Abstract
The angiopoietin-like protein 3 (ANGPTL3) is an important inhibitor of the endothelial and lipoprotein lipases and a promising drug target. ANGPTL3 undergoes proprotein convertase processing (RAPR224↓TT) for activation, and the processing site contains two potential GalNAc O-glycosylation sites immediately C-terminal (TT226). We developed an in vivo model system in CHO ldlD cells that was used to show that O-glycosylation in the processing site blocked processing of ANGPTL3. Genome-wide SNP association studies have identified the polypeptide GalNAc-transferase gene, GALNT2, as a candidate gene for low HDL and high triglyceride blood levels. We hypothesized that the GalNAc-T2 transferase performed critical O-glycosylation of proteins involved in lipid metabolism. Screening of a panel of proteins known to affect lipid metabolism for potential sites glycosylated by GalNAc-T2 led to identification of Thr226 adjacent to the proprotein convertase processing site in ANGPTL3. We demonstrated that GalNAc-T2 glycosylation of Thr226 in a peptide with the RAPR224↓TT processing site blocks in vitro furin cleavage. The study demonstrates that ANGPTL3 activation is modulated by O-glycosylation and that this step is probably controlled by GalNAc-T2.
Keywords: Glycoprotein, Glycosylation, Lipase, Lipoprotein Metabolism, Protein Processing, ANGPTL3, FGF23, Furin, O-Glycosylation, Mucin
Introduction
Site-specific O-glycosylation is emerging as a novel mechanism that regulates the action of proprotein convertase (PC)2 processing of proteins (1). PC processing is crucial in regulating many fundamental biological pathways (2). O-Glycans in or immediately adjacent to the recognition sites of PCs can interfere with proteolysis required for activation or inactivation of proteins normally undergoing processing. We recently presented the first example of O-glycosylation-mediated regulation of the PC processing of the phosphaturic factor FGF23 (1), which results in functional inactivation of FGF23 and direct inhibition of the FGFR receptor by the released C-terminal fragment (3). Deficiency in intact FGF23 leads to the rare syndromes familial tumoral calcinosis and hyperostosis-hyperphosphatemia associated with hyperphosphatemia (4). Deficiency in FGF23 can be caused by either mutations in the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase T3 (GalNAc-T3) that O-glycosylates the PC recognition site (RHTR179) of FGF23 or by mutations in FGF23 resulting in loss of secretion (5). Importantly, mutations in the PC recognition site of FGF23 that completely block cleavage lead to the mirror disease autosomal dominant hypophosphatemic Ricketts (6). Thus, O-glycosylation in the PC site is essential for preventing inactivation, whereas some degree of inactivation at the PC site is equally important. More recently, it was also found that furin processing of the brain natriuretic peptide precursor, pro-brain natriuretic peptide, was blocked by O-glycosylation (7, 8). These examples suggest that site-specific O-glycosylation directed by GalNAc-transferase isoforms play a direct regulatory role in PC processing.
ANGPTL3 (angiopoietin-like protein 3) expressed by the liver is an important inhibitor of the lipoprotein lipase (LPL) (9) and endothelial lipase (EL) (10), which both belong to the AB hydrolase superfamily (lipase family) and share 45% sequence identity. LPL is primarily expressed in heart, muscle, and adipose tissue, whereas the major EL expression is found in liver, placenta, and lung (11). Despite their homology, they exert their effect on lipoprotein metabolism quite differently. LPL primarily functions as a triglyceride (TG) lipase on chylomicrons and VLDL particles, whereas EL functions as a phospholipase with the phospholipid-rich high density lipoprotein (HDL) as its primary substrate (12). ANGPTL3 undergoes PC processing in the RAPR224↓TT sequence motif for activation (13), and the processing site contains two potential Thr O-glycosylation sites immediately C-terminal at the P1′- and P2′-positions. The mature processed N-terminal domain of ANGPTL3 is a considerably more potent inhibitor than the proprotein, especially in vivo (13, 14). The C-terminal fibrinogen-like domain induces endothelial cell adhesion and angiogenesis in vivo (15). Deficiency in ANGPTL3 in the KK/San mouse strain lowers serum TG, HDL-C, and HDL-phospholipid levels, and these can be restored by adenoviral supplementation with ANGPTL3 in agreement with its lipase-inhibitory function (9, 10, 16). Studies describing the relationship between human serum ANGPTL3 and parameters of lipid and cholesterol metabolism generally agree on a positive correlation between serum ANGPTL3 and HDL-C, but conflicting results on the correlation with TG levels exist (10, 17). Collectively, the results of these studies stress the involvement of ANGPTL3 in TG and HDL metabolism, albeit the precise role of ANGPTL3 and the consequences thereof are still not fully resolved.
Considerable evidence indicates that high LDL cholesterol, low HDL cholesterol, and high TG blood levels are independent factors predisposing for coronary artery disease and stroke, and these are strongly influenced by genetic variants in multiple genes, including ANGPTL3 and GALNT2 (18, 19). The latter gene encodes another GalNAc-transferase isoform, GalNAc-T2, involved in the initiation of O-glycosylation (20). In a recent study, the genetic association of GalNAc-T2 with altered plasma lipids was validated in a mouse model, and expression of murine GalNAc-T2 in the liver was shown to correlate inversely with plasma HDL cholesterol levels (21). We hypothesized that GalNAc-T2 could be involved in O-glycosylation of one or more proteins involved in lipid metabolism and thereby affect HDL and TG levels. We screened a panel of proteins known to affect lipid metabolism for potential O-glycosylation sites selectively glycosylated by the GALNT2 isoform and identified ANGPTL3. We demonstrated that ANGPTL3 was O-glycosylated at Thr226 immediately adjacent to a PC processing site (RAPR224↓TT) by GalNAc-T2. ANGPTL3 is exclusively expressed in the liver, and of the tested GalNAc-transferases capable of glycosylating ANGPTL3, only the GalNAc-T2 isoform is expressed in the liver. O-Glycosylation of ANGPTL3 expressed in CHO ldlD cells blocked PC processing. Furthermore, we demonstrated that glycosylation of ANGPTL3 at Thr226 in the PC RAPR224↓TT processing site by GalNAc-T2 blocks in vitro furin cleavage, further suggesting that GalNAc-T2-mediated O-glycosylation plays a regulatory role of activation of ANGPTL3 and hence lipid metabolism.
EXPERIMENTAL PROCEDURES
Glycosyltransferase Assays
All recombinant glycosyltransferases were expressed as soluble secreted truncated proteins in insect cells as described previously (22). Screening assays for GalNAc-transferases with peptides (Schafer-N, Sigma) were performed as product development assays in 25 μl of 25 mm cacodylic acid sodium, pH 7.4, 10 mm MnCl2, 0.25% Triton X-100, 1.5 mm UDP-GalNAc (Sigma), 10 μg of acceptor peptides, and 0.5 μg of purified enzyme.
Chemoenzymatic Synthesis of Glycopeptides
GalNAc O-glycosylation was performed in 25 mm cacodylic acid sodium, pH 7.4, 10 mm MnCl2, 0.25% Triton X-100, 1.5 mm UDP-GalNAc, 0.4 mg/ml acceptor peptides, and 15 μg/ml GalNAc-T2. Core 1 galactosylation was performed in 100 mm MES, pH 6.0, 20 mm MnCl2, 0.1% Triton X-100, 2.6 mm UDP-Gal (Sigma), 0.4 mg/ml of GalNAc-glycosylated ANGPTL3 peptide, and dC1Gal-T1 (22). Sialylation with ST6GalNAc-I was performed in 50 mm MES (pH 6.0), 20 mm EDTA, 2 mm dithiothreitol, 2 mm CMP-NeuAc, 0.4 mg/ml GalNAc-glycosylated ANGPTL3, and human ST6GalNAc-I (22). Synthesis was performed sequentially with HPLC purification between each step, and all glycopeptides were purified and analyzed by MALDI-TOF.
Characterization of O-Glycosylation Sites
The product of ANGPTL3 dodecapeptide with GalNAc-T2 was characterized by electrospray ionization-linear ion trap-Fourier transform mass spectrometry in an LTQ-Orbitrap XL hybrid spectrometer (Thermo-Scientific, Bremen, Germany), equipped with electron transfer dissociation (ETD) for peptide sequence analysis by MS/MS (MS2) with retention of glycan site-specific fragments. The sample was dissolved in methanol/water (1:1) containing 1% formic acid and introduced by direct infusion via a TriVersa NanoMate ESI-Chip interface (Advion BioSystems, Ithaca, NY), at a flow rate of ∼100 nl/min and 1.4 kV spray voltage. Mass spectra were acquired in positive ion Fourier transform mode at a nominal resolving power of 30,000; in addition to the normal calibration process, a polydimethylcyclosiloxane ion (m/z 445.1200) was used as an internal reference in MS1. Several ETD-MS2 spectra were acquired on the MH44+, MH33+, and MH22+ molecular precursor isotope clusters using an isolation width of 5 mass units; normalized collision energy, 35%; activation Q, 0.25; activation time of either 100, 150, or 200 ms; and supplemental activation, 20%. Experimental MS1 spectra were analyzed by comparison with exact m/z values calculated for the observed molecular ion charge states, using the known ANGPTL3 dodecapeptide sequence incremented with 1 HexNAc residue; ETD-MS2 spectra were analyzed by comparison with theoretical c- and z·-fragment m/z values calculated for both positional combinations of one HexNAc residue on the two potential glycosylation sites in the sequence. Calculations were performed using World Wide Web-based Protein Prospector MS-Isotope and MS-Product software routines.
Recombinant ANGPTL3 (Alexis Biochemicals) was GalNAc O-glycosylated with 20 μg/ml GalNAc-T2 and 2 μg of the protein as described for the peptide acceptors. The glycosylation reaction was stopped, and proteins were separated on a NuPage BisTris 4–12% gel (Invitrogen), followed by Coomassie protein staining. The ANGPTL3 band at 62 kDa was manually excised and then reduced and alkylated, followed by in-gel digestion with sequencing grade trypsin (Promega). 0.5 μl of the extract and 0.5 μl of 2,5-dihydrobenzoic acid (1 mg/ml) were mixed and applied to a 600-μm AnchorChip target (Bruker) with a peptide standard in dedicated calibration positions. Automatic acquisition of MALDI-TOF MS spectra was performed on a Bruker Reflex IV mass spectrometer. Spectra were recorded in the positive ion mode with external calibration using a pulsed UV laser beam (nitrogen laser, wavelength = 337 nm) and an acceleration voltage of 20 kV. The reflector voltage was set to 22.5 kV. Methods for automatic acquisition were defined in the autoexecute module of the FlexControl 1.3 software (Bruker). The raw spectra were processed by the FlexAnalysis 2.4 software, and the generated peak lists were transferred to the Biotools 3.0 module (Bruker), which triggered batch searches in the NCBI non-redundant protein data base using the MASCOT PMF search tools (Matrix Science, London, UK). Searches were restricted to human and trypsin specificity with one missed cleavage allowed. The maximum mass error was 100 ppm for externally calibrated spectra, and carbamidomethylation of cysteine and oxidation of methionine was considered.
Expression in CHO ldlD Cells
An expression construct of the entire coding sequence of human ANGPTL3 was cloned into pcDNA3.1/V5/His (C-terminal tags). Reporter constructs containing either 32 amino acids from ANGPTL3 or 30 amino acids from FGF23, furin cleavage sites and O-glycosylation sites, followed by 20 amino acids from the MUC1 tandem repeat containing O-glycosylation sites and the 5E5 antibody epitope were synthesized. Full-length monomeric EYFP (23) was amplified using primers 5′-GCGGGATCCAAATGGTGAGCAAGGGCGAGGAG-3′ and 5′-GCGAATTCCTTGTACAGCTCGTCCATGCCG-3′ (BamHI and EcoRI overhangs are underlined) and inserted into the BamHI/EcoRI site of a modified expression vector pcDNA-inf (GenBankTM accession number AJ131287). pcDNA-inf was modified by shifting the BamHI/EcoRI site one frame relative to the γ-interferon-encoded signal peptide sequence. The generated expression vector was designated pcDNA-γ-mEYFP. Two synthetic constructs encoding either His tag-flanked ANGLPTL3 or FGF23 fused to the MUC1 tandem repeat (Eurofins MWG Operon) were inserted into the EcoRI/NotI site of pcDNA-γ-mEYFP, generating pcDNA-γ-mEYFP-ANGLPTL3 and pcDNA-γ-mEYFP-FGF23.
CHO ldlD cells with defective UDP-Gal/UDP-GalNAc 4-epimerase (24) were grown in Ham's F-12/Dulbecco's modified Eagle's medium (1:1, v/v) supplemented with 3% fetal bovine serum and 1% glutamine. Cells were either transfected with 2 μg of pcDNA3.1 full coding ANGPTL3 or 2 μg of ANGPTL3 and FGF23 reporter constructs using Lipofectamine 2000 according to the manufacturer's instructions. Transient expression was analyzed 48 h post-transfection, and stable transfectants were selected in 0.4 mg/ml Zeocin (Invitrogen) by monitoring anti-V5 (R960-25, Invitrogen) staining or YFP tag fluorescence. To evaluate the effects of O-glycosylation, CHO ldlD transfectant cells were grown in medium supplemented with 0.1 mm galactose (Gal), 1 mm GalNAc (Gn), or a combination.
SDS-PAGE Western Blot Analysis
Cell culture medium (10 μl) was separated on NuPage Novex BisTris 4–12% gels (Invitrogen) and blotted onto nitrocellulose membrane for 60 min, followed by incubation in Western Breeze blocking solution (Invitrogen). Following wash in Tris-buffered saline (TBS) plus 0.05% Tween 20, the membranes were incubated in primary antibody (mouse anti-V5-AP (Invitrogen), rabbit anti-EYFP (Living Colors, Clontech), mouse anti-Tn-MUC1 mAb 5E5 (22), or mouse anti-ANGPTL3 1D10 (Alexis Biochemicals)) overnight at 4 °C. Blots were developed with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate after incubation with secondary alkaline phosphatase-conjugated antibodies for 1 h at room temperature.
Furin Cleavage of Peptides and Glycopeptides
A modification of an in vitro cleavage assay with a human furin protease (Sigma) was used to analyze furin cleavage of FGF23- and ANGPTL3-related peptides and glycopeptides covering the -RHTR- and -RAPR- sites, respectively (1). The assay was performed in 100 mm Hepes (pH 7.5), 1 mm CaCl2, 0.5% Triton X-100, 2 mm DTT using 10 μg of peptide or glycopeptide substrate and 1 unit of enzyme in a total volume of 50 μl. The mixture was incubated at 37 °C, and product development was evaluated after 0.5, 1.5, and 5 h by MALDI-TOF.
Inhibition of Furin-mediated Cleavage
CHO ldlD cells stably expressing FGF23rep or ANGPTL3rep were grown to 50% confluence in 6-well dishes, and the medium was replaced with fresh medium containing a 100, 50, 25, 10, or 0 μm concentration of the cell-permeable Furin inhibitor Dec-RVKR-CMK (EMD Chemicals) in DMSO. After 24 and 48 h, growth cell culture supernatant was sampled and assayed by SDS-PAGE Western blotting.
RESULTS
Identification of O-Glycoproteins Involved in HDL Metabolism as Targets for Selective GalNAc-T2-mediated O-Glycosylation
Two genome-wide association studies recently identified several novel putative genes associated with plasma levels of LDL-cholesterol, HDL-cholesterol, and triglycerides (18, 19). Of interest for the present study was the identification of SNPs in the first intron of the GalNAc-T2 gene associated with both HDL and triglyceride TG plasma levels. Subsequent studies on other populations have confirmed this association and validated that GalNAc-T2 plays a role in HDL cholesterol in a mouse model (21, 25). Based on these findings, we hypothesized that GalNAc-T2 could be involved in O-glycosylation of a protein involved in HDL and TG metabolism. We therefore searched for potential O-glycoproteins using literature information as well as the NetOGlyc algorithm for prediction of O-glycosylation sites (26). Table 1 summarizes known and potential O-glycosylation target sequences from 13 proteins identified. Four of these proteins were selected for further analysis based on known or potential O-glycosylation sites and the effect of genetic deficiency in these proteins. Apolipoprotein AII (apoA-II) is a major apolipoprotein in HDL, and the O-glycosylation level of apoA-II affects its association with HDL (27). Lecithin-cholesterol acyltransferase (LCAT) transacylates cholesterol in HDL particles and is O-glycosylated in two positions (28). Phospholipid transfer protein (PLTP) transfers phospholipids from TG-rich lipoproteins to HDL, and deficiency in PLTP is associated with selective decrease in HDL lipids (29). PLTP has several potential O-glycosylation sites. ANGPTL3 inhibits EL and LPL, resulting in inhibition of lipolysis and altered plasma TG and HDL levels (9, 30). ANGPTL3 has several potential O-glycosylation sites with some adjacent to a PC-processing site (Table 1).
TABLE 1.
Proteins involved in HDL and/or TG metabolism with established or predicted O-glycosylation sites
| Gene | Ensembl | ORF | Potential O-glycosylation sitesa | Established O-glycosylation sites | Target sequenceb | T1 | T2 | T3 |
|---|---|---|---|---|---|---|---|---|
| ABCA1 | ENSP00000363868 | 2261 | 0 | NDc | 251SKELAEATKTLLHSLG267 | NAd | NA | NA |
| APOA1 | ENSP00000364478 | 245 | 2 | ND | 99YPPTSMSRSPPTVLVI115 | NA | NA | NA |
| APOA2 | ENSP00000356969 | 61 | 1 | Yese | 46NFLSYFVELGTQPATQ61 | 1/2f | 0/2 | 0/2 |
| ANGPTL3 | ENSP00000360170 | 460 | 1 | ND | 216LSSKPRAPRTTPFLQL231 | 0/4 | 1/4 | 1/4 |
| BMP-1 | ENSP00000305714 | 986 | 0 | ND | 748DHKVTSTSGTITSPNW763 | NA | NA | NA |
| CETP | ENSP00000200676 | 493 | 1 | ND | 261EDLPLPTFSPTLLGDS277 | NA | NA | NA |
| LCAT | ENSP00000264005 | 440 | 3 | Yese | 424QGPPASPTASPEPPPP439 | 3/3 | 1/3 | 2/3 |
| LDLR | ENSP00000252444 | 860 | 6 | Yesg | 734TAVRTQHTTTRPVPDT749 | NA | NA | NA |
| LIPG | ENSP00000261292 | 500 | 1 | ND | 485DGWRMKNETSPTVELP500 | NA | NA | NA |
| PLTP | ENSP00000335290 | 441 | 10 | ND | 426RPADVRASTAPTPSTA439 | 3/5 | 2/5 | 3/5 |
| VLDLR | ENSP00000371532 | 873 | 2 | Yesg | 777PGGINVTTAVSEVSVP792 | NA | NA | NA |
| VNN-1 | ENSP00000356905 | 513 | 1 | ND | 458GRLFSLKPTSGPVLTV473 | NA | NA | NA |
| SCARB1 | ENSP00000261693 | 509 | 1 | ND | 489AYSESLMTSAPKGSVL504 | NA | NA | NA |
a Sites predicted by NetOGlyc 3.1 (available on the World Wide Web).
b Peptide sequence containing all or some of the potential O-glycosylation sites. Ser and Thr potential O-glycosylation sites are underlined. Sites predicted by NetOGlyc are shown in boldface type, and experimentally verified sites are shown in underlined boldface type.
c ND, not determined.
d NA, not analyzed.
e O-glycosylation-identified, determined sites in underlined boldface type.
f Sites incorporated in vitro by GalNAc-transferases in this study (number of GalNAc/total Ser/Thr sites).
g O-glycosylation identified, but sites not determined.
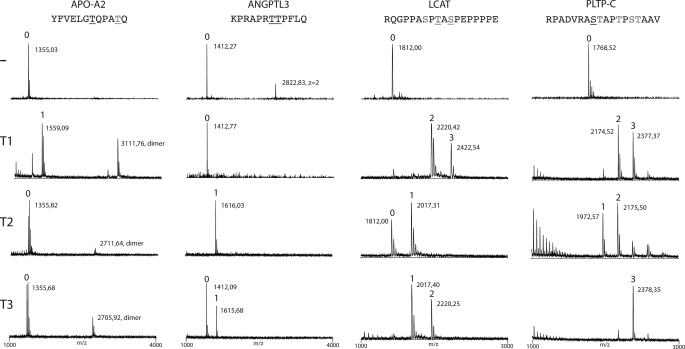
In order to assess if these four proteins where glycosylated and which GalNAc-transferase isoforms could be involved, we designed short peptides covering putative glycosylation sites and tested these as substrates for human recombinant GalNAc-transferases (Table 1). Our current knowledge of O-glycosylation of especially less abundant proteins and proteins with single O-glycosylation sites is quite limited, and sequence-based prediction models generally fail to predict isolated O-glycosylation sites in proteins (26). Considerable data indicate that in vitro GalNAc-transferase enzyme analysis of short peptide substrates reasonably reflects where the corresponding proteins are glycosylated in cells (31). We first tested the performance of three human GalNAc-transferase isoforms, GalNAc-T1, -T2, and -T3, in in vitro assays with peptide substrates designed from the putative O-glycosylation regions of the four identified proteins using 24-h reactions and characterization of products by MALDI-TOF mass spectrometry (Fig. 1). These isoforms are representative of three subfamilies of GalNAc-transferases with peptide specificities (31), and they are the most widely expressed and well characterized isforms. GalNAc-T2 was found to glycosylate three of the peptide targets, ANGPTL3, LCAT, and PLTP, but not the APO-A2 peptide. GalNAc-T1, in contrast, glycosylated APO-A2, LCAT, and PLTP but not ANGPTL3. GalNAc-T3 glycosylated mainly LCAT and PLTP, but low incorporation was also found with APO-A2 and ANGPTL3. ANGPTL3 was the only target that exhibited selective O-glycosylation by GalNAc-T2. Although GalNAc-T3 also exhibited weak activity with this substrate, GalNAc-T3 is not a candidate for glycosylation of ANGPTL3 in vivo because ANGPTL3 is almost exclusively expressed in the liver, where neither GalNAc-T3 nor its close homolog GalNAc-T6 are expressed (32, 33). We also tested GalNAc-T11, which is only expressed in the kidney and representative of a fourth GalNAc-T family, and this enzyme did not glycosylate ANGPTL3 (not shown). We therefore focused on ANGPTL3 as a candidate substrate for GalNAc-T2. A summary of the results is presented in Table 1.
FIGURE 1.
ANGPTL3 is selectively O-glycosylated by GalNAc-T2. In vitro screening of substrate specificities of recombinant human GalNAc-transferases, GalNAc-T1, -T2, and -T3, with peptides covering potential O-glycosylation sites identified in four proteins involved in HDL and TG metabolism (APO-A2, ANGPTL3, LCAT, and PLTP-C) (Table 1). Glycosylation of peptides was monitored by MALDI-TOF analysis, and products formed after 24 h incubation with enzymes are shown. Masses of peptides and glycopeptides are indicated with the predicted number of incorporated GalNAc residues indicated above the peaks. APO-A2 was only glycosylated by GalNAc-T1 (one GalNAc residue), ANGPTL3 was glycosylated by GalNAc-T2 (one GalNAc residue) and only partially by GalNAc-T3, and LCAT and PLTP-C were glycosylated by all three tested GalNAc-transferases with a varying number of GalNAc residues incorporated. Sequences of peptide substrates shown above with potential Ser and Thr O-glycosylation sites indicated by underlining with black type, sites predicted by the NetOGlyc server are shown with gray type, and sites experimentally verified are shown with underlining with gray type. The APO-A2 peptide partly formed dimers as indicated.
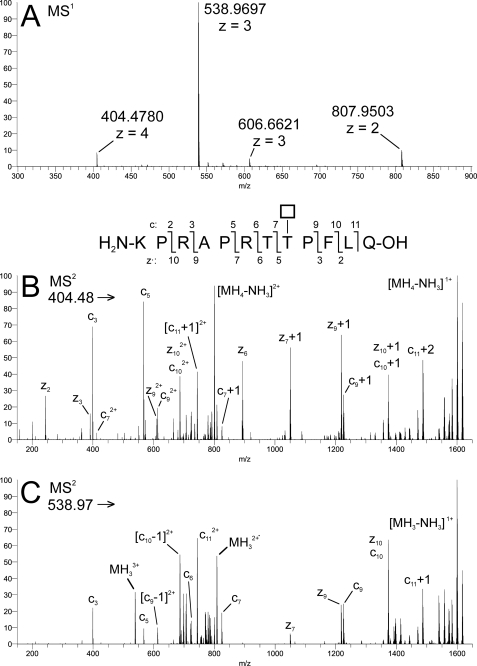
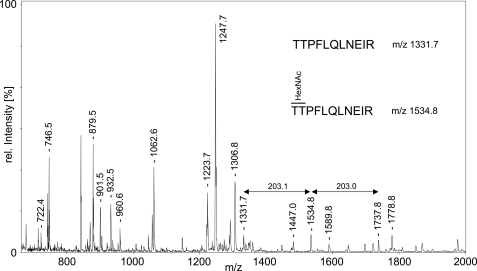
Mass spectrometric analysis of the GalNAc-glycopeptide product formed by GalNAc-T2 with the ANGPTL3 peptide showed that GalNAc-T2 specifically glycosylated the Thr226 residue immediately adjacent to the RAPR224↓ TT226 furin cleavage site in the ANGPTL3 peptide (Fig. 2). We therefore hypothesized that GalNAc-T2 was involved in regulation of PC processing of ANGPTL3 in a similar way as we previously demonstrated that the GalNAc-T3 isoform regulates processing of FGF23 (1). We further found that a commercially available recombinant ANGPTL3 proprotein (full coding except signal peptide) was not glycosylated at Thr225/226 and used this to demonstrate that GalNAc-T2 glycosylation of the entire protein substrate resulted in specific incorporation of at least one GalNAc residue into the Thr225/226 residues (Fig. 3). Interestingly, the supplied ANGPTL3 proprotein was partially cleaved with a C-terminal fragment of ∼39 kDa (Fig. 4C). These in vitro studies suggested that GalNAc-T2-mediated glycosylation potentially could play a role in the furin processing of ANGPTL3 that leads to activation of its in vivo functions (13).
FIGURE 2.
GalNAc-T2 glycosylated Thr226 of the ANGPTL3 peptide. Characterization of the product formed by GalNAc-T2 with the ANGPTL3 peptide by ETD in the LTQ-Orbitrap. A, MS1 of ANGPTL3 + 1Tn; B, ETD-MS2 of precursor MH44+ ion (200 ms activation time); C, ETD-MS2 of precursor MH33+ ion (150 ms activation time). Inset, fragmentation scheme for ANGPTL3 + 1Tn, illustrating the numbering of product ions generated by c/z• cleavage. The fragmentation pattern is consistent with glycosylation of Thr8 with a HexNAc residue (*) (i.e. abundant z·5+, z·6+, c7+, c9+, and related ions (cn ± 1 mass unit and/or doubly charged) were detected at m/z values calculated for HexNAc at T8. Corresponding fragments consistent with glycosylation at T7 were not detected. All detected fragment ions are listed in supplemental Table 1.
FIGURE 3.
GalNAc-T2 glycosylates Thr225/Thr226 of recombinant ANGPTL3. Shown is MALDI-TOF MS analysis of in-gel-digested ANGPTL3 after O-glycosylation with GalNAc-T2. m/z values of ANGPTL3 peptides are indicated above the peaks. Unglycosylated TTPFLQLNEIR (residues 225–235) and +1Tn glycoform are visible at m/z 1331.7 and 1534.8. A peak observed at m/z 1737.8 may correspond to the +2Tn glycoform, but it is also isobaric with the theoretical mass of another, potentially oxidized peptide (residues 446–460; STKM(Ox)LIHPTDSESFE); its identity could not be confirmed without MS/MS data. The peak intensity of the unglycosylated peptide at 1331.7 appeared strongly reduced as compared with the reference spectrum of untreated ANGPTL3 (not shown). Authenticity of ANGPTL3 in the gel slice was confirmed by PMF analysis (63% sequence coverage by 32 peptides matched, Mascot score 254).
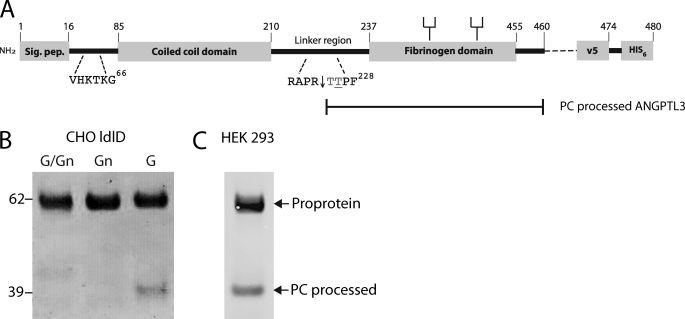
FIGURE 4.
PC processing of ANGPTL3 is blocked by O-glycosylation in CHO ldlD cells. A, schematic depiction of the full coding sequence of ANGPTL3 and design of the expression constructs used. Heparin binding site (VHKTKG), furin cleavage site (RAPR), and N-glycans are indicated. B, transient expression of full coding ANGPTL3 in CHO ldlD cells grown with Gal and GalNAc (G/Gn), with GalNAc (Gn) (allowing GalNAc O-glycosylation only), and with Gal (G) (allowing complete N-glycosylation but no O-glycosylation). Medium from transfected 6-well plates were harvested after 72 h, and His-tagged ANGPTL3 was affinity-purified on Talon beads (Co2+) and analyzed by Western blotting with anti-V5 antibody. The protein load was normalized to expression of ANGPTL3 evaluated by capture ELISA (not shown). The ANGPTL3 proprotein migrating at 62 kDa and the mature protein migrating at 39 kDa are indicated by arrows. C, Western blot analysis of 50 ng of commercial recombinant human ANGPTL3 with mAb 1D10 to ANGPTL3.
O-Glycosylation Blocks Processing of ANGPTL3 in Vivo
To evaluate the in vivo role of O-glycosylation in a cell system, we utilized the CHO ldlD cells in which O-glycosylation can be controlled (24). We first tested expression of a full coding ANGPTL3 construct transiently expressed in cells either capable or not capable of performing O-glycosylation (i.e. grown with and without exogenous sugars in medium, respectively). As shown in Fig. 4B, the ANGPTL3 protein produced in cells allowed to O-glycosylate (GalNAc/Gal or GalNAc added to medium) migrated to the expected molecular mass of ∼62 kDa without evidence of processing to the mature 236-amino acid product (molecular mass ∼39 kDa with tags and N-glycans). However, when cells were grown without capacity for O-glycosylation, a minor fraction (10–20%) of the ANGPTL3 proprotein was converted to the mature 39-kDa protein. The ratio of proprotein and mature protein was very similar to that found for the commercially available unglycosylated ANGPTL3 protein (Fig. 4C).
We next designed a reporter model construct for rapid screening of PC processing sites and to determine the role of O-glycosylation in CHO ldlD cells (Fig. 5A). The chimeric reporter protein consisted of the PC target sequence (32 and 30 amino acids from ANGPTL3 and FGF23, respectively, with the PC site in the middle) flanked by an N-terminal EYFP module for easy visualization of expression, a C-terminal MUC1 tandem repeat sequence for easy evaluation of O-glycosylation efficiency, and His tags for purification. We used the RHTR179↓ PC processing site in FGF23 to validate the model system. PC processing of FGF23 is only blocked by GalNAc-T3-mediated O-glycosylation of Thr178, and this enzyme activity is not present in CHO cells (1). Thus, as expected, the FGF23 reporter construct was efficiently cleaved (N-terminal fragment migrating at 39 kDa) with or without the capacity for O-glycosylation in CHO ldlD cells. A minor amount of the intact construct migrating at 48 kDa was rescued in cells grown in GalNAc alone and capable of O-glycosylation (Fig. 5B). The ANGPTL3 reporter construct was considerably more resistant to processing, in agreement with the findings with the full coding construct (Fig. 4). With O-glycosylation, the product migrated at 48 kDa, and without O-glycosylation, it migrated at 44 kDa. However, without O-glycosylation, a minor fraction (10–20%) of the reporter construct was cleaved and migrated at 38 kDa (Fig. 5B).
FIGURE 5.
Analysis of PC processing of FGF23 and ANGPTL3 reporter constructs in CHO ldlD cells. A, schematic depiction of design of reporter constructs. Vertical arrows indicate sites of furin cleavage, and filled circles indicate sites of O-glycosylation. All potential O-glycosylation sites are shown in gray. B, SDS-PAGE Western blot analysis of a representative clone expressing ANGPTL3 (6F7) or FGF23 (7B2) reporter constructs. Due to extensive O-glycosylation in the MUC1 tandem repeat region, the reporter construct migrates at 48 kDa compared with 44 kDa for the construct without O-glycosylation. The PC-processed N-terminal fragments with EYFP migrate at 38 kDa (ANGPTL3 reporter) and 39 kDa (FGF23 reporter). The upper panels are labeled with anti-EYFP, whereas the lower panels are labeled with mAb 5E5 specifically recognizing the GalNAc O-glycosylated MUC1 tandem repeat sequence in the C-terminal fragment (22). Migration of O-glycosylated, non-glycosylated, and processed proteins are indicated by arrows, and the additions of Gal and GalNAc to growth medium are indicated as described in the legend to Fig. 4.
ANGPTL3 Is Processed by a Furin-like Proprotein Convertase
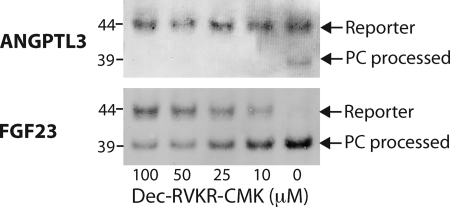
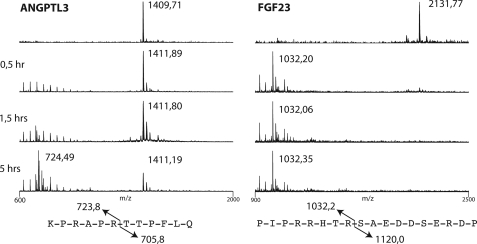
The processing of ANGPTL3 is probably catalyzed by the proprotein convertase furin as previously proposed (14), so we next tested the effect of the addition of the intracellular furin inhibitor Dec-RVKR-CMK for processing of the reporter constructs (34) (Fig. 6). Processing of the FGF23 control reporter construct was inhibited by Dec-RVKR-CMK in a dose-dependent way, with >50% inhibition at 50 mm concentration or above, as shown by the appearance of the uncleaved product migrating at 44 kDa (Fig. 6A). The partial processing of the ANGPTL3 product was completely blocked with a 10 mm concentration of the inhibitor, where the 38-kDa N-terminal fragment disappeared (Fig. 6B). These results combined with the previous results indicate that the PC processing site in ANGPTL3 is a substrate albeit a poor one for furin and possibly other PCs. To further validate this finding, we tested in vitro cleavage reactions of short peptides with furin in a time course monitored by MALDI-TOF analysis (Fig. 7). The ANGPTL3 peptide NH2-KPRAPR224TTPFLQ and the FGF23 peptide NH2-PIPRRHTR179SAEDDSERDP were treated with recombinant furin, and cleavage was monitored by MALDI-TOF in a time course assay. In agreement with our findings, the FGF23 peptide was a very efficient substrate for furin, with complete cleavage after 30 min, whereas the ANGPTL3 peptide was a considerably poorer substrate that was only partially cleaved after 5 h. It is likely that the Pro residue in P5 (PRHTR179), which is conserved in mammals, disfavors this furin substrate site, in agreement with previous analysis of consensus motifs for PC processing (35).
FIGURE 6.
The furin inhibitor Dec-RVKR-CMK blocks processing of FGF23 and ANGPTL3 reporter constructs in CHO ldlD cells. Shown is anti-EYFP SDS-PAGE Western blot analysis of culture medium of cells grown with inhibitor as indicated. Cells were grown in Gal alone without capacity for O-glycosylation. Stable CHO ldlD clones were grown to 50% confluence in 6-well dishes, and the medium was replaced with medium containing a 100, 50, 25, 10, or 0 μm concentration of the cell-permeable Furin-inhibitor Dec-RVKR-CMK in DMSO. After 24 and 48 h of growth, cell culture supernatant was sampled. The arrows indicate the non-processed reporter and the cleaved N-terminal fraction that migrates at ∼48 and 39 kDa, respectively.
FIGURE 7.
Furin time course digestion of ANGPTL3 and FGF23 peptides monitored by MALDI-TOF analysis. Furin cleavage reactions were sampled at 0.5, 1.5, and 5 h. The dodecapeptide ANGPTL3 was cleaved in the RAPR↓ motif after 5 h, yielding an N-terminal fragment of 724 Da as expected. The FGF23 peptide was readily cleaved in the RHTR↓ motif, yielding the N-terminal fragment of 1032 Da after 30 min.
GalNAc-T2-mediated O-Glycosylation of the ANGPTL3 PC Site Blocks Furin Processing
GalNAc-T2 is widely expressed in humans, and in fact, no human cell lines without GalNAc-T2 expression have been identified so far. We therefore used in vitro O-glycosylation of peptides with GalNAc-T2 to confirm the protective effect that this isoform has on processing. We have previously used this assay to demonstrate that GalNAc-T3-mediated O-glycosylation of the FGF23 PC site confers protection against furin cleavage (1). We prepared different glycoforms of ANGPTL3 glycopeptides with one O-glycan attached by GalNAc-T2 at Thr226, and time course MALDI analysis of furin digestion confirmed that all glycoforms provided almost complete protection (Fig. 8). In agreement with our studies of FGF23, the simple GalNAc glycoform appeared to provide the least protection.
FIGURE 8.
Furin time course digestion of ANGPTL3 glycopeptides monitored by MALDI-TOF analysis. An ANGPTL3 17-mer peptide was glycosylated at Thr226 with GalNAc (Tn) by GalNAc-T2 and further extended to core 1 (T, Galβ1-3GalNAcα1-O-Ser/Thr) or STn (NeuAcα2-3GalNAcα1-O-Ser/Thr). The furin cleavage reactions were sampled after 0.5, 1.5, and 5 h. The non-glycosylated ANGPTL3 peptide was cleaved in the RAPR↓ motif to completion after 5 h, giving rise to peaks with the expected masses of the C-terminal (933 Da) and N-terminal peptides (1,011 Da). The glycopeptides (Tn, T, and STn ANGPTL3) were almost completely protected against proteolytic cleavage. Only trace amounts of the cleaved C- and N-terminal peptides were visible after 5 h of incubation.
DISCUSSION
ANGPTL3 plays an important role in regulating HDL and TG metabolism and is regarded as a novel promising target for therapeutic intervention (36). ANGPTL3 is proposed to function through inhibition of the LPL and EL lipases, and the activity of ANGPTL3 is regulated partly through proprotein convertase activation (14). The present results suggest that PC activation of ANGPTL3 is further regulated by specific O-glycosylation performed by the GalNAc-T2 isoform in close proximity of the PC site. In addition to FGF23 and the brain natriuretic peptide, this is the third identified example of an apparent interplay between PC processing and O-glycosylation in regulating function of PC-processed proteins (1, 8). Differential processing of pro-opiomelanocortin has also been found to be affected by O-glycosylation (37). A survey of known PC processing sites in human proteins suggests that this may be a more common event.3 It is important to provide insight into the molecular mechanisms underlying the duality of regulation of processing by PCs on one hand and O-glycosylation mediated by GalNAc-transferase isoforms on the other. Defects in both the PC recognition site and GalNAc-transferase function are known to lead to diseases (4, 6), and strategies for therapeutic intervention may need to consider underlying defects.
The human GalNAc-transferase gene family consists of 20 genes, of which 15 have been cloned and functionally expressed to date (38). This is an unusually large number of isoforms catalyzing a single step in the biosynthesis of glycoproteins, and given that many of these isoforms have different albeit partly overlapping peptide substrate specificities and different cell and tissue expression patterns, it is clear that the initiation of mucin-type O-glycosylation is the single step in glycosylation of proteins that at least theoretically allows for the highest degree of differential regulation (31). The findings that defects in a single gene isoform result in disease in humans (1) or phenotypes in animal model systems, including Drosophila (39, 40) and mouse (41, 42), strongly argue that the large gene family serves more than functional redundancy and that the differential regulation provided by multiple GalNAc-transferase isoforms is important. A major challenge is to identify and backtrack a disease or phenotype to molecular understanding and identify defects in O-glycosylation. We previously identified one mechanism for disease-causing defects in GalNAc-transferase genes that involves the GalNAc-T3 isoform-specific O-glycosylation of the PC processing site RHTR179↓ in FGF23 and regulation of the processing and function of FGF23 (1). The PC processing of FGF23 is an essential inactivating step regulated by GalNAc-T3 O-glycosylation. In the CHO cell model system, GalNAc-T3 is required for secretion of intact FGF23 (1), and more recently we have found by metabolic labeling studies that the GalNAc-T3-mediated O-glycosylation mainly functions to modulate PC processing after secretion rather than during secretion.4 GalNAc-T3-mediated O-glycosylation appears to provide a physiological balance of active intact FGF23 in circulation, and studies suggest that the expression of GalNAc-T3 may be regulated by factors such as inorganic phosphate and calcium that regulate phosphate hemostasis (43). It is our hypothesis that O-glycosylation in or near the PC processing sites of proteins in fact not only regulates intracellular processing but also serves as a means to regulate activation or inactivation of proteins in circulation or at the site of action and hence plays a role in biodistribution and bioactivity.
The finding that the GalNAc-T2 isoform plays a role in regulating levels of plasma lipids poses a major challenge, which can ultimately only be conclusively addressed by identification and characterization of individuals with defects in GALNT2. Nevertheless, in the present study, we sought to utilize available methods to identify a glycoprotein that could play a role in HDL and TG metabolism and where the GalNAc-T2 isoform could modulate glycosylation. The identification of ANGPTL3 as a candidate target that has a PC processing site regulated by GalNAc-T2-mediated O-glycosylation provides a compelling scenario explaining a role for GalNAc-T2 in lipid metabolism. However, this model clearly has to be verified by analysis of PC processing and O-glycosylation of ANGPTL3 in GALNT2-deficient individuals and controls. Our results do provide strategies for future identification of individuals with defects in GALNT2 by analysis of levels of proprotein and mature ANGPTL3 in serum of subjects with low HDL and/or high TG and without other known genetic causes. Currently available ANGPTL3 serum assays, however, fail to distinguish the proprotein and mature protein (17).
We developed a simple model system for in vivo analysis of the effect of O-glycosylation on PC processing of specific sites. The reporter construct (Fig. 5) allows for rapid detection of expression and selection of clones by EYFP and antibody tags as well as monitoring of both O-glycosylation status and cleavage products. The “wild type” CHO ldlD cell line allows for evaluation of the effect of a complete lack of O-glycosylation, simple GalNAc (Tn) O-glycosylation, and core 1 sialylated O-glycosylation (ST; NeuAcα2–3Galβ1–3(±NeuAcα2–6)GalNAcα1-O-Ser/Thr) by growing cells in appropriate sugars. The repertoire of GalNAc-transferases in CHO cells is currently unknown but includes the rather ubiquitous GalNAc-T2 activity because the ANGPTL3 reporter construct was O-glycosylated (Fig. 5). To the extent that CHO ldlD cells lack certain GalNAc-transferase activities, such as GalNAc-T3, the model system can be used to demonstrate the requirement for a particular GalNAc-T isoform by co-transfection. We are currently successfully using the system to screen furin PC sites in proteins with prediction of adjacent O-glycosylation sites.
Our study demonstrated a role of O-glycosylation in PC activation of ANGPTL3. Perhaps surprisingly, the PC processing step in CHO cells as well as in vitro with furin was found to be quite ineffective, with only an estimated 10–20% processing. Although this can be explained by the PC sequence motif with an unfavored flanking Pro residue (KPRAPR224↓TT) (which interestingly is not conserved in mouse ANGPTL3 but substituted with a potential additional Ser O-glycosylation site (KSRAPR224↓TT)), it is possible that processing in cells with another repertoire of PCs or at specific sites of bioactivation is more efficient. Alternatively, activation of ANGPTL3 has to be carefully regulated, and both the sequence motif and the adjacent O-glycosylation are means to ensure this. Our results clearly warrant further studies of GalNAc-T2 and ANGPTL3 in relation to altered plasma lipids and enhanced risks for cardiovascular disease.
Supplementary Material
Acknowledgments
We kindly acknowledge Dr. Helen Hobbs for generously sharing the ANGPTL3 expression construct and Dr. Monte Krieger for generously sharing the CHO ldlD cell line.
This work was supported by the Carlsberg Foundation, the Alfred Benzon Foundation, The Danish Research Councils, and a Program of Excellence from the University of Copenhagen.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
K. Schjoldager, M. Vester-Christensen, and H. Clausen, unpublished results.
K. Kato and H. Clausen, unpublished results.
- PC
- proprotein convertase
- LPL
- lipoprotein lipase
- EL
- endothelial lipase
- TG
- triglyceride
- ETD
- electron transfer dissociation
- YFP
- yellow fluorescent protein
- EYFP
- enhanced YFP
- LCAT
- lecithin-cholesterol acyltransferase
- PLTP
- phospholipid transfer protein
- Dec-RVKR-CMK
- Decanoyl-Arg-Val-Lys-Arg-chloromethylketone.
REFERENCES
- 1.Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 2.Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 [DOI] [PubMed] [Google Scholar]
- 3.Goetz R., Nakada Y., Hu M. C., Kurosu H., Wang L., Nakatani T., Shi M., Eliseenkova A. V., Razzaque M. S., Moe O. W., Kuro-o M., Mohammadi M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. (2004) Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 5.Benet-Pagès A., Orlik P., Strom T. M., Lorenz-Depiereux B. (2005) Hum. Mol. Genet. 14, 385–390 [DOI] [PubMed] [Google Scholar]
- 6.White K. E., Evans W. E., O'Riordan J. L., Speer M. C., Econs M. J., Lorenz-Depiereux B., Grabowski M., Meitinger T., Strom T. M. (2000) Nat. Genet. 26, 345–34811062477 [Google Scholar]
- 7.Jiang J., Pristera N., Wang W., Zhang X., Wu Q. (2010) Clin. Chem. 56, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semenov A. G., Postnikov A. B., Tamm N. N., Seferian K. R., Karpova N. S., Bloshchitsyna M. N., Koshkina E. V., Krasnoselsky M. I., Serebryanaya D. V., Katrukha A. G. (2009) Clin. Chem. 55, 489–498 [DOI] [PubMed] [Google Scholar]
- 9.Shimizugawa T., Ono M., Shimamura M., Yoshida K., Ando Y., Koishi R., Ueda K., Inaba T., Minekura H., Kohama T., Furukawa H. (2002) J. Biol. Chem. 277, 33742–33748 [DOI] [PubMed] [Google Scholar]
- 10.Shimamura M., Matsuda M., Yasumo H., Okazaki M., Fujimoto K., Kono K., Shimizugawa T., Ando Y., Koishi R., Kohama T., Sakai N., Kotani K., Komuro R., Ishida T., Hirata K., Yamashita S., Furukawa H., Shimomura I. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 366–372 [DOI] [PubMed] [Google Scholar]
- 11.Hirata K., Dichek H. L., Cioffi J. A., Choi S. Y., Leeper N. J., Quintana L., Kronmal G. S., Cooper A. D., Quertermous T. (1999) J. Biol. Chem. 274, 14170–14175 [DOI] [PubMed] [Google Scholar]
- 12.Jaye M., Krawiec J. (2004) Curr. Opin. Lipidol. 15, 183–189 [DOI] [PubMed] [Google Scholar]
- 13.Ono M., Shimizugawa T., Shimamura M., Yoshida K., Noji-Sakikawa C., Ando Y., Koishi R., Furukawa H. (2003) J. Biol. Chem. 278, 41804–41809 [DOI] [PubMed] [Google Scholar]
- 14.Jin W., Wang X., Millar J. S., Quertermous T., Rothblat G. H., Glick J. M., Rader D. J. (2007) Cell Metab. 6, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camenisch G., Pisabarro M. T., Sherman D., Kowalski J., Nagel M., Hass P., Xie M. H., Gurney A., Bodary S., Liang X. H., Clark K., Beresini M., Ferrara N., Gerber H. P. (2002) J. Biol. Chem. 277, 17281–17290 [DOI] [PubMed] [Google Scholar]
- 16.Koishi R., Ando Y., Ono M., Shimamura M., Yasumo H., Fujiwara T., Horikoshi H., Furukawa H. (2002) Nat. Genet. 30, 151–157 [DOI] [PubMed] [Google Scholar]
- 17.Robciuc M. R., Tahvanainen E., Jauhiainen M., Ehnholm C. (2010) J. Lipid Res. 51, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., Albai G., Swift A. J., Morken M. A., Narisu N., Bennett D., Parish S., Shen H., Galan P., Meneton P., Hercberg S., Zelenika D., Chen W. M., Li Y., Scott L. J., Scheet P. A., Sundvall J., Watanabe R. M., Nagaraja R., Ebrahim S., Lawlor D. A., Ben-Shlomo Y., Davey-Smith G., Shuldiner A. R., Collins R., Bergman R. N., Uda M., Tuomilehto J., Cao A., Collins F. S., Lakatta E., Lathrop G. M., Boehnke M., Schlessinger D., Mohlke K. L., Abecasis G. R. (2008) Nat. Genet. 40, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., Wahlstrand B., Hedner T., Corella D., Tai E. S., Ordovas J. M., Berglund G., Vartiainen E., Jousilahti P., Hedblad B., Taskinen M. R., Newton-Cheh C., Salomaa V., Peltonen L., Groop L., Altshuler D. M., Orho-Melander M. (2008) Nat. Genet. 40, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White T., Bennett E. P., Takio K., Sørensen T., Bonding N., Clausen H. (1995) J. Biol. Chem. 270, 24156–24165 [DOI] [PubMed] [Google Scholar]
- 21.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., König I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Cecile J. W., Janssens A., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Döring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y. D., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Jr., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarp M. A., Sørensen A. L., Mandel U., Paulsen H., Burchell J., Taylor-Papadimitriou J., Clausen H. (2007) Glycobiology 17, 197–209 [DOI] [PubMed] [Google Scholar]
- 23.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 24.Kingsley D. M., Kozarsky K. F., Segal M., Krieger M. (1986) J. Cell Biol. 102, 1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama K., Bayasgalan T., Yamanaka K., Kumada M., Gotoh T., Utsumi N., Yanagisawa Y., Okayama M., Kajii E., Ishibashi S., Iwamoto S. (2009) J. Med. Genet. 46, 370–374 [DOI] [PubMed] [Google Scholar]
- 26.Julenius K., Mølgaard A., Gupta R., Brunak S. (2005) Glycobiology 15, 153–164 [DOI] [PubMed] [Google Scholar]
- 27.Remaley A. T., Wong A. W., Schumacher U. K., Meng M. S., Brewer H. B., Jr., Hoeg J. M. (1993) J. Biol. Chem. 268, 6785–6790 [PubMed] [Google Scholar]
- 28.Schindler P. A., Settineri C. A., Collet X., Fielding C. J., Burlingame A. L. (1995) Protein Sci. 4, 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X. C., Bruce C., Mar J., Lin M., Ji Y., Francone O. L., Tall A. R. (1999) J. Clin. Invest. 103, 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lichtenstein L., Kersten S. (2010) Biochim. Biophys. Acta 1801, 415–420 [DOI] [PubMed] [Google Scholar]
- 31.Hassan H., Bennett E. P., Mandel U., Hollingsworth M. A., Clausen H. (2000) in Carbohydrates in Chemistry and Biology: A Comprehensive Handbook (Ernst B., Hart G. W., Sinay P. eds) pp. 273–292, Wiley-VCH, New York [Google Scholar]
- 32.Bennett E. P., Hassan H., Clausen H. (1996) J. Biol. Chem. 271, 17006–17012 [DOI] [PubMed] [Google Scholar]
- 33.Bennett E. P., Hassan H., Mandel U., Hollingsworth M. A., Akisawa N., Ikematsu Y., Merkx G., van Kessel A. G., Olofsson S., Clausen H. (1999) J. Biol. Chem. 274, 25362–25370 [DOI] [PubMed] [Google Scholar]
- 34.Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., Shaw E., Klenk H. D. (1994) Biochimie 76, 217–225 [DOI] [PubMed] [Google Scholar]
- 35.Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C. (2009) Curr. Opin. Lipidol. 20, 357–359 [DOI] [PubMed] [Google Scholar]
- 37.Seger M. A., Bennett H. P. (1986) J. Steroid Biochem. 25, 703–710 [DOI] [PubMed] [Google Scholar]
- 38.Tabak L. A. (2010) Semin. Cell Dev. Biol. 21, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., Hollingsworth M. A., Clausen H. (2002) J. Biol. Chem. 277, 22623–22638 [DOI] [PubMed] [Google Scholar]
- 40.Zhang L., Tran D. T., Ten Hagen K. G. (2010) J. Biol. Chem. 285, 19491–19501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., Marth J. D. (2007) Mol. Cell Biol. 27, 8783–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miwa H. E., Gerken T. A., Jamison O., Tabak L. A. (2010) J. Biol. Chem. 285, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chefetz I., Kohno K., Izumi H., Uitto J., Richard G., Sprecher E. (2009) Biochim. Biophys. Acta 1792, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.