed] [Google Scholar]
- 32.Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Kortum RL, Lewis RE. The molecular scaffold ksr1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razidlo GL, Kortum RL, Haferbier JL, Lewis RE. Phosphorylation regulates ksr1 stability, erk activation, and cell proliferation. J Biol Chem. 2004;279:47808–47814. doi: 10.1074/jbc.M406395200. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen A, Burack WR, Stock JL, Kortum R, Chaika OV, Afkarian M, Muller WJ, Murphy KM, Morrison DK, Lewis RE, McNeish J, Shaw AS. Kinase suppressor of ras (ksr) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cell Biol. 2002;22:3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W, Cannon R, Kolesnick R. Deficiency of kinase suppressor of ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63:4232–4238. [PubMed] [Google Scholar]
- 37.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominantnegative caveolin inhibits h-ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 38.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. Gtp-dependent segregation of h-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 39.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for ras-induced raf-mek-erk activation and epithelial cell migration. Nat Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 40.Sitrin RG, Emery SL, Sassanella TM, Blackwood RA, Petty HR. Selective localization of recognition complexes for leukotriene b4 and formyl-met-leu-phe within lipid raft microdomains of human polymorphonuclear neutrophils. J Immunol. 2006;177:8177–8184. doi: 10.4049/jimmunol.177.11.8177. [DOI] [PubMed] [Google Scholar]
- 41.Harrison RE, Sikorski BA, Jongstra J. Leukocyte-specific protein 1 targets the erk/map kinase scaffold protein ksr and mek1 and erk2 to the actin cytoskeleton. J Cell Sci. 2004;117:2151–2157. doi: 10.1242/jcs.00955. [DOI] [PubMed] [Google Scholar]
- 42.Mineo C, James GL, Smart EJ, Anderson RG. Localization of epidermal growth factor-stimulated ras/raf-1 interaction to caveolae membrane. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- 43.Carey KD, Watson RT, Pessin JE, Stork PJ. The requirement of specific membrane domains for raf-1 phosphorylation and activation. J Biol Chem. 2003;278:3185–3196. doi: 10.1074/jbc.M207014200. [DOI] [PubMed] [Google Scholar]
- 44.Hallberg B, Rayter SI, Downward J. Interaction of ras and raf in intact mammalian cells upon extracellular stimulation. J Biol Chem. 1994;269:3913–3916. [PubMed] [Google Scholar]
- 45.Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH., Jr. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with fusarium moniliforme. J Biol Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 46.Muller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. Pkc zeta is a molecular switch in signal transduction of tnf-alpha, bifunctionally regulated by ceramide and arachidonic acid. Embo J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gulbins E, Coggeshall KM, Baier G, Telford D, Langlet C, Baier-Bitterlich G, Bonnefoy-Berard N, Burn P, Wittinghofer A, Altman A. Direct stimulation of vav guanine nucleotide exchange activity for ras by phorbol esters and diglycerides. Mol Cell Biol. 1994;14:4749–4758. doi: 10.1128/mcb.14.7.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fox TE, Houck KL, O'Neill SM, Nagarajan M, Stover TC, Pomianowski PT, Unal O, Yun JK, Naides SJ, Kester M. Ceramide recruits and activates protein kinase c zeta (pkc zeta) within structured membrane microdomains. J Biol Chem. 2007;282:12450–12457. doi: 10.1074/jbc.M700082200. [DOI] [PubMed] [Google Scholar]
- 49.Hajduch E, Turban S, Le Liepvre X, Le Lay S, Lipina C, Dimopoulos N, Dugail I, Hundal HS. Targeting of pkczeta and pkb to caveolin-enriched microdomains represents a crucial step underpinning the disruption in pkb-directed signalling by ceramide. Biochem J. 2008;410:369–379. doi: 10.1042/BJ20070936. [DOI] [PubMed] [Google Scholar]
- 50.Powell DJ, Hajduch E, Kular G, Hundal HS. Ceramide disables 3-phosphoinositide binding to the pleckstrin homology domain of protein kinase b (pkb)/akt by a pkczeta-dependent mechanism. Mol Cell Biol. 2003;23:7794–7808. doi: 10.1128/MCB.23.21.7794-7808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharkey NA, Leach KL, Blumberg PM. Competitive inhibition by diacylglycerol of specific phorbol ester binding. Proc Natl Acad Sci U S A. 1984;81:607–610. doi: 10.1073/pnas.81.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]