Herpes simplex virus/Sleeping Beauty (HSV/SB) amplicon vectors are being examined as a means to efficiently and stably deliver transgenes to neurons within the mammalian brain. De Silva et al. assess the in vivo functionality of an HSV/SB amplicon vector compared to a hyperactive SB following in utero delivery in mice.
Abstract
The Sleeping Beauty (SB) transposon system has been successfully used as a gene delivery tool in nonviral and viral vector platforms. Since its initial reconstruction, a series of hyperactive mutants of SB have been generated. Questions remain as to whether the enhanced in vitro activities of these SB transposase mutants translate to the in vivo setting, and whether such increased integration efficiencies will ultimately compromise the safety profile of the transposon platform by raising the risk of genomic insertional mutagenesis. Herein, we compared the in vivo impact of a herpes simplex virus (HSV) amplicon-vectored “wild-type” SB transposase (SB10) and a “hyperactive” SB mutant (HSB5), codelivered in utero with the HSVT-βgeo transposable reporter amplicon vector to embryonic day 14.5 C57BL/6 mice. The SB10 and HSB5 transposases do not disparately affect the viability and development of injected mouse embryos. Quantitation of brain-resident βgeo expression on postnatal day 21 revealed that mice receiving HSB5 exhibited only a trending increase in transgene expression compared with the SB10-infused group, an outcome that did not mirror the marked enhancement of HSB5-mediated transposition observed in vitro. These findings indicate that in vivo application of hyperactive SB mutants, although not differentially genotoxic to the developing mouse embryo, does not necessarily provide a significant therapeutic advantage over the employment of a lesser active SB when delivered in the context of the HSV/SB amplicon platform.
Introduction
Herpes simplex virus (HSV)-1 amplicon vectors are capable of efficiently transducing cells comprising the central nervous system and, thus, have been extensively engineered for gene-based therapeutic modalities targeting neurological disorders. The utility of the HSV amplicon vector has been enhanced by the generation of integration-competent, hybrid versions that facilitate long-term expression of the delivered transgene (e.g., HSV/AAV, HSV/Sleeping Beauty). Our laboratory previously demonstrated that the integration-competent, bipartite HSV/Sleeping Beauty (SB) amplicon vector system specifically transduced neuronally restricted precursor cells within the fetal mouse brain on embryonic day 14.5 (E14.5) after intraventricular delivery of vector particles (Bowers et al., 2006; Peterson et al., 2007). After transduction, the SB transposase, which is expressed from an “effector” amplicon (termed HSV-SB), efficiently catalyzes excision of the inverted repeat/direct repeat (IR/DR)-flanked reporter transgene (T-βgeo) from the “integrator” amplicon (termed HSVT-βgeo) and stably integrates it at a TA dinucleotide site within the cellular genome. These series of events have been shown to result in long-term expression of the reporter transgene in various regions of the brain, specifically the cortex, hippocampus, and dentate gyrus, due to subsequent proliferation and migration of the stably transduced neuronal precursor cells. Accordingly, the HSV/SB amplicon vector system is being developed as a fetal gene delivery vector platform capable of targeting the prevalent neuronal precursor cell population for widespread dissemination of therapeutic genes to treat early-onset neurological disorders.
Although initial studies have provided proof-of-principle concerning the feasibility of using the HSV/SB amplicon vector for stable integration of transgenes in vivo, the impact of in utero manipulation and stable transduction of neuronal precursor cells on postnatal survival rates has not been systematically determined. The HSV/SB amplicon vector platform employed in our initial investigations was designed with the “wild-type” version of the SB transposase (termed SB10). Since then, a series of “hyperactive” mutants of SB has been generated by selectively changing multiple residues within the SB transposase open reading frame. These mutants exhibit enhanced transposition activity compared with SB10 in cell line-based transposition assays (Geurts et al., 2003; Yant et al., 2004; Baus et al., 2005). Consequently, these hyperactive mutants have been used in gene delivery applications to enhance the efficiency of transposition as a means to overcome the limitations associated with plasmid-based gene delivery in vivo (Yant et al., 2000; Liu et al., 2004, 2006; Aronovich et al., 2007). Questions remain as to whether the enhanced activity of these SB transposase mutants observed in vitro in standard immortalized cell lines translates to the in vivo setting in specific target cell types, and whether such increased integration efficiencies will compromise the safety profile of the transposon platform by unduly introducing a higher risk of genomic instability.
In this paper we employed the hyperactive HSB5 mutant transposase, which has been shown to display an ∼10-fold higher transposition efficiency than SB10 during plasmid-based transposition (Yant et al., 2007), as a means to optimize the current HSV/SB vector platform for more efficient stable gene transfer and long-term expression of therapeutic genes in vivo. We sought to determine whether the hyperactivity observed in vitro with the HSB5 transposase would be exhibited when codelivered in utero with the HSVT-βgeo transposable reporter amplicon to the developing brain of E14.5 mouse embryos. In addition, we sought to assess the safety of using a hyperactive SB transposase to mediate transposition during mouse brain development and whether the increased transposition levels observed in vitro would be detrimental to the survival of the mouse embryo. Our results indicate that the hyperactive HSB5 mutant transposase marginally enhanced transgene expression levels in vivo compared with SB10. Importantly, HSB5 transposase-mediated transposition in the E14.5 mouse brain did not unduly affect the viability and development of the embryo compared with the wild-type SB transposase, which may help to mitigate safety concerns associated with stable integration of therapeutic transgenes via HSV/SB amplicons in the developing brain.
Materials and Methods
Cell culture
The human embryonic kidney (HEK 293) cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) and 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA) at 37°C and 5% CO2.
Amplicon plasmid construction and helper virus-free packaging
To generate the HSB5 open reading frame (Yant et al., 2007), the “wild-type” SB10 transposase open reading frame was codon optimized and synthesized for high-level protein expression along with four mutations at positions K13A, K33A, T83A, and S270A (GENEART, Regensburg, Germany). The HSB5 open reading frame was amplified by PCR from the pGA4-HSB5 plasmid, using primers containing a 5′ KpnI site in the forward primer and a 5′ SacI site in the reverse primer. The primer sequences were as follows: HSB5.Fwd, 5′-GCCGCCGGTACCATGGGCAAGAGCAAAGAGATCAGCCAGGAC-3′; HSB5.Rev, 5′-GCGCCGAGCTCTCAGTACTTGGTGGCGTTGCCCTTGAAC-3′.
The PCR product was ethanol precipitated, digested sequentially with KpnI and SacI, and cloned into the pHSVPrPUC amplicon plasmid using the corresponding sites to create the final pHSV-HSB5 vector (Fig. 1A). The construction of pHSV-SB10 and pHSVT-βgeo has been previously described (Bowers et al., 2006). The HSV-SB10, HSV-HSB5, HSVPrPUC, and HSVT-βgeo amplicons were packaged using a helper virus-free packaging system, which has been described previously (Bowers et al., 2001).
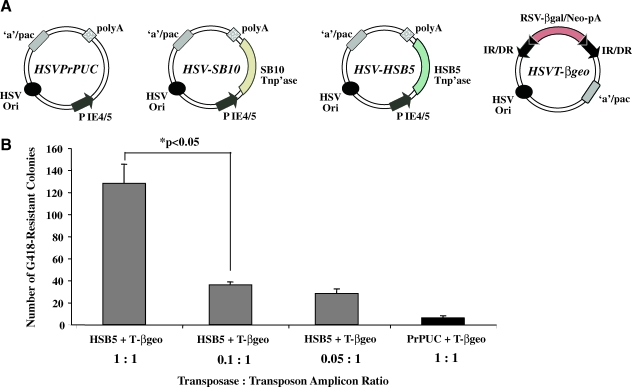
FIG. 1.
Determination of the optimal ratio of HSV-HSB5 to HSVT-βgeo amplicon to achieve maximal SB-mediated transposition in vitro. (A) Schematic representation of the amplicons used in the in vivo testing of the HSV/SB amplicon vector platform. The HSVPrPUC amplicon contains a multiple cloning site, and serves as an empty vector control. The HSV-SB10 amplicon expresses the “wild-type” SB transposase, whereas the HSV-HSB5 amplicon expresses the “hyperactive” HSB5 transposase. HSVT-βgeo serves as a substrate vector for the transposase, and contains an IR/DR-flanked Rous sarcoma virus (RSV) promoter-driven β-galactosidase–neomycin phosphotransferase fusion reporter gene unit (β-geo; adapted from Bowers et al., 2006). (B) Assessment of the optimal transposase-to-transposon (HSV-HSB5:HSVT-βgeo) amplicon ratio required to achieve maximal transposition efficiency. HEK 293 cell monolayers were individually cotransduced with various ratios of HSV-HSB5 and HSVT-βgeo viral particles at an MOI of 0.5. HSVPrPUC was used as a “balance” virus to ensure that equal numbers of viral particles were transduced under each condition. Two days posttransduction, the cells were seeded onto 100-mm dishes at a 1:3 dilution and placed under G418 selection for 2 weeks. G418-resistant colonies were enumerated by X-Gal histochemistry. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test. Color images available online at www.liebertonline.com/hum.
Cell line-based transposition assays
Transposition efficiency of HSB5 transposase at various transposase-to-transposon amplicon ratios
HEK 293 cells (4 × 105 cells per well in a 24-well dish) were individually cotransduced with increasing amounts of HSV-HSB5 viral particles together with a fixed amount of HSVT-βgeo amplicon to achieve various transposase-to-transposon amplicon ratios (0.01:1, 0.05:1, 0.1:1, and 1:1) while maintaining an overall multiplicity of infection (MOI) of 1. The HSVPrPUC amplicon served as a “balance” virus to maintain an MOI of 1 under each condition. On incubation for 1 hr at 37°C, the virus-containing medium was removed and cells were washed once and replenished with fresh medium. At 48 hr posttransduction, cells were trypsinized and seeded at a 1:3 dilution in 10-cm dishes containing medium supplemented with G418 antibiotic (600 μg/ml; Invitrogen). Cells were refed every 3–4 days with fresh medium supplemented with G418 antibiotic. After a 2-week selection period, the resulting G418-resistant colonies were fixed with 1% glutaraldehyde and enumerated after 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining (Fig. 1B).
Transposition efficiency of the hyperactive HSB5 transposase
HEK 293 cells (4 × 105 cells per well in a 24-well dish) were individually cotransduced for 1 hr at 37°C with a 1:1 viral particle ratio of either HSV-SB10 or HSV-HSB5 together with the HSVT-βgeo amplicon at an MOI of 1. The HSVPrPUC amplicon served as an empty vector control. A colony formation assay was performed as described previously (Fig. 2).
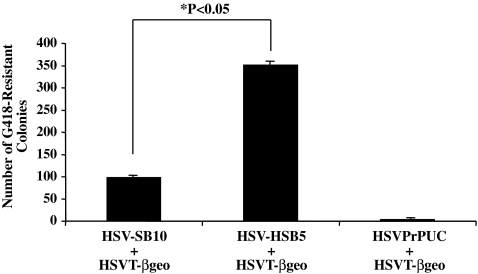
FIG. 2.
Comparison of the transposition efficiency of HSB5 transposase and wild-type SB10 transposase in human embryonic kidney 293 (HEK 293) cells. HEK 293 cells were cotransduced with HSV-SB10, HSV-HSB5, or HSVPrPUC amplicon together with the HSVT-βgeo reporter amplicon at a 1:1 amplicon ratio (MOI of 1). Two days later the cells were trypsinized and seeded at a 1:3 dilution onto dishes containing medium supplemented with G418 antibiotic (600 μg/ml). Two weeks later G418-resistant colonies were fixed, stained with X-Gal, and enumerated. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test.
In utero amplicon transduction
Under the surgical plane of anesthesia (Avertin, 0.5 mg/ml), the maternal abdomen of postcoitum 14.5 C57BL/6 mice was shaved and cleansed with povidone-iodine solution USP (Aplicare, Branford, CT). A laparotomy was performed and the uterus was gently exteriorized onto a sterile disposable drape. Two microliters of HSV-SB10, HSV-HSB5, or the control amplicon HSVPrPUC was mixed at a viral particle ratio of 1:1 (2 μl of total volume of virus, 2 × 104 total transducing units) with HSVT-βgeo and delivered via a glass-pulled pipet to each embryo intracranially, using an IM300 programmable microinjector (Narishige International, East Meadow, NY). Once born, the pups were allowed to develop to 21 days of age, killed, and brains were processed for immunohistochemical and integration site analyses.
Immunohistochemistry
β-Galactosidase-specific staining
The left hemisphere of each brain cotransduced with the HSV amplicon combinations (HSV-SB10 plus HSVT-βgeo, HSV-HSB5 plus HSVT-βgeo, and HSVPrPUC plus HSVT-βgeo) was analyzed 21 days postpartum by immunocytochemistry. Processing of tissue and mounting on glass slides were performed as described previously (Bowers et al., 2006). Double immunohistochemistry was performed on coronal mouse brain sections (30 μm) using anti-β-galactosidase, rabbit IgG fraction (diluted 1:2000; Biodesign, Saco, ME), with anti-NeuN mouse monoclonal antibody (diluted 1:200; Millipore, Billerica, MA) or anti-GFAP–Cy3 conjugate monoclonal antibody clone G-A-5 (diluted 1:2000; Sigma-Aldrich, St. Louis, MO). An Alexa 488 anti-rabbit IgG (H+L, diluted 1:200; Invitrogen) fluorescent secondary antibody was subsequently used. Immunopositive cells were visualized with a FV1000 Olympus laser scanning confocal microscope (Olympus, Center Valley, PA) at a magnification of ×40. Quantification of the average pixel density of β-galactosidase-specific cell staining in the visual motor cortex region (VM) was performed by diaminobenzidine (DAB) staining as described previously (Bowers et al., 2006). β-Galactosidase-specific cell staining in the VM region was determined with an Olympus AX-70 microscope equipped with a motorized stage and MCID 6.0 Elite software (Interfocus imaging subsidiary of GE Healthcare, Cambridge, UK). An average of eight equivalent sections of the VM region per mouse were analyzed at a magnification of × 20.
Nissl staining
Brain sections were mounted on Superfrost Plus slides (VWR International, West Chester, PA) and allowed to dry completely. Thereafter, the slides were hydrated in distilled H2O for 5 min, and stained with 0.02% cresyl violet acetate in 0.25% acetic acid for 30 min. Destaining was conducted by washing the slides in three changes of distilled H2O and placed in 50% ethanol and 70% ethanol for 1 min each, consecutively. The slides were allowed to dry and dipped in xylene before being coverslipped.
Microglial/macrophage staining
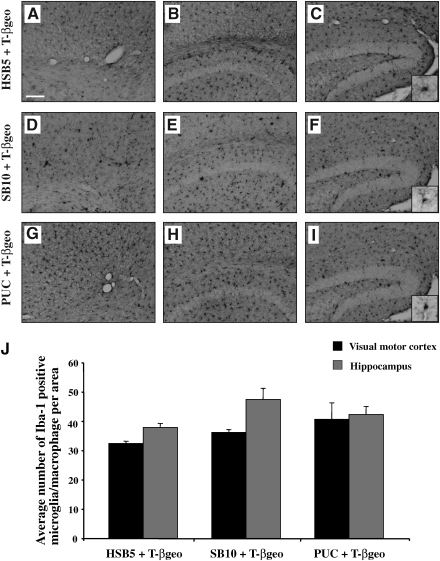
Immunohistochemical staining of brain-resident microglia/macrophages was performed with an anti-Iba-1 rabbit polyclonal antibody (1:750 dilution; Wako, Richmond, VA) in combination with a biotin-conjugated secondary antibody (1:1000 dilution; Vector Laboratories, Burlingame, CA). DAB staining was conducted according to the manufacturer's recommendations, using an avidin–biotin complex and nickel-enhanced diaminobenzidine (VECTASTAIN ABC system; Vector Laboratories). Quantification of the number of Iba-1-positive microglial/macrophage cells was conducted with an Olympus AX-70 microscope equipped with a motorized stage and MCID 6.0 Elite imaging software (Interfocus imaging subsidiary of GE Healthcare). Six equivalent sections of the VM cortex and hippocampus region were separately counted and averaged per mouse at a magnification of ×20 (n = 3 mice per group).
Statistical analysis
Statistical analysis was performed by Student t test and one-way analysis of variance.
Integration site mapping and analysis
Transposon–genomic DNA junctions were determined by linear amplification-mediated PCR (LAM-PCR) as described earlier (Mátés et al., 2009), with the following modifications. Genomic DNA (500 ng) was subjected to 100 cycles of linear amplification PCR in a final volume of 50 μl with a 5′-biotin-labeled, transposon-specific primer, under the following conditions: 94°C for 1 min, 59°C for 30 sec, and 72°C for 1 min 30 sec, and the products were captured on magnetic streptavidin beads (Dynabeads kilobase binder kit; Invitrogen). After second-strand synthesis, the DNA samples were digested with MboI (New England BioLabs, Ipswich, MA) to create compatible ends for linker ligation. Ligation reactions were performed with 50 pmol of the corresponding linkers overnight at room temperature. The ligation products were collected on a magnetic separator and resuspended in 10 μl of Tris-EDTA (TE) buffer. Two microliters of ligation product was amplified in the primary PCR in a 50-μl total volume, using a nested transposon-specific primer with the MboI linker-specific primer, under the following conditions: 94°C for 3 min; 35 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 1 min; followed by 72°C for 5 min. One microliter of the PCR product was further amplified in a nested round of PCR using a nested transposon-specific primer in combination with an MboI linker-specific nested primer. Conditions for the nested PCRs were as follows: 94°C for 3 min; 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; followed by 72°C for 5 min. PCR products were purified with a PCR purification kit (Qiagen, Hilden, Germany), and characterized by electrophoresis on a 1.25% agarose gel. The purified PCR product was cloned into the pGEM-T Easy vector (Promega, Madison, WI), and the DNA sequence was determined by standard sequencing technology (ABI 3730xl; Applied Biosystems, Foster City, CA). The identity and location of mouse chromosomal sequences that flank the integrated T-βgeo transposon were mapped by mouse BLAT genomic alignment (July 2007, NCBI37/mm9) on the University of California, Santa Cruz (UCSC) genome browser (available at http://genome.ucsc.edu).
Results
Cell line-based assessment of HSB5 transposase hyperactivity in the context of an HSV/SB amplicon vector
Previous plasmid-centric studies have established that, because of the stoichiometric nature of the transposition reaction, an optimal transposase-to-transposon plasmid ratio must be used to achieve maximal transposition, and to obviate the effect of “overproduction inhibition” (Zayed et al., 2004). The latter effect is common to a number of transposons representing different families, and is a result of high transposase levels relative to the transposon within the host cell that leads to dampening of the transposition (Hartl et al., 1997). We initially sought to test various transposase-to-transposon amplicon ratios to determine the optimal ratio that would facilitate efficient HSB5-mediated transposition from the HSV amplicon vector. A colony formation assay was performed by cotransducing HEK 293 cells with a fixed number of HSVT-βgeo amplicon particles and various amounts of the “hyperactive” HSB5 transposase expressing HSV amplicon. The HSVPrPUC amplicon served as an empty vector control and also as a “balance” virus to ensure that equal numbers of viral particles were being transduced under each condition. Schematic diagrams of the empty vector control (HSVPrPUC), effectors (HSV-SB10 and HSV-HSB5), and integrator (HSVT-βgeo) amplicons used in this study are presented in Fig. 1A. Cotransduction of HSV-HSB5 and HSVT-βgeo at a 1:1 amplicon ratio proved to be optimal, giving rise to the highest number of G418-resistant colonies (Fig. 1B).
Next, we compared the transposition efficiency of the hyperactive HSB5 transposase to that catalyzed by the wild-type SB10 transposase, using the optimized 1:1 transposase-to-transposon amplicon ratio. Cotransduction of HSV-SB10 plus HSVT-βgeo resulted in an ∼25-fold increase in the number of G418-resistant HEK 293 colonies compared with the HSVPrPUC plus HSVT-βgeo negative control group (Fig. 2). The cotransduction of HSV-HSB5 together with HSVT-βgeo resulted in a further increase in the number of G418-resistant colonies, which corresponded to a 3.5-fold increase in transposition efficiency compared with HSV-SB10. This result confirmed that the HSB5 transposase was significantly more active than SB10 in the context of an HSV/SB amplicon vector, but this gap in transposition activity is lower than the ∼10-fold difference observed between HSB5 and SB10 during plasmid-based transposition (Yant et al., 2007).
Comparison of mouse survival rates, brain architecture, and brain-resident inflammation after in utero vector infusion
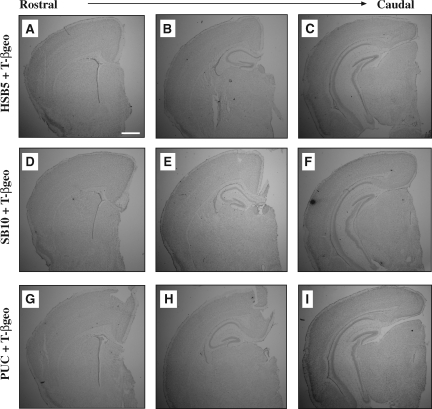
To determine whether the increased transposition activity of the HSB5 transposase observed in vitro would be preserved in an in vivo experimental setup, a total of ∼2–4 × 104 HSV amplicon vector particles containing an optimized 1:1 amplicon ratio of HSV-SB10 plus HSVT-βgeo, HSV-HSB5 plus HSVT-βgeo, or HSVPrPUC plus HSVT-βgeo was infused intraventricularly into E14.5 C57BL/6 embryos (Fig. 3). After surgery, the transduced embryos were allowed to reach full term and the numbers of live births were recorded to ascertain the survival rate of the mice on postnatal day 0 (P0) and P21. Mice injected with the HSV-HSB5 plus HSVT-βgeo amplicon combination exhibited a slightly higher survival rate (65.9%) than the wild-type HSV-SB10 plus HSVT-βgeo group (47%) and the HSVPrPUC plus HSVT-βgeo group (45%) on P0, although these differences did not reach statistical significance (Table 1). In addition, the numbers of live births from each cohort reaching P21 were not significantly different (Table 1). Mice surviving to P21 were subsequently killed and perfused, and brains were processed for histochemical analyses. Nissl staining was performed to assess overall brain architecture. Rostral-to-caudal assessment of 30-μm coronal brain sections did not reveal any gross abnormalities at the anatomical level (Fig. 4), thus indicating that brain development was not significantly affected after in utero HSV/SB vector transduction. In aggregate, these data indicate that the use of a hyperactive form of the SB transposase within the HSV/SB amplicon vector platform context does not deleteriously affect the viability or brain development of the embryo as compared with the wild-type SB10 transposase. Furthermore, because the empty vector control group (HSVPrPUC plus HSVT-βgeo) exhibited a survival rate similar to that of the HSV/SB amplicon vector groups on P0 and P21, it suggests that the observed impact on mouse survival is not due to SB-mediated transposition, but is most likely due to complications arising from in utero surgical manipulation and/or the exposure of the embryo to helper virus-free HSV amplicons.
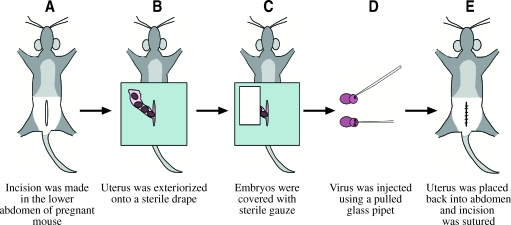
FIG. 3.
Schematic representation of the in utero surgical paradigm used for the delivery of HSV/SB amplicon vector platform to the developing mouse brain. Under the surgical plane of anesthesia (Avertin, 0.5 mg/ml), the maternal abdomen of postcoitum 14.5 C57BL/6 mice was shaved and an incision was made to exteriorize the uterus onto a sterile disposable drape. Sterile gauze was placed over the exposed uterus and hydrated with lactated Ringer solution. The appropriate virus combination was mixed at a ratio of 1:1 (total volume, 2 μl; total transducing units, 2 × 104) and delivered to each embryo intraventricularly, using a microinjector. The uterus was placed back into the abdomen and the incision was sutured by interrupted stitching. The virus-infused embryos were allowed to reach full term and left under maternal care for 21 days postpartum, at which time they were killed and the brains were processed for immunohistochemistry (left hemisphere) and integration site analysis (right hemisphere). Color images available online at www.liebertonline.com/hum.
Table 1.
Comparison of Survival Rates for C57BL/6 E14.5 Embryos Coinjected In Utero with HSV-HSB5, HSV-SB10, or HSVPrPUC Along with HSVT-βgeo Reporter Amplicon
| Group | Amplicon combination | No. of E14.5 embryos injected | No. of live births (P0) | Postsurgery survival rate at P0 (%) | No. of P0 Pups that lived to P21 | Postbirth survival rate (P21) (%) |
|---|---|---|---|---|---|---|
| 1 | HSV-HSB5 + HSVT-βgeo | 44 | 29 | 65.9 | 20 | 68.9 |
| 2 | HSV-SB10 + HSVT-βgeo | 59 | 28 | 47 | 16 | 57 |
| 3 | HSVPrPUC + HSVT-βgeo | 31 | 14 | 45 | 9 | 64.3 |
E14.5, embryonic day 14.5.
FIG. 4.
Representative Nissl-stained brain sections from P21 mice after in utero, intraventricular vector transduction. At 21 days of age, vector-transduced animals were killed and perfused with 4% paraformaldehyde, and brain sections (30 μm) were processed for Nissl staining to assess overall brain architecture, using an inverted microscope. A rostral-to-caudal display of representative P21 mouse brain sections corresponding to each of the treatment groups [HSV-HSB5 plus HSVT-βgeo (A–C), HSV-SB10 plus HSVT-βgeo (D–F), and HSVPrPUC plus HSVT-βgeo (G–I)] is presented. Images were captured at about 0.6 mm bregma (A, D, and G), about –1.3 mm bregma (B, E, and H), and about –3.1 bregma (C, F, and I) coordinates. Photomicrographs were obtained at an original magnification of × 1.25. Scale bar (A): 2 mm.
We subsequently assessed whether in utero delivery of the HSV/SB vector system and transposition within the fetal mouse brain would trigger differential inflammatory responses. We postulated that such inflammatory responses would be revealed by increases in microglia/macrophage activation marker staining patterns in the injected mouse brain on P21. Microglia are brain-resident macrophages that are activated and recruited to sites of inflammation within the brain as a primary participant in innate responses (Gehrmann et al., 1995). Brain sections were subjected to Iba-1-specific immunostaining to determine the overall presence of microglia/macrophages. Equivalent levels of Iba-1-specific microglial/macrophage staining were observed throughout the brain in all three groups of mice injected with HSV-SB10, HSV-HSB5, or HSVPrPUC plus the HSVT-βgeo reporter amplicon (Fig. 5). Quantitative analysis of the pixel density of Iba-1-specific microglia/macrophage staining in the visual motor (VM) cortex and hippocampal regions of the mice revealed comparable levels across the three groups of mice on P21 (n = 3 mice per group; Fig. 5J), thus confirming that SB-mediated transposition and resultant βgeo transgene expression did not lead to differential effects on microglial activation amongst the treatment groups.
FIG. 5.
Microglial/macrophage staining pattern in mouse brain on P21 after in utero HSV/SB vector transduction. Immunostaining of brain-resident microglia/macrophage was performed according to an Iba-1-specific diaminobenzidine (DAB) protocol. A rostral-to-caudal display of representative P21 mouse brain sections corresponding to each of the treatment groups [HSV-HSB5 plus HSVT-βgeo (A–C), HSV-SB10 plus HSVT-βgeo (D–F), and HSVPrPUC plus HSVT-βgeo (G–I)] is presented. Photomicrographs (A), (D), and (G) correspond to the M1 region of the motor cortex; (B), (E), and (H) correspond to the CA1 region of the hippocampus (about –1.58 mm bregma); and (C), (F), and (I) correspond to the dentate gyrus region (about –2.46 mm bregma). Photomicrographs were obtained at an original magnification of × 10. Scale bar (A): 500 μm. Each inset [in (C), (F), and (I)] represents a digitally magnified ( × 20) image of the photomicrograph for better visualization of stained cell morphology. The number of Iba-1-positive microglia was counted within the visual motor (VM) cortex and hippocampal region of the brain, using an Olympus AX-70 microscope equipped with a motorized stage and MCID 6.0 Elite software (J). Six equivalent sections of the VM cortex and hippocampus region were separately counted and averaged per mouse at a magnification of × 20 (n = 3 mice per group). Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test.
Both HSV-HSB5 and HSV-SB10 lead to neuronally restricted transgene expression after intraventricular coinfusion with HSVT-βgeo into fetal mouse brain
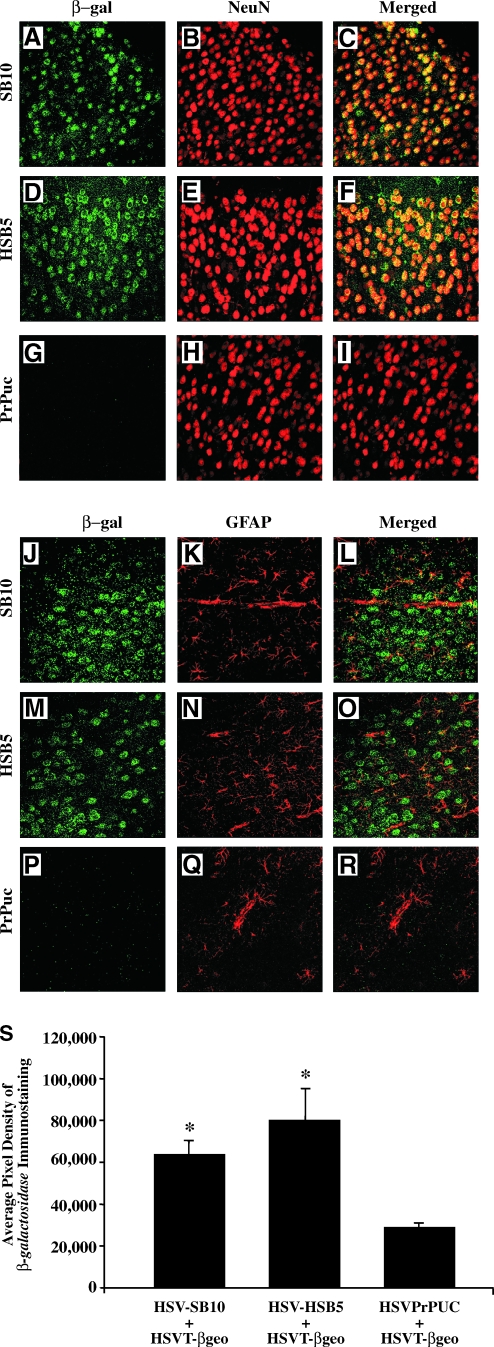
Previous studies using HSV-SB10 had established that the presence of β-galactosidase (β-Gal) transgene expression in the mouse brain on P21 was a result of SB-mediated integration of the T-βgeo reporter transgene in the mouse genome, because β-Gal expression from the episomally retained HSVT-βgeo amplicon was undetectable in the absence of SB transposase (Bowers et al., 2006). Furthermore, β-Gal expression on P21 was restricted to NeuN-positive mature neurons, which was subsequently shown to be a result of the HSV amplicon-specific transduction of neuronal precursor cells present in the subventricular zone of the developing fetal brain at E14.5 (Peterson et al., 2007). To determine whether use of the hyperactive HSB5 mutant altered the cell type specificity of transgene expression, we performed immunohistochemistry on brain sections of killed P21 mice. As depicted by the representative photomicrographs in Fig. 6, expression of β-Gal (green) was readily observed in cortical neurons that stained positive for the neuron-specific marker NeuN (red) in mice infused with HSV-SB10 plus HSVT-βgeo (Fig. 6A–C) or HSV-HSB5 plus HSVT-βgeo (Fig. 6D–E). Transgene-positive neurons were also detected in the hippocampus and dentate gyrus of these cohorts (data not shown). As expected, β-Gal expression was not detected in mice injected with HSVPrPUC plus HSVT-βgeo (Fig. 6G–I). Immunohistochemical costaining for a glia-specific marker, glial fibrillary acidic protein (GFAP), and β-Gal revealed no apparent colocalization of β-Gal expression (green) with GFAP-positive astrocytes (red; Fig. 6J–R). Overall, these results are consistent with previous findings, and indicate that the cell specificity of transgene expression on P21 is not influenced by the in vitro-ascribed hyperactivity of HSB5.
FIG. 6.
Immunohistochemical analysis and quantification of the cell type-specific expression of the T-βgeo transgenon (i.e., a transposable transgene) on P21 after in utero delivery. At 21 days of age, transduced animals were killed and perfused with 4% paraformaldehyde, brain sections were processed for dual β-galactosidase (β-Gal)/NeuN immunohistochemistry, and sections were imaged by confocal microscopy. Representative brain sections corresponding to the cortex are depicted. β-Gal-specific staining resulting from T-βgeo transgenon-mediated expression appears in the green channel (A, D, and G), NeuN-positive mature neurons in the red channel (B, E, and H), and colocalized staining (Merged) appears as yellow (C, F, and I). Similarly, brain sections were processed for dual β-Gal/GFAP immunohistochemistry (J–R). The GFAP-positive astrocytes appear in the red channel. Photomicrographs were obtained at an original magnification of ×40. Scale bar (A): 200 μm. β-Gal-specific diaminobenzidine (DAB) immunostaining was conducted on coronal brain sections of the left hemisphere to quantify the average pixel density of β-galactosidase-positive neurons in the visual motor cortex (VM) region of mice that were coinjected with HSV-SB10, HSV-HSB5, or HSVPrPUC along with the HSVT-βgeo amplicon (S). β-Gal-positive cells in the VM region were enumerated with an Olympus AX-70 microscope equipped with a motorized stage and MCID 6.0 Elite software. An average of eight equivalent sections of the VM region per mouse were analyzed at a magnification of × 20. Error bars represent the standard error of the mean and statistical analysis was conducted by Student t test. *Statistically significant difference between the two HSV-SB amplicon (SB10 and HSB5) plus HSVT-βgeo-injected groups and the HSVPrPUC plus HSVT-βgeo control group (p < 0.05).
In utero codelivery of HSV-HSB5 or HSV-SB10 with HSVT-βgeo results in comparable levels of transgene expression on P21
We subsequently quantified the level of β-Gal expression in the three groups of mice to assess whether the hyperactive HSB5 transposase was more efficient in generating transgene-positive neurons compared with SB10. The level of β-Gal expression was quantified by determining the average pixel density of β-Gal-specific diaminobenzidine (DAB) immunostaining present in the VM cortex region of each brain. The VM cortex was selected, because it consistently revealed the highest level of β-Gal staining in the brain on P21. Mice intraventricularly infused with either HSV-SB10 plus HSVT-βgeo or HSV-HSB5 plus HSVT-βgeo exhibited significantly higher average pixel density of β-Gal/DAB staining than the HSVPrPUC plus HSVT-βgeo control group (Fig. 6S). Mice that were infused with HSV-HSB5 plus HSVT-βgeo did show a trending increase in the level of β-Gal/DAB immunostaining as compared with the HSV-SB10 plus HSVT-βgeo group, although these differences did not reach statistical significance. Our in vivo findings are in contrast to what was shown in cell line-based tests (Fig. 2), indicating that the hyperactive HSB5 transposase mutant does not confer a marked advantage in the context of the HSV/SB amplicon vector platform when delivered in utero.
Identification of chromosomal integration sites of the T-βgeo transposable transgene in intraventricularly infused mouse brain
To confirm bona fide SB-mediated transposition in the mouse brain, linear amplification-mediated PCR (LAM-PCR) analysis was used to map the flanking mouse chromosomal sequences at T-βgeo integration sites in a subset of mice infused with HSV-SB10 plus HSVT-βgeo or with HSV-HSB5 plus HSVT-βgeo. Bona fide SB-mediated transposition is characterized by integration of the transposon into chromosomal TA dinucleotide sites, which are duplicated after integration (Ivics et al., 1997). Transposon–chromosome junction sequences containing the molecular signature of SB-mediated transposition were identified (Table 2). The flanking mouse chromosomal sequences were subjected to BLAT genomic alignment against the UCSC mouse genome database to map the chromosomal locations of the integrated T-βgeo transposons. As indicated in Table 2, we were able to recover two or three insertion sites per mouse in both treatment groups. In aggregate, these results indicate that intraventricular delivery of both the wild-type and hyperactive versions of the HSV/SB amplicon vector platforms at E14.5 leads to bona fide SB-mediated transposition within the mouse brain genome.
Table 2.
Chromosomal Integration Sites of T-βgeo Transposon in Mouse Brain Genome After Intraventricular Infusion with HSV-HSB5 or HSV-SB 10 Together with HSVT-βgeo Amplicon
| Mouse no. | Amplicon combination | Flanking chromosomal sequences | Mouse chromosome location |
|---|---|---|---|
| SB10_15C | HSV-SB10 + HSVT-βgeo | 1)…cttcaaCTGTA-TTTGCCAGTGACTTGGA | Chr2: 169384664-169384689 |
| 2)…cttcaaCTGTA-AACGCTTAAGAGTACCC | Chr1: 123400272-123400310 | ||
| SB10_15E | HSV-SB10 + HSVT-βgeo | 1)…cttcaaCTGTA-AGTTCAAGTATGAACAG | Chr4: 148872819-148873108 |
| 2)…cttcaaCTGTA-AATGCCAGTGACTTCGG | Chr10: 92289486-92289531 | ||
| 3)…cttcaaCTGTA-TGTGTATGTGGGGTGTG | Unknown | ||
| HSB5_A2 | HSV-HSB5 + HSVT-βgeo | 1)…cttcaaCTGTA-TATATACATATATATAT | Chr17: 91176203-91176234 |
| 2)…cttcaaCTGTA-TCTAGCCTTCACTGCAA | Unknown | ||
| 3)…cttcaaCTGTA-AGTGCCCGTGGTTTCAT | ChrX: 164815384-164815403 | ||
| HSB5_C | HSV-HSB5 + HSVT-βgeo | 1)…cttcaaCTGTA-TTCCCTTATGCCCAGCA | Unknown |
| 2)…cttcaaCTGTA-TTCTACAGTGTCCTGGA | Chr11: 9876902-9876922 |
Discussion
In this study we assessed the in vivo impact of using a “hyperactive” mutant of SB transposase (HSB5) to optimize the HSV/SB vector for efficient transgene integration in the developing mouse brain. Interestingly, we found that the extent of hyperactivity exhibited by the HSB5 transposase in vitro is not maintained in vivo when used in the context of the HSV/SB amplicon vector, and appeared to be marginally advantageous over the use of the “wild-type” SB transposase. Further in vivo evaluation confirmed that HSB5-mediated transposition within neuronal precursor cells of the embryonic brain did not unduly affect the viability of the embryo and allowed for normal brain development without eliciting an adverse immune response; outcomes that help to mitigate the safety concerns of in utero gene therapy.
Hyperactive mutants of SB transposase were initially developed with the intention of increasing the in vivo transposition efficiency of SB-based nonviral vector platforms, because in vivo testing of the first-generation SB vector system did not exhibit therapeutically beneficial levels of transgene integration (Yant et al., 2000; Baus et al., 2005). Although the enhanced transposition efficiency of these mutants has been validated in cell culture, their efficiency in vivo has not been fully tested. HSB5 transposase is one such mutant that has been shown to enhance plasmid-based transposition by ∼10-fold compared with the first-generation wild-type SB10 transposase in cell line-based testing (Yant et al., 2007). However, the testing of HSB5 in the context of the HSV/SB vector resulted in an ∼3.5-fold increase in transposition efficiency relative to SB10 in HEK 293 cells (Fig. 2). We speculate that the disparity between our result and that of Yant and colleagues might be due to the following: (1) intrinsic differences in plasmid-based transposition versus amplicon-based transposition (de Silva et al., 2010); (2) the use of different reporter transposable units, with T-βgeo approximately three times the sequence length of the T-neo transposon used by Yant and colleagues; and (3) inherent differences between the two human cell lines used to assess transposition efficiency.
The mutations resident in the HSB5 transposase have been postulated to stabilize the synaptic complex during transposition, thereby increasing the efficiency of the reaction (Yant et al., 2007). Thus, the failure to observe an increase in β-Gal-specific DAB immunostaining in the presence of the HSB5 transposase (Fig. 6S) in vivo may be due to the following: (1) the in vitro-optimized ratio of HSV-HSB5 to HSVT-βgeo amplicon used in our study may be suboptimal in neuronal precursor cells in vivo, because Yant and colleagues and Liu and colleagues had previously shown that different target tissue types (e.g., mouse liver and lung) required diverse transposon-to-transposase plasmid ratios for efficient transposition (Yant et al., 2000; Liu et al., 2004); (2) the neuronal precursor cell environment in the fetal mouse brain may lack an unknown cofactor normally present in immortalized cell lines that facilitates enhanced transposition by HSB5; and (3) the absence of selective pressure in vivo may differentially affect SB-mediated transposition via hyperactive mutants compared with a cell culture setting in which antibiotic selection is used to select for cells that have undergone transposition.
During its initial characterization, SB was shown to catalyze transposition in mouse embryonic stem cells (Luo et al., 1998), which provided the opportunity to match the transgene-integrating power of SB with the multipotent, proliferative nature of stem cells for widespread transgene delivery. Accordingly, the HSV/SB amplicon vector platform has proven to be capable of mediating transposition in neuronal precursor cells in vivo as evidenced by the widespread expression of β-Gal observed in the brains of mice on P21. The identification of transposon integration sites within the mouse genome revealed precise integration of the entire T-βgeo transposon into TA dinucleotide sites mediated by the wild-type SB transposase as well as the hyperactive HSB5 transposase, which is a testament to the efficiency of this integrating system (Table 2). It should be noted that we were able to recover only two or three integration sites from each mouse because of technical difficulties associated with mapping integration sites in the mouse brain genome. However, we speculate that more integration sequences, below the limit of detection, may be present in these mouse genomes. On the basis of previous studies by our laboratory it is possible that clonal expansion of the stably transduced neuronal precursors occurred during mouse brain development, which would have increased the probability of recovering integration sites from those specific clonal populations by LAM-PCR. These limited numbers of flanking mouse chromosomal sequences revealed that integration had occurred in nontranscriptionally active regions (data not shown). Even though many more integration sites will need to be mapped to characterize SB transposition in the context of the HSV/SB system, SB transposition out of plasmids clearly established a fully random insertional profile at the genome level (Vigdal et al., 2002; Yant et al., 2005; Mates et al., 2009), suggesting that SB might be safer than lentiviral and retroviral vectors, which have been shown to integrate into transcriptionally active genomic sites.
Another aim of this study was to assess the in vivo impact of using a hyperactive SB transposase in the context of the HSV/SB amplicon vector platform delivered in utero on E14.5. Our findings confirm that SB-mediated transposition did not unduly affect the viability of the mouse embryo (Table 1), and appeared to be innocuous during subsequent mouse brain development (Fig. 4). As seen in Table 1, the invasiveness of the surgical procedure itself resulted in fatalities in all three groups of mice on P0 injected with the HSVT-βgeo amplicon in the presence or absence of SB transposase. We observed that the survival rate of the transduced embryos on P0 was also dependent on the number of embryos present in the pregnant female mouse. Pregnant female mice bearing 6 or 7 embryos exhibited a higher birth rate after in utero surgery compared with those bearing 9 or 10 embryos (data not shown). Unlike stereotactic surgery, by which the viral vector can be delivered precisely into the adult brain on the basis of bregma coordinates, unguided delivery of the HSV amplicon virus into the ventricular space of the developing embryo on E14.5 is associated with a certain degree of inaccuracy, which could also have contributed to a subset of the observed fatalities. Thus, as described by Punzo and colleagues, ultrasound-guided in utero gene transfer can be adopted to minimize the fatalities that occur during surgery (Punzo and Cepko, 2008). Besides the survival rate on P0 after virus transduction, we were also interested in the survival rate of these mice on P21 to assess whether SB-mediated transposition during fetal development would cause any detrimental effects that manifest themselves after birth. On the basis of our results, we did not observe an increased fatality rate when the HSV/SB platform was transduced compared with the empty vector control group (HSVPrPUC plus HSVT-βgeo), which suggested that SB-mediated transposition was not a significant contributing factor in the deaths that occurred after birth. The fatalities that did occur after birth (i.e., within 24–48 hr) in all three groups of mice appeared to be due mainly to rejection and neglect of the pups by the dam. It remains unknown whether these deaths were due to developmental defects caused by the introduction of the virus during embryo development. Although the integration profile of SB has been shown to have no significant bias toward transcriptionally active regions within the human genome (Yant et al., 2005), the possibility still exists for insertional mutagenesis and the development of malignancies. The current study did not assess the potential for the development of brain tumors, but it should be noted that we have not observed the presence of any brain tumors in mice even after 90 days after stable transduction of neuronal precursor cells by the first-generation HSV/SB vectors (W.J. Bowers and H.J. Federoff, unpublished observation). Careful characterization of newly generated hyperactive mutants of SB transposase is warranted in an in vivo setting to ensure that the integration profile has not been skewed toward transcriptionally active sites. Current efforts have also focused on using hyperactive mutants of the SB transposase to generate site-specific chimeric versions to target “safe” chromosomal sites within the human genome for transgene integration (Yant et al., 2007).
Mátés and colleagues developed the SB100X hyperactive mutant transposase, which was shown to be 100-fold more active than the wild-type SB10 transposase in HeLa cells under conditions in which the amount of transposon template was a limiting factor (Mátés et al., 2009). SB100X was also shown to support 35–50% stable gene transfer in primary human CD34+ cells enriched in hematopoietic stem or progenitor cells that had previously proven difficult to transfect, and thus, unsupportive of transposition using the first-generation SB transposon system. In addition, it was demonstrated that the hyperactive SB100X transposase did not create any detectable genomic instability in the stably transfected CD34+ cells, which allowed them to differentiate into distinct lymphohematopoietic lineages similar to the nontransfected controls. Of particular interest to the present study, SB100X promoted far more robust, long-lasting transgene expression than early-generation hyperactive transposases in vivo after delivery into the mouse liver by hydrodynamic injection (Mátés et al., 2009). These results have highlighted the possibility of using SB-mediated, nonviral approaches to genetically modify hematopoietic stem cells ex vivo and hepatocytes in vivo for the treatment of human diseases. Thus, employing the appropriate hyperactive SB transposase in specific gene transfer paradigms may provide a means to enhance transposition efficiency, which in turn could improve a given therapeutic outcome. The efficiency of the SB100X hyperactive system in the context of the HSV amplicon delivery paradigm remains to be assessed.
In 2008, after a decade of extensive research, a human clinical trial was proposed for the treatment of CD19+ B-lymphoid malignancy by means of autologous transplantation of T cells that have been genetically modified via the nonviral SB transposon vector system to express a CD19-specific chimeric antigen receptor (CAR). In this protocol, investigators will use the SB11 hyperactive mutant transposase (Geurts et al., 2003) for stable integration of the CD19-specific CAR-encoding genetic unit in primary T cells (Singh et al., 2008). Notably, the hyperactive SB11 transposase has been used in in vivo studies to enhance plasmid-based transposition for the treatment of mucopolysaccharidosis I (MPSI; Aronovich et al., 2007) and also in chromosomal-based transposition for the discovery of cancer genes (Collier et al., 2005; Dupuy et al., 2005). Although limited studies conducted in human cell lines have not revealed genomic instability or insertional mutagenesis associated with the SB11 hyperactive transposase, one of the recommendations by the National Institutes of Health Recombinant DNA Advisory Committee (NIH RAC) has been to perform extended evaluations of SB11 transposase-modified T cells for possible clonal expansion and for insertional mutagenesis that could lead to T cell malignancies (Williams, 2008). Thus, it is imperative to conduct careful preclinical assessments of the in vivo impact of using hyperactive SB transposase mutants in specific tissue types to rule out the possibility of such detrimental outcomes.
In utero gene therapy, although invasive in nature, provides a therapeutic option for early-onset genetic conditions that manifest themselves during early development. As observed in our study, neuronal stem cells present in the developing CNS provide an excellent carrier to widely distribute a therapeutic gene by the HSV/SB amplicon vector platform. In addition, immune tolerance to the transgene product and possibly the viral vector can be achieved via in utero gene transfer because of the naive nature of the host immune system. This allows for vector persistence and possibly long-term expression of the delivered transgene compared with gene transfer into the adult CNS. Hence, further research in animal models of early-onset neurological diseases is warranted to assess the safety and efficacy of HSV/SB amplicon vector-based in utero gene therapy.
Acknowledgments
The authors thank Dr. Howard Federoff (Georgetown University) for helpful discussions, Anna Dalda (MDC) for assistance in transposon insertion site analysis, Dr. Deborah Ryan (University of Rochester) for conducting statistical analyses of data, and Dr. Keigan Park (University of Rochester) for setting up timed pregnant mice. NIH R01-AG023593 (W.J.B.) and a Bundesministerium für Forschung und Bildung NGFNplus research grant (Z.Iv.) supported this work.
Author Disclosure Statement
The authors declare that no conflicts of interest exist.
References
- Aronovich E.L. Bell J.B. Belur L.R. Gunther R. Koniar B. Erickson D.C. Schachern P.A. Matise I. McIvor R.S. Whitley C.B. Hackett P.B. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: Implications for non-viral gene therapy of mucopolysaccharidoses. J. Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baus J. Liu L. Heggestad A.D. Sanz S. Fletcher B.S. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- Bowers W.J. Howard D.F. Brooks A.I. Halterman M.W. Federoff H.J. Expression of vhs and VP16 during HSV-1 helper virus-free amplicon packaging enhances titers. Gene Ther. 2001;8:111–120. doi: 10.1038/sj.gt.3301340. [DOI] [PubMed] [Google Scholar]
- Bowers W.J. Mastrangelo M.A. Howard D.F. Southerland H.A. Maguire-Zeiss K.A. Federoff H.J. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol. Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Collier L.S. Carlson C.M. Ravimohan S. Dupuy A.J. Largaespada D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- De Silva S. Mastrangelo M.A. Lotta L.T., Jr. Burris C.A. Federoff H.J. Bowers W.J. Extending the transposable payload limit of Sleeping Beauty (SB) using the herpes simplex virus (HSV)/SB amplicon-vector platform. Gene Ther. 2010;17:424–431. doi: 10.1038/gt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy A.J. Akagi K. Largaespada D.A. Copeland N.G. Jenkins N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Gehrmann J. Matsumoto Y. Kreutzberg G.W. Microglia: Intrinsic immuneffector cell of the brain. Brain Res. Brain Res. Rev. 1995;20:269–287. doi: 10.1016/0165-0173(94)00015-h. [DOI] [PubMed] [Google Scholar]
- Geurts A.M. Yang Y. Clark K.J. Liu G. Cui Z. Dupuy A.J. Bell J.B. Largaespada D.A. Hackett P.B. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hartl D.L. Lozovskaya E.R. Nurminsky D.I. Lohe A.R. What restricts the activity of mariner-like transposable elements. Trends Genet. 1997;13:197–201. doi: 10.1016/s0168-9525(97)01087-1. [DOI] [PubMed] [Google Scholar]
- Ivics Z. Hackett P.B. Plasterk R.H. Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Liu L. Sanz S. Heggestad A.D. Antharam V. Notterpek L. Fletcher B.S. Endothelial targeting of the Sleeping Beauty transposon within lung. Mol. Ther. 2004;10:97–105. doi: 10.1016/j.ymthe.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Liu L. Mah C. Fletcher B.S. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol. Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Luo G. Ivics Z. Izsvak Z. Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L. Chuah M.K. Belay E. Jerchow B. Manoj N. Acosta-Sanchez A. Grzela D.P. Schmitt A. Becker K. Matrai J. Ma L. Samara-Kuko E. Gysemans C. Pryputniewicz D. Miskey C. Fletcher B. Vandendriessche T. Ivics Z. Izsvak Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Peterson E.B. Mastrangelo M.A. Federoff H.J. Bowers W.J. Neuronal specificity of HSV/Sleeping Beauty amplicon transduction in utero is driven primarily by tropism and cell type composition. Mol. Ther. 2007;15:1848–1855. doi: 10.1038/sj.mt.6300267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C. Cepko C.L. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev. Dyn. 2008;237:1034–1042. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H. Manuri P.R. Olivares S. Dara N. Dawson M.J. Huls H. Hackett P.B. Kohn D.B. Shpall E.J. Champlin R.E. Cooper L.J. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal T.J. Kaufman C.D. Izsvak Z. Voytas D.F. Ivics Z. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- Williams D.A. Sleeping Beauty vector system moves toward human trials in the United States. Mol. Ther. 2008;16:1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- Yant S.R. Meuse L. Chiu W. Ivics Z. Izsvak Z. Kay M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Yant S.R. Park J. Huang Y. Mikkelsen J.G. Kay M.A. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: Critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S.R. Wu X. Huang Y. Garrison B. Burgess S.M. Kay M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S.R. Huang Y. Akache B. Kay M.A. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed H. Izsvak Z. Walisko O. Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]