Abstract
Influenza has been circulating in the human population and has caused three pandemics in the last century (1918 H1N1, 1957 H2N2 and 1968 H3N2). The 2009 A(H1N1) was classified by World Health Organization as the fourth pandemic. Influenza has a high evolution rate, which makes vaccine design challenging. We here consider an approach for early detection of new dominant strains. By clustering the 2009 A(H1N1) sequence data, we found two main clusters. We then define a metric to detect the emergence of dominant strains. We show on historical H3N2 data that this method is able to identify a cluster around an incipient dominant strain before it becomes dominant. For example, for H3N2 as of 30 March 2009, the method detects the cluster for the new A/British Columbia/RV1222/2009 strain. This strain detection tool would appear to be useful for annual influenza vaccine selection.
Keywords: clustering, H1N1, H3N2, influenza
Introduction
The recent outbreak of 2009 A(H1N1) caused immediate international attention (Deem and Pan, 2009; Fraser et al., 2009; Garten et al., 2009; Smith et al., 2009). This new 2009 A(H1N1) virus contains a combination of gene segments from swine and human influenza viruses (Fraser et al., 2009; Garten et al., 2009). Confirmed infections reached 270 000 globally as of September 2009 (World Health Organization, 2009b). The novel 2009 A(H1N1) strain was defined as a pandemic strain by the World health Organization (WHO) in 2009 (World Health Organization, 2009d), and was the epidemic strain in the 2009 Northern winter.
Influenza viruses are hyper-mutating viruses. It has been estimated that the nucleotide mutation rate per genome per replication is approximately 0.76 (Drake and Holland, 1999). Influenza viruses escape the human immune system by continual antigenic drift and shift (Fitch et al., 1997; Gupta et al., 1998; Webster, 1998; Ferguson et al., 2003; Ghedin et al., 2005; Nelson and Holmes, 2007). The quasispecies nature of influenza viruses makes the strain structure complex (Domingo et al., 2002). Usually, there is one or a few dominant influenza strains circulating in the population for each flu season. The flu vaccine is most effective when it matches this dominant circulating strain (Hak et al., 2002; Gupta et al., 2006). The degree to which immunity induced by a vaccine protects against a different viral strain is determined by the antigenic distance between the vaccine and the virus. Due to evolution of the antigenic regions of the influenza virus, the composition of the flu vaccine is typically modified annually (Russell et al., 2008a). However, since the influenza strains used in the flu vaccine are decided 6 months before the flu season, a mismatch between the vaccine strain and dominant circulating strain may occur if the virus evolves significantly. Such a situation arose for the H3N2 virus in the 2009–2010 flu season, when A/British Columbia/RV1222/2009 emerged in the early spring (Seasonal influenza, 2009; Skowronski, 2009). Accurate early prediction of the dominant circulating strain is an essential and important task in influenza research.
There are several ways to estimate the flu vaccine effectiveness. Gupta et al. (2006) proposed pepitope as a measure of antigenic distance between influenza A vaccine and circulating strains. The hemagglutinin protein has five epitopes. The dominant epitope for a particular circulating strain in a particular season was taken as that which had the largest fractional change in amino acid sequence relative to the vaccine strain. The value of pepitope is defined as the fraction of number of amino acid differences in the dominant epitope to total number of amino acids in the dominant epitope. The antigenic distance between the vaccine strain and the circulating strain is quantified by pepitope. By a metaanalysis of historical vaccine efficacy data from over 50 publications, Gupta et al. (2006) showed in a metaanalysis that the pepitope between vaccine strain and circulating strain correlates well with the vaccine efficacy, with R2 > 0.8.
Understanding the evolution of influenza viruses has benefited from phylogenetic reconstructions of the hemagglutinin protein evolution (Ferguson et al., 2003; Russell et al., 2008b). In an alternative approach, Lapedes and Farber (2001), followed by Smith et al. (2004), applied a technique called multidimensional scaling to study antigenic evolution of influenza. Plotkin et al. (2002) clustered hemagglutinin protein sequences using the single-linkage clustering algorithm and found that influenza viruses group into clusters.
Here, we present a low-dimensional clustering method that can detect the cluster containing an incipient dominant strain for an upcoming flu season before the strain becomes dominant. The method builds upon the dimensional projection technique used by Lapedes and Farber (2001) and Smith et al. (2004) to characterize hemagglutination inhibition (HI) data. Importantly, the present method requires only sequence data, unlike the approach of Lapedes and Farber (2001) and Smith et al. (2004), which require ferret HI assay data animal data. In this paper, we first study the evolution of 2009 A(H1N1) by an evolutionary path map which leads to a suggestion for the H1N1 vaccine strain. Then, we introduce the low-dimensional protein sequence clustering method. We propose an influenza vaccine selection procedure based on this sequence clustering. The procedure is demonstrated and tested in detail using historical data. We show the performance of the method to predict the dominant H3N2 strain in an upcoming flu season using data solely from before the flu season, on data since 1996. We compare the results to those from existing methods since 1996. In the discussion section, we discuss the relationship between the protein sequence clustering method and previous approaches. We discuss the false positive rate, as well as other challenges.
Results
Evolutionary path of 2009 A(H1N1) influenza
We first construct the directional evolutionary path for the 2009 A(H1N1) influenza. We use high-resolution data in sequence, time and world spatial coordinate to construct this evolutionary relationship.
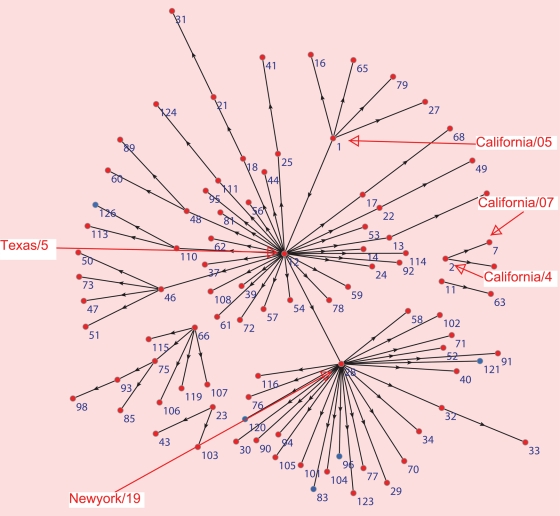
Since its first detection, the 2009 A(H1N1) virus has been extensively sequenced (Fraser et al., 2009; Garten et al., 2009). By 1 May 2009, the number of confirmed cases reported by WHO was 333 (World Health Organization, 2009b). At the same time, the sequenced hemagglutinin proteins (HA) available in NCBI Influenza Resources Database were 312 (Bao et al., 2008); that is to say most of the confirmed cases at that time were sequenced. On 1 July 2009, the ratio of sequenced HA protein to confirmed cases by WHO was 1039/77201 (World Health Organization, 2009b), a number which is still much larger than that for seasonal flu. In addition, the Influenza Resources Database contains the date of collection of each 2009 A(H1N1) virus strain. We reconstruct the evolutionary history of swine flu viruses with the following procedure. If strain B is mutated from strain A, we term strain A ‘founder’ and strain B ‘F1’. We align the HA proteins of all 2009 A(H1N1) strains. Then, for each strain, we find its founder strain based on the following four criteria: (i) the founder strain should appear earlier than the strain, as judged by collection date; (ii) the founder strain should have only one amino acid difference in the HA1 protein relative to the F1 strain; (iii) the founder should also have the most similar nucleotide sequence relative to F1; and (iv) the founder strain should have a large number of identical copies circulating in human population, as approximated by the number of different strains with identical HA sequences in the Influenza Resources Database. By applying these four criteria to 2009 A(H1N1) influenza, we construct the directional evolutionary path map, as shown in Fig. 1. We can see two clusters: one around A/New York/19/2009 (#28) and another one around A/Texas/05/2009 (#12). Most new strains are from the Northern hemisphere, and strains from the Southern hemisphere are mainly located at the edge of the map, such as strains #96, #120 and #126. That the Southern hemisphere strains appear at the boundary of the figure provides a self-consistency check of the validity of the assumptions entering the construction of this figure. Geographically, we see many founder to F1 links are from the USA and Mexico to other countries, but we rarely see founder to F1 links that are from other countries to the USA and Mexico, or from other countries to other countries except the USA and Mexico (see Materials and methods). We also found that strains with more F1 in Fig. 1 are more frequently seen in the human population. For example, in the Influenza Resources Database, we found 153 strains to be identical with A/New York/19/2009, which has 29 F1 strains, and 120 strains to be identical with A/Texas/05/2009, which has 24 F1 strains. We can see in Fig. 1 that A/Texas/05/2009 is at the very upstream of the map, with downward connections to most of the other strains by direct or two-step links. This result agrees with the US Food and Drug Administration (FDA, 2009) recommendation of A/Texas/05/2009 as a vaccination strain. The alternative vaccine strain A/California/7/2009 (#7) has fewer F1 strains and it is not located at the center of the network.
Fig. 1.
The evolutionary path of 2009 A(H1N1) influenza. Strain #1: A/California/05/2009. Strain #2: A/California/04/2009. Strain #7: A/California/07/2009. Strain #12: A/Texas/05/2009. Strain #28: A/New York/19/2009. For complete strain names, see Supplementary data. Strains from the Northern and Southern hemisphere are shown as red dots and blue dots, respectively. One branch represents one substitution in the amino acid sequence.
Low-dimensional clustering
We use a low-dimensional clustering method to visualize the antigenic distance matrix of the viruses. We use a statistical tool called ‘multidimensional scaling’ (Everitt et al., 2001). This method was used by Lapedes and Farber (2001) and Smith et al. (2004) to project ferret HI assay data to low dimensions. The influenza viral surface glycoprotein hemagglutinin is a primary target of the protective immune response. Here, we project the hemagglutinin protein sequence data, rather than animal model data, to low dimensions. The HA1 protein of influenza with 329 residues can be considered as a 329-dimension space. The multidimensional scaling method is applied to rescale the 329-dimension space to a two-dimensional space, so that we can plot and visualize it. First, we do a multialignment of the HA1 proteins. Then, the distance between any two proteins is calculated as
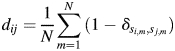 |
(1) |
where 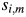 is the amino acid of protein i at position m. The term
is the amino acid of protein i at position m. The term 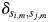 is 1 if amino acids of protein
is 1 if amino acids of protein 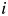 and
and 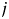 at position m are the same. Otherwise, it is 0. For the 2009 H1N1 viruses, we consider the entire HA protein, and N = 566. For H3N2 viruses, we consider only the HA1 protein, and N = 329, because the entire HA proteins are not completely sequenced in many cases. Thus,
at position m are the same. Otherwise, it is 0. For the 2009 H1N1 viruses, we consider the entire HA protein, and N = 566. For H3N2 viruses, we consider only the HA1 protein, and N = 329, because the entire HA proteins are not completely sequenced in many cases. Thus, 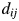 is the number of amino acid differences between HA proteins normalized by length. The multidimensional scaling produces a protein distance map, for example, Fig. 2b. In this map, each data point represents a flu strain isolate. The Euclidean distance between two points in the map approximates the protein distance in Equation (1) between these two flu strains (see Materials and methods for details of this distance approximation procedure). Two closely located points imply two strains with similar HA protein sequences.
is the number of amino acid differences between HA proteins normalized by length. The multidimensional scaling produces a protein distance map, for example, Fig. 2b. In this map, each data point represents a flu strain isolate. The Euclidean distance between two points in the map approximates the protein distance in Equation (1) between these two flu strains (see Materials and methods for details of this distance approximation procedure). Two closely located points imply two strains with similar HA protein sequences.
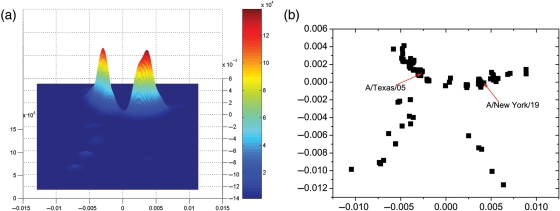
Fig. 2.
(a) Kernel density estimation for the protein distance map of 2009 A(H1N1) influenza as of 5 December 2009. (b) The protein distance map of 2009 A(H1N1) influenza. The vertical and horizontal axes of both figures represent protein distance as defined in Equation (1). A 0.0018 unit of protein distance equals one substitution in the HA protein sequence of H1N1. The height and colors in (a) both represent the density of isolates.
We apply the low-dimensional clustering method to study 2009 A(H1N1). We plot the protein distance map in Fig. 2b. Both A/Texas/05/2009 and A/New York/19/2009 are located near the center of the cluster, in good agreement with the observation from Fig. 1 that they are the founder strains for many F1 strains. To detect the clusters in the protein distance map, we use a statistical method known as kernel density estimation (Everitt et al., 2001). Kernel density estimation is a non-parametric method to estimate the probability density function from which data come. The kernel density figure is produced from the protein distance map, and it shows the density of influenza strains in sequence space. We plot the kernel density as the three-dimensional shaded surface. For example, the kernel density surface Fig. 2a is produced from Fig. 2b. The x and y axes in Fig. 2a are the same as that in Fig. 2b and are protein distance coordinates. The z dimension measures the density of flu strains around point (x, y). We use the surface height and the colors to represent z values, and the color is proportional to surface height. A peak in kernel density Fig. 2a indicates a cluster of related flu strains in the protein distance map Fig. 2b.
There are two significant clusters in Fig. 2a, as two peaks are observed. The cluster on the left side contains A/Texas/05/2009. Another cluster on the right side contains A/New York/19/2009. The 2009 A(H1N1) virus has evolved slowly to date. The greatest pepitope antigenic distance between A/Texas/05/2009 and all sequenced strains is measured to be <0.08. Values of pepitope less than 0.45 for H1N1 indicate positive expected vaccine efficacy (Pan et al., 2009), and so a vaccine is expected to be efficacious. All of the amino acids in all five epitopes of a strain of A/Texas/05/2009 and a strain of A/New York/19/2009 are the same. Multidimensional scaling predicts that A/Texas/05/2009 will be the dominant strain in the 2009–2010 season, and that A/Texas/05/2009 is a suitable strain for vaccination. Our focus is on the expected vaccine effectiveness, as it can be judged from antisera HI assay or sequence data alone. We do not consider other aspects such as growth in hen's eggs or other manufacturing constraints. Laboratory growth and passage data are needed to address these aspects.
H3N2 virus evolution for 38 years
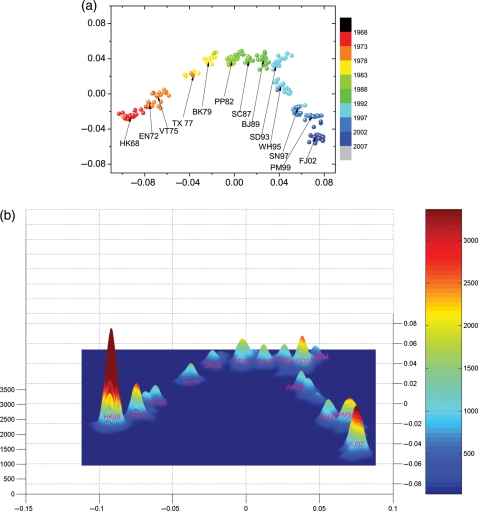
We construct the protein distance map to determine the evolution of influenza A(H3N2) virus from 1969 to 2007. Sequences of HA1 proteins were downloaded from the Influenza Virus Resources database (Bao et al., 2008). We use the multidimensional clustering method (Lapedes and Farber, 2001) to generate the protein distance map and corresponding kernel density estimation in Fig. 3. Smith et al. (2004) produced a similar graph using ferret antisera HI assay data. The figure presented here has a higher resolution, and more clusters are observed, because protein sequences data are more abundant and accurate than antisera HI assay data. The evolution of influenza tends to group strain into clusters. In Fig. 3, we identified 14 major clusters by setting a cutoff value of kernel density for the past 38 years from 1969 to 2007. The average duration time for a cluster is therefore 2.7 years, which is also the approximate duration of a vaccine. We marked each cluster by the first vaccine strain in the cluster. There are apparent gaps between clusters. The antigenic distance between two strains in two separate clusters is larger than the distances within the same cluster. The influenza virus evolves within one cluster before jumping from one cluster to another cluster. This dynamics occurs because small antigenic drift by one or a few sequential mutations does not lead the virus to completely escape from cross-immunity induced by vaccine protection or prior exposure.
Fig. 3.
(a) The protein distance map and (b) corresponding Kernel density estimation of influenza from 1968 to 2007. The vertical and horizontal axes of both figures represent protein distance as defined in Equation (1). A 0.0030 unit of protein distance equals one substitution in the HA1 protein sequence of H3N2. The colors in (a) represent the time of collection of the isolates. The colors and height in (b) represent the density of isolates. Each cluster is named after the first vaccine strain in the cluster. HK68: Hongkong/1/68, EN72: England/42/72, VT75: Victoria/3/75, TX77: Texas/1/77, BK79: Bangkok/1/79, PP82: Philippines/2/82, SC87: Sichuan/2/87, BJ89: Beijing/32/92, SD93: Shandong/9/93, JB94: Johannesburg/33/94, WH95: Wuhan359/95, SN97: Sydney/5/97, PM99: Panama/2007/99, FJ02: Fujian/411/2002.
For vaccine design, when the viruses evolve as a quasispecies in the same cluster, the vaccine that is targeted to the cluster provides protection. This protection decreases with antigenic distance. When the viruses jump to a new cluster by antigenic drift or shift, one would want to update the vaccine to provide protection against strains in the new cluster. In Fig. 3a, the arrows point to the exact position of vaccine strains. It can be seen that the positions of vaccine strains are near the center of clusters. It can be shown mathematically that choosing the consensus strain of a cluster as vaccine strain minimizes the pepitope antigenic distance between vaccine strain and cluster strains, and thus maximizes expected vaccine efficacy (Gupta et al., 2006).
Influenza vaccine strain selection
We now use the low-dimensional sequence clustering method in an effort to detect a new flu strain before it becomes dominant. A question of interest in influenza research is whether we can predict which strain will be dominant in the next flu season based on the information we have at present. WHO gathers together every February to make a recommendation for influenza strains to be used in vaccine for next flu season in the Northern hemisphere. The vaccine is expected to have high efficacy if the chosen strain is dominant in the next flu season. The recommendation is especially challenging to make when the dominant strain in next flu season has not been dominant before February of that year. For example, in mid-March 2009, a new H3N2 strain appeared (Seasonal influenza, 2009; Skowronski, 2009), which infected a significant fraction of the population in the Southern hemisphere.
The current accepted influenza vaccine strain selection procedure is as follows (Russell et al., 2008a). Isolates samples are collected by WHO GISN and are characterized antigenically using the HI assay. About 10% of samples are also sequenced in HA1 domain of HA gene. Antigenic maps are constructed from the HI assay data using the dimensional projection technique. Examination of HI data is not dependent on analysis using dimensional projection, but rather, the primary HI data may carry the most weight. If the vaccine does not match the current circulating strains, the vaccine is updated to contain one representative of the circulating strains. The emerging variant strains are identified. If the antigenically distinct emerging variants are judged to be the dominant strains in the upcoming season, the vaccine is updated to include one representative of emerging variants. The key issue and major difficulty is how to judge whether emerging variants will be the dominant variants in the next season. If a fourfold difference in antisera HI titer between the vaccine strain and the emerging strains is observed, the emerging strain is to be determined to be dominant strains in the upcoming season, and an updated vaccine is recommended to include the emerging strains (Russell et al., 2008a).
Here, we propose a modified vaccine selection process based on clustering detection. First, we apply the multidimensional scaling to make a protein distance map from HA1 sequences, instead of constructing an antigenic map from HI assay data. Then, we use kernel density estimation to determine the clusters of strains. If the vaccine does not match the current circulating cluster, the vaccine is updated to contain the current circulating strain. If the vaccine matches the current circulating cluster, but an emerging cluster is judged likely to be the major cluster in the upcoming season, the vaccine is updated to contain the consensus strain of the emerging cluster. We judge whether a cluster is an emerging dominant cluster by two criteria. The first criterion is that this cluster can be detected by kernel density estimation, and is separate from the cluster that contains the current circulating strain or vaccine strain. A cluster that can be detected by kernel density estimation usually contains a central strain that has multiple identical copies and some F1 strains that are closely related to the central strain. An example is the cluster of A/Texas/05/2009(H1N1) in Fig. 1. A/Texas/05/2009(H1N1) is the central strain, which has 120 strains with identical HA protein sequences in the Influenza Virus Resource database (Bao et al., 2008). A/Texas/05/2009(H1N1) also has 29 F1 strains with one amino acid different. So, A/Texas/05/2009(H1N1) and the surrounding strains form a cluster as we detected in Fig. 2 by kernel density estimation.
The second criterion is that the current vaccine strain does not match the consensus strain of the cluster and is estimated to provide low protection against strains in the cluster. That is, is the new strain sufficiently different so that an immune response stimulated by the current vaccine is not expected to be effective. The consensus strain is a protein sequence that shows which residues are most abundant in the multialignment at each position. The efficacy of current vaccine to the new cluster can be estimated from ferret antisera HI assay data. However, the antisera data have low resolution and has an imperfect correlation to vaccine effectiveness in humans (Gupta et al., 2006; Zhou et al., 2010). Instead, we use pepitope, which is calculated as the fraction of mutations in dominant epitope, to estimate vaccine efficacy and which has a more robust correlation to vaccine effectiveness in human than do ferret HI data (Gupta et al., 2006). When the pepitope between the current vaccine strain and consensus strain of the new cluster is larger than 0.19, expected vaccine efficacy decreases to 0 for H3N2 influenza, and the current vaccine cannot be expected to provide protection from new strains. As the examples shown below, our method can detect an incipient dominant strain at its very early stage, and the method appears to require about 10 sequences in the new cluster for detection.
Demonstration of low-dimensional sequence clustering method
We demonstrate the method of detecting the A/Fujian/411/2002(H3N2) strain. The A/Panama/2007/1999 had been the vaccination strain for four flu seasons between 2000 and 2004 in the Northern hemisphere.
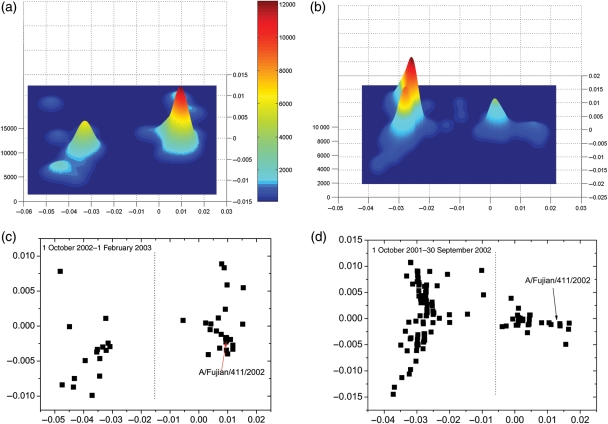
The vaccine strain was replaced by A/Fujian/411/2002(H3N2) in the 2004–2005 flu season, as described in Table I. The vaccine strain in the 2003–2004 season was A/Panama/2007/1999, while the dominant circulating strain became A/Fujian/411/2002(H3N2). This mismatch resulted in a large decrease in vaccine efficacy in the 2003–2004 flu season (Gupta et al., 2006). The vaccine efficacy is estimated to be only 12% (MMWR Morb, 2004). We test whether our method can detect A/Fujian/411/2002(H3N2) as an incipient dominant strain before it actually became dominant. We use only virus sequence data before 1 October 2003. We did not use any virus data collected in the 2003–2004 season. Therefore, our prediction and results are made without any knowledge from what happened in the 2003–2004 season. We plot the protein distance map of the 2001–2002 flu season in Fig. 4d. To detect the clusters, we plot the kernel density in Fig. 4b for the data in Fig. 4d. There are two separate significant clusters. The one with the largest kernel density on the left contains the current dominant strain A/Panama/2007/1999 and the widespread A/Moscow/10/1999 strain. The smaller one on the right is a new cluster, which contains A/Fujian/411/2002. Using the data as of 30 September 2002, we seek to determine whether the new cluster on the right in Fig. 4b and d will be the next dominant strain after A/Panama/2007/1999. We determine whether this cluster fulfills the two criteria above. First, this new cluster can be significantly detected by kernel density estimation. This cluster is separate from the current dominant strain, as we can see in the figure. Second, we calculated the average pepitope of the new cluster on the right with regard to A/Moscow/10/1999, A/Panama/2007/1999 and A/Fujian/411/2002 to be 0.214, 0.1214 and 0.083, respectively. This means the current vaccine contains A/Moscow/10/1999 is expected to provide little protection against viruses in the new cluster. This result makes the new cluster fulfill the second criterion. Thus, we predict based on the data as of 30 September 2002, that the cluster on the right in Fig. 4d will be the next dominant cluster. This prediction was made on data collected 1 year earlier than when the A/Fujian/411/2002 became dominant in the 2003–2004 season. To further support our prediction, in Fig. 4c, we plot the protein distance map from 1 October 2002 to 1 February 2003, right before the WHO selected the vaccine strain for the 2003–2004 season. To detect the clusters, we plot the kernel in Fig. 4a for the data in Fig. 4c. There are two separate major clusters observed in the kernel density estimation in Fig. 4a. The left cluster has the current dominant strain of A/Panama/2007/1999 and also A/Moscow/10/1999. The right cluster has the A/Fujian/411/2002. We calculated the average pepitope of the right new cluster with regard to A/Moscow/10/1999, A/Panama/2007/1999 and A/Fujian/411/2002 to be 0.2725, 0.1811 and 0.0367, respectively. This result further supports the prediction that the new cluster will become dominant, and A/Fujian/411/2002, which is the most frequent strain in the new cluster, will be or is very close to the next dominant strain. This suggestion proceeds the vaccine component switch by 1–2 years, as shown in Table I.
Table I.
Summary of results
| Flu season | Vaccine strain from WHO (World Health Organization, 2009c) | Our prediction | Circulating H3N2 strain | Circulating subtype |
|---|---|---|---|---|
| 1996–1997 | Wuhan/359/95 | Wuhan/359/95 | Wuhan/359/95 | H3 |
| 1997–1998 | Wuhan/359/95 | Wuhan/359/95 | Sydney/5/97 | H3 |
| 1998–1999 | Sydney/5/97 | Sydney/5/97 | Sydney/5/97 | H3 |
| 1999–2000 | Sydney/5/97 | Sydney/5/97 | Sydney/5/97 | H3 |
| 2000–2001 | Panama/2007/1999 | Panama/2007/1999 | N/A | H1 |
| 2001–2002 | Panama/2007/1999 | Panama/2007/1999 | Panama/2007/1999 | H3 |
| 2002–2003 | Panama/2007/1999 | Fujian/411/2002 | N/A | H1 |
| 2003–2004 | Panama/2007/1999 | Fujian/411/2002 | Fujian/411/2002 | H3 |
| 2004–2005 | Fujian/411/2002 | Fujian/411/2002 | Fujian/411/2002 | H3 |
| 2005–2006 | California/7/2004 | California/7/2004 | California/7/2004 | H3 |
| 2006–2007 | Wisconsin/67/2005 | Wisconsin/67/2005 | Wisconsin/67/2005 | H3 |
| 2007–2008 | Wisconsin/67/2005 | Wisconsin/67/2005 | N/A | H1 |
| 2008–2009 | Brisbane/10/2007 | Brisbane/10/2007 | Brisbane/10/2007 | H3 |
| 2009–2010 | Brisbane/10/2007 | BritishColumbia/RV1222/09 | BritishColumbia/RV1222/09 | H1 |
| 2010–2011 | Perth/16/2009 | BritishColumbia/RV1222/09 | N/A | N/A |
This table includes the H3N2 vaccine strains, our prediction of dominant strains, the reported dominant circulating H3N2 strains (World Health Organization, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002, 2003, 2004, 2005a, 2006, 2007, 2008, 2009a, 2010) and the circulating subtypes in the northern hemisphere (World Health Organization, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002, 2003, 2004, 2005a, 2006, 2007, 2008, 2009a, 2010). Circulating H3N2 strains are absent if the dominant subtype is H1 or influenza B. The reported dominant H3N2 strains and circulating subtypes data are from WHO Weekly Epidemiological Record (http://www.who.int/wer/en/).
Fig. 4.
(a) Kernel density estimation and (c) protein distance map for H3N2 viruses between 1 October 2002 and 1 February 2003. (b) Kernel density estimation and (d) protein distance map for H3N2 viruses between 1 October 2001 and 9 September 2002. We plot a dotted line to separate the two clusters. The vertical and horizontal axes of all figures represent protein distance as defined in Equation (1). A 0.0030 unit of protein distance equals one substitution of the HA1 protein sequence of H3N2.
Prediction for H3N2 influenza in 2009–2010
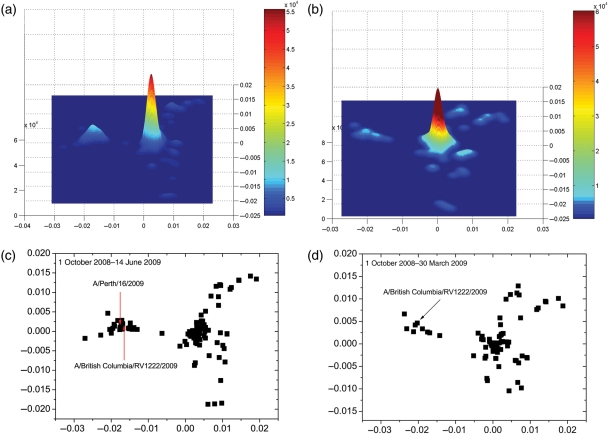
By applying our method to the 2008–2009 flu season, we predict that the dominant H3N2 strain in the 2009–2010 flu season may switch. Based on the flu activity in the 2008–2009 flu season, the WHO made the recommendation in February 2009 that A/Brisbane/10/2007(H3N2) should be used as the vaccine (World Health Organization, 2009c). However, a new strain evolved just after the recommendation was published. The British Columbia Center for Disease Control detected a new virus strain (Seasonal influenza, 2009; Skowronski, 2009) with 3 mutations in antigenic sites (two in epitope B and one in epitope D). Since this new strain is relatively far from the vaccine strain, with pepitope = 0.095, vaccine efficacy is expected to decrease to 20% (Gupta et al., 2006; Deem and Pan, 2009). However, since the mutations in this new strain ‘do not fulfill the criteria proposed by Cox as corresponding to meaningful antigenic drift’ (Cox and Bender, 1995; Skowronski, 2009), and this strain still remained the minority of H3N2 viruses in July 2009, health authorities were not certain that this new strain would replace the current dominant strain in the 2009–2010 flu season. We use our method to investigate whether this new strain will be the next dominant strain. We construct the protein distance map as shown in Fig. 5(c). We plot the kernel density estimation in Fig. 5(a) for data in Fig. 5(c). By the data up to June 14, 2009, we see two major clusters in Fig. 5(a). The larger one on the right contains the current dominant strain A/Brisbane/10/2007, and the left one is a new cluster which contains A/British Columbia/RV1222/2009. It is apparent that this new cluster is separate from the current dominant cluster. Thus, this cluster fulfills the first criterion. We calculated the average of pepitope of strains in the left new cluster with regards to A/Brisbane/10/2007 and A/British Columbia/RV1222/2009 to be 0.103 and 0.042 respectively. The vaccine that contains A/Brisbane/10/2007 has an expected efficacy of 20% to the virus strains in the new cluster. Thus, this new cluster satisfies both two criteria, and so we predict that this cluster which contains A/British Columbia/RV1222/2009 will be the dominant cluster in the 2009–2010 season. The earliest time for us to make this prediction is 30 March 2009. In Fig. 5d and b, we already see this new cluster on the left side of figure, though since there are only about 10 sequences in the new cluster, the kernel density of this new cluster is smaller than that in the dominant cluster. This strain was mentioned as a concern on 5 May 2009, although by conventional methods the strain was not considered a potentially new dominant strain in July 2009 (Skowronski, 2009). With the method of the present paper, this new cluster is suggested earlier using the data as of 30 March 2009.
Fig. 5.
(a) Kernel density estimation and (c) protein distance map for H3N2 viruses from 1 October 2008 to 14 June 2009. (b) Kernel density estimation and (d) protein distance map for H3N2 viruses between 1 October 2008 and 30 March 2009. The vertical and horizontal axes of all figures represent protein distance as defined in Equation (1). A 0.0030 unit of protein distance equals one substitution of the HA1 protein sequence of H3N2.
Comparison with previous results
Here we present a historical test of the method. For each flu season in the North Hemisphere from 1996, we use only the H3N2 sequences data until 1 February, before WHO published the recommendation for vaccine. We use the low-dimensional clustering to make the prediction for the dominant strain. The conventional method as used by WHO is phylogenetic analysis combined with ferret antisera HI assay. In Table I, we compare the method with the conventional method. In the most recent 14 flu seasons, influenza subtype H3 was dominant in 10. The WHO H3N2 vaccine component matches the circulating strains in eight seasons. Our predictions match the circulating strains in nine seasons. In the 1997–1998 season, a novel flu strain Sydney/5/97 was found in June 1997. Because no similar strains were collected before 1 February, neither of the two methods can predict it. In the 2003–2004 season, our method predicts Fujian/441/2002 as the dominant strain, while phylogenetic analysis combined with ferret antisera HI assay did not. For all other eight seasons dominated by influenza subtype H3, the predictions of both methods matched the dominant circulating strain. The 2009–2010 influenza season was dominated by H1N1. But data from local outbreaks of H3N2 infections (Seasonal influenza, 2009; Skowronski, 2009) showed that the dominant H3N2 strain was A/British Columbia/RV1222/2009, as predicted in Table I, rather than the vaccine strain A/Brisbane/10/2007. For the 2010–2011 season, we recommend A/British Columbia/RV1222/2009 as a vaccine strain, and the WHO recommended A/Perth/16/2009. These two strains are in the same cluster and antigenically similar with a small pepitope = 0.048. Although these two strains are slightly different, the vaccine is expected to be effective.
Detecting A/Wellington/1/2004 in the 2004 flu season in the Southern hemisphere
The low-dimensional clustering can also be applied to influenza in the Southern hemisphere. As an example, we test our method on the 2004 flu season. The recommended H3N2 vaccine strain by WHO used in the 2004 flu season in the Southern hemisphere was A/Fujian/411/2002. Data from the surveillance network suggested that the circulating dominant flu strain in the 2004 season in Southern hemisphere was A/Fujian/411/2002, and a late surge of A/Wellington/1/2004 was also observed. For example, in Argentina, a study showed that about 50% of infections were closely related to A/Fujian/411/2002 and another 50% were closely related to A/Wellington/1/2004 (Santamaria et al., 2008). In New Zealand, the dominant flu strain was A/Fujian/411/2002 which caused 78% of flu infections (Virology, 2004), and a late season surge of A/Wellington/1/2004 was also reported (Northern hemisphere, 2004). Therefore, the vaccine recommended by WHO matches the dominant strain and would be expected to have vaccine efficacy in the 2004 season in Southern hemisphere.
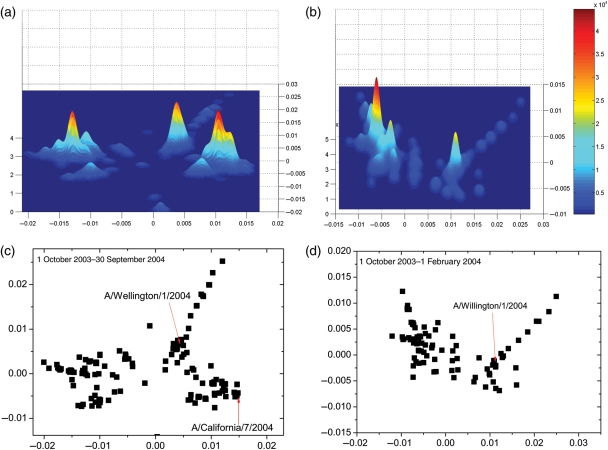
We here use the low-dimensional clustering method to detect the A/Wellington/1/2004 strain, which is not the major dominant strain but caused significant infections in the 2004 flu season. We plot the protein distance and kernel density estimation for the H3N2 viruses in Fig. 6d and b. We use the data only as of 1 February 2004, 3 months prior to the 2004 flu Southern hemisphere season, which is usually from May to September. We observed two clusters. The major cluster on the left side of Fig. 6d is A/Fujian/411/2002-like, which was the vaccine strain in the 2004 season. There is a new cluster in the right side of Fig. 6d which contains A/Wellington/1/2004. The pepitope of A/Wellington/1/2004 with regards to A/Fujian/411/2002 is 0.118. Therefore, we predict that A/Wellington/1/2004 will infect a large fraction of the population, and the A/Fujian/411/2002 vaccine is expected to provide only partial protection against the A/Wellington/1/2004 virus. However, since the appearance of A/Wellington/1/2004 was just before the 2004 flu season, it did not have sufficient time to spread out and become the dominant strain in the 2004 flu season. From our observation, it usually takes about 8 months or longer for a new strain to become dominant after its appearance in a new cluster. Therefore, the predominant flu strain in the 2004 season is expected to be A/Fujian/411/2002 based on the data as of 1 February 2004. This result agrees with the dominant flu strain in the 2004 flu season.
Fig. 6.
(a) Kernel density estimation for the protein distance map for H3N2 viruses between 1 October 2003 and 30 September 2004. (b) Kernel density estimation for the protein distance map for H3N2 viruses between 1 October 2003 and 1 February 2004. (c) Protein distance map for H3N2 viruses between 1 October 2003 and 30 September 2004. We plot a dotted line to separate the two clusters. (d) Protein distance map for H3N2 viruses between 1 October 2003 and 1 February 2004. The vertical and horizontal axes of all figures represent protein distance. A 0.0030 unit of protein distance equals one mutation of the HA1 protein sequence of H3N2.
Detecting A/California/4/2004 as a future dominant strain
As a further example of applying the low-dimensional clustering method to influenza in Southern hemisphere, we test the method on the 2005 flu season. The recommended H3N2 vaccine strain in the 2005 flu season in the Southern hemisphere was A/Wellington/1/2004. Data from HI assay tests and surveillance suggest that the dominant H3N2 strain in the 2005 season was A/California/7/2004. In HI tests with postinfection ferret sera, the majority of influenza A(H3N2) viruses from February 2005 to October 2005 were closely related to A/California/7/2004, as reported by WHO on 7 October 2005 (World Health Organization, 2005b). Surveillance data from Victoria, Australia, show that 45% of influenza A infections were A/California/7/2004-like(H3), 11% were A/Wellington/1/2004 (H3) and 44% were A/New Caledonia /20/99-like (H1), as collected in the 2005 flu season (Turner et al., 2006). Surveillance data from New Zealand also show that the dominant H3N2 strain in the 2005 flu season was A/California/7/2004 (Influenza Weekly, 2005).
We plot the protein distance for the H3N2 viruses in the 2003–2004 flu season in Fig. 6c. We only use the data as of 30 September 2004, earlier than the October 2004 date when the WHO published the influenza vaccine recommendation for Southern hemisphere. We plot the kernel density estimation in Fig. 6a for the data in Fig. 6c. There are three major clusters in Fig. 6a. The one on the left is the current dominant cluster which are mostly A/Fujian/422/2002-like viruses. There is a middle cluster centered on A/Wellington/1/2004. The one on the right contains A/California/7/2004. Both the A/California/7/2004 cluster and the A/Wellington/1/2004 cluster are antigenically novel from A/Fujian/411/2002.
When the protein distance map and kernel estimation as of 1 February, 2004 is plotted in Fig. 6d and b, we still see the A/Wellington/1/2004 cluster. With these data, the A/California/7/2004 cluster is no longer observed. Thus, A/California/7/2004 cluster is a newly appearing cluster and we consider it to be the emerging strain. The new cluster which contains A/California/7/2004 is separate from the current dominant cluster. We calculated the average pepitope of the new cluster that contains A/California/7/2004 with regard to A/Fujian/411/2002 to be 0.112. This makes the new cluster fulfill both criteria for an incipient dominant strain cluster. So we predict based on the information as of 30 September 2004 that A/California/7/2004 will be the next dominant strain after A/Fujian/411/2002 in Southern hemisphere. We further predict from these data that A/California/7/2004 will be the dominant strain in the following flu season in the Northern hemisphere. These predictions agree with the observed dominant strain in the 2005 flu season.
Discussion
The evolution of influenza virus is driven by cell receptor distributions, non-specific innate host defense mechanisms, cross-immunity (Gupta et al., 1998; Ferguson et al., 2003) and other contributions to viral fitness. In this paper, we focussed on HA protein evolution under antibody selection pressure. The degree to which the immunity induced by one strain protects against another strain depends on their antigenic distance (Gupta et al., 2006). Because the human immune response to viral infection is not completely cross-protective, natural selection favors amino acid variants of the HA protein that allow the virus to evade immunity, infect more hosts and proliferate. Mutant strains surround the dominant strain and group into a cluster rather than evolve in a defined direction (Plotkin et al., 2002; Smith et al., 2004). After the virus has circulated in population for one or more years, effective vaccines and cross-immunity of the population drive the evolution of influenza by mutation and reassortment. This evolution increases the immune-escape component of the fitness of new strains, and eventually causes a new epidemic. These new immune-escape strains will form a new cluster, and the old clusters will die out, thus starting a new cycle. This process of creating of new clusters is what our method detects. The low-dimensional clustering can be used not only in genetic sequences but also on distances calculated from inhibition assays of antibody and antigens, as first shown by Lapedes and Farber (2001) and Smith et al. (2004). The inhibition assay provides an approximation of antigenic distance and is broadly used as a marker for vaccine efficacy. The inhibition assay suffers from low resolution of data, which multidimensional scaling improves, and is less able to predict the vaccine efficacy than the pepitope method (Gupta et al., 2006). The genetic sequences used here are a direct description of the evolution of pathogen and the antigenic distance of influenza. To aid vaccine selection, the low-dimensional clustering on genetic sequences appears informative.
Challenges may arise in application of the method described here. If two or more new clusters appear in one season, additional information is needed to decide which cluster should be chosen for vaccine. Fortunately, it has been shown that the evolution of influenza is typically in one direction (Ferguson et al., 2003; Smith et al., 2004). It is rare to have two or more new clusters in the protein distance map in one season. As experience with the low-dimensional sequence clustering is gained, it may be that cluster structure will allow more precise prediction of vaccine efficacy. Despite these issues, the method described here can assist the design of vaccines, and it provides a new tool to analyze influenza viral dynamics. We did not see any false positive results in Table I.
The current WHO method works quite well in many years. The method discussed here appears to offer an additional tool which may provide additional utility.
Materials and methods
Data sources
Influenza hemagglutinin A(H3N2) sequences before 1 October 2008, and A(H1N1) sequences as of 5 December 2009, were downloaded from NCBI Influenza Virus Resources (Bao et al., 2008). All hemagglutinin sequences used in our study are filtered by removing identical sequences, Thus, all groups of identical sequences in the data set are be represented by the oldest sequence in each group. This approach reduces the number of sequences by keeping only the unique sequences in the data set. The hemagglutinin proteins of 2009 A(H1N1) used in our work are listed in Supplementary data, Table S3. The numerical labels in Figs 1 and 2 are the same as the labels in the first column of Supplementary data, Table S3. Influenza A(H3N2) sequences after 1 October 2008 were downloaded from GISAID database, see Supplementary data, Table S6. GISAID has the latest H3N2 sequence data.
Geographical spread pattern of 2009 A(H1N1)
It is believe that the 2009 A(H1N1) virus was most likely originated from Mexico (Fraser et al., 2009). It first spread to the neighboring country USA and then to other countries. We display this geographical spread pattern in Fig. 1. We take the founder–F1 relationship from Fig. 1, and assume the virus spreads from location of founder to the location of F1. We consider three regions: the USA, Mexico and other countries except the USA and Mexico. Then we count the cases of spreading from one region to another region. In Supplementary data, Table S2, we show that we observed many more paths of spreading from the USA to other countries than from other countries to the USA. The major path of spreading is from the USA to other countries. This result indicates our directional evolutionary map of Fig. 1 is in good agreement with the pattern of geographical spread.
Multidimensional scaling
The goal of multidimensional scaling is to represent the distance of proteins by a Euclidean distance in coordinate space. We calculate the distance between proteins 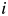 and
and 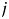 ,
, 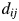 , by the number of amino acid residue differences divided by the total number of amino acid residues, as defined by Equation (1) in the main text. To do multidimensional scaling, we start with the distance of the proteins. The object of multidimensional scaling is to find the two, or p in general, directions that best preserve the distances
, by the number of amino acid residue differences divided by the total number of amino acid residues, as defined by Equation (1) in the main text. To do multidimensional scaling, we start with the distance of the proteins. The object of multidimensional scaling is to find the two, or p in general, directions that best preserve the distances 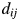 between the N proteins
between the N proteins
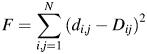 |
(2) |
Here, 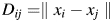 is the Euclidean distance between proteins
is the Euclidean distance between proteins 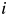 and
and 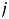 in the projected space, and
in the projected space, and  is the vector norm. The algorithm is as follows. Let the matrix
is the vector norm. The algorithm is as follows. Let the matrix 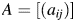 , where
, where 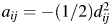 . The eigenvalues of A are
. The eigenvalues of A are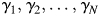 and
and 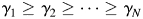 . Let
. Let 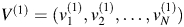 be the eigenvector of
be the eigenvector of 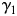 and
and 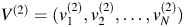 be the eigenvector of
be the eigenvector of 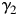 . Let
. Let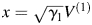 and
and 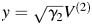 . The two coordinates in Figs 2–6 are
. The two coordinates in Figs 2–6 are 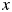 and
and 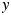 . The x-axis in the protein distance map is the largest eigenvector. We take the H3N2 2008–2009 season as an example. In Fig. 5c, we observe two clusters. One cluster is on the right side of figures with x value positive and another one has negative x values. We define the consensus sequence of a group of flu strains by taking the most frequent amino acid at each position. We calculate the consensus sequences both for the strains in the cluster on the right and on the left side of the figure. We found amino acids at four positions (76, 160, 172 and 203) are different for these two consensus H3N2 strains, see Supplementary data, Table S1. Interestingly, the Shannon entropy calculated from all 2008–2009 season sequences at these four positions (0.43, 0.67, 0.59 are 0.50) are the largest, which means the diversity at these four position are the largest.
. The x-axis in the protein distance map is the largest eigenvector. We take the H3N2 2008–2009 season as an example. In Fig. 5c, we observe two clusters. One cluster is on the right side of figures with x value positive and another one has negative x values. We define the consensus sequence of a group of flu strains by taking the most frequent amino acid at each position. We calculate the consensus sequences both for the strains in the cluster on the right and on the left side of the figure. We found amino acids at four positions (76, 160, 172 and 203) are different for these two consensus H3N2 strains, see Supplementary data, Table S1. Interestingly, the Shannon entropy calculated from all 2008–2009 season sequences at these four positions (0.43, 0.67, 0.59 are 0.50) are the largest, which means the diversity at these four position are the largest.
There is software available to run the multidimensional scaling. We use the Matlab function ‘CMD-SCALE’ to generate an 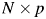 configuration matrix Y. Rows of Y are the coordinates of N points in p-dimensional space. The ‘CMDSCALE’ also returns a vector E containing the sorted eigenvalues of what is often referred to as the ‘scalar product matrix,’ which, in the simplest case, is equal to
configuration matrix Y. Rows of Y are the coordinates of N points in p-dimensional space. The ‘CMDSCALE’ also returns a vector E containing the sorted eigenvalues of what is often referred to as the ‘scalar product matrix,’ which, in the simplest case, is equal to 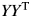 . If only two or three of the largest eigenvalues E are much larger than others, then the matrix D based on the corresponding columns of Y nearly reproduces the original distance matrix d. We used the influenza H3N2 in the 2001–2002 season as an example. The five largest of all 180 eigenvalues are 0.0361, 0.0032, 0.0024, 0.0020 and 0.0016. The first two largest eigenvalues contribute 70% to the sum of all 180 eigenvalues, which indicates p = 2. Then, we plot the N points in a two-dimensional graph. Each point represents a protein. The Euclidean distance between any two points
. If only two or three of the largest eigenvalues E are much larger than others, then the matrix D based on the corresponding columns of Y nearly reproduces the original distance matrix d. We used the influenza H3N2 in the 2001–2002 season as an example. The five largest of all 180 eigenvalues are 0.0361, 0.0032, 0.0024, 0.0020 and 0.0016. The first two largest eigenvalues contribute 70% to the sum of all 180 eigenvalues, which indicates p = 2. Then, we plot the N points in a two-dimensional graph. Each point represents a protein. The Euclidean distance between any two points 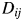 on the graph should be equal to or close to the distance of these two proteins. That is,
on the graph should be equal to or close to the distance of these two proteins. That is, 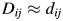 . As an example, in Supplementary data, Fig. S1, we show that
. As an example, in Supplementary data, Fig. S1, we show that 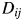 and
and 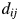 have a strong linear relationship. A short MATLAB program of multidimensional scaling is as follow.
have a strong linear relationship. A short MATLAB program of multidimensional scaling is as follow.
% Multidimensional scaling.
% alignment.aln is a sequence multialignment file
% generated by software ClustalW.
clear
Sequences = multialignread(’alignment.aln’);
distances = seqpdist(Sequences,’Method’,’p-distance’);
Y = cmdscale(distances);
scatter(Y(:,1), Y(:,2));
Biases in the data
There are two biases in the sequence data. First, more isolates are sequenced in recent years. Generally speaking, more sequences make the vaccine selection based on low-dimensional clustering methods more reliable. That is why we compared low-dimensional clustering methods with WHO results only since 1996 in Table I. To avoid these biases in the generation of the figure of evolution history of influenza for the 40 years (Fig. 3), we choose 20 random isolates for each season, even though the database contains more sequences in recent years. Second, most isolates are collected in the USA. We found that many isolates collected in the USA are identical, because of the high sampling rate in the USA. To reduce this bias, we collapse redundant strains, keeping only distinct strains.
Supplementary data
Funding
This research was supported by DARPA under the FunBio program.
Supplementary Material
References
- Bao Y., Bolotov P., Dernovoy D., et al. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N., Bender C. Semin. Virol. 1995;6:359–370. doi:10.1016/S0168-583X(01)00748-0. [Google Scholar]
- Deem M., Pan K. Protein Eng. Des. Sel. 2009;22:543–546. doi: 10.1093/protein/gzp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Holland J., Biebricher C. Quasispecies and RNA Virus Evolution: Principles and consequences. Austin, TX: Landes; 2002. doi:10.1016/j.radmeas.2003.12.023. [Google Scholar]
- Drake J.W., Holland J.J. Proc. Natl Acad. Sci. USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt B.S., Landau S., Leese M. Cluster Analysis. Oxford University Press; 2001. doi:10.1016/S1350-4487(96)00085-6. [Google Scholar]
- FDA. Regulatory considerations regarding the use of novel influenza A (H1N1) virus vaccines. 2009. 23 July.
- Ferguson N., Galvani A., Bush R. Nature. 2003;422:428. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- Fitch W.M., Bush R.M., Bender C.A., Cox N.J. Proc. Natl Acad. Sci. USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. doi:10.1097/00004032-197108000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C., Donnelly C.A., Cauchemez S., et al. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten R.J., Davis C.T., Russell C.A., et al. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Sengamalay N.A., Shumway M., et al. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. doi:10.1016/j.radmeas.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Gupta S., Ferguson N., Anderson R. Science. 1998;280:912. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- Gupta V., Earl D.J., Deem M. Vaccine. 2006;24:3881–3888. doi: 10.1016/j.vaccine.2006.01.010. doi:10.1016/j.radmeas.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hak E., Nordin J., Wei F., et al. Clin. Infect. Dis. 2002;35:370–377. doi: 10.1086/341403. doi:10.1016/j.radmeas.2009.10.089. [DOI] [PubMed] [Google Scholar]
- Lapedes A., Farber R. J. Theor. Biol. 2001;212:57–69. doi: 10.1006/jtbi.2001.2347. [DOI] [PubMed] [Google Scholar]
- Nelson M.I., Holmes E.C. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. doi:10.1016/j.radmeas.2009.10.075. [DOI] [PubMed] [Google Scholar]
- Northern hemisphere: Risk of A/Wellington/1/2004(H3N2)-like virus. ProMed. 2004, October 24. Available from http://www.promedmail.org. archive no. 20041024.2879.
- Pan K., Subieta K., Deem M. Protein Eng. Deg. Sel. doi: 10.1093/protein/gzq105. in press doi:10.1016/j.radmeas.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J.B., Dushoff J., Levin S.A. Proc. Natl Acad. Sci. USA. 2002;99:6263–6268. doi: 10.1073/pnas.082110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., et al. Vaccine. 2008a;26:D31–D34. doi: 10.1016/j.vaccine.2008.07.078. doi:10.1016/j.asr.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., et al. Science. 2008b;320:340–346. doi: 10.1126/science.1154137. doi:10.1093/rpd/ncp265. [DOI] [PubMed] [Google Scholar]
- Santamaria C., Urue A., Videla C., et al. Influenza and Other Respiratory Viruses. 2008;2:131–134. doi: 10.1111/j.1750-2659.2008.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seasonal influenza (H3N2) virus - potential vaccine mismatch. ProMed. 2009, July 24. Available from http://www.promedmail.org. archive no. 20090724.2623.
- Skowronski D. Influenza A (H1N1) - worldwide (11): Coincident H3N2 variation. ProMed. 2009, 5 May. Available from http://www.promedmail.org. archive no. 20090505.1679.
- Smith D.J., Lapedes A.S., de Jong J.C., et al. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Smith G., Vijaykrishna D., Bahl J., et al. Nature. 2009;459:1122. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Turner J.L., Fielding J.E., Clothier H.J., Kelly H.A. Commun. Dis. Intell. 2006;30:137. [PubMed] [Google Scholar]
- Webster R.G. Emerging Infect. Dis. 1998;4:436–441. doi: 10.3201/eid0403.980325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 1995;70:53. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 1996;71:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 1997;72:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 1998;73:56. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 1999;74:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2000;75:61. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2001;76:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2002;77:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2003;78:57. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2004;79:85. [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2005a;80:65. [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2005b;80:341. [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2006;81:81. [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2007;82:69. [PubMed] [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2008;83:77. [Google Scholar]
- World Health Organization. Wkly Epidemiol. Rec. 2009a;84:65. [Google Scholar]
- World Health Organization. Pandemic (H1N1) 2009 – update 64. 2009b cited 2010, 27 January Available from. [Google Scholar]
- World Health Organization. Recommendations for influenza viruses. 2009c. Available from http://www.who.int/csr/disease/influenza/vaccinerecommendations/en/index.html .
- World Health Organization. World now at the start of 2009 influenza pandemic. 2009d. 4 June Available from http://www.who.int/mediacentre/news/statements/2009. /
- World Health Organization. Wkly Epidemiol. Rec. 2010;85:81. [Google Scholar]
- Zhou H., Pophale R.S., Deem M.W. Wang Q., Tao Y.J. Influenza: Molecular Virology. Caister Academic Press; 2010. Chapter 10. [Google Scholar]
- MMWR Morb Mortal Wkly Rep. 2004;53:8–11. [PubMed] [Google Scholar]
- Virology quarterly report July–September 2004. 2004.
- Influenza Weekly Update. 2005:22–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.