Abstract
Runx2 is a critical transcription factor for osteoblast differentiation. Regulation of Runx2 expression levels and transcriptional activity is important for bone morphogenetic protein (BMP)-induced osteoblast differentiation. Previous studies have shown that extracellular signal-regulated kinase (Erk) activation enhances the transcriptional activity of Runx2 and that BMP-induced Runx2 acetylation increases Runx2 stability and transcriptional activity. Because BMP signaling induces Erk activation in osteoblasts, we sought to investigate whether BMP-induced Erk signaling regulates Runx2 acetylation and stability. Erk activation by overexpression of constitutively active MEK1 increased Runx2 transcriptional activity, whereas U0126, an inhibitor of MEK1/2, suppressed basal Runx2 transcriptional activity and BMP-induced Runx2 acetylation and stabilization. Overexpression of constitutively active MEK1 stabilized Runx2 protein via up-regulation of acetylation and down-regulation of ubiquitination. Erk activation increased p300 protein levels and histone acetyltransferase activity. Knockdown of p300 using siRNA diminished Erk-induced Runx2 stabilization. Overexpression of Smad5 increased Runx2 acetylation and stabilization. Erk activation further increased Smad-induced Runx2 acetylation and stabilization, whereas U0126 suppressed these functions. On the other hand, knockdown of Smad1 and Smad5 by siRNA suppressed both basal and Erk-induced Runx2 protein levels. Erk activation enhanced the association of Runx2 with p300 and Smad1. Taken together these results indicate that Erk signaling increases Runx2 stability and transcriptional activity, partly via increasing p300 protein levels and histone acetyltransferase activity and subsequently increasing Runx2 acetylation by p300. In addition to the canonical Smad pathway, a BMP-induced non-Smad Erk signaling pathway cooperatively regulates osteoblast differentiation partly via increasing the stability and transcriptional activity of Runx2.
Keywords: Bone Morphogenetic Protein (BMP), Erk, Histone Acetylase, p300, Ubiquitination
Introduction
The bone morphogenetic proteins (BMPs)3 are members of the transforming growth factor-β superfamily and are primary growth factors that induce formation of both cartilage and bone. Receptors for BMP are serine/threonine kinase receptors and consist of type I (BMPR-I) and II (BMPR-II) receptors. After ligand binding, BMPR-I kinases are activated by BMPR-II kinase-induced phosphorylation. R-Smad proteins are then recruited to activated receptors and play a role in transmitting the BMP signal from the receptor to target genes such as alkaline phosphatase (ALP), bone sialoprotein, osteocalcin (OC), Runx2, and Dlx5 (1, 2). In addition to the Smad pathway, diverse intracellular signaling molecules also participate in BMP-induced osteoblast differentiation. These are collectively called the non-Smad pathway of BMP signaling and include extracellular signal-regulated protein kinase (Erk), p38 mitogen-activated protein kinase, c-Jun N-terminal kinase, phosphatidylinositol 3-kinase, and protein kinases C and D (3–7). These cooperate with and/or regulate the Smad pathway (8).
Runx2 is a transcription factor that belongs to the runt-domain gene family. Runx2 acts as a master regulator for the commitment of mesenchymal stem cells to the osteoblastic lineage and regulates expression levels of osteogenic marker genes including ALP, osteopontin, type I collagen, and OC (9–11). Runx2-deficient mice do not form mineralized bone, whereas mice heterozygous for Runx2 show a phenotype similar to that in humans with cleidocranial dysplasia (9, 11). Loss-of-function mutations in human RUNX2 are responsible for the cleidocranial dysplasia phenotype (12). These reports imply that the appropriate gene dosage of Runx2 is crucial for bone development. Runx2 expression is regulated by various extracellular signals including BMP and fibroblast growth factors (FGFs) (13–15).
In addition to transcriptional regulation, Runx2 can be modified post-translationally by phosphorylation, acetylation, and ubiquitination, and these post-translational modifications seem to affect transcriptional activity and/or stability of Runx2 (16, 17). Phosphorylation of Runx2 by distinct kinases and signaling pathways results in differential effects on Runx2 function and osteoblast differentiation. Runx2 transcriptional activity is decreased by phosphorylation at Ser-104 and Ser-451 (18) or by glycogen synthase kinase 3β-dependent phosphorylation at Ser-369—Ser-373—Ser-377 (19). In contrast, Runx2 phosphorylation by Erk in its proline-, serine-, and threonine-rich domain increases transcriptional activity and osteoblast differentiation (20–22). Previous studies have shown that Erk is activated by various extracellular signals such as extracellular matrix, FGF, and BMP, which all stimulate osteoblast differentiation (13, 14, 23). A recent in vivo study has elucidated that transgenic mice overexpressing a constitutively active MEK (Mek-sp) in osteoblasts show enhanced bone formation and that crossing Mek-sp transgenic mice to Runx2 heterozygote mice partially rescues the phenotype of cleidocranial dysplasia (24). These reports suggest that the MEK/Erk pathway plays a pivotal role in osteogenesis, at least through stimulation of Runx2 phosphorylation and transcriptional activity. However, the molecular mechanism that links Erk activation and the resulting Runx2 phosphorylation to enhanced Runx2 transcriptional activity remains unclear.
Ubiquitination and subsequent proteasome-dependent protein degradation is an important regulatory mechanism for control of numerous cellular processes (25). Runx2 is also selectively targeted for ubiquitination of lysine residues and subsequent proteasomal degradation by specific ubiquitin E3 ligases such as Smad ubiquitin regulatory factor (Smurf) (26). Cyclin D1/CDK4-mediated phosphorylation of Runx2 at Ser-472 is known to stimulate ubiquitination and proteasomal degradation of Runx2 (27). In addition, p300 increases the Runx2 half-life as well as transcriptional activity through acetylation of specific lysine residues, precluding Smurf1-mediated ubiquitination (28). The p300 protein is a transcriptional coactivator with a histone acetyltransferase (HAT) domain (29, 30). In contrast, histone deacetylase 4 (HDAC4) and HDAC5 have been shown to deacetylate Runx2, allowing the protein to undergo Smurf-mediated ubiquitination and subsequent degradation (28).
Regulation of Runx2 transcriptional activity and stability is important to BMP-induced osteoblast differentiation. BMP-2 increases p300-mediated Runx2 acetylation, enhancing the stability and transcriptional activation of Runx2 and consequently osteoblast differentiation (28). Because Erk is also activated by BMP-2, we aimed to investigate the role of Erk activation in BMP-2-induced Runx2 stabilization and transcriptional activation. In this study we show that Erk activation is required for BMP-2-induced Runx2 acetylation and stabilization, which is at least in part achieved through increasing p300 protein levels and HAT activity.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human BMP-2 was purchased from Cytolab (Rehovot, Israel). The easy-BLUETM, i-star TaqTM, Maxime RT PreMix, and WEST-ZOL (plus) were purchased from iNtRON Biotechnology (Sungnam, Korea). The AccuPower RT-PreMix was purchased from Bioneer (Daejoen, Korea) and SYBR Premix Ex TaqTM was from TaKaRa (Otsu, Japan). The alkaline phosphatase staining kit and anti-FLAG M2 monoclonal antibody were purchased from Sigma. Anti-acetyl lysine, anti-phosphoserine for MAPK/CDK substrates ((PXS*P or S*PXR/K motif), anti-ubiquitin, anti-phospho-Erk, anti-Erk antibodies, and the MEK1/2 inhibitor U0126 were purchased from Cell Signaling Technology (Beverly, MA). Anti-HA monoclonal antibody was purchased from Covance (Berkley, CA), and anti-p300 antibody was from Upstate Biotechnology (Lake Placid, NY). Anti-Myc, anti-Runx2, and anti-actin antibodies and HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The Lipofectamine 2000 reagent was purchased from Invitrogen and WelFect-ExTM PLUS was from WelGENE (Seoul, Korea). The luciferase assay system was purchased from Promega (Madison, WI).
Cell Culture and ALP Staining
C2C12 and 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. To induce osteoblastic differentiation, C2C12 cells were cultured in DMEM supplemented with 5% FBS and BMP-2 (200∼300 ng/ml). To observe the effect of Erk activation on BMP-2-induced osteogenic differentiation, ALP activity was examined by ALP histochemical staining. When C2C12 cells became ∼90% confluent, the cells were pretreated with vehicle or 40 μm U0126 for 1 h and further incubated in the presence of 300 ng/ml BMP-2 for an additional 48 h. ALP staining was then performed using the alkaline phosphatase staining kit according to the manufacturer's instructions.
Plasmid Constructs and Site-directed Mutagenesis
Previously reported expression plasmids and reporter vectors were used: Myc- or FLAG-tagged Runx2 full-length type I isoform (MRIPV N-terminal sequence) and type II isoform (MASNS N-terminal sequence) expression vectors; pGL3–6XOSE2-luc reporter (15, 31); constitutively active BMP receptor IB (BMPR-IB), HA-p300, FLAG-Smad1, FLAG-Smad5, Myc-HDAC4, and Myc-HDAC5 expression vectors (28). The constitutively active MEK1 plasmid (pFC-MEK1) was purchased from Stratagene (La Jolla, CA). The type II Runx2-S301A/S319A mutant was generated by the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions (32). These mutants were confirmed by sequencing. The primers used were as follows: S301A forward, 5′-CCC AGG CAG GCA CAG TCT GCC CCA CCG TGG TCC TAT GAC-3′; S301A reverse, 5′-GTC ATA GGA CCA CGG TGG GGC AGA CTG TGC CTG CCT GGG-3′; S319A forward, 5′-TAT CTG AGC CAG ATG ACA GCC CCA TCC ATC CAC TCC ACC ACG-3′; S319A reverse, 5′-CGT GGT GGA GTG GAT GGA TGG GGC TGT CAT CTG GCT CAG ATA-3′.
RT-PCR and Real-time PCR
Expression levels of ALP, OC and p300 mRNA were examined by real-time PCR. Total RNA was isolated using easy-BLUETM RNA extraction reagent. cDNA was synthesized from 1 μg of total RNA using the Maxime RT PreMix kit. Real-time PCR analysis was performed using SYBR Premix Ex TaqTM and an AB 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, CA). Each sample was analyzed in quadruplicate, and target genes were normalized to the reference housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Fold differences were then calculated for each treatment group using normalized CT values for the control. Mouse genes and their primer sequences for real-time PCR were as follows: OC forward, 5′-ATCTCACCATTCGGATGAGTCT-3′; OC reverse, 5′-TCAGTCCATAAGCCAAGCTCTCA-3′; ALP forward, 5′-CCAACTCTTTTGTGCCAGAGA-3′, ALP reverse, 5′-GGCTACATTGGTGTTGAGCTTTT-3′; p300 forward, 5′-GAAGAACAGCCAAGCACCTC-3′; p300 reverse, 5′-CGGTAAAGTGCCTCCAATGT-3′; GAPDH forward, 5′-CATGTTCCAGTATGACTCCACTC-3′; GAPDH reverse 5′-GGCCTCACCCCATTTGATGT-3′.
Luciferase Reporter Assay
C2C12 cells were plated in 96-well plates, and transient transfection was performed with the Lipofectamine Plus reagent. In each transfection, 100 ng of expression plasmid (pcDNA3, pcDNA3-Runx2, and/or pFC-MEK1), 100 ng of reporter plasmid (pGL3-luc or pGL3–6XOSE2-luc), and Renilla luciferase plasmid were used as indicated. After overnight recovery from transfection, the cells were further incubated with vehicle or 40 μm U0126 for 24 h. The cells were then harvested, and luciferase activity was determined using a luciferase assay kit according to the manufacturer's instructions. The data were represented as activity relative to Renilla luciferase activity.
Immunoblot Analysis and Immunoprecipitation
After appropriate treatments, the cells were washed with ice-cold PBS twice and lysed in radioimmune precipitation assay buffer consisting of 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 150 mm NaCl, 1% Nonidet P-40, 2% SDS, 1% sodium deoxycholate, 50 mm NaF, 0.2 mm Na3VO4, 1 mm PMSF, and a protease inhibitor mixture. Protein concentrations were measured using BCA reagents. Each sample containing equal amounts of protein was subjected to SDS-PAGE and immunoblot analysis as described previously (33). For immunoprecipitation, cells were washed with ice-cold PBS twice and lysed in 1× HEPES lysis buffer consisting of 25 mm HEPES (pH 7.5), 150 mm NaCl, 10 mm sodium butyrate, 1% Nonidet P-40, 0.25% sodium deoxycholate, 10% glycerol, protease inhibitor mixture, and phosphatase inhibitor cocktails I and II. After measuring protein concentrations, 1 mg of protein from each sample was used for immunoprecipitation with appropriate primary antibodies and protein G-agarose beads. Binding reactions were performed for 16 h at 4 °C with continuous rotation. The beads were collected and washed three times with 1× HEPES lysis buffer. Bound proteins were eluted by boiling in 1× Laemmli sample buffer with 1 m DTT and then subjected to SDS-PAGE and immunoblot analysis.
Determination of Runx2 Stability
293T cells were transiently transfected with Myc-Runx2, BMPR-IB, and/or MEK1 expression plasmids. Thirty hours after transfection, cycloheximide (10 μg/ml) was added to cells in order to inhibit new protein synthesis. One hour after cycloheximide addition, cells were further incubated in the presence or absence of U0126 (40 μm) for the indicated times. Immunoblot analysis was then performed.
Ubiquitination Analysis
C2C12 cells were transiently transfected with pcDNA or MEK1 expression vectors using WelFECT. After overnight recovery, the cells were incubated in the presence or absence of MG132 (5 μm), a proteasome inhibitor, for an additional 18 h. For the U0126-treated group, U0126 (40 μm) was added to the culture for the final 2 h. The cells were then lysed in ice-cold lysis buffer consisting of 25 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.2% SDS, 5 mm N-ethylmaleimide, protease inhibitor mixture, and phosphatase inhibitor cocktails I and II. Immunoprecipitation with anti-Runx2 antibody and subsequent immunoblot analysis with anti-ubiquitin antibody were performed to detect ubiquitinated Runx2.
HAT Assay
The HAT assay was performed as described previously (34). C2C12 cells were transiently transfected with HA-p300 and MEK1 expression vectors and incubated for 48 h. For the U0126-treated sample, U0126 (40 μm) was added to the culture for the final 2 h. Cell lysates were prepared using 1× HEPES lysis buffer described above, and immunoprecipitation was performed with anti-p300 antibody or with control IgG overnight at 4 °C. The beads were then washed three times and incubated with 1 μg (50 pmol) histones and [14C]acetyl CoA (50 μCi/μl, 1000 pmol/μl, Amersham Biosciences) for 30 min at 37 °C. SDS-PAGE was then performed using 5 μl of each reaction mixture. The 14C-labeled acetylated histone level was detected using phosphorimaging, and Coomassie staining of gels was performed to check equal loading of histone.
Gene Knockdown by Small Interfering RNA (siRNA)
ON-TARGETplus SMARTpool siRNAs for human p300, SMAD1, and SMAD5 genes and a non-targeting siRNA (control siRNA) were purchased from Dharmacon (Chicago, IL). ON-TARGETplus SMARTpool siRNAs were a mixture of 4 siRNAs, providing advantages in both potency and specificity. Transfection of siRNA into 293T cells was performed according to the manufacturer's instructions. The second transfection with Myc-Runx2 and MEK1 expression vectors was performed 6 h later, and cells were incubated for an additional 32 h. Whole cell lysates were then prepared, and immunoprecipitation and/or immunoblot analysis was performed.
Statistical Analysis
All results were expressed as mean ± S.E. The statistical significance was analyzed by Student's t test. A p value < 0.01 was considered to be statistically significant.
RESULTS
Erk Activation Is Involved in BMP-2-induced Osteoblast Differentiation
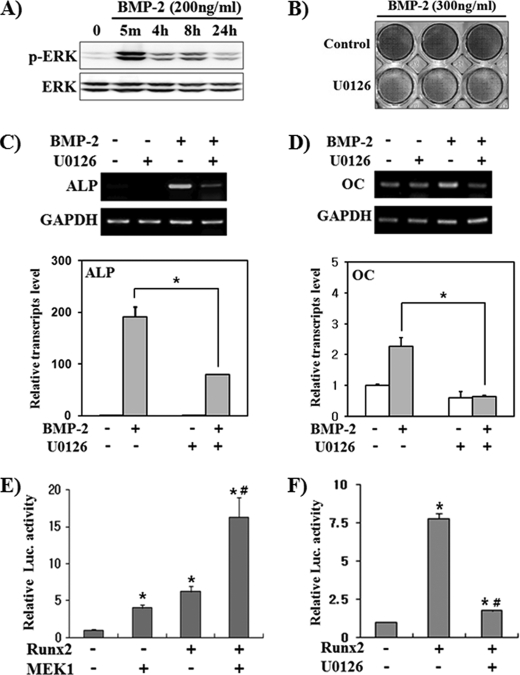
To investigate the role of activated Erk in BMP-2-induced osteoblast differentiation, we used pluripotent mesenchymal C2C12 cells that can be differentiated into osteoblasts by BMP-2 treatment (35). We first examined whether BMP-2 induces Erk activation in C2C12 cells (Fig. 1A). BMP-2-induced Erk phosphorylation was observed within 5 min and persisted for at least 24 h, although there was a fluctuation in Erk phosphorylation level. This result is similar to a previous report (36). We then investigated the role of Erk activation in BMP-2-induced osteoblast marker gene expression using U0126, a specific inhibitor of Erk upstream kinases MEK1/2. ALP and OC were used as phenotypic markers for early and late differentiation, respectively (37). ALP staining showed that blocking Erk activation by U0126 clearly suppressed ALP activity (Fig. 1B). Semiquantitative RT-PCR and real-time PCR results showed that the ALP mRNA level was greatly increased by BMP-2, whereas the OC mRNA level was weakly induced (Fig. 1, C and D). U0126 significantly suppressed BMP-2-induced expression of both ALP and OC. These results confirm previous reports that Erk activation is necessary for BMP-2-induced osteoblast marker gene expression (21, 36). We next examined whether the transcriptional activity of Runx2 is regulated by Erk activation. We performed reporter assays using the pGL3–6XOSE2-luc vector that contains six tandem repeats of a Runx2 binding element in the promoter of the mouse OC gene (15). MEK1 overexpression alone enhanced luciferase activity similar to Runx2 overexpression and synergistically increased Runx2-induced luciferase activity (Fig. 1E). On the other hand, U0126 significantly suppressed Runx2 transcriptional activity (Fig. 1F). MEK1-stimulated luciferase activity seems to be due to endogenous Runx2 in C2C12 cells, and this result is similar to the previous report showing that MEK1 stimulated OSE2-driven reporter activity only in Runx2-positive cells (38). These results confirm previous reports that Erk activation stimulates Runx2 transcriptional activity (38) and indicate that Erk activation is necessary for Runx2 transcriptional activation.
FIGURE 1.
Erk activation is involved in BMP-2-stimulated osteoblast differentiation. A, BMP-2 induced Erk activation. C2C12 cells were serum-starved for 16 h and treated with BMP-2 for the indicated times. Whole cell lysates were prepared and subjected to immunoblot analyses. B–D, Erk inhibition by U0126 suppressed BMP-2-induced ALP and OC expression. C2C12 cells were treated with BMP-2 and/or U0126 (40 μm) for 2 days and then ALP histochemical staining (B) and semiquantitative RT-PCR (C and D, upper panels) or real-time PCR (C and D, lower panel) were performed. Data represent the mean ± S.E. (n = 4). *, p < 0.01. E and F, Erk signaling increased Runx2 transcriptional activity. C2C12 cells were transfected with pGL3–6XOSE2-Luc and FLAG-Runx2 or a constitutively active MEK1 expression vector. After overnight recovery from transfection, cells were incubated in the presence or absence of U0126 for an additional 24 h. Data are shown as activity relative to Renilla luciferase activity and represent the mean ± S.E. (n = 6). * p < 0.01, compared with control; #, p < 0.01, compared with FLAG-Runx2 overexpressed group.
Erk Activation Increases Runx2 Protein Stability and Acetylation
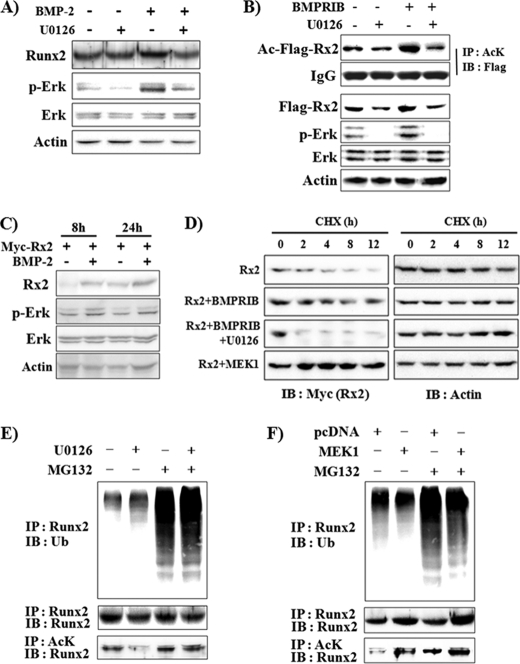
Because it has been reported that BMP-2 increases Runx2 protein stability as well as transcriptional activity (28), we next examined whether Erk activation is involved in BMP-2-induced Runx2 protein stabilization. C2C12 cells were incubated with BMP-2 and/or U0126 for 48 h (Fig. 2A). At this time point, Erk activation was still observed in the BMP-2-treated group. BMP-2 clearly increased endogenous Runx2 protein levels, and U0126 treatment completely abolished the effect of BMP-2 on Runx2 protein expression. To rule out a regulatory effect of Erk activation on Runx2 transcription, we observed the effect of Erk activation on exogenously expressed FLAG-Runx2 (Fig. 2B). Overexpression of constitutively active BMPR-IB increased the level of phosphorylated Erk, and U0126 suppressed both basal and BMPR-induced Erk activation. BMPR-IB increased FLAG-Runx2 protein levels, whereas U0126 decreased both basal and BMPR-IB-induced FLAG-Runx2 levels. Because a previous report had shown that BMP-2 increases Runx2 protein stability via Runx2 acetylation (28), we also looked for an effect of Erk activation on acetylated FLAG-Runx2 levels (Fig. 2B). Similar to FLAG-Runx2 protein levels, BMPR-IB significantly increased acetylated FLAG-Runx2, whereas U0126 decreased this effect. We also observed that BMP-2 treatment increased exogenously expressed Myc-Runx2 protein levels and that this increase could be detected 8 h after exposure to BMP-2 (Fig. 2C). We next examined whether the Erk-induced increase in Runx2 protein levels is due to increased protein stability (Fig. 2D). A previous report showed that the exogenously expressed Runx2 protein half-life was ∼5 h in 293T cells but that BMP signaling extended it to 50 h (28). Similar to that report, BMPR-IB overexpression significantly delayed Runx2 protein degradation. U0126, however, blocked BMPR-IB-induced stabilization and enhanced Runx2 degradation further than the control. In contrast, MEK1 overexpression dramatically increased Runx2 protein stability. These results suggest that Erk signaling is critical for the maintenance of Runx2 protein stability. Because the above data showed that Erk activation increased Runx2 acetylation and protein stability, we next analyzed whether Erk activation affects endogenous Runx2 ubiquitination (Fig. 2, E and F). In the presence of MG132, a proteasomal inhibitor, the polyubiquitinated Runx2 ladder increased, suggesting that proteasomal degradation of ubiquitinated Runx2 is continuously in progress in C2C12 cells. U0126 increased Runx2 ubiquitination, whereas MEK1 overexpression suppressed it (Fig. 2, E and F, upper panel). Both nonubiquitinated Runx2 and acetylated Runx2 protein levels were increased by MEK1 but decreased by U0126 (Fig. 2, E and F, middle and lower panels). These results indicate that Erk enhances Runx2 stability via stimulation of Runx2 acetylation and concomitant diminution of Runx2 ubiquitination.
FIGURE 2.
Erk activation increases Runx2 acetylation and stability, whereas it decreases Runx2 ubiquitination. A, U0126 decreased BMP-2-induced Runx2 protein expression. C2C12 cells were pretreated with vehicle or U0126 for 1 h and further incubated in the presence or absence of BMP-2 (200 ng/ml) for an additional 48 h. Endogenous Runx2 protein levels were determined by immunoprecipitation and immunoblot analysis. B, U0126 suppressed constitutively active BMP type IB (BMPR-IB)-induced increase in Runx2 protein levels and acetylation. C2C12 cells were transiently transfected with FLAG-Runx2 and BMPR-IB expression plasmids and incubated for 24 h. When indicated, the cells were incubated with U0126 for the last 3 h. Immunoprecipitation (IP) and immunoblot (IB) analysis were then performed. AcK, anti-acetylated lysine antibody. C, BMP-2 increased exogenously expressed Myc-Runx2 protein levels. C2C12 cells were transfected with Myc-Runx2, incubated for 8 or 24 h in the presence of BMP-2, and subjected to IB analysis. D, Erk activation by BMPR-IB or MEK1 expression stabilized Runx2 protein, whereas U0126 enhanced degradation of Runx2. 293T cells were transiently transfected with Myc-Runx2 and concomitantly with either BMPR-IB or MEK1 as indicated. Thirty hours after transfection the cells were treated with cycloheximide (CHX, 10 μg/ml) in the presence or absence of U0126 for the indicated times. The levels of Myc-Runx2 and actin were determined by IB analysis. E and F, U0126 increased, whereas Erk activation decreased Runx2 ubiquitination (upper panel). C2C12 cells were treated with vehicle or 5 μm MG132 for 18 h. When indicated, U0126 was added for the last 3 h (E). C2C12 cells were transfected with a MEK1 expression vector, incubated for 24 h, and treated with vehicle or MG132 for an additional 18 h (F). Ubiquitinated endogenous Runx2 was detected by IP with an anti-Runx2 antibody and subsequent IB with an anti-ubiquitin (Ub) antibody. Nonubiquitinated Runx2 and acetylated Runx2 were also examined using the same cell lysates (middle and lower panels).
p300-mediated Runx2 Acetylation Is Essential for Erk-induced Runx2 Stabilization
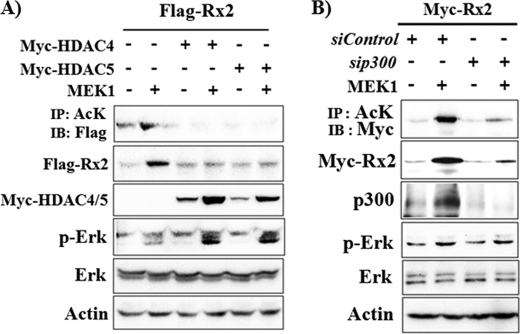
Previously it had been reported that overexpression of HDAC4 or HDAC5 results in deacetylation and destabilization of Runx2, whereas their knockdown by siRNA enhances Runx2 acetylation and stabilization (28). Therefore, to further verify that Runx2 acetylation is indispensible for Erk-mediated Runx2 stabilization, we induced deacetylation of Runx2 by overexpression of Myc-HDAC4 or Myc-HDAC5 in 293T cells (Fig. 3A). As expected, Runx2 acetylation was decreased in the presence of HDAC4 and HDAC5. Moreover, overexpression of HDAC4 or HDAC5 blocked MEK1-induced Runx2 acetylation and stabilization. These results indicate that Erk-induced Runx2 stabilization requires Runx2 acetylation. To further confirm the necessity for p300-mediated acetylation in Erk-induced Runx2 stabilization, we knocked down the endogenous p300 by siRNA (Fig. 3B). The efficiency of p300 siRNA was confirmed by immunoblot analysis. MEK1 overexpression also significantly increased endogenous p300 protein levels. siRNA for p300 decreased basal as well as Erk-induced p300 protein expression. Furthermore, knockdown of p300 suppressed Erk-induced Runx2 acetylation and stabilization. These results indicate that p300 is involved in Erk-induced Runx2 acetylation.
FIGURE 3.
Runx2 acetylation and p300 are required for Erk-induced Runx2 stabilization. A, overexpression of HDAC4 or HDAC5 blocked an Erk-mediated increase in Runx2 protein and acetylation. 293T cells were transiently transfected with FLAG-Runx2, MEK1, Myc-HDAC4, and Myc-HDAC5 expression vectors as indicated. Twenty-four hours after transfection, IP and IB analysis was performed. B, knockdown of p300 using siRNA suppressed Erk-mediated Runx2 acetylation and stabilization. 293T cells were transfected with siRNAs for p300 (sip300) and a non-targeting control siRNA (siControl). Six hours after transfection, a second transfection with Myc-Runx2 and MEK1 expression vector was performed, and the cells were incubated for an additional 32 h, then IP and IB analysis was performed. AcK, anti-acetylated lysine antibody.
Erk Activation Increases p300 Protein Levels and HAT Activity
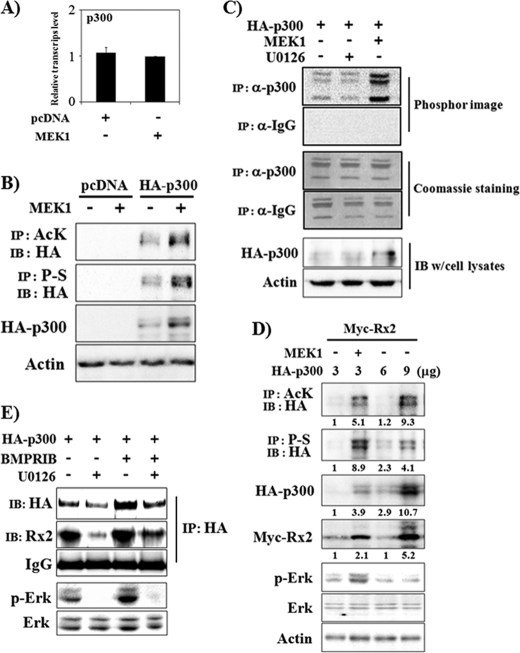
Because the above data showed that Erk activation increases p300 protein levels, we examined whether p300 mRNA levels are also changed by Erk activation. For this, C2C12 cells were transiently transfected with pcDNA or MEK1 expression plasmids and incubated for 13 h. The mRNA levels for p300 were then examined using real-time PCR (Fig. 4A). Dissimilar to the effect on p300 protein levels, Erk activation did not exert a significant effect on p300 mRNA levels. Because Erk activation increased exogenously expressed p300 protein as well as the endogenous form, it is likely that Erk regulates p300 at the post-transcriptional level. A previous study had demonstrated that Erk2 increases p300 HAT activity directly by phosphorylation (39). Therefore, we assayed whether overexpression of MEK1 increases p300 phosphorylation in 293T cells. Erk activation increased p300 phosphorylation as well as its total protein level (Fig. 4B). Because p300 has auto-acetylating activity (29), we also checked the acetylation level of p300 (Fig. 4B). Similar to phosphorylation, p300 acetylation is also increased by Erk activation. In accordance with the p300 acetylation data, MEK1 increased p300 HAT activity, whereas U0126 slightly decreased the activity (Fig. 4C). These results indicate that Erk activation increases p300 protein levels as well as HAT activity. We next asked whether increasing p300 expression can overcome the need for further Erk activation to increase Runx2 stability. Transient transfection of 293T cells with increasing amounts (3∼9 μg) of HA-p300 expression vector induced expression of HA-p300 in a dose-dependent manner (Fig. 4D). Concomitant transfection of 3 μg of HA-p300 with MEK1 induced total and acetylated HA-p300 more than with 6 μg of HA-p300. In addition, the Myc-Runx2 stabilization effect was stronger for 3 μg of HA-p300/MEK1-transfected cells than for 6 μg of HA-p300-transfected cells. However, transfection of 9 μg of HA-p300 induced higher levels of total and acetylated p300 protein expression and consequent stabilization of Runx2 than transfection of 3 μg of HA-p300/MEK1, suggesting that an increase in p300 protein levels is sufficient to increase Runx2 protein stability in the presence of basal Erk activation (Fig. 4D). We next examined whether BMPR signaling increases p300 protein levels and the association of p300 and Runx2 in C2C12 cells. Because it is difficult to detect endogenous p300 protein in C2C12 cells using immunoblot analysis, we examined the effect of BMPR activation using exogenously expressed HA-p300. Interactions between p300 and endogenous Runx2 were observed by co-immunoprecipitation with anti-HA antibody and immunoblot analysis with anti-Runx2 antibody (Fig. 4E). The results showed that BMPR-IB overexpression increased HA-p300 protein levels, whereas U0126 treatment decreased both basal and BMPR-IB-induced HA-p300 protein levels. In addition, the association of endogenous Runx2 and HA-p300 was increased by BMPR-IB, whereas it was suppressed by U0126. These results suggest that BMPR-IB-induced Erk activation increases Runx2 acetylation via increasing p300 protein and HAT activity and consequently increasing the association of p300 and Runx2.
FIGURE 4.
Erk activation increases protein levels and HAT activity of p300 and the association of Runx2 and p300. A, Erk activation did not significantly increase p300 mRNA levels. C2C12 cells were transiently transfected with a MEK1 expression vector and incubated for 13 h. Total RNA was then prepared, and real-time PCR was performed. B and C, Erk activation increases p300 protein levels and HAT activity. 293T cells were transiently transfected with HA-p300 and MEK1. Forty-eight hours after transfection, whole cell lysates were prepared for IP and/or IB analysis. When indicated, U0126 was used for the last 2 h (C). AcK, anti-acetylated lysine antibody. A HAT assay was performed using immunoprecipitates with anti-p300 antibody or with control IgG, and 14C-labeled acetylated histone levels were detected by phosphor-imaging (C, upper panel). Coomassie staining shows equal loading of histone mixture used for HAT assay (C, middle panel), and IB results represent HA-p300 expression levels of cell lysates (C, lower panel). D, Erk activation increased p300 acetylation, phosphorylation, and protein levels. The Runx2 protein level was related to the acetylated p300 protein level. 293T cells were transiently transfected with Myc-Runx2 and increasing amounts of HA-p300 expression vector. When indicated, MEK1 was also overexpressed. Forty-eight hours after transfection, whole cell lysates were prepared for IP and/or IB analysis. Densitometry analysis results were provided under the respective bands. P-S, anti-phosphoserine antibody. E, U0126 suppressed the physical association between Runx2 and p300. C2C12 cells were transiently transfected with HA-p300 and BMPR-IB expression vectors and incubated in the presence or absence of U0126 for 48 h. Whole cell lysates were then prepared, and IP and/or IB analysis was performed.
Runx2 Stabilization Requires Both Smad1/5 and Erk Activation
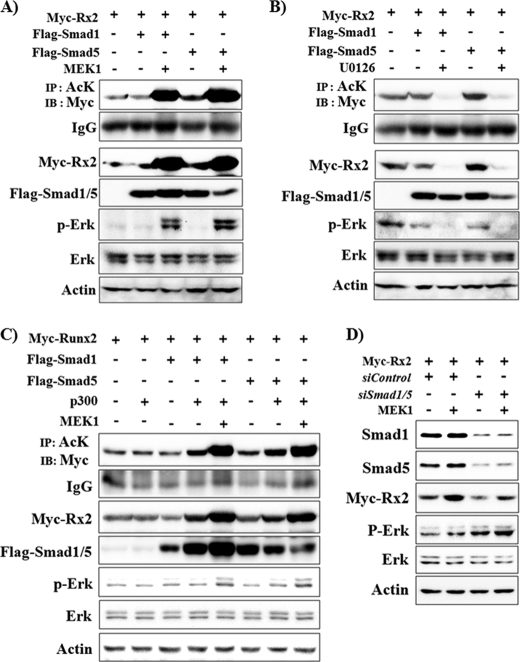
A previous report demonstrated that overexpression of Smad1 and Smad5 induces Runx2 acetylation, whereas knockdown of these by siRNA interferes with p300-mediated Runx2 acetylation, suggesting that Smad1/5 are necessary for BMP-mediated Runx2 acetylation (28). Because the above data showed that Erk activation is also essential for BMPR signal-induced Runx2 acetylation and stabilization, we next analyzed the effect of Erk activation on Smad1/5-induced Runx2 acetylation and stabilization. FLAG-tagged Smad1 and Smad5 were overexpressed in 293T cells, and the effect of Erk activation or inhibition was observed (Fig. 5, A and B). Smad1 overexpression alone did not show a significant effect on Runx2 acetylation and stability, whereas Smad5 noticeably increased Runx2 acetylation and stability. In the presence of Smad1/5 overexpression, MEK1 greatly enhanced Runx2 acetylation and stability, whereas U0126 decreased Runx2 acetylation and stability below basal levels. Because overexpressed Smad1 or 5 alone did not show a strong effect on Runx2 acetylation and stability, we overexpressed HA-p300 concomitantly with FLAG-Smad1/5. Overexpression of HA-p300 synergistically enhanced Smad1- and Smad5-induced Runx2 acetylation and stabilization (Fig. 5C). MEK1 activation further enhanced the effect of HA-p300, whereas U0126 suppressed the effect of HA-p300 and Smad1/5 (U0126 data not shown). These results strongly indicate that Erk signaling is necessary for BMP/Smad-induced Runx2 acetylation. To further define the requirement for Smad1/5 in Erk-induced Runx2 stabilization, we knocked down the endogenous SMAD1 and SMAD5 by siRNA. The efficiency of siRNA was confirmed by immunoblot analysis (Fig. 5D). Knockdown of SMAD1/5 decreased both the basal and Erk-induced Runx2 protein levels, but the Erk-induced increase in Runx2 stability was still observable. These results suggest that both Smad1/5 and Erk activation are required for full stabilization of Runx2 by BMP signaling.
FIGURE 5.
Both Smad1/5 and Erk activation are required for Runx2 stabilization. A–C, Erk activation further enhanced Smad1/5-induced Runx2 stabilization, whereas U0126 blocked it. 293T cells were transiently transfected with Myc-Runx2, FLAG-Smad1, FLAG-Smad5, HA-p300, and MEK1 expression vectors as indicated and incubated for 24 h. When indicated, cells were treated with U0126 for the last 2 h. IP and/or IB analysis was then performed. AcK, anti-acetylated lysine antibody. D, knockdown of Smad1/5 decreased both the basal and Erk-induced Runx2 protein levels. 293T cells were transfected with siRNAs for Smad1 and -5 (siSmad1/5) and a non-targeting control siRNA (siControl), and a second transfection with Myc-Runx2 and MEK1 expression vectors was performed 6 h later. After an additional incubation for 32 h, IP and IB analysis was performed.
Erk Activation Increases the Association of Runx2 with Smad1 and p300
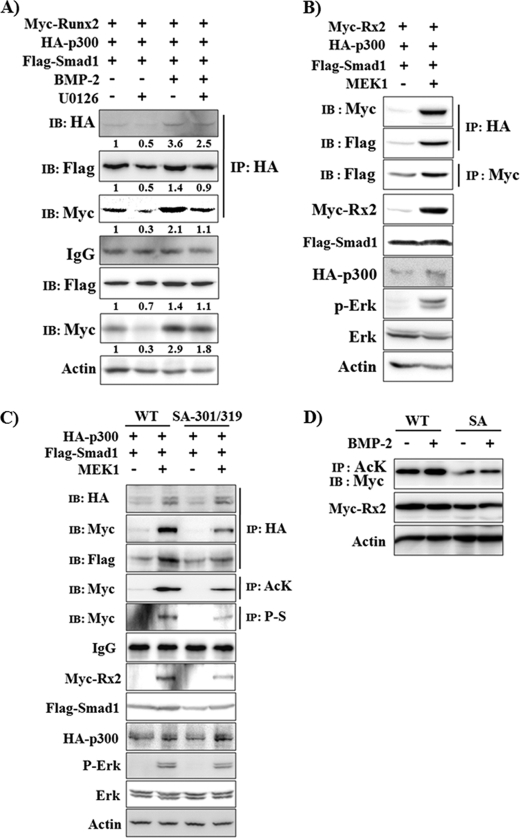
A previous study had demonstrated that the association of Runx2 and Smad1/5 is essential for BMP2-induced osteogenic induction of C2C12 cells and that the physical interaction between Runx2 and Smads is dependent on Erk-mediated phosphorylation of Runx2 (40). In addition, it had been demonstrated that Smad1/5 stimulates p300-mediated Runx2 acetylation by facilitating their physical interaction (28). Therefore, we examined whether BMP-2-induced Erk activation enhances the interaction between Runx2, Smad1, and p300 by co-immunoprecipitation. C2C12 cells were transfected with Myc-Runx2, FLAG-Smad1, and HA-p300 expression plasmids and treated with BMP-2 in the presence or absence of U0126 (Fig. 6A). BMP-2 increased protein levels of Runx2 and p300 as well as the association between them, whereas U0126 treatment diminished the BMP-2-induced effect. We also examined whether MEK1 overexpression enhances the interaction between Runx2, Smad1, and p300 in 293T cells (Fig. 6B). Overexpression of MEK1 increased the interaction between p300 and Smad1 as well as the interaction between p300 and Runx2. Furthermore, MEK1 enhanced interaction between Runx2 and Smad1, suggesting that Erk activation increases complex formation between Runx2, Smad1 and p300. Recently it has been reported that Ser-301 and Ser-319 of type II Runx2 are directly phosphorylated by Erk and that they are responsible for Erk-induced transcriptional activation of Runx2 (32). Therefore, we further looked for whether S301A/S319A mutation exerts any effect on Erk-induced Runx2 stabilization. To address this issue, 293T cells were transiently transfected with HA-p300, FLAG-Smad1, and MEK1 together with wild type Myc-Runx2 or Myc-Runx2-S301A/S319A expression vectors (Fig. 6C). As expected, MEK1-induced phosphorylation was decreased in Myc-Runx2-S301A/S319A mutant. Furthermore, S301A/S319A mutation diminished MEK1-induced association of p300, Runx2, and Smad1, resulting in reduced levels of Runx2 acetylation and protein stabilization. In addition, we examined the effect of Runx2 SA mutation on BMP-2-induced Runx2 acetylation and stabilization (Fig. 6D). Runx2 SA mutation reduced the basal Myc-Runx2 protein expression and acetylation levels as well as BMP-2-induced acetylation. Our data indicate that Erk-induced phosphorylation of Runx2 is necessary for complex formation between Runx2, Smad1, and p300 and consequent Runx2 acetylation and stabilization.
FIGURE 6.
Erk activation increases the association between Runx2, Smad1 and p300. A, C2C12 cells were transiently transfected with Myc-Runx2, FLAG-Smad1, and HA-p300 expression vectors and incubated for 24 h in the presence or absence of BMP-2 (200 ng/ml) and/or U0126 (40 μm). IP and/or IB analysis was then performed. Densitometry analysis results were provided under the respective band. B, 293T cells were transiently transfected with Myc-Runx2, FLAG-Smad1, HA-p300, and MEK1 expression vectors as indicated and incubated for 24 h. IP and/or IB analysis was then performed. C and D, S310A/S319A mutation reduces Erk activation-induced Runx2 acetylation and stabilization. C, 293T cells were transiently transfected with wild type Myc-Runx2 (WT) or Myc-Runx2-S301A/S319A (SA-301/319) concomitantly with FLAG-Smad1, HA-p300, and MEK1 expression vectors as indicated and incubated for 24 h. IP and/or IB analysis was then performed. AcK, anti-acetylated lysine antibody, P-S, anti-phosphoserine antibody. D, C2C12 cells were transiently transfected with WT or SA mutant Myc-Runx2 expression plasmids and were treated with BMP-2 (80 ng/ml) for 18 h as indicated. IP and/or IB analysis was then performed.
DISCUSSION
Runx2 is a master transcription factor for osteoblast differentiation and bone formation (11, 41). BMP signaling has been shown to be required for Runx2-dependent osteoblast differentiation (42). In addition to stimulation of Runx2 transcription, BMP-2-activated R-Smads enhance the expression of osteoblast differentiation marker genes partly via synergistic actions with Runx2 (14). Furthermore, BMP-2-activated Smad1/5 increases p300-mediated acetylation and stabilization of Runx2 as well as transcriptional activity of Runx2 (28). In this study we provide another clue for the role of BMP signaling in Runx2 regulation by showing that BMP-2-induced Erk increases acetylation and stability of Runx2 via increasing p300 protein levels and HAT activity. In this study we demonstrate that (i) BMP-2 induced Erk activation, (ii) BMP-2-induced expression of ALP and OC was suppressed by U0126 treatment, (iii) transcriptional activity of Runx2 was increased by Erk activation but suppressed by U0126 treatment, (iv) Erk activation increased acetylation and stability of Runx2, while decreasing ubiquitination of Runx2, (v) induction of Runx2 deacetylation by overexpression of HDAC4 or -5 or knockdown of p300 expression abolished Erk-mediated Runx2 stabilization, (vi) Erk activation increased p300 protein levels, HAT activity, and the association of Runx2 with p300, (vii) both Erk and Smad1/5 signaling are involved in Runx2 acetylation and stabilization, and (viii) Erk activation increases formation of a complex composed of Runx2, Smads, and p300.
The Erk signaling pathway is one of the major links between the cell surface and nucleus to control diverse functions including cell proliferation, differentiation, migration, and survival (43). Although many studies have reported that the Erk pathway is activated by various extracellular signals including growth factors (22, 44, 45), extracellular matrix (21), and mechanical force (46), it has been controversial whether Erk signaling plays an important role in the modulation of osteoblast differentiation in vivo. However, a recent study conclusively established a pivotal function for Erk signaling in osteoblast differentiation and skeletal development using transgenic mice expressing dominant negative or constitutively active MEK1 protein (24). Furthermore, the authors demonstrated that the effects of transgenes were maximized in the background of a Runx2 heterozygote, suggesting that activation of Erk signaling somehow helps overcome haploinsufficiency of Runx2 in Runx2+/− mice. Although Erk activation is known to increase transcriptional activity of Runx2 (38, 47), the molecular mechanism by which Erk signaling helps to overcome haploinsufficiency for Runx2 remains unclear.
Phosphorylation, acetylation, and ubiquitination are important post-translational modifications of Runx2 that regulate Runx2 stability and/or transcriptional activity (16, 17). Although previous reports showed that Runx2 phosphorylation and transcriptional activity were stimulated by the Erk signaling pathway (38, 48) and BMP-2-activated Smads increased Runx2 acetylation, stability, and transcriptional activity (28), there had been no study linking BMP-induced Erk signaling to the regulation of Runx2 acetylation and stability. In this study we provide evidence that Erk activation increases Runx2 acetylation and stability via increasing p300 protein levels and HAT activity as well as its association with p300. Because a previous report has clearly demonstrated that p300-mediated acetylation of Runx2 is essential not only for Runx2 stabilization but also for its transcriptional activity (28), our results imply that Erk activation increases the transcriptional activity of Runx2 by enhancing its association with p300 and subsequent acetylation and stabilization of Runx2.
Regulation of protein stability and/or transcriptional activity by cooperative post-translational modifications is also well known for transcription factors other than Runx2. For instance, stability of the tumor suppressor p53 is increased by p300/CBP-mediated acetylation, whereas it is negatively regulated by MDM2-HDAC1 complex-mediated deacetylation (49). The p300-dependent increase in Runx3 acetylation increased its stability and transcriptional activity (50). The ER81 protein, a transcription factor of the ETS family, is a downstream effector of the proto-oncoproteins HER2/Neu and Ras, which stimulate not only ER81 phosphorylation but also the HAT activity of p300 and P/CAF and thereby jointly regulate ER81 stability, DNA binding, and transactivation (51). Furthermore, Ras/Erk2-mediated phosphorylation of Ets-1 and Ets-2 activates them by increasing p300/CBP recruitment and binding (52).
In addition to Erk, several kinases are known to regulate Runx2 transcriptional activity and/or stability by phosphorylating Runx2. FGF2 stimulates Runx2 transcriptional activity via protein kinase Cδ-dependent phosphorylation of Ser-247 (53), and parathyroid hormone increases Runx2 transcriptional activity via protein kinase A-dependent phosphorylation of Ser-347 (54). In contrast, glycogen synthase kinase 3β inhibits Runx2 transcriptional activity via phosphorylation of Ser-369—Ser-373—Ser-377 (19), and the cyclin D1-CDK4 complex induces Runx2 degradation via phosphorylation of Ser-472 (27). Despite these reports, little information has been available concerning how Runx2 phosphorylation regulates its transcriptional activity and/or stability.
Regarding the regulation of Runx2 protein stability, it has been reported that ubiquitination of Runx2 by E3 ligases such as Smurf1 and WWP1 promotes its degradation, whereas acetylation by p300 stabilizes it via prevention of ubiquitination (28, 55, 56). In this study an Erk-induced increase in Runx2 acetylation was accompanied by enhanced binding of p300 to Runx2. Because total protein levels for both p300 and Runx2 were increased by Erk activation, it is not clear whether the increased association between Runx2 and p300 is simply a consequence of protein increase or of Erk-induced phosphorylation of Runx2 and p300 that affects their affinity for each other. Based on a recent report showing that Ser-301 and Ser-319 are the major phosphorylation target sites of Erk and are necessary for Erk-induced transcriptional activation of Runx2 (32), we examined the effect of S301A/S319A mutation on Erk-induced Runx2 acetylation and stabilization. Our data showed that S301A/S319A mutation diminishes Erk-induced association of p300 and Runx2 as well as Runx2 acetylation and stabilization. These results suggest that Erk-mediated phosphorylation of Runx2 enhances the association of Runx2 and p300 and consequent Runx2 acetylation and stabilization.
Although it is not clear whether p300 is the only HAT that regulates Runx2 acetylation, p300 knockdown data indicate that p300 plays an important role in Erk-induced Runx2 acetylation and stabilization. Recently, it was demonstrated that Erk2 directly phosphorylates p300 and increases its HAT activity (39). In addition, the study showed that phosphorylation of p300 confers a higher affinity for the Sp1 transcription factor. We also observed that MEK1 overexpression increased p300 protein levels as well as its phosphorylation and HAT activity. In addition, when we compared the effects on Runx2 protein levels, it appears that acetylated p300 protein levels influence Runx2 stabilization. Because p300 has auto-acetylating activity (29, 30), it is assumed that acetylated p300 levels reflect its HAT activity. Furthermore, knockdown of p300 using siRNA suppressed Erk-induced Runx2 acetylation and stabilization. Therefore, our results imply that an Erk-induced increase in p300 protein levels and HAT activity is involved in the Erk-mediated increase in Runx2 acetylation and stabilization. Because Erk activation increased both endogenous and exogenously expressed p300 protein but did not significantly affect p300 mRNA levels, it is likely that Erk increases p300 protein levels via post-transcriptional regulation, necessitating further studies.
Jeon et al. (28) have also demonstrated that Smad5 and BMPR-IB synergistically increase Runx2 acetylation. Similar to that report, our results show that Smad5-induced Runx2 acetylation was robustly stimulated by Erk activation, whereas knockdown of Smad1/5 suppressed Runx2 acetylation and stabilization. Because U0126 treatment almost completely abolished Smad5-induced Runx2 acetylation, basal Erk activation seems to be required for Smad5-induced Runx2 acetylation. Although overexpression of Smad1 or Smad5 led to self-activation by inducing C-terminal phosphorylation, significant activation of Erk was not observed by Smad1/5 overexpression, suggesting that basal Erk activation signals may come from the culture environment but not from Smad1/5 overexpression (data not shown). These results indicate that BMP-2-induced Runx2 acetylation requires both Smad activation and non-Smad Erk activation. The relative roles for Smad1/5 and Erk in Runx2 acetylation are not clear. Previous studies have demonstrated that the association of Runx2 and Smad1/5 is essential for BMP2-induced osteogenic induction of C2C12 cells and that the physical interaction between Runx2 and Smad depends on Erk-mediated phosphorylation of Runx2 (40). In addition, Jeon et al. (28) demonstrated that Smad1/5 stimulates p300-mediated Runx2 acetylation by facilitating their physical interaction. Our results also show that Erk activation by MEK1 overexpression greatly enhanced the association of Runx2, Smad1, and p300. Furthermore, S310A/S319A mutation reduced Erk-induced formation of a complex composed of p300, Smad1, and Runx2. Taking these reports and our results together, we can propose a model for the relative roles of BMP-activated Smads and Erk activation on Runx2 acetylation. Activated Erk increases p300 levels and its HAT activity as well as phosphorylation of Runx2. Smad1/5 then recruits p300 and binds to phosphorylated Runx2, thereby increasing the physical interaction between Runx2 and p300 and subsequent Runx2 acetylation.
In summary, we show here that BMP-activated Erk increases Runx2 acetylation and stability at least in part via up-regulating p300 protein levels and HAT activity and that both R-Smad and non-Smad Erk signaling is required for BMP-induced Runx2 acetylation and stabilization. Further study is necessary to verify whether Erk-mediated phosphorylation of Runx2 and/or p300 is necessary for direct binding between the two proteins and/or increases their binding affinity.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (Grants 311-2006-1-E00093 and 531-2007-1-E00070) and by the Korea Health 21 R&D Project, Ministry of Health and Welfare (Grant A085021).
- BMP
- bone morphogenetic protein
- BMPR
- BMP receptor
- ALP
- alkaline phosphatase
- HDAC
- histone deacetylase
- HAT
- histone acetyltransferase
- IP
- immunoprecipitation
- IB
- immunoblot
- OC
- osteocalcin.
REFERENCES
- 1.Nohe A., Keating E., Knaus P., Petersen N. O. (2004) Cell. Signal. 16, 291–299 [DOI] [PubMed] [Google Scholar]
- 2.Lee M. H., Kim Y. J., Kim H. J., Park H. D., Kang A. R., Kyung H. M., Sung J. H., Wozney J. M., Kim H. J., Ryoo H. M. (2003) J. Biol. Chem. 278, 34387–34394 [DOI] [PubMed] [Google Scholar]
- 3.Celil A. B., Campbell P. G. (2005) J. Biol. Chem. 280, 31353–31359 [DOI] [PubMed] [Google Scholar]
- 4.Yue J., Mulder K. M. (2000) Methods Mol. Biol. 142, 125–131 [DOI] [PubMed] [Google Scholar]
- 5.Yakymovych I., Ten Dijke P., Heldin C. H., Souchelnytskyi S. (2001) FASEB J. 15, 553–555 [DOI] [PubMed] [Google Scholar]
- 6.Aubin J., Davy A., Soriano P. (2004) Genes Dev. 18, 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nohe A., Hassel S., Ehrlich M., Neubauer F., Sebald W., Henis Y. I., Knaus P. (2002) J. Biol. Chem. 277, 5330–5338 [DOI] [PubMed] [Google Scholar]
- 8.Moustakas A., Heldin C. H. (2005) J. Cell Sci. 118, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 9.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 10.Banerjee C., McCabe L. R., Choi J. Y., Hiebert S. W., Stein J. L., Stein G. S., Lian J. B. (1997) J. Cell. Biochem. 66, 1–8 [DOI] [PubMed] [Google Scholar]
- 11.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 12.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y. X., Xu X., Chen L., Li C., Brodie S. G., Deng C. X. (2000) Hum. Mol. Genet 9, 2001–2008 [DOI] [PubMed] [Google Scholar]
- 14.Lee K. S., Kim H. J., Li Q. L., Chi X. Z., Ueta C., Komori T., Wozney J. M., Kim E. G., Choi J. Y., Ryoo H. M., Bae S. C. (2000) Mol. Cell. Biol. 20, 8783–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. J., Kim J. H., Bae S. C., Choi J. Y., Kim H. J., Ryoo H. M. (2003) J. Biol. Chem. 278, 319–326 [DOI] [PubMed] [Google Scholar]
- 16.Bae S. C., Lee Y. H. (2006) Gene 366, 58–66 [DOI] [PubMed] [Google Scholar]
- 17.Jonason J. H., Xiao G., Zhang M., Xing L., Chen D. (2009) J. Dental Res. 88, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wee H. J., Huang G., Shigesada K., Ito Y. (2002) EMBO Rep. 3, 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kugimiya F., Kawaguchi H., Ohba S., Kawamura N., Hirata M., Chikuda H., Azuma Y., Woodgett J. R., Nakamura K., Chung U. I. (2007) PLoS One 2, e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno T., Takahashi T., Tsujisawa T., Ariyoshi W., Nishihara T. (2007) J. Cell. Biochem. 101, 1266–1277 [DOI] [PubMed] [Google Scholar]
- 21.Xiao G., Gopalakrishnan R., Jiang D., Reith E., Benson M. D., Franceschi R. T. (2002) J. Bone Miner. Res. 17, 101–110 [DOI] [PubMed] [Google Scholar]
- 22.Xiao G., Jiang D., Gopalakrishnan R., Franceschi R. T. (2002) J. Biol. Chem. 277, 36181–36187 [DOI] [PubMed] [Google Scholar]
- 23.Otto F., Lübbert M., Stock M. (2003) J. Cell. Biochem. 89, 9–18 [DOI] [PubMed] [Google Scholar]
- 24.Ge C., Xiao G., Jiang D., Franceschi R. T. (2007) J. Cell Biol. 176, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershko A. (1983) Cell 34, 11–12 [DOI] [PubMed] [Google Scholar]
- 26.Huang G., Shigesada K., Ito K., Wee H. J., Yokomizo T., Ito Y. (2001) EMBO J. 20, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen R., Wang X., Drissi H., Liu F., O'Keefe R. J., Chen D. (2006) J. Biol. Chem. 281, 16347–16353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 29.Wolffe A. P., Pruss D. (1996) Cell 84, 817–819 [DOI] [PubMed] [Google Scholar]
- 30.Yang X. J. (2004) Nucleic Acids Res. 32, 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y. J., Lee M. H., Wozney J. M., Cho J. Y., Ryoo H. M. (2004) J. Biol. Chem. 279, 50773–50780 [DOI] [PubMed] [Google Scholar]
- 32.Ge C., Xiao G., Jiang D., Yang Q., Hatch N. E., Roca H., Franceschi R. T. (2009) J. Biol. Chem. 284, 32533–32543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H. L., Woo K. M., Ryoo H. M., Baek J. H. (2010) Biochem. Biophys. Res. Commun. 391, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 34.Chakravarti D., Ogryzko V., Kao H. Y., Nash A., Chen H., Nakatani Y., Evans R. M. (1999) Cell 96, 393–403 [DOI] [PubMed] [Google Scholar]
- 35.Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. (1994) J. Cell Biol. 127, 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallea S., Lallemand F., Atfi A., Rawadi G., Ramez V., Spinella-Jaegle S., Kawai S., Faucheu C., Huet L., Baron R., Roman-Roman S. (2001) Bone 28, 491–498 [DOI] [PubMed] [Google Scholar]
- 37.Franceschi R. T., Iyer B. S. (1992) J. Bone Miner. Res. 7, 235–246 [DOI] [PubMed] [Google Scholar]
- 38.Xiao G., Jiang D., Thomas P., Benson M. D., Guan K., Karsenty G., Franceschi R. T. (2000) J. Biol. Chem. 275, 4453–4459 [DOI] [PubMed] [Google Scholar]
- 39.Chen Y. J., Wang Y. N., Chang W. C. (2007) J. Biol. Chem. 282, 27215–27228 [DOI] [PubMed] [Google Scholar]
- 40.Afzal F., Pratap J., Ito K., Ito Y., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., Javed A. (2005) J. Cell Physiol. 204, 63–72 [DOI] [PubMed] [Google Scholar]
- 41.Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G. (1999) Genes Dev. 13, 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phimphilai M., Zhao Z., Boules H., Roca H., Franceschi R. T. (2006) J. Bone Miner. Res. 21, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebisuya M., Kondoh K., Nishida E. (2005) J. Cell Sci. 118, 2997–3002 [DOI] [PubMed] [Google Scholar]
- 44.Hurley M. M., Marcello K., Abreu C., Kessler M. (1996) J. Bone Miner. Res. 11, 1256–1263 [DOI] [PubMed] [Google Scholar]
- 45.Chen C., Koh A. J., Datta N. S., Zhang J., Keller E. T., Xiao G., Franceschi R. T., D'Silva N. J., McCauley L. K. (2004) J. Biol. Chem. 279, 29121–29129 [DOI] [PubMed] [Google Scholar]
- 46.You J., Reilly G. C., Zhen X., Yellowley C. E., Chen Q., Donahue H. J., Jacobs C. R. (2001) J. Biol. Chem. 276, 13365–13371 [DOI] [PubMed] [Google Scholar]
- 47.Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H. (1996) Mol. Cell. Biol. 16, 3967–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franceschi R. T., Xiao G. (2003) J. Cell. Biochem. 88, 446–454 [DOI] [PubMed] [Google Scholar]
- 49.Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., Yao T. P. (2002) EMBO J. 21, 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y. H., Jeon E. J., Li Q. L., Lee Y. H., Choi J. K., Kim W. J., Lee K. Y., Bae S. C. (2004) J. Biol. Chem. 279, 29409–29417 [DOI] [PubMed] [Google Scholar]
- 51.Goel A., Janknecht R. (2003) Mol. Cell. Biol. 23, 6243–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foulds C. E., Nelson M. L., Blaszczak A. G., Graves B. J. (2004) Mol. Cell. Biol. 24, 10954–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim B. G., Kim H. J., Park H. J., Kim Y. J., Yoon W. J., Lee S. J., Ryoo H. M., Cho J. Y. (2006) Proteomics 6, 1166–1174 [DOI] [PubMed] [Google Scholar]
- 54.Selvamurugan N., Pulumati M. R., Tyson D. R., Partridge N. C. (2000) J. Biol. Chem. 275, 5037–5042 [DOI] [PubMed] [Google Scholar]
- 55.Zhao M., Qiao M., Oyajobi B. O., Mundy G. R., Chen D. (2003) J. Biol. Chem. 278, 27939–27944 [DOI] [PubMed] [Google Scholar]
- 56.Jones D. C., Wein M. N., Oukka M., Hofstaetter J. G., Glimcher M. J., Glimcher L. H. (2006) Science 312, 1223–1227 [DOI] [PubMed] [Google Scholar]