In this brief report by de Silva and colleagues, the authors reveal that HSV/SB vectors contain a biologically active non-histone nuclear protein, called HMGB1, that facilitates SB-mediated transposition in vitro.
Abstract
The development of the integration-competent, herpes simplex virus/Sleeping Beauty (HSV/SB) amplicon vector platform has created a means to efficiently and stably deliver therapeutic transcription units (termed “transgenons”) to neurons within the mammalian brain. Furthermore, an investigation into the transposition capacity of the HSV/SB vector system revealed that the amplicon genome provides an optimal substrate for the transposition of transgenons at least 12 kb in length [de Silva, S., Mastrangelo, M.A., Lotta, L.T., Jr., Burris, C.A., Federoff, H.J., and Bowers, W.J. (2010). Gene Ther. 17, 424–431]. These results prompted an investigation into the factors that may contribute toward efficient transposition from the HSV/SB amplicon. One of the cellular cofactors known to play a key role during SB-mediated transposition is the high-mobility group DNA-binding protein-1 (HMGB1). Our present investigation into the role of HMGB1 during amplicon-based transposition revealed that transposition is not strictly dependent on the presence of cellular HMGB1, contrary to what had been previously demonstrated with plasmid-based SB transposition. We have shown for the first time that during amplicon preparation, biologically active HMGB1 derived from the packaging cell line is copackaged into amplicon vector particles. As a result, HSV/SB amplicon virions arrive prearmed with HMGB1 protein at levels sufficient for facilitating SB-mediated transposition in the transduced mammalian cell.
Introduction
With a packaging capacity of approximately 130 kb and an ability to efficiently transduce numerous cell types, the herpes simplex virus type 1 (HSV-1) amplicon provides an attractive modality for gene therapeutic applications (Spaete and Frenkel, 1982; Oehmig et al., 2004). The HSV amplicon contains two noncoding elements from the wild-type HSV-1 genome, which are the packaging/cleavage site (pac) and the HSV-1 origin of replication (oriS), and is devoid of all viral genes. Once packaged, the 150-kb vector consists of a concatemer of repeating amplicon units arranged in a head-to-tail configuration, which provides efficient expression of the harbored transgene(s). However, persistent therapeutic gene expression within the transduced cell is infeasible because of its episomal existence, which leads to loss of the HSV amplicon genome in actively dividing cells (Suzuki et al., 2007). The more recent development of “integration-competent” iterations of the HSV amplicon via the generation of HSV/adeno-associated virus (AAV) hybrid vectors or through the adaptation of the Sleeping Beauty (SB) transposon system (Ivics et al., 1997) has circumvented this hurdle by providing the means to deliver and stably integrate a transgene within the mammalian genome (Fraefel et al., 1997; Bakowska et al., 2003; Bowers et al., 2006; Peterson et al., 2007).
The current HSV/SB amplicon vector platform includes an “effector” amplicon designed to express the SB transposase under the transcriptional regulation of the HSV immediate-early 4/5 (IE4/5) promoter, and an “integrator” amplicon that harbors a transgene unit flanked by the inverted/direct repeat (IR/DR) elements of SB. On cotransduction of the two amplicons into a target cell, the SB transposase excises the IR/DR-flanked transgene unit from the integrator amplicon and integrates it into a TA dinucleotide site within the host cell genome. As a safety measure, the integrated transgene is transcriptionally regulated by the cytomegalovirus (CMV) IE promoter, which unlike long terminal repeat (LTR) sequences present in retrovirus-derived vectors, does not aberrantly influence the expression of genes located at distal sites within the host cell genome. Application of this new amplicon vector platform has resulted in prolonged expression and genomic retention of a reporter transgene unit in vivo (Bowers et al., 2006). The HSV/SB vector system integrates its IR/DR-flanked cargo in a precise manner without causing any chromosomal rearrangements, which is a result of the step-wise transposition process mediated by the SB transposase (Luo et al., 1998). This is an advantage over integration-competent HSV/AAV vectors, which have the potential to cause chromosomal rearrangements at the site of transgene integration due to the action of the Rep protein (Young and Samulski, 2001).
The transposition process catalyzed by the Sleeping Beauty (SB) transposase involves a series of highly coordinated events, which include the following: (1) the binding of SB transposase to its cognate inverted repeat/direct repeat (IR/DR) elements, (2) formation of the synaptic complex, (3) excision of transposon DNA from the donor locus, and (4) integration of the transposon into chromosomal DNA (Ivics et al., 1997; Hackett et al., 2005). Although the critical elements required for efficient transposition involve the SB transposase and its cognate flanking DNA elements, host cell-encoded factors have also been shown to aid the complex excision and integration processes (Yant and Kay, 2003; Zayed et al., 2003; Izsvak et al., 2004; Walisko et al., 2006). Among the identified cellular cofactors that assist in the transposition process, the high-mobility group DNA-binding protein-1 (HMGB1) has unequivocally been shown to facilitate transposition from a plasmid-based vector (Zayed et al., 2003). However, the role of HMGB1 in SB-mediated transposition from the HSV amplicon genome has yet to be determined.
HMGB1 is a nonhistone nuclear protein that is found abundantly in eukaryotic cells and is closely associated with chromatin (Bianchi and Beltrame, 2000). Extensive investigation into the functionality of HMGB1 has revealed its involvement in DNA replication, gene regulation, V(D)J recombination, and more recently, inflammation (Bustin, 1999; Scaffidi et al., 2002). Structurally, HMGB1 contains a conserved HMG-1 domain (also known as the HMG-1 box) that binds A/T-rich DNA in the minor groove, which causes local unwinding and bending of the DNA. During their investigation into the involvement of HMGB1 in SB transposition, Zayed and coworkers demonstrated that SB transposition from a plasmid-based vector was severely compromised in the absence of HMGB1 in a mouse embryonic fibroblast (MEF) cell line, which had been derived from an Hmgb1–/– mouse (Calogero et al., 1999). Transposition could be restored in this cell line by plasmid-based transient expression of HMGB1. They further demonstrated that with its ability to bend transposon DNA and physically interact with SB transposase, HMGB1 enhanced the binding affinity of SB transposase to the inner direct repeat (DR) element and facilitated the formation of the synaptic complex; a requisite of SB-mediated integration (Zayed et al., 2003). In a separate study, Zayed and colleagues also demonstrated that transient overexpression of HMGB1 during transposition of a modified plasmid-harbored transposon (termed “T-zeo322”) by a hyperactive mutant of the SB transposase (termed “SB12”) resulted in an increase in transposition efficiency as compared with when HMGB1 was not overexpressed (Zayed et al., 2004). Furthermore, our laboratory showed that significant HMGB1 expression was evident in neuronal precursor cells populating the ependymal and subventricular layers of the embryonic day 14.5 (E14.5) mouse embryo, wherein SB-mediated integration of a reporter transcription unit (a “transgenon”; β-galactosidase/neomycin phosphotransferase fusion gene) was shown to occur after in utero delivery of the HSV/SB amplicon vector platform (Peterson et al., 2007). On the basis of these prior reports we initiated a set of studies to determine the potential for optimizing transposition efficiencies of the current HSV/SB amplicon platform by systematically altering HMGB1 protein levels during mammalian cell transduction. Unexpectedly, we discovered that HSV amplicon virions harbor biologically active HMGB1 at levels sufficient to promote SB-mediated transposition.
Materials and Methods
Cell line maintenance
The baby hamster kidney (BHK) cell line used for helper virus-free amplicon packaging was obtained from the American Type Culture Collection (ATCC, Manassas, VA; cat. no. CCL-10) and maintained in Dulbecco's modified Eagle's medium (DMEM) and 10% fetal bovine serum at 37°C and 5% CO2 levels. The mouse embryonic fibroblast (MEF)-derived HMGB1 wild-type (VA1) and HMGB1-deficient (C1) cell lines were kindly provided by Z. Izsvák (Max Delbrúck Center for Molecular Medicine, Berlin, Germany), and were maintained in DMEM and 10% fetal bovine serum at 37°C and 5% CO2 levels.
Amplicon plasmid construction
To construct the pHSVT-Zeo transposable reporter amplicon plasmid, the Sh Ble open reading frame, which confers resistance to Zeocin, was amplified by PCR from pT-Zeo-R6K (kindly provided by M. Kay, Stanford University, Stanford, CA). The primer sequences were designed to contain a BamHI site in the forward primer and an EcoRI site in the reverse primer. The primer sequences are as follows: Zeo.Fwd, 5′-GCCGCGGATCCACCATGGCCAAGTTGACCAGTGCTGTCC-3′; Zeo.Rev, 5′-GCCGCGAATTCTCAGTCCTGCTCCTCTGCCACAAAGTG-3′. The PCR product was ethanol precipitated, digested with EcoRI and BamHI, and cloned into the corresponding sites in the pFBGR vector to generate the CMV-Zeo-SV40 poly(A) transcription unit. This unit was excised from the pFBGR-Zeo plasmid, using the flanking NotI sites, and cloned into the blunted ClaI site of the pHSVTmcs vector to generate the final pHSVT-Zeo amplicon plasmid. To construct pHSV-HMGB1, the HMGB1 open reading frame was amplified by PCR from the pEBB-HMGB1 plasmid (kindly provided by M. Bianchi, San Raffaele Scientific Institute, Milan, Italy). The primer sequences were designed to contain a 5′ KpnI site in the forward primer and a 5′ SacI site in the reverse primer. The primer sequences are as follows: HMGB1.Fwd, 5′-GCCGCCGGTACCATGGGCAAAGGAGATCCTAAGAAGC-3′; HMGB1.Rev, 5′-GCGCCGAGCTCCTATTATTCATCATCATCATCTTCTTCTTCATC-3′. The PCR product was ethanol precipitated, digested with EcoRI and BamHI, and cloned into the corresponding sites in the pHSVPrPUC vector downstream of the HSV IE4/5 promoter to generate the pHSV-HMGB1 plasmid.
Helper virus-free HSV-1 amplicon packaging
Helper virus-free amplicon vectors were packaged using the BHK cell line (cat. no. CCL-10; ATCC) as described previously (Bowers et al., 2001). The HSVlac amplicon vector was individually packaged using the HMGB1 wild-type and HMGB1-deficient MEF cell lines in a similar manner and titers ranged between 4 × 107 and 5 × 107 transducing units (TU)/ml.
Colony formation assays
For HSV amplicon plasmid-based transposition, HMGB1-deficient MEF cells (1.65 × 105 cells per well in a 35-mm dish) were individually triple-transfected with pHSV-SB10, pHSVT-CMV-Zeo, and pHSV-HMGB1 plasmids at a 1:1:1 mass ratio (333 ng per construct), using the FuGENE 6 transfection reagent (Roche, Mannheim, Germany). The pHSVPrPUC plasmid served as a negative control in the absence of pHSV-HMGB1 to ensure that equal amounts of DNA were being transfected under each condition. Forty-eight hours posttransfection, the cells were trypsinized and seeded at a 1:13 dilution onto 100-mm dishes containing medium supplemented with Zeocin (100 μg/ml; Invitrogen, Carlsbad, CA). Antibiotic selection was carried out for 14 days, at which point the Zeocin-resistant colonies were fixed with 4% paraformaldehyde, stained with 2% methylene blue, and enumerated.
To assess the amplicon-based transposition efficiency of SB transposase in the presence of HMGB1 overexpression, HMGB-deficient MEF cells (1.65 × 105 cells per well in a 35-mm dish) were individually transduced with increasing amounts of HSV-HMGB1 viral particles (multiplicities of infection [MOIs] of 0.25, 0.5, and 1). On incubation for 1 hr at 37°C, the virus-containing medium was removed and cells were washed once and replenished with fresh medium. Four hours after the initial transduction, the same cells were individually cotransduced at an MOI of 1 with HSV-SB10 and HSVT-CMV-Zeo at a 1:1 viral particle ratio. The HSVPrPUC amplicon served as a negative control in the absence of HSV-HMGB1 to ensure that equal amounts of viral particles were being transduced. The colony formation assay was performed as described previously.
To test transposition efficiency of SB in the presence of virion-associated HMGB1, HMGB1-deficient MEF cells (1.65 × 105 cells per well in a 35-mm dish) were transduced with HSVlac viral particles (MOI of 1) packaged from either wild-type MEF cells or HMGB1-deficient MEF cells. After incubation for 1 hr at 37°C, the virus-containing medium was removed and cells were washed once and replenished with fresh medium. Four hours after the initial transduction, the cells were cotransfected with pHSV-SB10 and pHSVT-CMV-Zeo at a 1:1 mass ratio, using the FuGENE 6 transfection reagent. The colony formation assay was performed as described previously.
Western blotting
To confirm expression of the HSV-HMGB1 amplicon in the HMGB1 wild-type and HMGB1-deficient MEF cell lines, 1 × 106 cells from each cell line were separately transduced with HSV-HMGB1 amplicon at an MOI of 0.5. Cell lysates were analyzed for the presence of HMGB1 via a primary anti-HMGB1 rabbit polyclonal antibody (diluted 1:2000; Abcam, Cambridge, MA) in combination with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L) secondary antibody (diluted 1:2000; Jackson ImmunoResearch Laboratories, West Grove, PA). Subsequently, the membrane was stripped and reprobed with an anti-β-actin monoclonal mouse antibody (diluted 1:3000; Sigma-Aldrich, St. Louis, MO) in combination with an HRP-conjugated goat anti-mouse IgG (H+L) secondary antibody (diluted 1:2000; Jackson ImmunoResearch Laboratories) to verify that equal amounts of protein were being loaded in each lane. To determine the presence of HMGB1 protein in each viral stock, 10 μl of virus was lysed with an equal volume of radioimmunoprecipitation assay (RIPA) buffer and subjected to Western blot analysis. After immunoblotting for HMGB1, the membrane was stripped and reprobed for the presence of the HSV VP16 tegument protein in each viral stock, using a primary anti-VP16 polyclonal rabbit antibody (diluted 1:2000; Sigma-Aldrich) in combination with an HRP-conjugated goat anti-rabbit IgG secondary antibody (diluted 1:2000; Jackson ImmunoResearch Laboratories).
Detergent treatment and sucrose gradient fractionation of amplicon virions
The detergent treatment of amplicon virions and sucrose gradient fractionation to determine the location of HMGB1 in the virion was carried out as described by Murphy et al. (2008). Briefly, helper virus-free HSVlac virions packaged in either the wild-type MEF or HMGB1-deficient MEF cell line were individually treated with 1% Nonidet P-40 (NP-40) at 4°C for 30 min. After treatment, the samples were overlaid on a 20–50% (w/w) linear sucrose gradient (10 ml) and subjected to rate zonal centrifugation at 24,500 rpm for 1 hr at 4°C in a Beckman SW-41 rotor (Beckman Coulter, Fullerton, CA). After centrifugation, 1-ml gradient fractions were collected from the bottom of the tube and proteins were precipitated overnight at 4°C, using 10% trichloroacetic acid (TCA). After centrifugation at 14,000 rpm for 5 min, the protein pellets were resuspended in 1× sample buffer and electrophoretically separated on a sodium dodecyl sulfate–10% polyacrylamide gel. Western blot analysis was performed to detect the presence of HMGB1 and VP16 in each fraction as described previously.
Results and Discussion
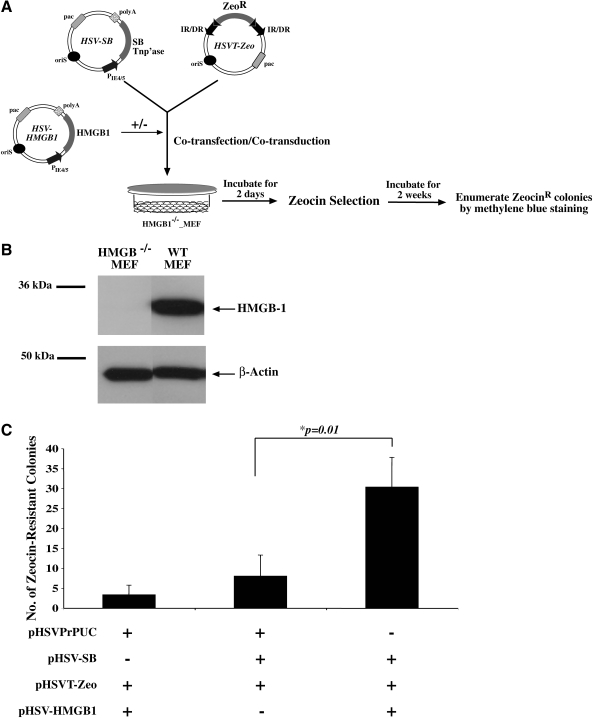
The requirement of HMGB1 during SB-mediated transposition from the plasmid version of the HSV amplicon was determined with an HMGB1-deficient mouse embryonic fibroblast (MEF) cell line (kindly provided by Z. Izsvák, and described previously [Calogero et al., 1999]). Western blot analysis confirmed that the HMGB1-deficient MEF cell line was indeed devoid of cellular HMGB1, as compared with the control wild-type MEF cell line (Fig. 1B). A new “integrator” amplicon plasmid was constructed that carried a Zeocin-resistant reporter gene (termed pHSVT-Zeo), and a colony formation assay was performed with the HMGB1-deficient MEF cell line in the presence or absence of a cotransfected pHSV-SB plasmid (Fig. 1A). Cotransfection of the pHSVT-Zeo amplicon plasmid together with the SB transposase-expressing amplicon plasmid (pHSV-SB) did not result in an increase in Zeocin-resistant colonies in the HMGB1-deficient cell line compared with when SB transposase was absent (Fig. 1C). However, we were able to rescue SB-mediated transposition via the cotransfection of an HMGB1-expressing amplicon plasmid (pHSV-HMGB1) (Fig. 1C). This result was consistent with the findings of Zayed and colleagues (2003), and confirmed that HMGB1 was required for SB transposition from the HSV amplicon plasmid.
FIG. 1.
HMGB1 facilitates Sleeping Beauty (SB)-mediated transposition from the context of an HSV-1 amplicon plasmid. (A) Schematic representation of the colony formation assay. All assays were performed at a minimum of n = 3 per condition. (B) Western blot analysis was conducted with an anti-HMGB1 rabbit polyclonal antibody to confirm the lack of HMGB1 protein expression in the HMGB1-deficient mouse embryonic fibroblast (MEF) cell line. Wild-type MEF cells served as a positive control. (C) HMGB1-deficient MEF cells were cotransfected with pHSVT-Zeo in combination with pHSV-SB and pHSV-HMGB1 at an equivalent mass ratio. pHSVPrPUC served as a negative control and was employed to ensure that equal amounts of DNA were transfected under each condition. After a 14-day antibiotic selection period, the numbers of Zeocin-resistant colonies were enumerated via methylene blue staining. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test. Values of p are indicated.
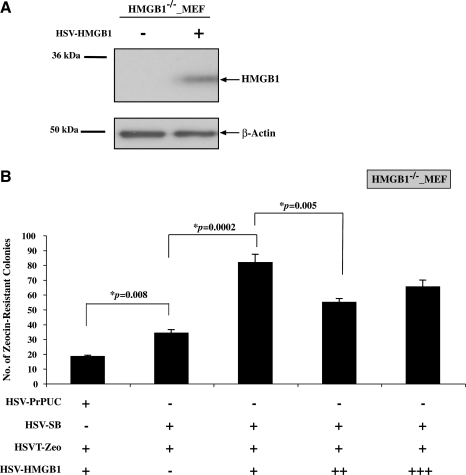
Next, the HSVT-Zeo, HSV-SB, and HSV-HMGB1 amplicons were packaged into virions in BHK cells, using helper virus-free packaging, and titered (Bowers et al., 2001; Bowers and Federoff, 2006). To confirm expression of the HSV-HMGB1 amplicon, HMGB1-deficient MEF cells were transduced and Western blot analysis of the cell lysates confirmed that the packaged HSV-HMGB1 amplicon virions express HMGB1 in transduced HMGB1-deficient MEF cells (Fig. 2A). To determine the effect of various HMGB1 levels on SB-mediated transposition from HSV/SB virions, we performed a colony formation assay (Bowers et al., 2006) in HMGB1-deficient cells. As shown in Fig. 2B, cotransduction of HSV-SB and HSVT-Zeo in the HMGB1-deficient MEF cell line resulted in a 1.86-fold increase in the number of Zeocin-resistant colonies compared with when HSVPrPUC (empty vector control) was cotransduced with the HSVT-Zeo amplicon. This was surprising because we had not observed such an increase in the number of Zeocin-resistant colonies in the absence of HMGB1 when plasmid versions of the same amplicons were used (Fig. 1C). This indicated that SB transposase was capable of mediating a modest level of transposition in the absence of target cell-expressed HMGB1 when delivered via the HSV/SB amplicon vector particles. Furthermore, transduction of HMGB1-deficient MEF cells with various numbers of HSV-HMGB1 viral particles 4 hr before cotransduction with HSV-SB and HSVT-Zeo virus resulted in a significant increase in the transposition efficiency compared with when an empty amplicon vector, HSVPrPUC, was transduced before SB-mediated transposition. Progressively higher HSV-HMGB1 MOIs did not further enhance transposition efficiency, demonstrating that an upper limit exists to the HMGB1-mediated potentiating effect.
FIG. 2.
Effect of HMGB1 overexpression on SB-mediated transposition from the HSV amplicon. (A) Detection of HMGB1 protein via Western blot analysis of cell lysates prepared from HMGB1-deficient MEF cells either nontransduced or transduced with HSV-HMGB1 amplicon at an MOI of 0.5 (top). β-Actin served as a loading control (bottom). (B) HMGB1-deficient MEF cells were cotransduced at an MOI of 1 with equal numbers of HSV-SB10 and HSVT-Zeo viral particles in combination with increasing numbers of HSV-HMGB1 viral particles [MOIs: 0.25 (+), 0.5 (++), and 1.0 (+++)]. The HSVPrPUC amplicon served as a negative control and was used as a “balance” virus to ensure that equal numbers of viral particles were transduced. Two days posttransduction, cells were placed under Zeocin selection for 14 days (n = 4 per condition), at which point Zeocin-resistant colonies were enumerated to determine the extent of transposition. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test. Values of p are indicated.
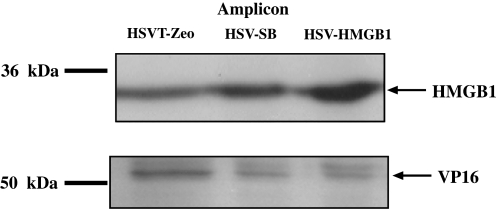
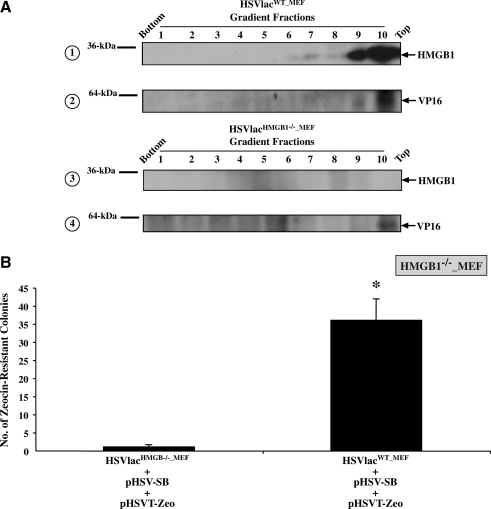
HSV/SB-mediated transposition in the absence of target cell HMGB1 expression suggests that an alternative source of the cofactor exists. Packaging of HSV amplicon genomes into particles results in the incorporation of viral and packaging cell-derived proteins (Bowers and Federoff, 2006). Loret and coworkers conducted a comprehensive proteomic analysis of extracellular wild-type HSV-1 virions and identified 49 cellular proteins, which included actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), but failed to identify HMGB1 as a detectable component of wild-type HSV-1 particles (Loret et al., 2008). The amplicon genome, although similar in size to the wild-type HSV-1 genome, is inherently different because of its concatemeric organization. Hence, it is possible that the multiple HSV-1 ori and pac sequences in the amplicon genome somehow promote retention of packaging cell-derived HMGB1 at the time of virion assembly/genome insertion. Thus, we employed Western blot analysis to assess whether HMGB1 copurifies with HSV-SB (8 × 108 TU/ml), HSVT-Zeo (1.1 × 108 TU/ml), and HSV-HMGB1 (5 × 108 TU/ml) amplicon stock. A substantial level of HMGB1 protein derived from the BHK packaging cell line was detected in each purified stock (Fig. 3). The presence of the HSV tegument protein, VP16, confirmed the presence of packaged HSV amplicon vector particles in each of the purified viral stocks. Next, we sought to identify the location of HMGB1 within the packaged amplicon particle. HSVlac virions packaged in either the wild-type MEF cell line (termed HSVlacWT_MEF) or the HMGB1-deficient MEF cell line (termed HSVlacHMGB1–/–_MEF) were treated with 1% NP-40 to de-envelope the virions and subsequently subjected to rate zonal centrifugation in a 20–50% sucrose gradient to isolate the tegument fraction from the nucleocapsid fraction (Murphy et al., 2008). Western blot analysis of the gradient fractions of HSVlacWT_MEF virus treated with NP-40 revealed that HMGB1 was restricted to fractions that contained the tegument protein VP16 (Fig. 4A). Conversely, HSVlacHMGB1–/–_MEF particles treated with NP-40 did not contain HMGB1 (Fig. 4A, panel 3), as expected. Given that we were able to detect HMGB1 in amplicon vector stocks packaged in two separate cell lines (i.e., BHK and MEF cells), irrespective of the type of transgene that was present in the amplicon, it is evident that cellular HMGB1 is preferentially incorporated into the tegument of the amplicon vector particle as evidenced by its absence in wild-type HSV-1 virions (Loret et al., 2008). Thus, the HSV/SB amplicon vector system arrives prearmed with tegument-associated HMGB1 derived from the packaging cell line, which could be advantageous for SB-mediated transposition under conditions in which HMGB1 is a limiting factor. This result also provides insight into the dose-dependent effect of HMGB1 on SB-mediated transposition from the HSV amplicon, because the presence of tegument-associated HMGB1 and the HMGB1 overexpressed from the HSV-HMGB1 vector (at the lowest MOI) might have inadvertently established a cellular level of HMGB1 that was optimally conducive to SB-mediated transposition (Fig. 2B). Furthermore, by transducing increasing amounts of HSV-HMGB1 vector particles, we may have exceeded the threshold level of HMGB1 required for SB-mediated transposition, and thus, either directly or indirectly inhibited the transposition process.
FIG. 3.
Packaging cell line-derived HMGB1 protein is associated with purified HSV amplicon viral particles. A 10-μl aliquot (equivalent to ∼5 × 106 transduction units) of each HSV-1 amplicon vector packaged from the BHK cell line was lysed in RIPA buffer and subjected to Western blot analysis to determine the presence of HMGB1 protein derived from the BHK cell line (top). On HMGB1 immunoblotting, the blots were stripped and subsequently analyzed for the presence of the HSV tegument protein VP16 (bottom). Molecular masses are represented as kilodaltons (kDa).
FIG. 4.
HMGB1 protein is incorporated into the viral tegument during helper virus-free amplicon packaging and facilitates SB-mediated transposition. (A) HSVlac amplicon vector stocks separately packaged in MEF-derived HMGB1 wild-type and HMGB1-deficient cell lines (designated HSVlacWT_MEF and HSVlacHMGB1–/–_MEF, respectively) were treated with 1% NP-40 and subjected to rate zonal centrifugation in a linear sucrose gradient. Gradient fractions were analyzed by Western blotting to determine the fractions containing HMGB1 (panels 1 and 3). Subsequently, the samples were analyzed for the presence of the HSV tegument protein VP16 (panels 2 and 4). (B) HMGB1-deficient MEF cells were separately transduced with HSVlacWT_MEF and HSVlacHMGB1–/–_MEF at an MOI of 1. Four hours posttransduction, the cells were cotransfected with pHSVT-Zeo and pHSV-SB amplicon plasmids at a 1:1 mass ratio. On incubation for 48 hr, the cells were trypsinized and seeded at a 1:13 dilution on 100-mm dishes containing medium supplemented with Zeocin (100 μg/ml) (n = 3 per condition) and incubated for 14 days. Zeocin-resistant colonies were enumerated via methylene blue staining. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test. *denotes p < 0.05.
To determine whether the virion-associated HMGB1 was biologically active, in terms of its ability to facilitate SB-mediated transposition, a modified colony formation assay was conducted. Briefly, HMGB1-deficient MEF cells were initially transduced with either HSVlacWT_MEF or HSVlacHMGB–/–_MEF. Of note, titering of the individual amplicon vector stocks revealed that both wild-type and HMGB1-deficient MEF cell lines packaged the HSVlac amplicon at ∼5 × 107 TU/ml in a reproducible manner, thus indicating that the amplicon genome is packaged to a similar degree independent of the presence of HMGB1. Four hours after transduction, the cells were cotransfected with pHSV-SB and pHSVT-Zeo amplicon plasmids. As illustrated in Fig. 4B, transduction of the HMGB1-deficient cell line with the HSVlacWT_MEF viral stock before introduction of the pHSV-SB and pHSVT-Zeo amplicon plasmids, resulted in a significant increase in the numbers of Zeocin-resistant colonies compared with the HSVlacHMGB1–/–_MEF-transduced control. This result confirmed that virion-incorporated HMGB1 derived solely from the wild-type MEF cell line was indeed sufficient for facilitating HSV/SB transposition in the absence of target cell HMGB1 expression. On the basis of previous studies by Zayed and coworkers and our current findings, we propose that after HSV/SB amplicon transduction, virion-associated HMGB1 might be recruited by SB transposase to bend the concatemeric amplicon genome into a malleable configuration that facilitates SB-mediated integration under conditions in which cellular HMGB1 is a limiting factor.
Our findings provide new evidence that the DNA-bending protein HMGB1 plays a significant role during SB-mediated transposition from the HSV amplicon, and demonstrate for the first time that HSV/SB particles arrive prearmed with HMGB1 derived from the packaging cell line, which serendipitously facilitates SB-mediated transposition. Indeed, it appears that specific levels of HMGB1 are required within the target cell at the time of SB transposition to achieve optimal levels of transposition. This may be a moot point in many cell types, as HMGB1 is thought to be a ubiquitously expressed protein (Bustin, 1999). However, Guazzi and coworkers revealed that whereas HMGB1 protein is present in certain cell types in the developing mouse brain, HMGB1 was undetectable in most cells in adult mouse brain except in areas of continuing neurogenesis (Guazzi et al., 2003). Furthermore, Izsvak and colleagues demonstrated that various cell types support various SB transposition efficiencies (Izsvak et al., 2000), which could be due to limitations imposed by the cellular levels of cofactors, including HMGB1. On the basis of our results, it is evident that titration of appropriate levels of HMGB1 before SB-mediated transposition can impart significant effects on transgene integration efficiency from the HSV/SB amplicon vector platform, which is being developed as a gene therapy vector for the treatment of neurological diseases. Such a feat in vivo could be achieved within the design of the HSV/SB amplicon vector platform via the use of specific promoters that temporally regulate the expression of SB transposase and HMGB1 to catalyze optimal SB transposition. Further characterization, development, and optimization of this newer HSV amplicon-based vector is warranted to ensure that the codelivery of HMGB1 and other packaging cell line-derived proteins does not adversely affect target cellular physiology and that this hybrid platform maintains host cell genomic integrity by integrating the transgene into “safe” chromosomal locations.
Acknowledgments
The authors thank Dr. Howard Federoff (Georgetown University) for helpful discussions, and Dr. Richard Courtney (Pennsylvania State University) for technical advice. NIH R01-AG023593 (W.J.B.) supported this work.
Author Disclosure Statement
The authors declare that no conflicts of interest exist.
References
- Bakowska J.C. Di Maria M.V. Camp S.M. Wang Y. Allen P.D. Breakefield X.O. Targeted transgene integration into transgenic mouse fibroblasts carrying the full-length human AAVS1 locus mediated by HSV/AAV rep+ hybrid amplicon vector. Gene Ther. 2003;10:1691–1702. doi: 10.1038/sj.gt.3302061. [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. Beltrame M. Upwardly mobile proteins. Workshop: The role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 2000;1:109–114. doi: 10.1093/embo-reports/kvd030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers W.J. Federoff H.J. Herpes simplex virus type 1-derived amplicon vectors. In Gene Transfer: Delivery and Expression of DNA and RNA, A Laboratory Manual. In: Rossi J., editor; Friedmann T., editor. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2006. pp. 227–254. [Google Scholar]
- Bowers W.J. Howard D.F. Brooks A.I. Halterman M.W. Federoff H.J. Expression of vhs and VP16 during HSV-1 helper virus-free amplicon packaging enhances titers. Gene Ther. 2001;8:111–120. doi: 10.1038/sj.gt.3301340. [DOI] [PubMed] [Google Scholar]
- Bowers W.J. Mastrangelo M.A. Howard D.F. Southerland H.A. Maguire-Zeiss K.A. Federoff H.J. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol. Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero S. Grassi F. Aguzzi A. Voigtlander T. Ferrier P. Ferrari S. Bianchi M.E. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- de Silva S. Mastrangelo M.A. Lotta L.T., Jr. Burris C.A. Federoff H.J. Bowers W.J. Extending the transposable payload limit of Sleeping Beauty (SB) using the herpes simplex virus (HSV)/SB amplicon-vector platform. Gene Ther. 2010;17:424–431. doi: 10.1038/gt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraefel C. Jacoby D.R. Lage C. Hilderbrand H. Chou J.Y. Alt F.W. Breakefield X.O. Majzoub J.A. Gene transfer into hepatocytes mediated by helper virus-free HSV/AAV hybrid vectors. Mol. Med. 1997;3:813–825. [PMC free article] [PubMed] [Google Scholar]
- Guazzi S. Strangio A. Franzi A.T. Bianchi M.E. HMGB1, an architectural chromatin protein and extracellular signalling factor, has a spatially and temporally restricted expression pattern in mouse brain. Gene Expr. Patterns. 2003;3:29–33. doi: 10.1016/s1567-133x(02)00093-5. [DOI] [PubMed] [Google Scholar]
- Hackett P.B. Ekker S.C. Largaespada D.A. McIvor R.S. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv. Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Ivics Z. Hackett P.B. Plasterk R.H. Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvak Z. Ivics Z. Plasterk R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- Loret S. Guay G. Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. Ivics Z. Izsvak Z. Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.A. Bucks M.A. O'Regan K.J. Courtney R.J. The HSV-1 tegument protein pUL46 associates with cellular membranes and viral capsids. Virology. 2008;376:279–289. doi: 10.1016/j.virol.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Oehmig A. Fraefel C. Breakefield X.O. Update on herpesvirus amplicon vectors. Mol. Ther. 2004;10:630–643. doi: 10.1016/j.ymthe.2004.06.641. [DOI] [PubMed] [Google Scholar]
- Peterson E.B. Mastrangelo M.A. Federoff H.J. Bowers W.J. Neuronal specificity of HSV/Sleeping Beauty amplicon transduction in utero is driven primarily by tropism and cell type composition. Mol. Ther. 2007;15:1848–1855. doi: 10.1038/sj.mt.6300267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P. Misteli T. Bianchi M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Spaete R.R. Frenkel N. The herpes simplex virus amplicon: A new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Suzuki M. Chiocca E.A. Saeki Y. Early STAT1 activation after systemic delivery of HSV amplicon vectors suppresses transcription of the vector-encoded transgene. Mol. Ther. 2007;15:2017–2026. doi: 10.1038/sj.mt.6300273. [DOI] [PubMed] [Google Scholar]
- Walisko O. Izsvák Z. Szabó K. Kaufman C.D. Herold S. Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S.R. Kay M.A. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol. Cell. Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S.M., Jr. Samulski R.J. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13525–13530. doi: 10.1073/pnas.241508998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed H. Izsvak Z. Khare D. Heinemann U. Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed H. Izsvak Z. Walisko O. Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]