Abstract
Ribosomes synthesizing secretory and membrane proteins are bound to the endoplasmic reticulum (ER) membrane and attach to ribosome-associated membrane proteins such as the Sec61 complex, which forms the protein-conducting channel in the membrane. The ER membrane-resident Hsp40 protein ERj1 was characterized as being able to recruit BiP to ribosomes in solution and to regulate protein synthesis in a BiP-dependent manner. Here, we show that ERj1 and Sec61 are associated with ribosomes at the ER of human cells and that the binding of ERj1 to ribosomes occurs with a binding constant in the picomolar range and is prevented by pretreatment of ribosomes with RNase. However, the affinity of ERj1 for ribosomes dramatically changes upon binding of BiP. This modulation by BiP may be responsible for the dual role of ERj1 at the ribosome, i.e. acting as a recruiting factor for BiP and regulating translation.
Keywords: Endoplasmic Reticulum (ER), Membrane Proteins, Protein Translocation, Ribosomes, Surface Plasmon Resonance (SPR), Chaperone, Hsp40, Hsp70
Introduction
In eukaryotic cells, protein secretion begins with the typically cotranslational translocation of presecretory proteins across the membrane of the rough endoplasmic reticulum (ER)3 (1). In general, translocation is mediated by a protein translocase that resides in the ER membrane and contains the heterotrimeric Sec61 complex as the central and pore-forming component and the main receptor for ribosomes in the ER membrane, respectively (2, 3). In yeast cells, the cotranslationally operating protein translocase comprises Sec71p, Sec72p, Sec63p (a membrane-integrated Hsp40 protein), and Kar2p (an ER lumenal Hsp70 protein) as additional components (4, 5). In mammalian cells, the Kar2p ortholog BiP (immunoglobulin heavy chain-binding protein) and an as yet unidentified Hsp40 protein of the ER are also involved in cotranslational protein import into the ER (6, 7). Furthermore, Sec62 and Sec63 are found in stoichiometric amounts compared with Sec61α in the mammalian ER and in association with the Sec61 complex in microsomal detergent extracts (8, 9). In addition, the mammalian ER membrane contains the Hsp40 protein ERj1 (ER-resident J-domain protein 1), which is related to Sec63 in providing a lumenal J-domain (10, 11) and can complement inactivation of the SEC63 gene in yeast (12).
ERj1 associates with the ribosomal tunnel exit via a positively charged oligopeptide in its cytosolic domain and recruits BiP to translating ribosomes as well as to nascent polypeptide chains via the lumenal J-domain, which is separated from the cytosolic domain by one transmembrane domain (10, 11, 13). Furthermore, ERj1 is able to modulate translation. In the absence of BiP, ERj1 inhibits initiation of protein synthesis. When BiP is present, ERj1 recruits BiP to ribosomes, and protein synthesis is not inhibited (11). Therefore, we proposed that the function of ERj1 is to allow communication between ER lumenal BiP and translating ribosomes, i.e. to recruit ER lumenal BiP to translating ribosomes as well as to nascent polypeptide chains to aid polypeptide translocation or folding and to play a regulatory function in protein synthesis. However, the mechanism of this differential activity has remained elusive.
ERj1 is structurally related to the cytosolic mammalian Hsp40 protein MPP11 (M-phase phosphoprotein 11), which forms an unusually stable complex with the cytosolic Hsp70 protein Hsp70L1 (14). The complexes have been designated ribosome-associated complexes: mRAC in mammals (15, 16) and RAC in yeast (17, 18). The function of yeast RAC is to recruit the cytosolic Hsp70 proteins, such as Ssb1p and Ssb2p, to nascent polypeptide chains and thus to aid cotranslational polypeptide folding. The function of mRAC is only poorly understood (15, 16).
Here, we employed quantitative assays (immunofluorescence microscopy and surface plasmon resonance (SPR) spectroscopy) to test various aspects of our hypothesis for the function and molecular mechanism of ERj1 at the cellular level as well as in a cell-free system. We determined the effect of BiP on the affinity of ERj1 for ribosomes, and we observed that ERj1 is in close proximity to ribosomes at the ER of mammalian cells. Furthermore, we compared the affinity of ERj1 for ribosomes with the structurally related but cytosolic MPP11 protein.
EXPERIMENTAL PROCEDURES
Materials
ERj1-His6, GST-ERj1, GST-ERj1C, GST-ERj1-H89Q, His6-BiP, His6-BiP-G227D, and His6-BiP-T229G were purified from Escherichia coli as described previously (11, 19, 20). N-terminally His6-tagged MPP11 was expressed from pET28a-HisMPP11 (16) in E. coli. For the purification of mRAC, an N-terminally His6-tagged version of Hsp70L1 was cloned in pACYC-DUET1 (Novagen) from pET28a-HisHsp70L1 (16) (both vectors were digested with NcoI and BamHI) and was coexpressed with His6-tagged MPP11. The harvested cells were resuspended in nickel-nitrilotriacetic acid binding buffer (40 mm potassium phosphate buffer (pH 7.8), 240 mm KCl, and 15 mm imidazole). In the first step, mRAC and MPP11 were purified via nickel-nitrilotriacetic acid (Qiagen) according to the manufacturer's protocol for native protein purification at pH 7.8. The bound proteins were eluted with elution buffer (40 mm potassium phosphate buffer (pH 7.8), 240 mm KCl, and 200 mm imidazole). Subsequently, the eluate was diluted with 10 mm potassium phosphate buffer (pH 7.8) and loaded onto a hydroxylapatite column (ceramic hydroxylapatite, Bio-Rad). Bound proteins were eluted with a 200–500 mm linear gradient of potassium phosphate buffer (pH 7.8). MPP11- or mRAC-containing fractions were pooled, diluted with 40 mm Hepes-KOH buffer (pH 7.8) to a final concentration of 50 mm potassium phosphate, and loaded onto a Mono Q HR 5/5 anion exchange column (GE Healthcare). Elution of bound proteins was performed with a 90–600 mm linear potassium acetate gradient in 40 mm Hepes-KOH buffer (pH 7.8). MPP11 was recovered in the flow-through fraction of the Mono Q column, whereas mRAC was eluted at a concentration of 280–400 mm potassium acetate. For further purification, MPP11 was directly loaded on a Mono S HR 5/5 cation exchange column (GE Healthcare). In the case of mRAC, fractions were pooled and diluted with 40 mm Hepes-KOH buffer (pH 7.8) to a final concentration of 50 mm potassium acetate and were also further purified via a Mono S HR 5/5 cation exchange column. Both proteins were eluted with an 80–800 mm linear potassium acetate gradient. Finally, the buffer was exchanged to 40 mm Hepes-KOH (pH 7.4), 240 mm potassium acetate, and 10% glycerol. Ribosomes were purified either from rabbit reticulocyte lysate (Roche Applied Science) or from canine pancreas by centrifugation and, where indicated, washed with puromycin (0.5 mm) and 500 mm potassium acetate. The molecular mass and hydrodynamic radius of the washed ribosomes were determined by a combination of asymmetric flow field-flow fractionation (Eclipse 2, Wyatt Technology) and light scattering analysis (miniDAWN TriStar and QELS, Wyatt Technology) as 3.62 ± 0.15 MDa and 18.25 ± 1.71 nm, respectively (21). Pretreatment of ribosomes with RNase A was carried out by incubation for 30 min at 30 °C.
Quantitative Fluorescence Microscopy
All steps were carried out as described by Müller et al. (21).
SPR Spectroscopy
SPR spectroscopy was performed in a BIAlite upgrade system (Biacore). The CM5 sensor chip was activated and loaded with antibodies according to the manufacturer's protocol. Purified proteins were immobilized on the chip-bound antibodies (anti-GST in the case of ERj1 and anti-His in the case of MPP11 and mRAC) at a flow rate of 10 μl/min in 50 mm Tris-HCl (pH 8), 150 mm KCl, 1 mm MgCl2, and 0.65% Chaps (ERj1) or in 20 mm Hepes-KOH (pH 7.4), 120 mm potassium acetate, and 1 mm magnesium acetate (MPP11 and mRAC). For interaction analysis with ribosomes, the chip was equilibrated with the same respective buffer at a flow rate of 30 μl/min. For interaction analysis of ERj1 and BiP and in the case of comparative interaction analysis of ribosomes and ERj1 in the presence or absence of BiP, 1 mm ATP was added to the running buffer. Subsequently, solutions containing increasing concentrations of ribosomes were passed over the chip surface. Unless stated otherwise, each ribosome application was followed by application of 1 m KCl in buffer. The analysis was carried out employing BIAevaluation Version 3.1 (Biacore) using 1:1 binding models and mass transfer. For analyte titrations, the regeneration steps were omitted, and the special model “titration kinetics 1:1 binding with drift” was employed.
Ribosome Binding Assay
Pretreatment of ribosomes with RNase A (80 μg/ml) was carried out by incubation for 30 min at 30 °C. The ribosomal complexes were formed as described previously (10), and the mixtures were then subjected to sucrose gradient centrifugation (linear sucrose gradient between 10 and 60% (w/v) in 20 mm Hepes-KOH (pH 7.5), 100 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, and 0.65% (w/v) Chaps, adjusted to 33 μg/ml BSA) at 54,000 rpm for 80 min at 2 °C (Beckman SW 55 Ti rotor). After fractionation of the gradients, proteins were precipitated and subjected to SDS-PAGE and subsequent protein Western blotting plus immunodetection (10).
RESULTS
ERj1 Is Protected from Antibody Access by Ribosomes in Cells
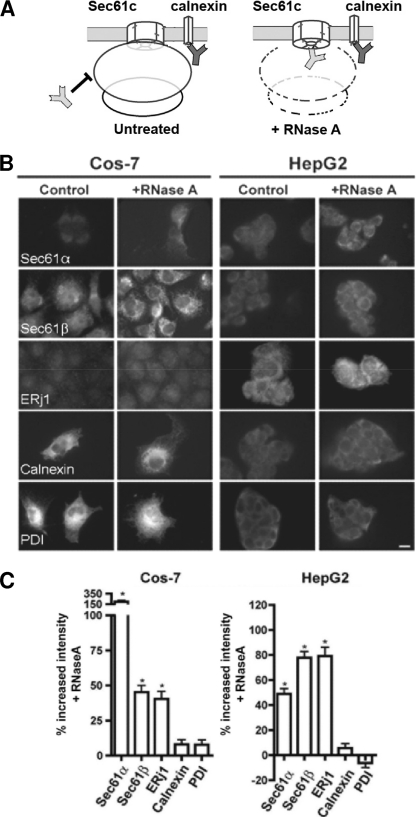
As a first step, we asked if ribosome association of ERj1 can be demonstrated in cells, and we used an established microscopic method to address this question in mammalian cells (21). The experimental strategy is to permeabilize formaldehyde-fixed cells with detergent, destroy ribosomes by RNase treatment or not, and incubate the cells with specific primary and fluorescently labeled secondary antibodies (Fig. 1A). In the subsequent quantitative fluorescence microscopy and image analysis, the data are compared with the minus-RNase control. The RNase treatment leads to a higher accessibility of antibodies against proteins like the Sec61 complex that are shielded by ribosomes, which results in an increased fluorescence signal, whereas the signal of unprotected proteins remains unaffected. Here, COS-7 and HepG2 cells were analyzed with respect to ERj1 (Fig. 1, B and C). The ribosome-associated membrane proteins Sec61α and Sec61β served as positive controls, whereas the ER membrane protein calnexin and the ER lumenal protein-disulfide isomerase served as negative controls. Although the extent of the differential effects varied between the two cell types, a significant increase in fluorescence intensity after RNase treatment was detected for Sec61α, Sec61β, and ERj1, but not for calnexin and protein-disulfide isomerase. Similar results were obtained for Madin-Darby canine kidney and HeLa cells (data not shown). Thus, like the Sec61 complex, ERj1 is associated with ribosomes at the intact ER. Because the Sec61 complex is the main receptor for ribosomes in the ER membrane, one could speculate that ERj1 is also in close proximity to the Sec61 complex.
FIGURE 1.
ERj1 is associated with ribosomes at the ER of mammalian cells. A, shown is the experimental strategy. B, COS-7 and HepG2 cells were fixed, left untreated or treated with RNase, and labeled with the indicated primary antibodies plus Alexa 555-conjugated secondary antibodies. Fluorescence was then recorded and quantified. PDI, protein-disulfide isomerase. C, the difference in fluorescence intensity between the RNase-treated and control samples is given as percent change, and the S.E. is indicated. *, p < 0.0001 relative to the control (n > 20 cells).
In a control experiment, we asked if the effect of RNase treatment can be mimicked by puromycin treatment, i.e. by the release of nascent polypeptides from translating ribosomes (data not shown). There was no intensity increase detected for any of the proteins after the addition of puromycin. These data demonstrate that the RNase experiments visualized translating as well as non-translating ribosomes.
ERj1 Interacts with Ribosomes with High Affinity
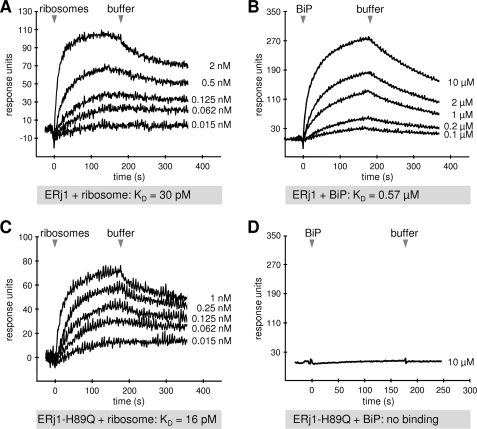
To further characterize the ribosome interaction of ERj1, SPR experiments were carried out. GST-ERj1 was immobilized in the measuring cell of a sensor chip. Mammalian ribosomes were then passed over the chip, and after washing with buffer, the association of the analyte and its dissociation were recorded (Fig. 2A). We determined an apparent affinity (KD) of ERj1 for ribosomes of 30 ± 3 pm.
FIGURE 2.
SPR analysis of the ERj1-ribosome and ERj1-BiP interaction. GST-ERj1 (A and B) or GST-ERj1-H89Q (C and D) was immobilized on an activated CM5 sensor chip in the measuring cell. A and C, increasing concentrations of canine pancreatic ribosomes were passed over the chip, followed by buffer application. B and D, increasing concentrations of BiP or 10 μm BiP, respectively, was passed over the chip, followed by buffer application.
The applied ribosomes were derived either from rabbit reticulocyte lysate or from dog pancreas. In addition, dog pancreas ribosomes were also treated with puromycin and high salt. There was no difference in the binding affinities of ERj1 for all tested ribosomes (supplemental Fig. S1).
Two experiments were carried out to demonstrate the specificity of ribosome binding in the SPR experiments. The interaction of ERj1C, a C-terminally truncated version of the cytosolic domain of ERj1 that had been characterized in complex with ribosomes by three-dimensional reconstruction after cryo-EM (13), has been suggested to involve electrostatic interactions with rRNA (10, 11). Therefore, GST-ERj1C was immobilized in the measuring cell of a sensor chip, and mammalian ribosomes were passed over the chip, followed by buffer and then by 500 mm KCl in buffer. According to the sensorgram, the interaction of ERj1C with ribosomes was salt-sensitive (supplemental Fig. S2B). Next, RNase-treated ribosomes were employed as the analyte (supplemental Fig. S2A). As expected, RNase-treated ribosomes showed a decreased binding to ERj1C, leading to a lower response difference compared with untreated ribosomes. Thus, the interaction of ERj1 with ribosomes is RNase-sensitive. Similar observations were made in ribosome binding assays and subsequent gradient centrifugation (supplemental Fig. S2, C and D).
BiP Modulates the Affinity of ERj1 for Ribosomes
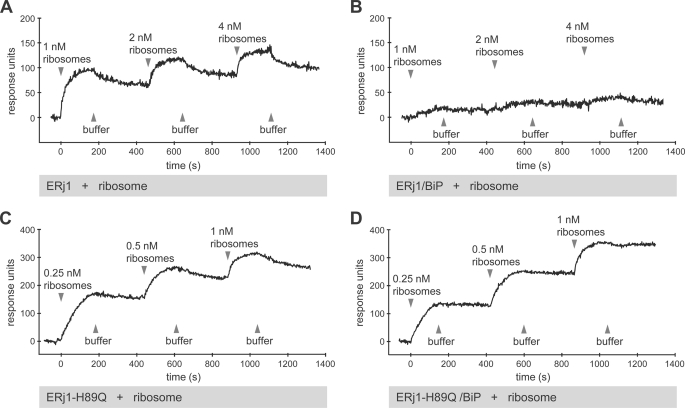
We addressed the question of whether the binding of ERj1 to ribosomes is modulated by BiP. GST-ERj1 was immobilized in the measuring cell of a sensor chip, equilibrated with 5 μm BiP, and analyzed via a titration analysis with respect to ribosomes in the presence of BiP (Fig. 3, A and B). Here, increasing concentrations of analyte were injected without regeneration of the chip between injections. In the presence of BiP, hardly any ribosome interaction was detected up to a ribosome concentration of 4 nm. Thus, binding of BiP reduces the affinity of ERj1 for ribosomes.
FIGURE 3.
SPR analysis of the ERj1-ribosome interaction in the presence of BiP. GST-ERj1 (A and B) or GST-ERj1-H89Q (C and D) was immobilized on an activated CM5 sensor chip in the measuring cell. Where indicated, the chip was saturated with BiP. Increasing concentrations of canine pancreatic ribosomes were then passed over the chip in the presence or absence of BiP, followed by buffer application. Note that the chips were not regenerated after each ribosome injection (titration experiment).
A control experiment for the effect of BiP was carried out with a mutant version of ERj1 (GST-ERj1-H89Q) in the same analysis. ERj1-H89Q is mutated in the conserved HPD motif of the J-domain, and in contrast to wild-type ERj1 (Fig. 2B), was not able to interact with BiP (Fig. 2D) but still interacted with ribosomes (Fig. 2C). GST-ERj1-H89Q was immobilized, equilibrated with 5 μm BiP, and analyzed via titration analysis with respect to ribosomes in the presence of BiP (Fig. 3, C and D). The binding of ribosomes to GST-ERj1-H89Q was not affected by the presence of BiP. Thus, the observed modulation of ribosome binding in the case of ERj1 involves the J-domain.
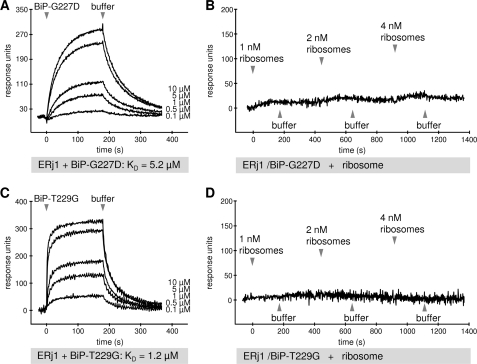
To further examine the BiP modulation of the ERj1-ribosome interaction, a BiP ATP-binding mutant (His6-BiP-G227D) and an ATP hydrolysis mutant (His6-BiP-T229G) (19) were employed. First, both mutants were analyzed for ERj1 binding by pulldown experiments (data not shown) and SPR measurements (Fig. 4, A and C). Both showed a decreased affinity for ERj1 in comparison with wild-type BiP (BiP, KD = 0.57 μm, ka = 4600 m−1 s−1, and kd = 0.003 s−1; BiP-G227D, KD = 5.2 μm, ka = 4200 m−1 s−1, and kd = 0.02 s−1; and BiP-T229G, KD = 1.2 μm, ka = 29,400 m−1 s−1, and kd = 0.04 s−1) (Fig. 2B). GST-ERj1 was then immobilized in the measuring cell of a sensor chip, equilibrated with 15 μm BiP-G227D or BiP-T229G, and assayed by titration analysis for binding to ribosomes (Fig. 4, B and D). Similar to the results obtained in the presence of wild-type BiP (Fig. 3B), there was no binding of ERj1 to ribosomes in the presence of the BiP mutants, indicating that neither ATP binding to BiP nor ATP hydrolysis is necessary for the modulation of the ERj1-ribosome interaction. This suggests that binding of BiP to ERj1 is sufficient for modulation of the affinity of ERj1 for ribosomes.
FIGURE 4.
SPR analysis of the ERj1-ribosome interaction in the presence of BiP mutants. GST-ERj1 was immobilized on an activated CM5 sensor chip in the measuring cell. A and C, increasing concentrations of the indicated BiP mutants were passed over the chip, followed by buffer application. B and D, the chip was saturated with the indicated BiP mutant. Increasing concentrations of canine pancreatic ribosomes were then passed over the chip, followed by buffer application. Note that the chips were not regenerated after each ribosome injection (titration experiment).
Affinity of MPP11 for Ribosomes Is Modulated by Its Hsp70 Partner, too
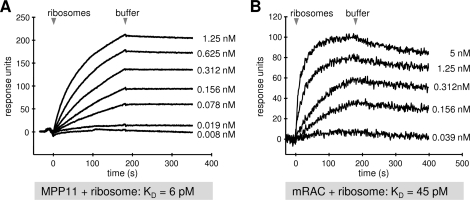
Another established ribosome interaction partner, human MPP11, which is structurally related to ERj1 but present in the cytosol, was analyzed. First, His6-MPP11 was immobilized in the measuring cell of a sensor chip. Mammalian ribosomes were then passed over the chip, followed by buffer, and the association of the analyte and its dissociation were recorded (Fig. 5A). We determined an apparent affinity (KD) of MPP11 for ribosomes of 6 pm. Thus, the affinity of MPP11 for ribosomes is about five times higher compared with that of ERj1. Next, His6-mRAC, which comprised His6-MPP11 plus His6-Hsp70L1, was analyzed in a similar manner (Fig. 5B). We determined a KD of mRAC for ribosomes of 45 pm. Remarkably, binding of Hsp70L1 reduced the affinity of MPP11 for ribosomes. However, the effect was not as dramatic as that caused by BiP on ERj1-ribosome binding.
FIGURE 5.
SPR analysis of the ribosome interactions of MPP11 and the mRAC complex. His6-MPP11 (A) or His6-mRAC (B) was immobilized on an activated CM5 sensor chip in the measuring cell. Increasing concentrations of canine pancreatic ribosomes were passed over the chip, followed by buffer application.
DISCUSSION
Previously, we observed that the ER membrane-resident Hsp40 protein ERj1 associates with the ribosomal tunnel exit and recruits BiP to translating ribosomes (11, 13). Furthermore, we found that ERj1 inhibits initiation of protein synthesis in the absence of BiP but does not inhibit translation when BiP is bound. Therefore, we proposed that the dual role of ERj1 in the ER membrane is to recruit ER lumenal BiP to translating ribosomes and to prevent initiation of translation on ER-associated ribosomes when BiP is not available. The latter appears to be relevant to ribosomes or 60 S ribosomal subunits that do not leave the ER surface after termination of protein synthesis/transport and initiate the translation of a new transport substrate directly at the ER membrane (22). Here, we have shown that ERj1 is associated with ribosomes in vivo in different cell types. The increased fluorescence intensity of ERj1 signals after RNase treatment varied between the cell types but was similar to the values obtained for the Sec61 complex. Thus, just like the Sec61 complex, ERj1 is a ribosome-associated ER membrane protein and therefore appears to be involved in protein biogenesis at the ER surface.
How do the affinities of ERj1 for ribosomes fit into the above model? ERj1 has a higher affinity for ribosomes (30 pm) than the Sec61 complex (∼20 nm) (23). However, the ERj1-BiP complex has a lower affinity for ribosomes compared with BiP-free ERj1 and, one could speculate, also the Sec61 complex. The inference is that BiP-free ERj1 binds to ribosomes and prevents initiation of presecretory protein synthesis. Under these conditions, ERj1 may occupy the ribosomal tunnel exit, as we have observed by cryo-EM for ERj1C (13). When BiP is available for binding to ERj1, however, initiation of translation should be allowed, and the simultaneous transfer of the tunnel exit to the Sec61 complex occurs. We postulate that the BiP-bound form of ERj1 binds to the ribosome in a different mode compared with the BiP-free form, allowing simultaneous binding of ERj1 and Sec61 to the ribosome. This mechanism is difficult to visualize on the basis of the reconstructions of the ribosome-Sec61 complex and ribosome-ERj1 (13, 24, 25). However, there are precedents for rearrangement of ligands of the ribosomal tunnel exit. During signal peptide recognition, SRP54 (the 54-kDa subunit of the signal peptide recognition particle) is positioned at the ribosomal tunnel exit. When SRP54 contacts the signal recognition particle receptor at the ER membrane, SRP54 is rearranged to allow the Sec61 complex to dock with the ribosome, leading to insertion of the nascent polypeptide chains into the Sec61 complex (26). Therefore, a similar situation is envisaged for the Sec61 complex and ERj1 at the ER surface. This scenario would be ideal for the recruitment of BiP to either incoming polypeptide chains (to act as a molecular ratchet) (7) or lumenal loops of the Sec61 subunits (to facilitate channel closure to maintain calcium homeostasis) (20, 27). Our current efforts are directed at the formation and cryo-EM analysis of ribosome-ERj1-BiP trimeric complexes and ribosome-ERj1-BiP-Sec61 quaternary complexes.
Furthermore, we studied a second Hsp40-Hsp70 pair (termed mRAC), MPP11 and Hsp70L1, with regard to its ribosome binding properties. MPP11 alone, but especially MPP11 in complex with Hsp70L1, interacted much more strongly with ribosomes compared with ERj1 or ERj1-BiP. The interaction between MPP11 and Hsp70L1 appears to be different from the ERj1-BiP interaction. In contrast to ERj1 and BiP, MPP11 and Hsp70L1 form a stable complex (16). Nevertheless, both Hsp40 proteins are affected in their ribosome affinity by the respective Hsp70 protein. The different binding affinities suggest that mRAC and ERj1 bind to different sites at the ribosome. This is supported by our observation that ERj1 and MPP11 do not compete for ribosome binding in SPR measurements (data not shown) and by findings obtained for the yeast complex. RAC binds the ribosome near the ribosomal protein Rpl31. Rpl31 is located at the ribosomal tunnel exit site on the site opposite to the binding site of ERj1 (28). Thus, the ER lumenal ERj1-BiP and cytosolic MPP11-Hsp70L1 complexes do not seem to represent compartment-specific Hsp40-Hsp70 pairs with an identical mechanism at the ribosome.
Remarkably, almost all of the features of ERj1 are shared by the mammalian Sec63-Sec62 complex: Sec63 provides an ER lumenal J-domain; Sec62 is also protected from antibody access by ribosomes in cells; the cytosolic N-terminal domain of Sec62 (Sec62N) binds to ribosomes with high affinity in a salt- and RNase-sensitive manner; Sec62N contains two positively charged oligopeptides that are essential for ribosome binding; like ERj1, Sec62N inhibits translation at the level of initiation; and Sec62N can be cross-linked to short nascent polypeptide chains at the surface of ribosomes. In addition, Sec62N competes with ERj1C for binding to ribosomes (21). Therefore, we propose that ERj1 and the Sec63-Sec62 complex have identical or overlapping roles in protein biogenesis and that this explains why the loss of Sec63 function associated with polycystic liver disease is not lethal in humans (in contrast to yeast) (29, 30). This view is also supported by the observation that expression of human ERj1 in yeast can complement the loss of Sec63 function (12).
Supplementary Material
Acknowledgments
We are grateful to Linda Hendershot for the kind gift of the BiP expression plasmids. We thank A. Müller, S. Amann, and M. Lerner for expert technical assistance.
This work was supported in part by grants from Homburger Forschungsförderungsprogramm (HOMFOR) and the Deutsche Forschungsgemeinschaft (FOR 967 and SFB530).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- mRAC
- mammalian ribosome-associated complex
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Palade G. (1975) Science 189, 347–358 [DOI] [PubMed] [Google Scholar]
- 2.Deshaies R. J., Sanders S. L., Feldheim D. A., Schekman R. (1991) Nature 349, 806–808 [DOI] [PubMed] [Google Scholar]
- 3.Görlich D., Rapoport T. A. (1993) Cell 75, 615–630 [DOI] [PubMed] [Google Scholar]
- 4.Brodsky J. L., Goeckeler J., Schekman R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young B. P., Craven R. A., Reid P. J., Willer M., Stirling C. J. (2001) EMBO J. 20, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dierks T., Volkmer J., Schlenstedt G., Jung C., Sandholzer U., Zachmann K., Schlotterhose P., Neifer K., Schmidt B., Zimmermann R. (1996) EMBO J. 15, 6931–6942 [PMC free article] [PubMed] [Google Scholar]
- 7.Tyedmers J., Lerner M., Wiedmann M., Volkmer J., Zimmermann R. (2003) EMBO Rep. 4, 505–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer H. A., Grau H., Kraft R., Kostka S., Prehn S., Kalies K. U., Hartmann E. (2000) J. Biol. Chem. 275, 14550–14557 [DOI] [PubMed] [Google Scholar]
- 9.Tyedmers J., Lerner M., Bies C., Dudek J., Skowronek M. H., Haas I. G., Heim N., Nastainczyk W., Volkmer J., Zimmermann R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudek J., Volkmer J., Bies C., Guth S., Müller A., Lerner M., Feick P., Schäfer K. H., Morgenstern E., Hennessy F., Blatch G. L., Janoscheck K., Heim N., Scholtes P., Frien M., Nastainczyk W., Zimmermann R. (2002) EMBO J. 21, 2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudek J., Greiner M., Müller A., Hendershot L. M., Kopsch K., Nastainczyk W., Zimmermann R. (2005) Nat. Struct. Mol. Biol. 12, 1008–1014 [DOI] [PubMed] [Google Scholar]
- 12.Kroczynska B., Evangelista C. M., Samant S. S., Elguindi E. C., Blond S. Y. (2004) J. Biol. Chem. 279, 11432–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blau M., Mullapudi S., Becker T., Dudek J., Zimmermann R., Penczek P. A., Beckmann R. (2005) Nat. Struct. Mol. Biol. 12, 1015–1016 [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto-Taniura N., Pirollet F., Monroe R., Gerace L., Westendorf J. M. (1996) Mol. Biol. Cell 7, 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hundley H. A., Walter W., Bairstow S., Craig E. A. (2005) Science 308, 32–34 [DOI] [PubMed] [Google Scholar]
- 16.Otto H., Conz C., Maier P., Wölfle T., Suzuki C. K., Jenö P., Rücknagel P., Stahl J., Rospert S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10064–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautschi M., Lilie H., Fünfschilling U., Mun A., Ross S., Lithgow T., Rücknagel P., Rospert S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3762–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautschi M., Mun A., Ross S., Rospert S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J., Gaut J. R., Hendershot L. M. (1995) J. Biol. Chem. 270, 26677–26682 [DOI] [PubMed] [Google Scholar]
- 20.Hamman B. D., Hendershot L. M., Johnson A. E. (1998) Cell 92, 747–758 [DOI] [PubMed] [Google Scholar]
- 21.Müller L., de Escauriaza M. D., Lajoie P., Theis M., Jung M., Müller A., Burgard C., Greiner M., Snapp E. L., Dudek J., Zimmermann R. (2010) Mol. Biol. Cell 21, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter M. D., Seiser R. M., Nicchitta C. V. (2001) Trends Cell Biol. 11, 112–115 [DOI] [PubMed] [Google Scholar]
- 23.Prinz A., Behrens C., Rapoport T. A., Hartmann E., Kalies K. U. (2000) EMBO J. 19, 1900–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. (1997) Science 278, 2123–2126 [DOI] [PubMed] [Google Scholar]
- 25.Beckmann R., Spahn C. M.., Eswar N., Helmers J., Penczek P. A., Sali A., Frank J., Blobel G. (2001) Cell 107, 361–372 [DOI] [PubMed] [Google Scholar]
- 26.Pool M. R., Stumm J., Fulga T. A., Sinning I., Dobberstein B. (2002) Science 297, 1345–1348 [DOI] [PubMed] [Google Scholar]
- 27.Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. (2005) J. Cell Biol. 168, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peisker K., Braun D., Wölfle T., Hentschel J., Fünfschilling U., Fischer G., Sickmann A., Rospert S. (2008) Mol. Biol. Cell 19, 5279–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davila S., Furu L., Gharavi A. G., Tian X., Onoe T., Qian Q., Li A., Cai Y., Kamath P. S., King B. F., Azurmendi P. J., Tahvanainen P., Kääriäinen H., Höckerstedt K., Devuyst O., Pirson Y., Martin R. S., Lifton R. P., Tahvanainen E., Torres V. E., Somlo S. (2004) Nat. Genet. 36, 575–577 [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann R., Müller L., Wullich B. (2006) Trends Mol. Med. 12, 567–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.