Abstract
Fibroblast growth factor 21 (FGF21) is a potent antidiabetic and triglyceride-lowering hormone whose hepatic expression is highly responsive to food intake. FGF21 induction in the adaptive response to fasting has been well studied, but the molecular mechanism responsible for feeding-induced repression remains unknown. In this study, we demonstrate a novel link between FGF21 and a key circadian output protein, E4BP4. Expression of Fgf21 displays a circadian rhythm, which peaks during the fasting phase and is anti-phase to E4bp4, which is elevated during feeding periods. E4BP4 strongly suppresses Fgf21 transcription by binding to a D-box element in the distal promoter region. Depletion of E4BP4 in synchronized Hepa1c1c-7 liver cells augments the amplitude of Fgf21 expression, and overexpression of E4BP4 represses FGF21 secretion from primary mouse hepatocytes. Mimicking feeding effects, insulin significantly increases E4BP4 expression and binding to the Fgf21 promoter through AKT activation. Thus, E4BP4 is a novel insulin-responsive repressor of FGF21 expression during circadian cycles and feeding.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Expression, Insulin, Liver, Metabolism, Transcription Repressor, E4BP4, FGF21, Feeding, Circadian Rhythm
Introduction
The mammalian circadian rhythm system plays a fundamental role in coordinating various physiological processes, which are manifested by a precise 24-h cycle and responsiveness to light or food cues (1–4). Recent genetic and biochemical studies of mammals, Drosophila, and bacteria have provided a general model of the circadian clock that is based on a transcriptional-translational feedback loop consisting of both positive and negative circadian clock proteins (1, 4). Besides controlling the core circadian oscillation loop, the clock proteins also actively participate in rhythmic expression of various output genes, which may account for the rhythmic activities in peripheral tissues (1, 3). As demonstrated in various microarray studies, genes important for gluconeogenesis, lipogenesis, and cholesterol synthesis are potential targets of clock proteins (5–8). Therefore, for drug administration and drug design, it becomes critical to understand how the cycling of individual metabolic genes is regulated in a 24-h rhythm (9, 10).
E4BP4 (E4-binding protein 4), also called NFIL3, is a b-ZIP (basic leucine zipper) transcription factor initially identified as an IL-3-inducible factor in pro-B lymphocytes (11–13). The biological function of E4BP4 has been largely explored in the immune system, in which E4BP4 knock-out mice are defective in natural killer cell development and IgE class switch (14–16). E4BP4 was first identified as a clock-controlled gene in mouse liver (17, 18). Its mRNA and protein levels oscillate in a circadian fashion, which is anti-phase to DBP (D-site of albumin promoter-binding protein), another clock-controlled output gene (19). The mRNA of E4BP4 peaks at circadian time (CT)2 0 and troughs at CT 12 (17, 20). Although the role of E4BP4 in the mammalian circadian system is unclear, its homologue in Drosophila, vrille, serves as a key component of the core circadian network via a negative feedback loop (21–23). E4BP4 binds a D-box cis-element of the promoter region of target genes to repress transcription (24–27). So far, only a few circadian targets, including Cyp7A, Mdr1, and PPARγ (peroxisome proliferator-activated receptor γ), have been identified as targets of E4BP4 (28–30). Nevertheless, the circadian function of E4BP4 in the liver metabolism remains unknown.
FGF21 (fibroblast growth factor 21) belongs to a superfamily of FGF peptides without mitogenic effects (31–34). FGF21 induction is critical to lipolysis, gluconeogenesis, and ketogenesis during the prolonged adaptive response to fasting (35, 36). It has been observed that chronic injection of FGF21 improves insulin sensitivity, lowers blood glucose and lipid levels, and stimulates glucose uptake in adipose tissue in several animal models (36, 37). However, in human studies, FGF21 level is also increased in patients with obesity and diabetes, raising a possibility of FGF21 resistance in those patients (38–40). Nuclear receptor PPARα is the major transcription activator for FGF21 induction during fasting (35, 36, 41). A recent report suggests that thyroid hormone T3 induces FGF21 expression through thyroid hormone receptor β, retinoid X receptor, and PPARα (42). Two major components of the circadian network, RORα (retinoic acid receptor-related receptor α) and Rev-erbα, have also been found to regulate FGF21 expression (43, 44). Given that FGF21 expression is sensitive to the feeding state (45–47), we speculate that the FGF21 level oscillates during a circadian cycle, which is intimately coupled with a feeding and fasting cycle.
In this report, we present evidence that Fgf21 is a direct target of E4BP4. Manipulation of E4BP4 level inversely changes the expression and secretion of FGF21 in the mouse liver cell line and primary mouse hepatocytes. Knockdown of E4BP4 alters the circadian oscillation of Fgf21 mRNA in synchronized hepatoma cells. On the physiological level, the E4BP4-mediated repression of FGF21 is downstream of the insulin-induced AKT signaling and correlated to the fasting-feeding cycling.
EXPERIMENTAL PROCEDURES
Plasmids and Adenovirus Generation
The E4BP4 expression construct was made by cloning the full-length E4BP4 cDNA into pQCXIP (Clontech). The E4BP4 shRNA knockdown construct was made by ligating the targeting oligonucleotide sequence into the RNAi-Ready pSIREN-Retro-Q vector (Clontech). The targeting sequence for mouse E4bp4 is 5′-ACGUAUUCCACCUCCAUCU-3′. The FGF21 promoter-driven luciferase reporter construct was a generous gift from Dr. Kliewer (University of Texas Southwestern Medical Center). The QuikChange site-directed mutagenesis kit (Agilent-Stratagene) was used to introduce targeted deletions to the Fgf21 promoter luciferase construct. pAdEasy FLAG-E4BP4 virus was made by using the Agilent AdEasy adenoviral vector system according to the user's manual. The constitutive active AKT expression vector was kindly provided by Dr. Liangyou Rui (University of Michigan).
Animal Experiments
Wild-type C57BL6 mice aged 6–10 weeks were maintained on 12 h/12 h light/dark with free access to food and water for 2 weeks before being transferred to the dark/dark condition for an additional 48 h. Mice (n = 3) were sacrificed every 4 h for 2 circadian cycles. The liver tissues were harvested and prepared for mRNA analysis. For the fasting and feeding study described in Fig. 3, wild-type C57BL6 mice aged 6–8 weeks (n = 5/group) were either fed ad libitum or restricted from food for 24 h (from 5 p.m. to 5 p.m.). After sacrifice, liver tissues were collected for both mRNA and protein analysis. All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee at the University of Pennsylvania.
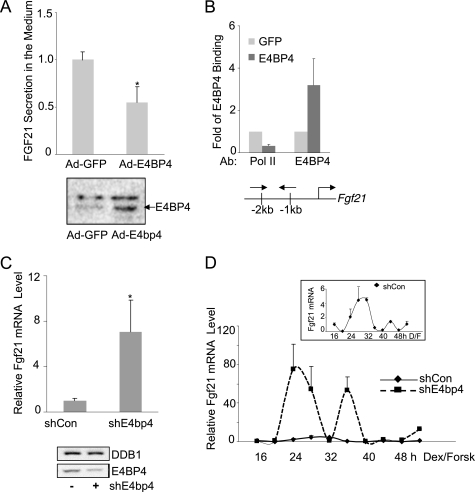
FIGURE 3.
E4BP4 directly represses Fgf21 and is required for its circadian oscillation in mouse liver cells. A, overexpression of E4BP4 suppresses the secretion of FGF21 in culture medium of primary mouse hepatocytes. Cells were infected with either Ad-GFP or Ad-E4BP4 for 48 h and the culture medium was then collected for ELISA analysis of FGF21. Results are expressed as mean ± S.D. (error bars) of triplicates. A representative result of three individual experiments is shown. The levels of total E4BP4 protein in primary hepatocytes transduced with either Ad-GFP or Ad-E4BP4 were detected by Western blot. B, chromatin immunoprecipitation was performed to detect E4BP4 binding to the mouse Fgf21 promoter in Hepa1c1c-7 cells transfected with either GFP or E4BP4 expression plasmid. The recruitment of RNA polymerase II was examined as well. Results are expressed as mean ± S.D. of three experiments. The locations of ChIP primers are shown as well. C, Hepa1c1c-7 cells were transfected with the control or E4bp4 shRNA for 72 h. Cells were synchronized by 50% horse serum for 2 h and incubated in serum-free medium for another 16 h. Cells then were harvested for mRNA analysis of the Fgf21 gene by Q-PCR. Results are expressed as mean ± S.D. of at least three independent experiments. *, p < 0.05. Knockdown efficiency of E4BP4 protein knockdown is shown below. D, knockdown of E4BP4 alters the circadian oscillation of mouse Fgf21 in synchronized Hepa1c1c-7 cells. After synchronization treatment with both dexamethasone (100 nm) and forskolin (10 μm), the Hepa1c1c-7 cells stably expressing either control shRNA or E4bp4 shRNA were collected for mRNA analysis at the indicated time points. The value given for the amount of mRNA present at the 16 h time point was set as 1. Error bars, ±range (n = 2). Inset, circadian oscillation of Fgf21 in the shRNA control group.
Cell Cultures and Transfection
Hepa1c1c-7 cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. Cells were plated in a 24-well plate overnight before transfection with the Fgf21 promoter luciferase reporter alongside either GFP control or various circadian clock gene expression constructs using Lipofectamine 2000 (Invitrogen). 48 h post-transfection, cells were lysed for a luciferase activity assay on BioTek Synergy 2. The β-galactosidase construct was also cotransfected in each well for normalizing luciferase activity. Insulin was purchased from Invitrogen. LY294002 was purchased from Promega.
Generation of Stable Cell Lines
A Hepa1c1c-7-derived stable cell clone expressing E4bp4 shRNA was generated for the circadian study. Hepa1c1c-7 cells were transiently transfected with pSiren-shNC (control vector) or pSiren-shE4 (E4bp4 shRNA) vector and subsequently were selected in the medium containing puromycin (1.5 μg/ml) for 3 weeks. The positive clones were confirmed by both RT-PCR and immunoblotting for a decreased level of E4BP4 mRNA and protein.
Primary Mouse Hepatocyte Isolation and Culture
Hepatocytes were isolated from male mice (9–10 weeks in C57BL/6 background). The liver was perfused with 0.5 mg/ml type II collagenase (Worthington) via the inferior vena cava to isolate hepatocytes. Cells were seeded for 2 h on collagen-coated 6-well plates in Williams Medium E (catalogue number W4125, Sigma) supplemented with 2% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (5 × 105 cells/well).
cDNA Synthesis and Q-PCR
Total cellular RNA extraction was performed with TRIzol reagent (Invitrogen) and chloroform. cDNA was synthesized with the Verso cDNA kit (Thermo Fisher Scientific, Surrey, UK) and subjected to Q-PCR using Absolute Blue SYBR Green ROX Mix (Thermo Fisher Scientific) on an ABI 7900 HT thermal cycler (Applied Biosystems, Foster City, CA). The value of each cDNA was calculated using the ΔCt method and normalized to the value of a housekeeping gene control, arbp. The data were plotted as -fold change. The primer sequences used in this study are as follows: mFgf21, 5′-gctgctggaggacggttaca-3′ (forward) and 5′-cacaggtccccaggatgttg-3′ (reverse); mArbp, 5′- ccgatctgcagacacacact-3′ (forward) and 5′-accctgaagtgctcgacatc-3′ (reverse); mE4bp4, 5′- cggagcttgaatcgcgcccc (forward) and 5′-gggttatcgtggttctgctccctg (reverse).
ELISA
Primary mouse hepatocyte cells were treated with either Ad-GFP or Ad-E4BP4 viral stock overnight before switching to serum-free medium. 72 h later, the cell culture medium was used for measuring FGF21 secretion by the ELISA kit (MF2100, R&D Systems) according to the manufacturer's instructions.
Immunoblotting
48 h post-transfection, Hepa1c1c-7 cells were washed once in 1× PBS buffer and lysed in lysis buffer supplemented with 1× protease inhibitor (Roche Applied Science). After whole cell lysates were precleared at maximal speed at 4 °C in a microcentrifuge, the protein concentration of each supernatant was measured by a Bio-Rad protein assay. Equal amounts of protein samples were separated in 10% SDS-polyacrylamide gels and transferred to PVDF membrane (Millipore). The membranes were incubated in antibodies against FLAG (Sigma), E4BP4 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), AKT (Cell Signaling), phosphorylated AKT (Cell Signaling), GSKβ (Santa Cruz Biotechnology, Inc.), phosphorylated GSKβ (Cell Signaling), DDB1 (Abcam), and RAN (BD Biosciences). HRP-conjugated secondary antibody against mouse IgG or rabbit IgG and Western Lightning ECL substrate (PerkinElmer Life Sciences) were used to detect chemiluminescence signals on a Fluorchem HD2 AlphaImager (Cell Biosciences).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay in culture cells was performed as described previously (48). Hepa1c1c-7 cells were transfected with either GFP or E4bp4 expression vector and harvested 72 h later for the ChIP assay. The liver ChIP assay was performed according the protocol described previously (49, 50). The frozen liver tissues were minced in the presence of liquid nitrogen and cross-linked in 1% formaldehyde-PBS buffer. The resulted nuclei pellets were then sonicated to generate soluble chromatin materials with sizes between 1 and 2 kb. The chromatin materials were immunoprecipitated with the following antibodies at 4 °C overnight: anti-E4BP4, RNA polymerase II (Santa Cruz Biotechnology, Inc.), and anti-acetyl H4 (Abcam). The resulting DNA fragments were purified and subjected to PCR analysis using primers encompassing the D-box region of the mouse Fgf21 promoter. The sequences of the PCR primers are as follows: mFgf21–5UTR, 5′-agtccttgctcagggttcct-3′ (forward) and 5′-acaggtgctctccagatgct-3′ (reverse); mFgf21-TSS, 5′-tggtatttctgcgttcacca-3′ (forward) and 5′-atgggtcaggttcagactgg-3′ (reverse); 18 S RNA, 5′-ttgacggaagggcaccaccag-3′ (forward) and 5′-gcaccaccacccacggaatgg-3′ (reverse).
Statistical Analysis
All of the luciferase assays were done in triplicates. Luciferase readings were normalized by the β-galactosidase activity for each well. Three individual experiments were performed for each treatment in FGF21 ELISA. The results are indicated as the mean ± S.E. or mean ± S.D. To compare two groups, p < 0.05 was considered statistically significant for Student's t test.
RESULTS
Fgf21 Is a Circadian-regulated Gene
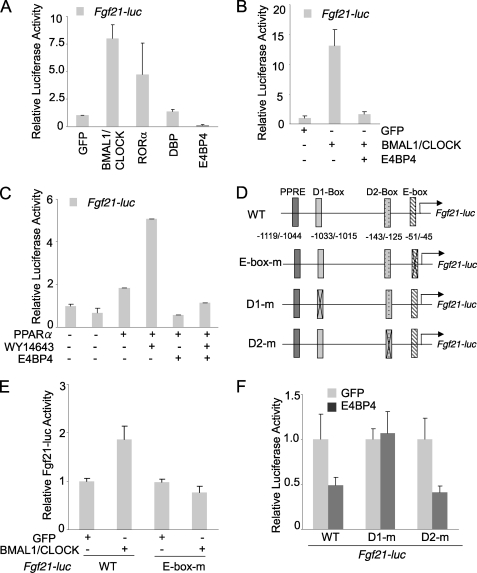
The genome-wide expression analysis reveals about 5–10% of liver transcripts oscillate in a circadian fashion, suggesting that circadian clock proteins directly regulate the expression of those circadian output genes (7, 8, 18, 51). One of the key features of circadian output genes is the presence of one or more of the three classical cis-elements, including E-box, ROR-response element, and D-box sites (25, 26, 52). To determine whether the critical regulator of liver metabolism, FGF21, is under the direct control of the circadian network, we used Genomatrix software to examine the mouse Fgf21 promoter and indeed located a canonical E-box (−51 to −45) and two putative D-box sites (−1033 to −1015 and −143 to −125) (Fig. 1B). To test whether the circadian proteins directly regulate the Fgf21 promoter, we co-transfected a mouse hepatoma cell line, Hepa1c1c-7, with both the Fgf21 promoter-driven luciferase reporter construct and a panel of clock protein expression vectors. Our results (Fig. 1A) clearly show that both the BMAL1-CLOCK complex and RORα activated the Fgf21 promoter, whereas E4BP4 potently repressed it. Our results are consistent with a recent report showing that RORα regulates the FGF21 expression and secretion (44). Additionally, we tested the cross-talk between E4BP4 and other circadian activators of the Fgf21 promoter. E4BP4 not only suppressed the BMAL1-CLOCK-mediated activation of Fgf21-luc (Fig. 1B) but also abolished the PPARα-induced activation of Fgf21-luc (Fig. 1C). To determine the function of the putative circadian binding elements, we generated both E-box and D-box deletion mutants of the Fgf21-luc construct (Fig. 1D). The E-box mutant Fgf21-luc was no longer responsive to BMAL1-CLOCK activation (Fig. 1E), whereas loss of the distal D-box element (−1033 to −1015) abolished the suppression of Fgf21 promoter by E4BP4 (Fig. 1F), strongly supporting our hypothesis that the mouse Fgf21 is indeed a circadian target gene.
FIGURE 1.
Mouse Fgf21 is a circadian output gene regulated by the circadian protein E4BP4. A, mouse Fgf21 promoter is differentially regulated by clock proteins. A luciferase vector driven by the mouse Fgf21 promoter was cotransfected with the expression vectors encoding various clock proteins in Hepa1c1c-7 cells. The relative luciferase activities were calculated based on the value of GFP group set as 1. Results are expressed as mean ± S.D. (error bars) (n = 3). B, E4BP4 overexpression blocks transcription activation of the Fgf21-luc by BMAL1-CLOCK. The mouse Fgf21 promoter-driven luciferase reporter was co-transfected with expression vectors of various clock proteins in Hepa1c1c-7 cells. The relative luciferase activities were calculated based on the value of GFP group set as 1. Results are expressed at mean ± S.D. (n = 3). C, E4BP4 abolishes the activation of the Fgf21-luc by PPARα ligand WY14643. 24 h after co-transfection with the mouse Fgf21-luc and expression vectors of E4BP4 and PPARα, Hepa1c1c-7 cells were treated with either DMSO or PPARα ligand WY14643 (10 μm) for 16 h. The relative luciferase activities are expressed as mean ± S.D. (n = 3). D, schematic diagram shows the putative binding sites (PPAR-response element (PPRE), E box, and D box) within the 1.5 kb Fgf21 promoter upstream of the transcription starting site. The E-box and D-box deletion mutants are also indicated. E, mouse Fgf21 promoter contains a functional E-box element. Deletion of the E-box binding site (−51 to −45) abolished the BMAL1-CLOCK-mediated activation of the Fgf21 promoter. F, a D-box-binding element is required for E4BP4-mediated repression of the Fgf21 promoter. The Fgf21-luc mutant D1m (−1033 to −1015 deleted) was not repressed by E4BP4 in the co-transfection assay. Both D2m mutant (−143 to −125 deleted) and wild-type Fgf21-luc respond to E4BP4 in similar manners.
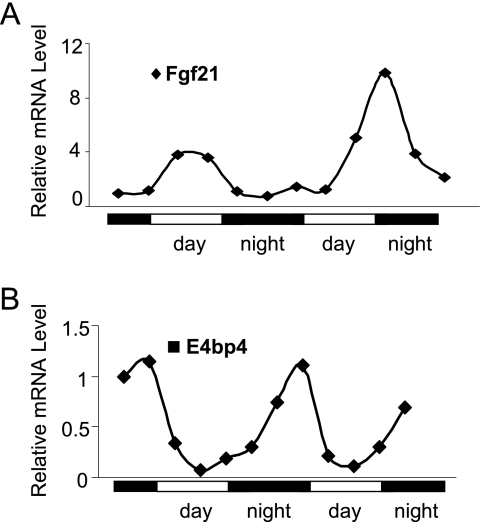
We are particularly interested in the repression of FGF21 by E4BP4 in the context of circadian regulation because such transcription repressors of FGF21 have not been reported. In the mouse liver samples harvested during a circadian cycle, the Fgf21 mRNA peaked around CT 6 in a 24-h rhythm (Fig. 2A). In contrast, the liver E4bp4 mRNA peaks at CT 0, completely opposite to that of Fgf21, suggesting that E4BP4 represses the Fgf21 gene expression during a circadian cycle.
FIGURE 2.
The circadian expression of Fgf21 and E4bp4 in the mouse liver. The liver mRNA levels of Fgf21 (A) and E4bp4 (B) are shown during two circadian cycles. The cDNA of each time point was derived from the pooled liver samples of three mice.
To elucidate the detailed mechanisms of how E4BP4 represses the Fgf21 promoter, we manipulated the E4BP4 function by using either an E4BP4 repression domain-truncated mutant (12) or E4BP4 shRNA. In both cases, repression of the Fgf21 promoter by E4BP4 is lost (data not shown). Taken together, our data demonstrate that E4BP4 is a repressor of Fgf21 promoter via a D-box binding element.
E4BP4 Represses FGF21 Transcription and Secretion
To test whether E4BP4 directly regulates Fgf21 expression, we transduced primary mouse hepatocytes with either Ad-GFP or Ad-E4BP4 and detected a significant decrease of the FGF21 level in the culture medium by ELISA (Fig. 3A). The expression of FLAG-E4BP4 was confirmed by Western blot in adenovirus-transduced primary mouse hepatocyte cells (Fig. 2A). To further test how E4BP4 regulates the endogenous Fgf21 gene expression in liver cells, we used a ChIP assay to confirm the E4BP4 occupancy on the Fgf21 promoter in cells transfected with either GFP or E4BP4 expression construct. An increase in E4BP4 binding to the D-box-containing Fgf21 promoter region was detected, whereas the binding of RNA polymerase II, a marker of active transcription, diminished drastically (Fig. 3B). Both results indicate that E4BP4, when expressed in abundance, can directly bind to the Fgf21 promoter and inhibit FGF21 expression and secretion.
E4BP4 Regulates Circadian Oscillation of FGF21
Next, we examined the effects of E4BP4 knockdown on the FGF21 circadian oscillation. The basal level of the endogenous FGF21 mRNA increased about 7-fold in the cells transiently transfected with E4bp4 shRNA versus control shRNA (Fig. 3C). We previously established a protocol to synchronize Hepa1c1c-7 cells, which display the rhythmic expression of the Dbp gene for two continuous cycles (53). To determine the extent of E4BP4 contribution to the circadian oscillation of Fgf21, we generated a Hepa1c1c-7 stable cell line expressing E4bp4 shRNA. Knockdown of the E4BP4 protein was confirmed by immunoblotting (data not shown). To validate the circadian samples, we first examined the Dbp oscillation in Hepa1c1c-7 stable cells (shCon versus shE4bp4). The Dbp mRNA oscillation was impaired in shE4bp4-expressing cells, suggesting that a chronic depletion of E4bp4 interferes with the clock function (data not shown). In contrast, E4bp4 knockdown drastically increased the amplitude of the Fgf21 mRNA oscillation without affecting its phase (Fig. 3D), consistent with the notion that E4BP4 functions as a circadian repressor of FGF21 in liver cells. Based on these results, we conclude that E4BP4 controls the circadian oscillation of Fgf21 mRNA.
Feeding Suppresses FGF21 Expression through Up-regulating E4BP4
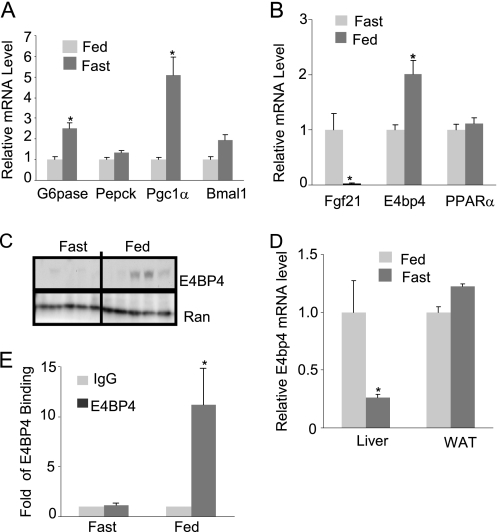
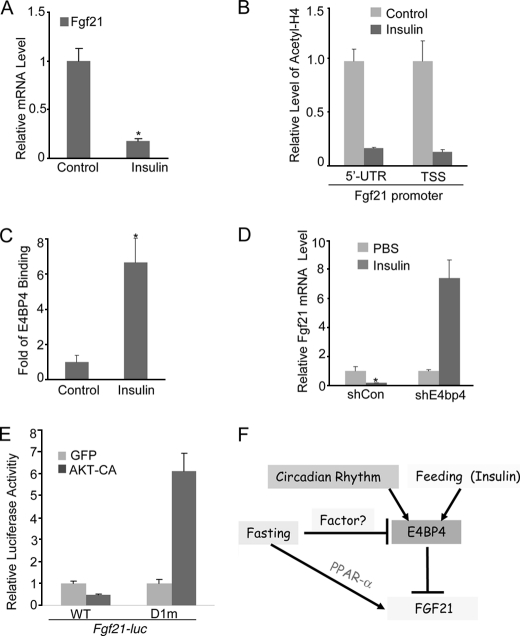
Circadian rhythms control the daily cycle of feeding and fasting (2–4, 47). The Fgf21 mRNA is significantly induced by fasting but potently repressed by feeding (35, 36). Although Fgf21 induction has been attributed to the PPARα-mediated activation in the adaptive response during fasting (35, 36, 54), the underlying mechanism for feeding-induced Fgf21 repression is unknown. To determine whether E4BP4 also plays a role in feeding-induced Fgf21 repression, we measured the E4bp4 mRNA and protein levels in the mouse liver tissues after fasting and ad libitum feeding for 24 h. Fasting, but not feeding, induced the expression of G6pase, Pepck, and Pgc1α in the mouse livers (Fig. 4A). In contrast, the E4BP4 mRNA and protein levels were increased upon feeding but decreased in the fasting condition (Fig. 4, B and C). It is noteworthy that only E4bp4 expression in the liver rather than adipose tissues is affected by feeding (Fig. 4D), indicating an existence of a tissue-specific regulation for E4bp4 expression. To examine whether E4BP4 is involved in the Fgf21 repression during the feeding state, we performed a ChIP assay to detect the occupancy of E4BP4 binding on the Fgf21 promoter. In liver tissues from fasted mice, there was no E4BP4 binding on the Fgf21 promoter. However, a strong signal of E4BP4 binding was detected on the Fgf21 promoter in the fed mouse liver tissues (Fig. 4E). Combined with in vitro evidence, these results allow us to conclude that E4BP4 is likely to be the transcription repressor mediating the feeding-induced suppression of Fgf21.
FIGURE 4.
E4BP4 mediates feeding-induced FGF21 gene suppression. A, the mRNA levels of gluconeogenic genes, including G6pase, Pepck, and Pgc1α, in the fasted or fed liver tissues isolated from wild-type C57BL6 mice (n = 5). Data are presented as mean ± S.E. (error bars). *, p < 0.05. B, Q-PCR analysis for the mRNA level of E4bp4 in the same set of liver tissues (n = 5). The expression levels of Fgf21 and PPARα were measured as well. Data are presented as mean ± S.E. *, p < 0.05. C, immunoblotting analysis for the liver E4BP4 protein level from the same tissue samples. D, Q-PCR analysis of the E4bp4 mRNA levels in both the liver and adipose tissues isolated from fasted or fed wild-type C57BL6 mice (n = 5). Data are presented as mean ± S.E. *, p < 0.05. E, ChIP assay for the occupancy of E4BP4 on the Fgf21 promoter in both fasting and feeding conditions. Liver tissues (n = 3) harvested after fasting or feeding for 24 h were used as input materials. Data are presented as mean ± S.E. *, p < 0.05.
E4BP4 Mediates Suppression of FGF21 by Insulin
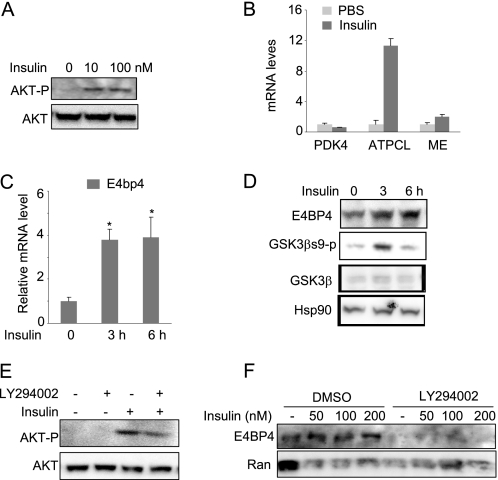
Upon food intake, the insulin level increases in response to glucose surge. Although the insulin action on the liver and adipose tissue has been well documented in SREBP-mediated lipogenesis (55–57), its inhibition on lipolysis is largely unexplored. Because the E4bp4 mRNA level is higher during feeding, we speculate that insulin regulates its transcription. To test this possibility, we treated serum-starved Hepa1c1c-7 cells with insulin and then analyzed the E4PB4 expression at both the mRNA and protein levels. Hepa1c1c-7 cells respond to a wide range of insulin dosage with an increase in AKT Ser473 phosphorylation, ATP citrate lyase expression (58) and a down-regulation of PDK4 (59) (Fig. 5, A and B). Indeed, we found that a 200 nm insulin treatment for 3 h was sufficient to induce E4BP4 at both mRNA and protein levels in Hepa1c1c-7 cells and primary mouse hepatocytes (Fig. 5, C and D). Because insulin activates multiple signaling pathways, including PI3K-AKT, ERK, and PKC (60–62), we tested whether the PI3K-AKT pathway is involved in E4BP4 induction by insulin. Specifically, we used a PI3K inhibitor, LY294002, to block insulin-induced AKT activation (confirmed by the reduced level of phosphorylated AKT in Fig. 5E). In this condition, we observed that E4BP4 induction by insulin is completely abrogated when the PI3K-AKT pathway is inhibited (Fig. 5F). In contrast with the insulin induction of E4bp4, the same dose of insulin caused a significant down-regulation of Fgf21 expression in Hepa1 cells after overnight treatment (Fig. 6A). The insulin effect on Fgf21 expression is mediated through a transcriptional event because insulin treatment resulted in a significant decrease in the levels of acetyl-H4 binding around the 5′-untranslated region (UTR) as well as the transcription start site (TSS) region of the Fgf21 promoter (Fig. 6B). To examine whether the Fgf21 down-regulation by insulin treatment is mediated through E4BP4, we performed ChIP analysis on the Fgf21 promoter following a 12-h insulin treatment. The result shows that insulin stimulated the E4BP4 recruitment onto the Fgf21 promoter (Fig. 6C). To ultimately test whether E4BP4 is required for this regulation, we examined the Fgf21 expression in insulin-treated cells after E4BP4 depletion. E4BP4 knockdown completely abolished insulin suppression of the Fgf21 expression (Fig. 6D), indicating that E4BP4 is the major mediator of insulin action on Fgf21 regulation. Moreover, we addressed the potential role of AKT activation in the insulin-induced repression of the Fgf21 promoter activity. Overexpression of a constitutively active form of AKT (AKT-CA in Fig. 6) was sufficient to repress the Fgf21 promoter activity in Hepa1C1C-7 cells. On the other hand, constitutively active AKT failed to repress the distal D-box-deleted Fgf21 promoter mutant (D1m), indicating that such effects are likely to be mediated through E4BP4 (Fig. 6E).
FIGURE 5.
Insulin up-regulates the expression of E4BP4 through AKT activation in liver cells. A, insulin-induced AKT phosphorylation in Hepa1c1c-7 cells. Both total and phosphorylated AKT proteins were detected by immunoblotting. B, Q-PCR analysis of the mRNA levels of Pdk4, ATP citrate lyase, and malic enzyme (ME) in the insulin-treated Hepa1C1C-7 cells. C, insulin up-regulates the gene expression of E4bp4 in a time-dependent manner in Hepa1c1c-7 cells. Confluent Hepa1c1c-7 cells were treated with insulin at 200 nm for 0, 3, and 6 h before harvest. The mRNA level of E4bp4 was determined by Q-PCR. Results are expressed as mean ± S.D. (error bars) (n = 3). *, p < 0.05. D, insulin up-regulates the E4BP4 protein levels in freshly isolated primary mouse hepatocytes. Cells were treated with insulin at 200 nm for 0, 3, and 6 h. The protein levels of E4BP4, GSK3β, GSK3βs9p, and loading control Ran were detected by specific antibodies. E, pretreatment with PI3K inhibitor LY294002 (50 μm) blocks AKT phosphorylation by insulin in Hepa1c1c-7 cells. The control was treated with DMSO. F, AKT inhibition blocks the insulin-induced E4BP4 protein expression in Hepa1C1C-7 cells. Cells were treated with either DMSO or LY294002 for 2 h before insulin treatment at various concentrations for 12 h. An immunoblot was used for detecting E4BP4 protein levels.
FIGURE 6.
Insulin suppresses Fgf21 via the induction of E4BP4 in liver cells. A, insulin suppresses the Fgf21 expression in Hepa1c1c-7 cells. Confluent Hepa1c1c-7 cells were first synchronized by 50% horse serum for 2 h and incubated at serum-free medium for another 6 h. Cells were then treated with 200 nm insulin for 12 h before Fgf21 Q-PCR. Results are expressed as mean ± S.D. (error bars) (n = 3). *, p < 0.05. B, insulin treatment increases the levels of acetylated H4 around the Fgf21 promoter regions. Confluent Hepa1c1c-7 cells were treated as in A before ChIP analysis with specific antibodies. Results are expressed as mean ± S.D. (n = 3). C, insulin treatment stimulates the recruitment of E4BP4 onto the Fgf21 promoter. Confluent Hepa1c1c-7 cells were treated as in A before a ChIP assay with anti-E4BP4 antibody. D, E4BP4 is required for the insulin-induced suppression of Fgf21 in liver cells. After synchronization by 50% horse serum for 2 h and incubation in serum-free medium for another 6 h, Hepa1c1c-7 cells stably expressing either shRNA or E4bp4 shRNA were then treated with insulin at 200 nm for 12 h before Fgf21 mRNA analysis. Results are expressed at mean ± S.D. (n = 3). *, p < 0.05. E, AKT activation represses the luciferase activity driven by the wild type Fgf21 promoter but not by the E4BP4 binding site-deleted mutant. A constitutively active AKT was co-transfected into Hepa1c1c-7 cells along with either WT Fgf21-luc or D1m Fgf21-luc. The relative luciferase activities 48 h post-transfection are presented as mean ± S.D. (n = 3). F, model for the regulation of hepatic FGF21 expression by E4BP4. Both insulin and feeding induce E4bp4 to regulate the Fgf21 circadian oscillation upon food intake. E4BP4 directly binds to a distal D-box element of the Fgf21 promoter to suppress its expression. As a first order circadian output gene, E4BP4 links the circadian rhythm to liver metabolism.
DISCUSSION
In this report, we identified a novel regulatory mechanism for the circadian protein E4BP4 to control the Fgf21 expression during a circadian cycle and upon food intake (Fig. 6F). As a transcription repressor, E4BP4 mediates the repressive state of Fgf21 expression during the circadian night as well as during the feeding phase. Our study has identified a functional D-box element in the Fgf21 promoter region. E4BP4 directly binds to this site within the promoter region of the Fgf21 gene and suppresses its transcription. Depletion of E4BP4 derepresses Fgf21 expression and shifts its oscillation pattern in the synchronized mouse hepatoma cells. Overexpression of E4BP4 suppresses secretion of FGF21 in primary mouse hepatocytes. Mimicking the feeding effect, insulin stimulates the E4BP4 expression and increases the E4BP4 binding to the D-box-containing Fgf21 promoter. More importantly, we found that E4BP4 mediates the insulin-induced down-regulation of Fgf21 because insulin fails to suppress Fgf21 expression in cells depleted of E4BP4. In summary, we uncovered a transcriptional pathway linking a critical metabolic regulator with the circadian clock through the circadian-controlled transcription repressor, E4BP4.
As a circadian output gene, the E4BP4 mRNA was found to be robustly cycling in the liver and suprachiasmatic nucleus but out of phase with DBP (17). The mammalian E4BP4 has been implicated in the circadian regulation of a number of circadian output genes, including Per2 (63), Cyp7A (28), and Cyp3A4 (64), suggesting that E4BP4 functions as a mediator of the circadian clock to control its output gene expression. In this study, we discovered a novel link between the circadian protein E4BP4-dependent transcription repression and the circadian oscillation of a key metabolic regulator in liver metabolism. We identified Fgf21 as another E4BP4-regulated circadian output gene and provided new insights into the circadian function of E4BP4 in energy homeostasis. Because FGF21 displays diverse effects on lipolysis, gluconeogenesis, and mitochondrial biogenesis in the liver (32–34), it is very likely that the E4BP4-FGF21 regulatory axis could influence the diurnal fluctuation of these metabolic pathways. The recently developed E4BP4 knock-out mice (14–16) will be a very useful model to address the circadian biology and metabolic relevance of E4BP4, in particular on FGF21 regulation.
Along with the work of others (43, 44), we showed that the Fgf21 promoter contains three circadian-responsive elements, including an E-box, a ROR-response element site, and a D-box, all of which are hallmarks of a classical circadian-regulated gene (25, 26). An E-box mutant of Fgf21-luc completely abolished the BMAL1-CLOCK activation of the promoter activity. It will be of great interest to investigate how the circadian pattern of Fgf21 is altered in the circadian clock mutant mice, such as Bmal1 null mice (65). Several recent human studies show that PPARα-dependent transcription activation could not fully explain the FGF21 dysregulation in metabolic diseases, such as obesity (40, 66), non-alcoholic fatty liver disease (66, 67), and diabetes (68, 69). It is notable that none of the previously mentioned studies has examined the diurnal rhythm of the circulating FGF21, which is critical to the temporal changes of FGF21 in diseased conditions. Liver circadian clocks are usually perturbed in the condition of obesity and diabetes (20, 70–72). Although the role of FGF21 in metabolism is still being clarified (35–37, 73), it is conceivable that dysregulation of FGF21 by clock proteins contributes to the pathogenesis of diabetes and obesity.
Herein, we first reported that both E4bp4 mRNA and protein could be induced by insulin in a mouse hepatoma cell line and primary mouse hepatocytes. At this stage, it remains unclear how insulin induces the E4BP4 expression. Both SREBP and ChREBP are the two major transcription factors mediating transcription activation of lipogenesis upon insulin treatment (55, 56, 74, 75). As a circadian output protein, the E4bp4 promoter contains a canonical binding site for nuclear receptor RORα and Rev-erbα as well as an E-box for BMAL1-CLOCK (25, 28). E4bp4 circadian expression is altered in Rev-erbα null mice and Clock mutant mice. No report about the E4bp4 expression in RORα null mice has been published yet. Future work is needed to pin down the transcription factors involved in the insulin-induced expression of E4BP4. Besides transcriptional regulation, phosphorylation of the E4BP4 protein by casein kinase 1ϵ was also reported (76). Therefore, it is also possible that the insulin-signaling cascade leads to a change of the phosphorylation status of E4BP4, which in turn enhances its repression activity on the target genes.
We show in this study that E4BP4 is important for maintaining a repressive state of the Fgf21 gene during feeding and in response to insulin in hepatocytes. Several studies have presented data showing that the levels of E4BP4 and its circadian counterpart DBP are correlated to body mass index in humans, and their levels change in high fat diet-fed animals (77, 78). Interestingly, Fgf21 also shows dysregulation in those conditions. A future study will test whether dysregulation of E4BP4 accounts for abnormal levels of Fgf21 in circadian knock-out mice or high fat diet-induced obese mice.
Acknowledgments
We thank Dr. John Hogenesch (University of Pennsylvania) for providing the liver circadian samples and for critical reading of the manuscript. We thank Dr. Bishr Omar (University of Michigan) for insightful comments on the manuscript. We also thank Dr. Steven Kliewer (University of Texas Southwestern) for providing the Fgf21-luciferase vector.
This work was supported, in whole or in part, by National Institutes of Health Grants K99/R00 DK 077449 (to L. Y.) and R01 DK45586 (to M. A. L.).
- CT
- circadian time
- Q-PCR
- quantitative PCR
- UTR
- untranslated region.
REFERENCES
- 1.Dibner C., Schibler U., Albrecht U. (2010) Annu. Rev. Physiol. 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 2.Green C. B., Takahashi J. S., Bass J. (2008) Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schibler U. (2009) J. Biol. Rhythms 24, 3–15 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) Nat. Rev. Genet. 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceriani M. F., Hogenesch J. B., Yanovsky M., Panda S., Straume M., Kay S. A. (2002) J. Neurosci. 22, 9305–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., Panda S., Hogenesch J. B. (2009) PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 8.Panda S., Hogenesch J. B. (2004) J. Biol. Rhythms 19, 374–387 [DOI] [PubMed] [Google Scholar]
- 9.Innominato P. F., Lévi F. A., Bjarnason G. A. (2010) Adv. Drug Deliv. Rev. 62, 979–1001 [DOI] [PubMed] [Google Scholar]
- 10.Paschos G. K., Baggs J. E., Hogenesch J. B., FitzGerald G. A. (2010) Annu. Rev. Pharmacol. Toxicol. 50, 187–214 [DOI] [PubMed] [Google Scholar]
- 11.Cowell I. G. (2002) BioEssays 24, 1023–1029 [DOI] [PubMed] [Google Scholar]
- 12.Cowell I. G., Hurst H. C. (1994) Nucleic Acids Res. 22, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowell I. G., Hurst H. C. (1996) Nucleic Acids Res. 24, 3607–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gascoyne D. M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H. J. (2009) Nat. Immunol. 10, 1118–1124 [DOI] [PubMed] [Google Scholar]
- 15.Kamizono S., Duncan G. S., Seidel M. G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E. F., Haight J. P., Ohashi P. S., Look A. T., Mak T. W. (2009) J. Exp. Med. 206, 2977–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashiwada M., Levy D. M., McKeag L., Murray K., Schröder A. J., Canfield S. M., Traver G., Rothman P. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsui S., Yamaguchi S., Matsuo T., Ishida Y., Okamura H. (2001) Genes Dev. 15, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oishi K., Amagai N., Shirai H., Kadota K., Ohkura N., Ishida N. (2005) DNA Res. 12, 191–202 [DOI] [PubMed] [Google Scholar]
- 19.Ripperger J. A., Schibler U. (2006) Nat. Genet. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 20.Zvonic S., Ptitsyn A. A., Conrad S. A., Scott L. K., Floyd Z. E., Kilroy G., Wu X., Goh B. C., Mynatt R. L., Gimble J. M. (2006) Diabetes 55, 962–970 [DOI] [PubMed] [Google Scholar]
- 21.Cyran S. A., Buchsbaum A. M., Reddy K. L., Lin M. C., Glossop N. R., Hardin P. E., Young M. W., Storti R. V., Blau J. (2003) Cell 112, 329–341 [DOI] [PubMed] [Google Scholar]
- 22.Glossop N. R., Houl J. H., Zheng H., Ng F. S., Dudek S. M., Hardin P. E. (2003) Neuron 37, 249–261 [DOI] [PubMed] [Google Scholar]
- 23.Hardin P. E., Krishnan B., Houl J. H., Zheng H., Ng F. S., Dryer S. E., Glossop N. R. (2003) Novartis Found. Symp. 253, 140–150; discussion 150–160 [PubMed] [Google Scholar]
- 24.Kotaka M., Onishi Y., Ohno T., Akaike T., Ishida N. (2008) Neurosci. Res. 60, 307–313 [DOI] [PubMed] [Google Scholar]
- 25.Kumaki Y., Ukai-Tadenuma M., Uno K. D., Nishio J., Masumoto K. H., Nagano M., Komori T., Shigeyoshi Y., Hogenesch J. B., Ueda H. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda H. R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S. (2005) Nat. Genet. 37, 187–192 [DOI] [PubMed] [Google Scholar]
- 27.Yang F., Nakajima Y., Kumagai M., Ohmiya Y., Ikeda M. (2009) Biochem. Biophys. Res. Commun. 380, 22–27 [DOI] [PubMed] [Google Scholar]
- 28.Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Baugé E., Havinga R., Bloks V. W., Wolters H., van der Sluijs F. H., Vennström B., Kuipers F., Staels B. (2008) Gastroenterology 135, 689–698 [DOI] [PubMed] [Google Scholar]
- 29.Murakami Y., Higashi Y., Matsunaga N., Koyanagi S., Ohdo S. (2008) Gastroenterology 135, 1636–1644 [DOI] [PubMed] [Google Scholar]
- 30.Takahashi S., Inoue I., Nakajima Y., Seo M., Nakano T., Yang F., Kumagai M., Komoda T., Awata T., Ikeda M., Katayama S. (2010) J. Atheroscler. Thromb. 17, 73–83 [DOI] [PubMed] [Google Scholar]
- 31.Cuevas-Ramos D., Almeda-Valdes P., Aguilar-Salinas C. A., Cuevas-Ramos G., Cuevas-Sosa A. A., Gomez-Perez F. J. (2009) Curr. Diabetes Rev. 5, 216–220 [DOI] [PubMed] [Google Scholar]
- 32.Kharitonenkov A. (2009) Curr. Opin Pharmacol. 9, 805–810 [DOI] [PubMed] [Google Scholar]
- 33.Kharitonenkov A., Shanafelt A. B. (2009) Curr. Opin. Investig. Drugs 10, 359–364 [PubMed] [Google Scholar]
- 34.Kliewer S. A., Mangelsdorf D. J. (2010) Am. J. Clin. Nutr. 91, 254S-257S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badman M. K., Pissios P., Kennedy A. R., Koukos G., Flier J. S., Maratos-Flier E. (2007) Cell Metab. 5, 426–437 [DOI] [PubMed] [Google Scholar]
- 36.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., Elmquist J. K., Gerard R. D., Burgess S. C., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. (2007) Cell Metab. 5, 415–425 [DOI] [PubMed] [Google Scholar]
- 37.Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., Gromada J., Brozinick J. T., Hawkins E. D., Wroblewski V. J., Li D. S., Mehrbod F., Jaskunas S. R., Shanafelt A. B. (2005) J. Clin. Invest. 115, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavez A. O., Molina-Carrion M., Abdul-Ghani M. A., Folli F., Defronzo R. A., Tripathy D. (2009) Diabetes Care 32, 1542–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dushay J., Chui P. C., Gopalakrishnan G. S., Varela-Rey M., Crawley M., Fisher F. M., Badman M. K., Martinez-Chantar M. L., Maratos-Flier E. (2010) Gastroenterology 139, 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Yeung D. C., Karpisek M., Stejskal D., Zhou Z. G., Liu F., Wong R. L., Chow W. S., Tso A. W., Lam K. S., Xu A. (2008) Diabetes 57, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 41.Lundåsen T., Hunt M. C., Nilsson L. M., Sanyal S., Angelin B., Alexson S. E., Rudling M. (2007) Biochem. Biophys. Res. Commun. 360, 437–440 [DOI] [PubMed] [Google Scholar]
- 42.Adams A. C., Astapova I., Fisher F. M., Badman M. K., Kurgansky K. E., Flier J. S., Hollenberg A. N., Maratos-Flier E. (2010) J. Biol. Chem. 285, 14078–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estall J. L., Ruas J. L., Choi C. S., Laznik D., Badman M., Maratos-Flier E., Shulman G. I., Spiegelman B. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22510–22515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Solt L. A., Burris T. P. (2010) J. Biol. Chem. 285, 15668–15673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Challet E., Caldelas I., Graff C., Pévet P. (2003) Biol. Chem. 384, 711–719 [DOI] [PubMed] [Google Scholar]
- 46.Escobar C., Cailotto C., Angeles-Castellanos M., Delgado R. S., Buijs R. M. (2009) Eur. J. Neurosci. 30, 1665–1675 [DOI] [PubMed] [Google Scholar]
- 47.Feillet C. A., Albrecht U., Challet E. (2006) J. Physiol. Paris 100, 252–260 [DOI] [PubMed] [Google Scholar]
- 48.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 49.Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C. P., Mackay S., Talianidis I., Flicek P., Odom D. T. (2010) Science 328, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas A. M., Hart S. N., Kong B., Fang J., Zhong X. B., Guo G. L. (2010) Hepatology 51, 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oishi K., Miyazaki K., Kadota K., Kikuno R., Nagase T., Atsumi G., Ohkura N., Azama T., Mesaki M., Yukimasa S., Kobayashi H., Iitaka C., Umehara T., Horikoshi M., Kudo T., Shimizu Y., Yano M., Monden M., Machida K., Matsuda J., Horie S., Todo T., Ishida N. (2003) J. Biol. Chem. 278, 41519–41527 [DOI] [PubMed] [Google Scholar]
- 52.Bozek K., Kiełbasa S. M., Kramer A., Herzel H. (2007) Genome Inform. 18, 65–74 [PubMed] [Google Scholar]
- 53.Yin L., Joshi S., Wu N., Tong X., Lazar M. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11614–11619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oishi K., Uchida D., Ishida N. (2008) FEBS Lett. 582, 3639–3642 [DOI] [PubMed] [Google Scholar]
- 55.Mounier C., Posner B. I. (2006) Can. J. Physiol. Pharmacol. 84, 713–724 [DOI] [PubMed] [Google Scholar]
- 56.Shimano H. (2007) J. Mol. Med. 85, 437–444 [DOI] [PubMed] [Google Scholar]
- 57.Utzschneider K. M., Kahn S. E. (2006) J. Clin. Endocrinol. Metab. 91, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 58.Fukuda H., Noguchi T., Iritani N. (1999) FEBS Lett. 464, 113–117 [DOI] [PubMed] [Google Scholar]
- 59.Kwon H. S., Huang B., Unterman T. G., Harris R. A. (2004) Diabetes 53, 899–910 [DOI] [PubMed] [Google Scholar]
- 60.Fritsche L., Weigert C., Häring H. U., Lehmann R. (2008) Curr. Med. Chem. 15, 1316–1329 [DOI] [PubMed] [Google Scholar]
- 61.Taguchi A., White M. F. (2008) Annu. Rev. Physiol. 70, 191–212 [DOI] [PubMed] [Google Scholar]
- 62.Whiteman E. L., Cho H., Birnbaum M. J. (2002) Trends Endocrinol. Metab. 13, 444–451 [DOI] [PubMed] [Google Scholar]
- 63.Ohno T., Onishi Y., Ishida N. (2007) Biochem. Biophys. Res. Commun. 354, 1010–1015 [DOI] [PubMed] [Google Scholar]
- 64.Takiguchi T., Tomita M., Matsunaga N., Nakagawa H., Koyanagi S., Ohdo S. (2007) Pharmacogenet. Genomics 17, 1047–1056 [DOI] [PubMed] [Google Scholar]
- 65.Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A. (2000) Cell 103, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duguay D., Cermakian N. (2009) Chronobiol. Int. 26, 1479–1513 [DOI] [PubMed] [Google Scholar]
- 67.Yilmaz Y., Eren F., Yonal O., Kurt R., Aktas B., Celikel C. A., Ozdogan O., Imeryuz N., Kalayci C., Avsar E. (2010) Eur. J. Clin. Invest. 40, 887–892 [DOI] [PubMed] [Google Scholar]
- 68.Eto K., Tumenbayar B., Nagashima S., Tazoe F., Miyamoto M., Takahashi M., Ando A., Okada K., Yagyu H., Ishibashi S. (2010) Diabetes Res. Clin. Pract. 89, 52–57 [DOI] [PubMed] [Google Scholar]
- 69.Mai K., Bobbert T., Groth C., Assmann A., Meinus S., Kraatz J., Andres J., Arafat A. M., Pfeiffer A. F., Möhlig M., Spranger J. (2010) Am. J. Physiol. Endocrinol. Metab. 299, E126–E130 [DOI] [PubMed] [Google Scholar]
- 70.Ando H., Oshima Y., Yanagihara H., Hayashi Y., Takamura T., Kaneko S., Fujimura A. (2006) Biochem. Biophys. Res. Commun. 346, 1297–1302 [DOI] [PubMed] [Google Scholar]
- 71.Ando H., Takamura T., Matsuzawa-Nagata N., Shima K. R., Eto T., Misu H., Shiramoto M., Tsuru T., Irie S., Fujimura A., Kaneko S. (2009) Diabetologia 52, 329–335 [DOI] [PubMed] [Google Scholar]
- 72.Herichová I., Zeman M., Stebelová K., Ravingerová T. (2005) Mol. Cell Biochem. 270, 223–229 [DOI] [PubMed] [Google Scholar]
- 73.Badman M. K., Koester A., Flier J. S., Kharitonenkov A., Maratos-Flier E. (2009) Endocrinology 150, 4931–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Denechaud P. D., Dentin R., Girard J., Postic C. (2008) FEBS Lett. 582, 68–73 [DOI] [PubMed] [Google Scholar]
- 75.Postic C., Dentin R., Denechaud P. D., Girard J. (2007) Annu. Rev. Nutr. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 76.Doi M., Okano T., Yujnovsky I., Sassone-Corsi P., Fukada Y. (2004) Curr. Biol. 14, 975–980 [DOI] [PubMed] [Google Scholar]
- 77.Hsieh M. C., Yang S. C., Tseng H. L., Hwang L. L., Chen C. T., Shieh K. R. (2010) Int. J. Obes. 34, 227–239 [DOI] [PubMed] [Google Scholar]
- 78.Wu X., Xie H., Yu G., Hebert T., Goh B. C., Smith S. R., Gimble J. M. (2009) Int. J. Obes. 33, 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]