Abstract
The 26 S proteasome, composed of the 20 S core and 19 S regulatory particle, plays a central role in ubiquitin-dependent proteolysis. Disruption of this process contributes to the pathogenesis of the various diseases; however, the mechanisms underlying the regulation of 26 S proteasome activity remain elusive. Here, cell culture experiments and in vitro assays demonstrated that apoptosis signal-regulating kinase 1 (ASK1), a member of the MAPK kinase kinase family, negatively regulated 26 S proteasome activity. Immunoprecipitation/Western blot analyses revealed that ASK1 did not interact with 20 S catalytic core but did interact with ATPases making up the 19 S particle, which is responsible for recognizing polyubiquitinated proteins, unfolding them, and translocating them into the 20 S catalytic core in an ATP-dependent process. Importantly, ASK1 phosphorylated Rpt5, an AAA ATPase of the 19 S proteasome, and inhibited its ATPase activity, an effect that may underlie the ability of ASK1 to inhibit 26 S proteasome activity. The current findings point to a novel role for ASK1 in the regulation of 26 S proteasome and offer new strategies for treating human diseases caused by proteasome malfunction.
Keywords: Cell Death, JNK, Proteasome, Protein Degradation, Protein Phosphorylation, ASK1, Rpt5
Introduction
The 26 S proteasome is the major proteolytic complex carrying out ubiquitin-mediated substrate degradation in eukaryotic cells. This multiprotein complex is central to the regulation of many basic cellular processes including cell cycle progression, apoptosis, stress response, and the regulation of immune and inflammatory responses (1). The 26 S proteasome is an ATP-dependent complex that is made up of the functionally and structurally distinct 20 S core particle and 19 S regulatory complex. The 20 S core particle contains three distinct peptidase activities including trypsin-like, chymotrypsin-like, and peptidyl glutamyl-peptide-hydrolyzing activity (2). To reach the active sites of the 20 S core particle, native substrate proteins must first be unfolded by the 19 S regulatory particles and then be threaded into the 20 S core. The 19 S regulatory complex can be subdivided into two subcomplexes, known as the “base” and “lid.” The base is primarily composed of three non-ATPase subunits (known as regulatory particle non-ATPases or Rpns)3 and six ATPases (known as regulatory particle AAA ATPases or Rpts), whereas the lid contains at least nine non-ATPase subunits (3). Aberrations in the 26 S proteasome have been implicated in the pathogenesis of several human diseases in which some proteins are lost or accumulated due to their accelerated or decreased degradation (e.g. cancer, AIDS, and neurodegenerative diseases) (4). Therefore, understanding the mechanisms that regulate proteasomal substrate digestion and identifying the molecules that modulate 26 S proteasome function are of great importance.
Apoptosis signal-regulating kinase 1 (ASK1) is a member of the mitogen-activated protein kinase (MAPK) kinase kinase family and activates JNK/SAPK and p38 cascades (5). ASK1 is activated in response to diverse stresses and apoptotic stimuli that participate in the pathogenesis and exacerbation of various human diseases. These include oxidative stress, death receptor ligands, lipopolysaccharide, and endoplasmic reticulum stress (6, 7). Activated ASK1 directly phosphorylates MKK3/MKK6 and MKK4/MKK7, resulting in the activation of p38 and JNK, respectively. Importantly, ASK1 has been shown to play an essential role in stress- and cytokine-induced apoptosis, a finding underscored by the resistance of ASK1-deficient mice to such forms of apoptosis (8). In addition to phosphorylating the MKKs, ASK1 phosphorylates JNK/stress-activated protein kinase-associated protein 1, allowing it to regulate the function of this JNK scaffolding protein (9). ASK1 also phosphorylates cardiac troponin T to regulate cardiac contractile function (10).
Nishitoh et al. (11) reported that ASK1 deficiency reduces apoptosis in cells incubated with various proteasomal inhibitors, suggesting that ASK1 is required for proteasomal dysfunction-induced cell death. This led us to investigate the possible functional link between ASK1 and the proteasome. Here, we demonstrate that ASK1 is a novel negative regulator of 26 S proteasome. We also provide evidence that this negative regulatory effect is attributable to the ability of ASK1 to phosphorylate and inhibit Rpt5, a 19 S proteasomal ATPase.
EXPERIMENTAL PROCEDURES
Materials and cDNA Constructs
Peroxidase-conjugated anti-rabbit and anti-mouse antibodies were purchased from Zymed Laboratories Inc..(San Francisco, CA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and Lipofectamine PLUS reagents were obtained from Invitrogen. Clasto-lactacystin β-lactone was purchased from A. G. Scientific (San Diego, CA). λ-Protein phosphatase (P0753S) was purchased from New England Biolabs (Beverly, MA). Fluorogenic peptides for proteasomal degradation (Bz-VGR-AMC, Suc-LLVY-AMC, Z-LLE-AMC,), fluorogenic peptide for caspase-3 substrate (AC-DEVD-AMC), purified 26 S proteasome, anti-α1, -2, -3, -5, -6, and -7 subunits of 20 S proteasome, anti-Rpt2, anti-Rpt5, and anti-Rpn10 were purchased from Enzo Life Science (Plymouth Meeting, PA). Epoxomicin and Z-VAD(Ome)-FMK were purchased from Calbiochem. Anti-HA, anti-GFP, anti-α-tubulin, anti-Hsp90, anti-ubiquitin, anti-IκBα, and anti-Myc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-JNK and anti-phospho-JNK antibodies were purchased from Cell Signaling Technology (Beverly, MA), and anti-ASK1 antibody was from Abcam (Cambridge, MA). Cycloheximide, anti-FLAG®, anti-actin, and anti-FLAG M2 affinity gel (A2220) were purchased from Sigma-Aldrich. Rabbit polyclonal anti-T7 antibody was purchased from KOMA Biotech (Seoul, Korea), and mouse monoclonal anti-T7 antibody was from Novagen (Madison, WI).
Plasmid encoding UbG76VGFP (12) was a kind gift from N. P. Dantuma (Karolinska Institutet, Stockholm, Sweden). Plasmids encoding FLAG-tagged Rpn1, Rpn10, Rpt6, Rpt5, α1 of the 20 S core, and β4 of the 20 S core as well as plasmids encoding for T7-Rpt1, T7-Rpt2, T7-Rpt3, T7-Rpt6, T7-Rpn6, Tet-Off transactivator, GFP-fused ornithine decarboxylase (GFP-ODC), and GFPu were prepared as described previously (13). Human Rpt5 was amplified using following primers: forward primer 5′-GCACAACGCGTATGAATCTGCTGCCGAATATTG(MluI)-3′ and reverse primer 5′-GCACAGCGGCCGCCTAGGCGTAGTATTGTAGGTTGGCTTT(NotI)-3′, and subcloned into pRK5-FLAG-vector using MluI and NotI. Plasmids encoding HA-tagged full-length ASK1 and HA-tagged ASK1 deletion mutants (ΔN-ASK1 (ASK1649–1375), ΔC-ASK1 (ASK11–936), or NT-ASK1 (ASK11–656)) were kindly provided by E. J. Choi (Korea University, Seoul, Korea). Plasmids encoding HA-tagged ASK1-K709R mutant (ASK1-KM) and Δ1–277-ASK1 (ASK1278–1375) were kindly provided by H. Ichijo (The University of Tokyo, Tokyo, Japan). Plasmid encoding HA-tagged ΔC-ASK1-K709R mutant was generated through the mutagenesis of HA-ΔC-ASK1 using QuikChange® XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and amplified using the following primers: forward primer 5′-AACCAAGTCAGAATTGCTATTAGGGAAATCCCAGAGAGAGACAGC-3′ and reverse primer 5′-GCTGTCTCTCTCTGGGATTTCCCTAATAGCAATTCTGACTTGGTT-3′. For purification of recombinant GST-tagged human Rpt5 truncated mutant proteins, human Rpt5 gene was amplified by PCR using the following primers: for Rpt51–198, forward primer 5′-GAATTCATGAATCTGCTGCCGAATATTGA-3′ and reverse primer 5′-CTCGAGCTACAGCTCCTGGATCTGCTTGTC-3′; for Rpt5199–439, forward primer 5′-GAATTCGTGGAGGCCATTGTCTTGC-3′ and reverse primer 5′-CTCGAGCTAGGCGTAGTATTGTAGGTTGGCTTT-3′; for Rpt5199–355, the same forward primer used for Rpt5199–439 and reverse primer 5′-CTCGAGCGGGAACTCTATCTTGCG-3′; for Rpt5356–439, forward primer 5′-GAATTCATGCCCAATGAGGAGGCC-3′ and the same reverse primer used for Rpt5199–439; for Rpt5199–275, the same forward primer used for Rpt5199–439 and reverse primer 5′-CTCGAGCTAGGCCAGGGCAAAGGC-3′; for Rpt5276–439, forward primer 5′-GAATTCAAGGAGAAAGCGCCCTCTATC-3′ and the same reverse primer used for Rpt5199–439. The PCR products were subcloned into pGEX-4T-1 vector using EcoRI and XhoI sites.
Cell Culture and Preparation of Cell Lysates
HeLa Tet-Off (Clontech) cells, human embryonic kidney 293 (HEK293) cells, mouse neuroblastoma N2a cells, and rat C6 glioma cells were maintained in DMEM containing 10% FBS and 100 units/ml penicillin-streptomycin. Cell lysates were prepared by rinsing cells twice with ice-cold phosphate-buffered saline (PBS) and solubilizing them in lysis buffer containing 10 mm Tris (pH 7.4), 1.0% Nonidet P-40, 150 mm NaCl, 10% glycerol, 1 mm Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 10 mm NaF, and 0.2 mm phenylmethylsulfonyl fluoride (PMSF). For detection of endogenous ASK1 protein, cell lysates were prepared in lysis buffer containing 8 m urea, 50 mm Tris (pH 8.0), 1.0% Triton X-100, and 0.2 mm PMSF.
Immunoprecipitation
One microgram of appropriate antibody was incubated with 1 mg of cell extracts prepared in lysis buffer overnight at 4 °C. The mixture was then incubated with 30 μl of a 1:1 suspension of protein A-Sepharose beads for 2 h at 4 °C with gentle rotation. The beads were pelleted and washed five times with cell lysis buffer. The immunocomplexes were dissociated by boiling the mixture in SDS-PAGE sample buffer. Whole protein samples were then separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in TBST buffer containing 20 mm Tris, pH 7.6, 137 mm NaCl, 0.05% Tween 20, and 5% nonfat dry milk for 1 h at the room temperature and then incubated overnight at 4 °C in 3% nonfat dry milk containing the appropriate primary antibodies. The membrane was washed several times in TBST and incubated with horseradish peroxidase-coupled secondary IgG. After 60 min, the membrane was washed several times with TBST, and bands were visualized using ECL reagents (PerkinElmer Life Sciences).
RNA Interference
The small interference RNAs (siRNA) for human ASK1 were purchased from Invitrogen (Stealth Select RNAi, 5′-CCUGUGCUAACGACUUGCUUGUUGA-3′ (sense), 5′-UCAACAAGCAAGUCGUUAGCACAGG-3′ (antisense)). Stealth RNAi negative control (Invitrogen) was used as a control. HEK293 cells seeded into 12-well plates were transfected with 100 nm siRNA using Lipofectamine RNAiMAX (Invitrogen).
RT-PCR
Total RNAs from ASK1+/+ mouse embryonic fibroblasts (MEFs) or ASK1−/− MEFs were isolated with TRIzol reagents (Invitrogen) and reverse-transcribed using SuperScript® III reverse transcriptase (Invitrogen) according to the manufacturer's guide. PCR assays were performed with Taq polymerase (BIONEER, Seoul, Korea). Sequences of oligonucleotide primers used in RT-PCR amplifications are as follows: mASK1 (NM_008580, 506–905 bp) forward primer 5′-GGACTTCGGGGAAACCGCCG-3′, reverse primer 5′-GGAACTCGCTTGCGCCACCT-3′ (400 bp); mGAPDH (NM_008084, 570–928 bp) forward primer 5′-ACCACAGTCCATGCCATCAC-3′, reverse primer 5′-AAGGTGGAAGAGTGGGAGTTG-3′ (359 bp).
Measurement of 26 S Proteasome Catalytic Activity
HEK293 or MEFs cells were mock-transfected or transfected with HA-tagged wild type ASK1, ΔC-ASK1, or ASK1-KM, and cell lysates were incubated for 1 h at 37 °C with reaction buffer containing 20 mm Tris-HCl (pH 7.4), 2 mm MgCl2, and 80 μm fluorogenic peptide (Suc-LLVY-AMC, Z-LLE-AMC, or Bz-VGR-AMC) in the presence of 1 mm ATP. The amount of cleaved AMC fragment was quantified using a Victor® X3 multilabel plate reader (PerkinElmer Life Sciences) at λEX = 380/λEM = 460.
Measurement of in Vitro 26 S Ubiquitin-dependent Proteasomal Activity
Proteasome activity was assayed using ubiquitin-conjugated lysozyme as a substrate. Ubiquitin-conjugated lysozyme was generated by ubiquitinating lysozyme in vitro, using a ubiquitin protein-conjugation kit (Calbiochem). The resulting ubiquitinated proteins were flash-frozen in aliquots and thawed just before use. Proteasome activity was assayed using the 26 S proteasome degradation kit (Calbiochem) according to the manufacturer's instructions. Briefly, after HEK293 cells were mock-transfected or transfected with HA-tagged ASK1-WT or ASK1-KM, immunoprecipitation of cell lysates was performed with anti-HA antibody. Purified 26 S proteasome (10 nm) and the anti-HA immunocomplexes were preincubated with 10 μm lactacystin for 30 min at 37 °C as a positive control. An equal amount of polyubiquitinated lysozyme (10 nm) was added and incubated for 60 min at 37 °C in buffer containing Mg2+/ATP, purified 26 S proteasome, and the anti-HA immunoprecipitates. The reaction was terminated by adding an equal volume of 2× sample buffer. The reaction products were analyzed by Western blot using anti-ubiquitin or anti-lysozyme antibodies.
Native Gel Immunoblotting
After HEK293 cells were transfected with HA-tagged wild type ASK1 or mutant ASK1, cell lysates were separated on 4–12% Novex® Tris-glycine gels (Invitrogen) in 1× NativePAGE running buffer (Invitrogen) for 3 h at 150 V. Proteins were then transferred to a PVDF membrane (Amersham Biosciences) at 25 V for 2 h in 1× NativePAGE transfer buffer (Invitrogen). The PVDF membrane was incubated for 10 min in methanol to fix the proteins, blocked in 5% milk for 1 h, and incubated overnight with mouse monoclonal antibody to α1, -2, -3, -5, -6, and -7 subunits of 20 S proteasome antibody (BIOMOL) or antibody to the Rpn10 (19 S) subunit (BIOMOL) in TBST. The blot was incubated with horseradish peroxidase-coupled secondary IgG for 1 h, washed several times with TBST, and processed using ECL reagents.
In Vitro Kinase Assay
After HEK293 cells were mock-transfected or transfected with HA-tagged ASK1, immunoprecipitation was performed with anti-HA antibody. The resulting immunoprecipitates were rinsed with lysis buffer and with kinase buffer containing 20 mm Tris (pH 7.4), 20 mm MgCl2. The samples were then incubated for 30 min at 30 °C in kinase reaction buffer containing 1 μCi of [γ-32P]ATP and 1 μg of GST-fused recombinant protein as a substrate, such as GST, GST-Rpt51–198, GST-Rpt5199–439, GST-Rpt5199–355, GST-Rpt5356–439, GST-Rpt5199–275, or GST-Rpt5276–439. The reaction mixtures were subjected to SDS-PAGE, and the phosphorylation of substrate proteins was observed by autoradiography.
ATPase Activity Assay
Rpt5 ATPase activity was determined using an ATP determination kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Briefly, FLAG-immunoprecipitates from cells overexpressing FLAG-tagged Rpt5 and/or HA-tagged wild type ASK1, ΔN-ASK1, NT-ASK1, nonspecific control siRNA (nc siRNA), or ASK1 siRNA were incubated for 15 min with reaction buffer containing 1 μm ATP and 1.25 μg/ml firefly luciferase. The initial ATP concentration in the reaction mixtures was 1 μm, and the ATP remaining after ATP hydrolysis was measured using the ATP determination kit. The reaction was initiated by adding 0.5 mm d-luciferin. Ten seconds later, luminescence detection was performed using a Victor X3 multilabel plate reader. A standard curve was generated by measuring the luminescence of reaction mixtures containing varying concentrations of ATP ranging from 1 nm to 1 μm. Background luminescence was subtracted from all samples.
Caspase Assay
Cell lysates were prepared by rinsing cells twice with ice-cold PBS and solubilizing in lysis buffer containing 50 mm Tris (pH 7.0), 2 mm EDTA, 1% Triton X-100, 1 mm Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 10 mm NaF, and 0.2 mm PMSF. Cell lysates were incubated with 25 μm AC-DEVD-AMC and reaction buffer (100 mm HEPES, pH 7.4, 10% sucrose, 5 mm DTT, 0.1% CHAPS) at 37 °C for 1 h. The amount of cleaved AMC fragment was quantified using a Victor X3 multilabel plate reader (PerkinElmer Life Sciences) at λEX = 380/λEM = 460.
Gel Filtration Analysis
HEK293 cells transiently overexpressing wild type ASK1 were harvested in PBS, washed once with PBS, and homogenized with an ultrasonicator in 1 ml of buffer A (25 mm Tris-HCl, pH 7.5, 5 mm DTT, 2 mm MgCl2, 2 mm ATP), and centrifuged at 10,000 × g for 15 min. The resulting supernatant was applied to a gel filtration column (Superdex 200 10/300 GL, Amersham Biosciences) after equilibration with buffer A. One hundred-microliter aliquots of fractions were collected and subjected to immunoblotting with anti-20 S core, anti-Rpt2, and anti-HA antibodies.
Generation of ASK1 Null Mouse Embryonic Fibroblast Culture
A knock-out mouse harboring the first coding exon and a part of the intron deletion in ASK1 gene (ASK1−/− mice) was kindly provided by H. Ichijo (The University of Tokyo). MEFs were collected from day 12.5 mouse embryos and cultured in DMEM supplemented with 10% fetal calf serum and 100 units/ml penicillin G in a 5% CO2 atmosphere at 37 °C. Experiments were performed on MEFs between passages 2 and 5. For reconstitution of ASK1 into ASK1−/− MEFs, ASK−/− MEFs cells were transfected with HA-tagged wild type ASK1 or its kinase-inactive mutant using Lipofectamine PLUS reagents (Invitrogen).
Statistical Analysis
Significant differences in fluorescence intensity of 26 S proteasomal activity and ATPase activity were determined with an unpaired Student's t test using Sigma Plot 9.0. Values were expressed as the mean ± S.E.
RESULTS
ASK1 Modulates the 26 S Proteasome Activity in Cultured Cells
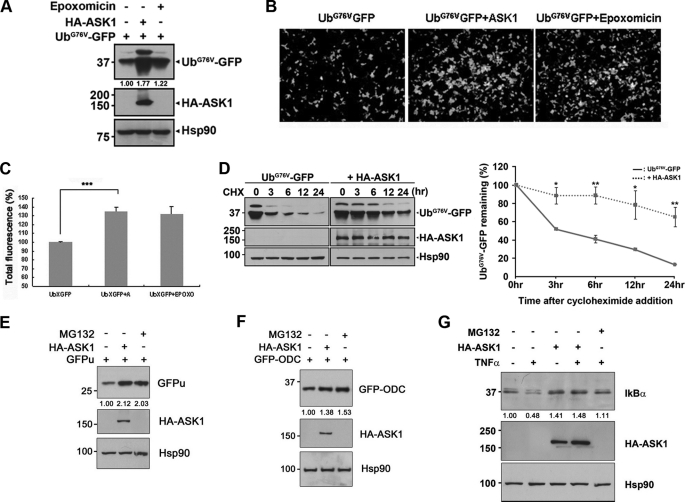
First, we investigated the effect of ASK1 overexpression on the 26 S proteasome activity in HEK293 cells. To measure the activity of proteasome, we used a reporter plasmid encoding a ubiquitin-fused GFP-based substrate (UbG76VGFP). In this substrate, the polyubiquitin chain is covalently attached to GFP to prevent its removal by deubiquitination machinery (12). As UbG76VGFP is only degraded solely by the 26 S proteasome, the steady-state levels of UbG76VGFP are inversely proportional to the intracellular activity of the 26 S proteasome. Accordingly, Western blot analysis of HEK293 cell lysates revealed that UbG76VGFP levels were increased in the presence of epoxomicin, a potent and selective proteasomal inhibitor (Fig. 1A). Interestingly, the level of UbG76VGFP was higher in cells expressing both ASK1 and UbG76VGFP than in those expressing UbG76VGFP alone (Fig. 1A), whereas the GFP level was not affected by ASK1 (supplemental Fig. S1). This finding was confirmed by fluorescence microscopy (Fig. 1B) and quantification of fluorescence intensity (Fig. 1C), the latter of which revealed that ASK1 increased UbG76VGFP fluorescence by more than 35%. In addition, we investigated whether the effect of ASK1 on the level of UbG76VGFP was induced by an inhibition of UbG76VGFP degradation using cycloheximide chase experiments. As shown in Fig. 1D, UbG76VGFP was degraded significantly slower in the presence of ASK1 when compared with the absence of ASK1 (Fig. 1D).
FIGURE 1.
ASK1 reduces 26 S proteasome activity. A–C, HEK293 cells were co-transfected with HA-tagged ASK1 and UbG76VGFP and then cultured in the presence or absence of 100 nm epoxomicin for 6 h. Total cell lysates were subjected to Western blot analysis using anti-GFP or anti-HA antibodies. Hsp90 served as a loading control (A). Alternatively, UbG76VGFP degradation was measured at λEX = 480/λEM = 510 using a fluorescence microplate reader (***, p < 0.001) (C) or observed by fluorescent microscopy (B). D, HEK293 cells were transfected with UbG76VGFP alone or together with HA-tagged ASK1 and then treated with 25 μg/ml cycloheximide (CHX) for the indicated time periods. Total cell lysates were subjected to Western blot analysis with anti-GFP or anti-HA antibodies. Hsp90 served as a loading control. Western blots are representative of three independent experiments. Left panel, quantification of relative UbG76VGFP levels from images of three independent experiments (**, p < 0.05, *, p < 0.5). E and F, HEK293 cells were transfected with HA-ASK1 and GFPu (E) or GFP-ODC (F) in the presence or absence of 10 μm MG132 for 6 h. Total cell lysates were subjected to Western blot analysis using anti-GFP or anti-HA antibodies. Hsp90 served as a loading control. G, N2a cells were mock-transfected or transfected with HA-tagged ASK1. After C6 glioma cells were treated with 5 ng/ml TNFα for 24 h, the cell culture media were transferred to the N2a cells overexpressing HA-ASK1. Thirty minutes later, N2a lysates were collected for Western blotting with anti-IκBα or anti-HA antibodies. Hsp90 served as a loading control. The values at the bottom of the top panel in each figure indicate the relative intensities of UbG76VGFP (A), GFPu (E), GFP-ODC (F), or IκBα (G) bands measured using the ImageJ program (NIH). Western blot images and quantification values in E–G are representative of three independent experiments.
As ASK1-induced apoptotic pathways include the activation of caspase-3 (14, 15), we next examined whether downstream caspase activity may have an effect on the level of UbG76VGFP. After cells were co-transfected with UbG76VGFP and ASK1, the level of UbG76VGFP was measured in the absence or presence of pan-caspase inhibitor, Z-VAD-FMK. Although the addition of Z-VAD-FMK led the blockade of ASK1-induced casapse-3 activity to the control level, the amount of UbG76VGFP was comparable between the cells treated and not treated with Z-VAD-FMK (supplemental Fig. S2). This finding suggests that ASK1-induced caspase-3 activation has no effect on the regulation of 26 S proteasome.
The effect of ASK1 on the 26 S proteasome was also measured using two other GFP-based constructs, GFP-ODC and GFPu. The C-terminal 37 amino acids of ODC act as a degradation signal by allowing ODC to bind to and to be degraded by the 26 S proteasome without polyubiquitination or the action of any other target protein (16, 17). GFPu carries a 16-amino acid consensus degradation signal known as degron CL1 that targets GFPu for polyubiquitination and subsequent proteasomal degradation (18). Similar to UbG76VGFP, ODC or GFPu was more abundant in cells expressing both ASK1 and ODC-GFPu than in those expressing ODC/GFPu alone (Fig. 1, E and F). These data indicate that ASK1 overexpression inhibits 26 S proteasomal activity.
To further investigate the inhibitory effect of ASK1 on the 26 S proteasome, we determined whether ASK1 affects the level of endogenous IκBα, a well known target of proteasome-mediated turnover. Inflammatory cytokines such as TNFα induce targeted phosphorylation and subsequent degradation of IκBα, leading to the activation of and nuclear translocation of NF-κB (19, 20). Consistent with the previous reports, TNFα treatment caused a significant reduction in endogenous IκBα (Fig. 1G). Pretreatment of cells with MG132 abrogated this reduction in IκBα (Fig. 1G), indicating that the steady-state level of IκBα is controlled by proteasomal degradation. This TNFα-induced decrease in IκBα was also prevented by overexpression of ASK1 (Fig. 1G). These data lend further support for ASK1 inhibition of the 26 S proteasome in cultured cells.
ASK1 Regulates 26 S Proteasome through Two Separate Regions, C-terminal and Kinase Domain
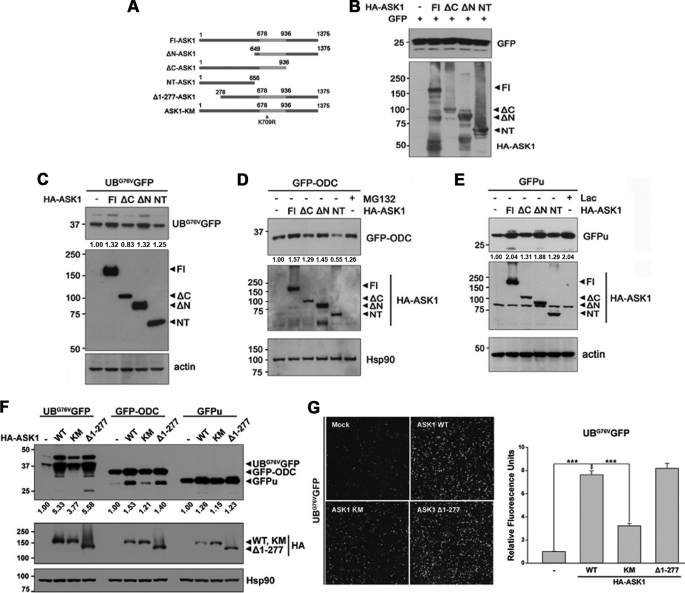
ASK1 is composed of an inhibitory N-terminal domain, an internal kinase domain, and a C-terminal regulatory domain (5, 21, 22). To determine which domain within ASK1 is crucial for 26 S proteasome inhibition, we expressed UbG76VGFP along with full-length ASK1 (amino acids 1–1375), ASK11–656 (NT-ASK1), ASK11–936 (ΔC-ASK1), or ASK1649–1375 (ΔN-ASK1) in HEK293 cells (Fig. 2A). To rule out any confounding effects of differences in transfection efficiency between the vectors encoding ASK1 and GFP-based vectors, we first analyzed GFP expression in cells transfected with these different forms of ASK1 and an EGFP vector. We found that GFP level did not differ between cells expressing GFP and those expressing each of the various forms of ASK1 (Fig. 2B). Analysis of UbG76VGFP levels in HEK293 cells revealed that full-length ASK1 and ASK-ΔN increased UbG76VGFP but that NT-ASK1 and ΔC-ASK1 did not (Fig. 2C). ASK-ΔN also increased the levels of ODC/GFPu (Fig. 2, D and E). These results indicate that ASK1 reduces 26 S proteasome activity via its C-terminal domain.
FIGURE 2.
The C-terminal domain of ASK1 is critical for the down-regulation of proteasome function. A, schematic diagram of full-length ASK1 (Fl-ASK1), truncation mutants, and catalytically inactive mutant. The ASK1-kinase domain located at 676–936 amino acids is marked in gray. B–E, HEK293 cells were transfected with HA-tagged full-length ASK1 (Fl) or ASK1 deletion mutants (ΔC-ASK1, ΔN-ASK1, and NT-ASK1) along with GFP vector (B), UbG76VGFP (C), GFP-ODC (D), or GFPu (E). Total cell lysates were probed with anti-GFP or anti-HA antibodies. Actin or Hsp90 served as a loading control. The values at the bottom of the top panel indicate the relative intensities of UbG76VGFP (C), GFP-ODC (D), or GFPu (E) bands measured using MultiGauge version 3.1. Lac, lactacystin. F, HEK293 cells were transfected with HA-tagged full-length ASK1 (Fl), its kinase-inactive K709R mutant (ASK1-KM), or constitutive active mutant (Δ1–277-ASK1) along with UbG76VGFP, GFP-ODC, or GFPu. Total cell lysates were probed with anti-GFP or anti-HA antibodies. Western blots are representative of three independent experiments. Hsp90 served as a loading control. The values at the bottom of the top panel indicate the relative intensities of UbG76VGFP, GFPu, or GFP-ODC bands measured using MultiGauge version 3.1. Western blot images and quantification values in B–F are representative of three independent experiments. G, after HEK293 cells were transfected with HA-tagged full-length ASK1, its kinase-inactive K709R mutant (ASK1-KM), or constitutive active mutant (Δ1–277-ASK1) along with UbG76VGFP, UbG76VGFP degradation was observed by fluorescent microscopy (left) or measured at λEX = 485/λEM = 535 using a VICTOR X3 (***, p < 0.001) (right).
Next, to determine whether ASK1-induced reduction of 26 S proteasome activity is dependent on ASK1 kinase activity, we expressed UbG76VGFP, GFP-ODC, or GFPu along with catalytically inactive ASK1-K709R mutant (ASK1-KM) or the constitutive active form of ASK1 (Δ1–277-ASK1). As a result, full-length ASK1 and Δ1–277-ASK1 increased the levels of UbG76VGFP, GFP-ODC, and GFPu, but ASK1-KM did not (Fig. 2F). This finding was confirmed by fluorescence microscopy and quantification of fluorescence intensity of UbG76VGFP (Fig. 2G) or GFP-ODC (data not shown). Taken together, these results suggest that ASK1 reduces 26 S proteasome activity in its kinase activity-dependent manner.
Because the K709R point mutation is located in the kinase domain, but not in C-terminal region (amino acids 937–1375), we then determined the effect of ΔC-ASK1-K709R mutant that has C-terminal truncation plus K709R point mutation on the level of UbG76V-GFP (supplemental Fig. S3). Western blot analysis of GFP demonstrated that the level of UbG76V-GFP in cells transfected with ΔC-ASK1-K709R was much lower than that in cells overexpressing ΔC-ASK1 or ASK1-KM, suggesting that two distinct motifs of ASK1, C-terminal and kinase domain, additively contribute to its repressive activity on 26 S proteasome.
ASK1 Inhibits the 26 S Proteasome in Vitro
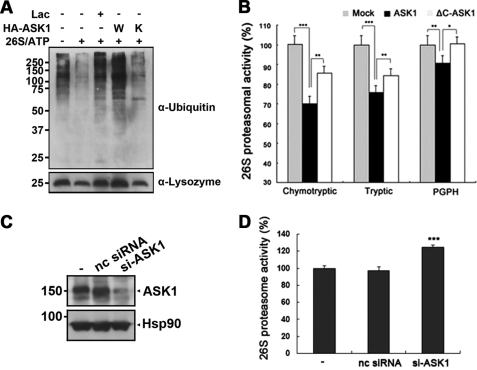
Next, we determined the effect of ASK1 on ubiquitin-dependent 26 S proteasome activity by performing in vitro assays with polyubiquitinated lysozyme as substrate. Anti-HA immunocomplexes from the cells transfected with HA-ASK1-WT or HA-ASK1-KM were incubated in the reaction buffer containing 26 S proteasome and polyubiquitinated lysozyme. The degree of lysozyme degradation by the 26 S proteasome was analyzed by Western blot analysis using anti-ubiquitin or anti-lysozyme antibodies. As shown in Fig. 3A, the amount of polyubiquitinated lysozyme was reduced in the presence of 26 S proteasome. However, the addition of wild type ASK1 led to a dramatic increase in the amount of polyubiquitinated lysozyme, which is comparable with that arising with lactacystin as a positive control (Fig. 3A). Moreover, the addition of ASK1-KM did not enhance the amount of polyubiquitinated lysozyme (Fig. 3A), suggesting that ASK1 inhibits 26 S proteasomal activity in vitro in a kinase-dependent fashion.
FIGURE 3.
ASK1 inhibits ubiquitin-dependent 26 S proteasomal activity in vitro. A, where specified, HEK293 cells were mock-transfected or transfected with HA-ASK1-WT (W) or HA-ASK1-KM (K), and the cell lysates were immunoprecipitated with anti-HA IgG. The immunocomplexes alone or together with purified 26 S proteasome were preincubated with 10 μm lactacystin and incubated with polyubiquitinated lysozyme, as indicated. Reaction products were probed with anti-lysozyme or anti-ubiquitin antibodies. Lac, lactacystin. B, HEK293 cells were mock-transfected (Mock) or transfected with HA-ASK1 (ASK1) or ΔC-ASK1 for 24 h. Lysates were then analyzed for chymotryptic activity using Suc-LLVY-AMC, peptidyl-glutamyl hydrolytic (PGPH) activity using Z-LLE-AMC, or tryptic activity using Bz-VGR-AMC. The amount of cleaved AMC fragment was normalized to values obtained from the mock-transfected group. Experiments were repeated four times (***, p < 0.001, **, p < 0.01). C, after HEK293 cells were mock-transfected (−) or transfected with either ASK1-siRNA (si-ASK1; 100 nm) or nonspecific control siRNA (nc siRNA; 100 nm), Western blot analysis was performed with anti-ASK1 antibodies. Hsp90 served as a loading control. D, the 26 S proteasomal activity in each sample was determined by assaying Suc-LLVY-AMC cleavage. The amount of cleaved AMC fragment was normalized to values obtained from the mock-transfected group. Experiments were repeated four times (***, p < 0.001).
The effect of ASK1 on the three individual ATP-dependent protease activities associated with the 26 S proteasome was also assessed. For this purpose, chymotryptic activity was measured with Suc-LLVY-AMC peptide, peptidyl glutamyl hydrolytic (PGPH) activity with Z-LLE-AMC peptide, and tryptic activity with Bz-VGR-AMC peptide. We found that all three major catalytic activities of the 26 S proteasome were significantly lower in cells overexpressing full-length ASK1 than in control cells or cells overexpressing ΔC-ASK1 (Fig. 3B). On the contrary, the knockdown of endogenous ASK1 increased the steady-state activity of 26 S proteasome by ∼20% (Fig. 3, C and D).
ASK1 Does Not Alter the Steady-state Levels of Proteasomal Subunits or the Natively Assembled 26 S Proteasome
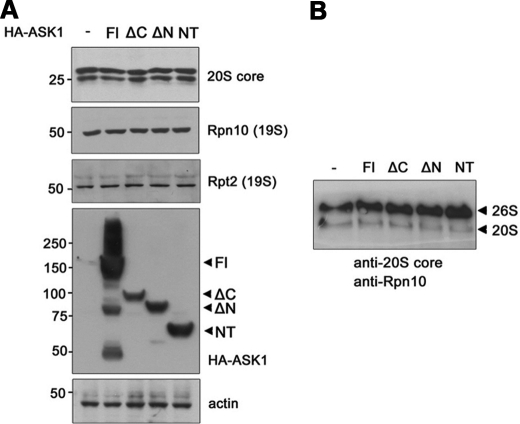
To investigate how ASK1 inhibits 26 S proteasome activity, we tested whether ASK1 affects steady-state levels of the components of the 20 S or 19 S proteasome subunits. Western blot analysis revealed that levels of the 20 S proteasome α subunits and the 19 S regulatory particle Rpn10 and Rpt2 were not changed by overexpression of wild type ASK1 or the ASK1 deletion mutants in HEK293 cells (Fig. 4A). These data suggest that ASK1 does not inhibit 26 S proteasome activity by decreasing steady-state levels of the 19 S or 20 S proteasomal subunits.
FIGURE 4.
ASK1 does not affect the levels of the 20 S or 26 S proteasome components or the assembly/disassembly of the 26 S proteasome. A, HEK293 cells were transfected with plasmids encoding HA-tagged full-length ASK1 (Fl) or one of the ASK1 deletion mutants (ΔC, ΔN, or NT). Cell lysates were then subjected to Western blot analysis using antibodies to the 20 S proteasome α subunits, Rpn10, Rpt2, or HA. Actin served as a loading control. B, the samples prepared in A were resolved on a NativePAGE gel and subjected to Western blot analysis using anti-20 S core and anti-Rpn10 antibodies.
Dissociation of the 26 S proteasome into the 20 S core and 19 S regulatory particle inhibits proteolysis (23). Thus, we used non-denaturing gel electrophoresis to determine whether ASK1 inhibits the 26 S proteasome by enhancing its dissociation into two parts. NativePAGE using 19 S and 20 S antibodies showed that wild type ASK1 and the ASK1 deletion mutants did not affect the structure of the native 26 S proteasome (Fig. 4B).
ASK1 Interacts with ATPases of 19 S Regulatory Particles and Phosphorylates Rpt5 Subunit
To further investigate how ASK1 reduces 26 S proteasome activity, we tried to identify the individual subunits(s) of the 19 S or 20 S proteasomal complex with which ASK1 associates. To this end, plasmid encoding HA-tagged ASK1 and a plasmid encoding one of the 20 S proteasomal subunits (α1 or β4) or one of the 19 S proteasomal subunits (Rpn1, Rpn6, Rpn10, Rpt1, Rpt2, Rpt3, Rpt5, or Rpt6) were co-transfected into HeLa cells. Co-immunoprecipitation/Western blot analysis of HeLa cell lysates revealed that HA-ASK1 associated with neither α1 nor β4 (Fig. 5A). Moreover, the endogenous α subunits of the 20 S core complex did not bind to ASK1 in HEK293 cells (Fig. 5B). The same was true of Rpn1, Rpn6, and Rpn10, the non-ATPases of the 19 S regulatory subunit (Fig. 5, C and D). On the other hand, HA-tagged ASK1 interacted well with Rpt1, Rpt2, Rpt3, Rpt5, and Rpt6, the ATPases subunits of this complex (Fig. 5, D and E). These results suggest that ASK1 associates with the ATPases of the 19 S regulatory complex but not with the 20 S proteasomal subunits.
FIGURE 5.
ASK1 interacts with the ATPases of the 19 S proteasome complex. A, HeLa cells were transfected with HA-ASK1, FLAG-tagged α1, or FLAG-tagged β4. Whole lysates (Input) and anti-HA immunoprecipitates (IP) were then probed with anti-HA or anti-FLAG antibodies. Asterisks indicate IgG heavy chains. B, HEK293 cells were transfected with HA-tagged ASK1. Whole lysates and anti-HA immunoprecipitates were probed with anti-20 S core or anti-HA antibodies. C, HeLa cells were transfected with HA-ASK1 alone or together with FLAG-tagged Rpn1 or Rpn10 for 24 h. Total cell lysates were immunoprecipitated with anti-FLAG antibody, and the immunocomplexes were then probed with anti-HA antibody. D, HeLa cells were transfected with HA-ASK1 alone or together with T7-tagged Rpn6, Rpt1, Rpt2, Rpt3, or Rpt6 for 24 h. Total cell lysates were immunoprecipitated with anti-T7 antibody, and the immunocomplexes were probed with anti-HA antibody. E, HeLa cells were transfected with plasmids encoding HA-tagged ASK1 (full-length (Fl) ASK1 or one of the ASK1 deletion mutants (ΔC, ΔN, or NT)) and/or FLAG-Rpt5. Whole lysates and FLAG-immunoprecipitates were probed with anti-HA or anti-FLAG antibodies. F, HEK293 cells transiently expressing wild type ASK1 were homogenized, and soluble proteins were loaded into a Superdex 200 column. Fractions were subjected to the immunoblot analysis with anti-HA, anti-19 S Rpt2, and anti-20 S core antibodies. G, the high (fraction numbers 13–25) molecular weight fractions prepared in panel F were immunoprecipitated with either preimmune IgG or anti-HA antibodies followed by immunoblot analysis with anti-Rpt2, anti-20 S core, or anti-HA antibodies.
To determine which domain within ASK1 is critical for binding to the 19 S proteasomal subunit, we co-expressed FLAG-tagged Rpt5 with full-length ASK1 (amino acids 1–1375), NT-ASK1 (amino acids 1–656), ΔC-ASK1 (amino acids 1–936), or ΔN-ASK1 (amino acids 649–1375). In addition to binding to full-length ASK1, Rpt5 bound to ΔN-ASK1, indicating that Rpt5 can interact with ASK1 through the C-terminal ASK1937–1375 domain (Fig. 5E). These data account for the finding that among the three ASK1 deletion mutants, only ΔN-ASK1 had the same effect on the 26 S proteasome as wild type ASK1 (Fig. 2).
Next, to investigate whether ASK1 binds to 19 S proteasomal subunits within the context of proteasomal complexes, gel filtration of lysates from HEK293 cells overexpressing HA-ASK1 was performed, and subsequently, the high and low molecular weight protein fractions containing 26 S proteasome and ASK1 were resolved. Immunoblot analyses of the eluted fractions with anti-20 S core, anti-19 S Rpt2, and anti-HA antibodies revealed that the components of the 20 S and 19 S proteasome co-eluted over the same range of high molecular weight fractions (fraction numbers 13–25), whereas ASK1 was present in the lower molecular weight fraction (fraction number 43–55) (Fig. 5F). Interestingly, a considerable portion of ASK1 co-eluted with the high molecular weight fractions containing the 19 S and 20 S proteasome components (Fig. 5F), raising the possibility that ASK1 binds to 19 S proteasomal subunits. A co-immunoprecipitation assay of the high molecular weight fraction revealed that ASK1 interacts with Rpt2, one of the 19 S proteasomal subunits, not with the 20 S core (Fig. 5G). Taken together, these findings suggest that ASK1 binds to the ATPases of the 19 S regulatory complex, but not to the 20 S proteasomal subunits, in the context of proteasomal complexes.
ASK1 Phosphorylates the Rpt5 Subunit of the 19 S Proteasomal Complex
The finding that ASK1 reduces 26 S proteasome activity raises the possibility that ASK1 may phosphorylate components of the 26 S proteasome. Interestingly, when wild type ASK1 or ΔN-ASK1 was co-expressed with Rpt5, Rpt5 appeared as a doublet on Western blots, suggesting that Rpt5 is phosphorylated by ASK1. Both mouse Rpt5 (Fig. 6A) and human Rpt5 (Fig. 6B) appeared as doublets following co-expression with wild type ASK1 or ΔN-ASK1. Although Rpt6 can also bind to ASK1 (Fig. 5C), Rpt6 did not appear as a doublet in the presence of wild typeASK1 or ΔN-ASK1 (Fig. 6C). Observation of the Rpt doublet was not limited to where Rpt5 was overexpressed as endogenous Rpt5 also appeared as a doublet in cells overexpressing ASK1 (Fig. 6D). In addition, the upper band of Rpt 5 induced by ASK1 was reduced by ASK1 siRNA, but not by nonspecific control siRNA (Fig. 6E). To investigate whether the upper band of the Rpt5 doublet resulted from ASK1-mediated phosphorylation, we incubated cell lysates or anti-FLAG immunoprecipitates with λ-protein phosphatase. As shown in Fig. 6F, the upper bands of Rpt5 induced by wild type ASK1 disappeared after the treatment with protein phosphatase (Fig. 6F). To verify this result, we conducted in vitro kinase assays using several truncated Rpt5 forms fused with GST such as Rpt51–198, Rpt5199–439, Rpt5199–355, Rpt5356–439, Rpt5199–275, Rpt5276–439 (Fig. 6G), and GST as a negative control (supplemental Fig. S4). The in vitro kinase assay revealed that ASK1 phosphorylates the N-terminal 1–198 amino acids and C-terminal 356–439-amino acid region of Rpt5 (Fig. 6H), but not GST protein (supplemental Fig. S4). These results demonstrate that ASK1 phosphorylates the N-terminal and C-terminal regions of Rpt5 in vitro.
FIGURE 6.
ASK1 phosphorylates Rpt5 in vivo and in vitro. A–C, HeLa cells were transfected with HA-tagged ASK1 (full-length (Fl) ASK1 or one of the ASK1 deletion mutants (ΔC, ΔN, or NT)) and FLAG-tagged mouse Rpt5 (mRpt5) (A), FLAG-tagged human Rpt5 (hRpt5) (B), or FLAG-tagged Rpt6 (C). Total cell lysates were subjected to Western blot analysis using anti-FLAG or anti-HA antibodies. Actin and Hsp90 served as a loading control. D, HEK293 cells were transfected with HA-tagged wild type ASK1, and lysates were probed with anti-HA or anti-Rpt5 antibodies. α-Tubulin served as a loading control. Mock, mock-transfected. E, HEK293 cells were transfected with FLAG-Rpt5, Tet-Off transactivator, and HA-ASK1 or together with ASK1 siRNA (100 nm) or nonspecific control siRNA (nc siRNA) (100 nm). Total cell lysates were analyzed by Western blot using anti-FLAG or anti-HA antibodies. α-Tubulin served as a loading control. F, HeLa cells were transfected with FLAG-tagged human Rpt5 (Rpt5) and either HA-tagged Fl ASK1 or HA-tagged ΔN-ASK1. Total cell lysates and FLAG-immunoprecipitates were then probed with anti-FLAG or anti-HA antibody. A subset of samples was incubated with λ-protein phosphatase (λ-PPase) for 1 h at 37 °C prior to Western blot analysis. Actin served as a loading control. The values at the bottom of the top and middle panel indicate the intensity ratio of upper Rpt5 bands to lower ones measured using the ImageJ program (NIH). G, the schematic diagrams of GST-fused Rpt51–198, Rpt5199–439, Rpt5199–355, Rpt5356–439, Rpt5199–275, and Rpt5276–439 used as a substrate in panel H. H, HEK293 cells were transfected with HA-ASK1. In vitro kinase assays were performed by incubating HA immunoprecipitates from these cells with various GST-fused truncated forms of Rpt5 shown in panel G. Phosphorylated GST-fused Rpt5 was visualized by autoradiography, and proper expression of GST-fused Rpt5 was confirmed by Coomassie Brilliant Blue (CBB) staining. Asterisks indicate the bands of various truncated forms of GST fused Rpt5.
ASK1 Inhibits the Rpt5 ATPase Activity
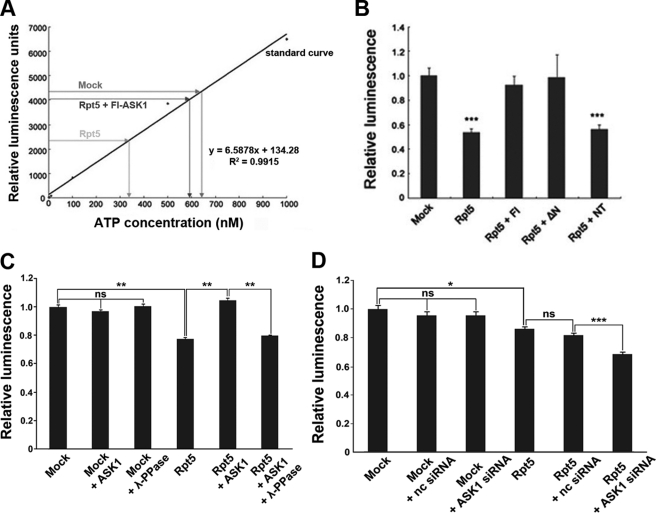
Next, we investigated whether ASK1-mediated phosphorylation of Rpt5 alters the ATPase activity of this 19 S subunit. ATPase activity was determined by measuring residual ATP after ATP hydrolysis by Rpt5. Thus, ATP levels (i.e. luminescence intensity) are inversely proportional to Rpt5 ATPase activity. As shown in Fig. 7A, residual ATP concentrations in assays containing Rpt5 immunoprecipitates from FLAG-Rpt5-expressing cells were ∼300 nm, whereas this value was almost doubled in control cells performed with corresponding immunoprecipitates from mock-transfected cells. Importantly, co-expression of ASK1 with Rpt5 significantly enhanced residual ATP, returning it to a level comparable with that seen in control assays (Fig. 7, A and B). The ATPase activity of Rpt5 was also almost completely blocked when Rpt5 was co-expressed with ΔN-ASK1, but not with NT-ASK1 (Fig. 7B). In addition, when the cell lysates overexpressing Rpt5 plus ASK1 were incubated with λ-protein phosphatase, decreased ATPase activity of Rpt5 by ASK1 was restored to a level comparable with that seen in the absence of ASK1 (Fig. 7C). Moreover, ATPase activity of Rpt5 was significantly increased when the level of ASK1 was knocked down by ASK1 siRNA (Fig. 7D). These results suggest that ASK1 down-regulates 26 S proteasome activity possibly by phosphorylating Rpt5 and consequently inhibiting its ATPase activity.
FIGURE 7.
ASK1 inhibits the ATPase activity of Rpt5. A and B, in vitro ATPase activity assays were performed with FLAG immunoprecipitates collected from HeLa cells expressing FLAG-Rpt5 and/or HA-tagged ASK1 (wild type or mutant ASK1 (ΔN or NT)). A standard curve (A) was generated using an ATP determination kit. The luminescence of each sample was plotted (***, p < 0.001 versus control) (B). Fl Ask1, full-length Ask1; Mock, mock-transfected. C, HEK cells were mock-transfected or transfected with pRK5-FLAG-Rpt5 or together with HA-ASK1. Immunoprecipitation was performed with anti-FLAG M2 affinity gel. A subset of samples was incubated with λ-protein phosphatase (λ-PPase) for 30 min at 37 °C prior to in vitro ATPase activity assay. The luminescence of each sample was plotted (**, p < 0.001; ns, not significant). D, HEK293 cells were mock-transfected or transfected with pRK5-FLAG-Rpt5 alone or together with either 100 nm nonspecific control siRNA (nc siRNA) or 100 nm ASK1 siRNA. Anti-FLAG M2 affinity gel immunoprecipitates were used for in vitro ATPase activity assay. The luminescence of each sample was plotted (*, p < 0.05, ***, p < 0.0005; ns, not significant).
Increased 26 S Proteasomal Activity in MEFs Derived from ASK1 Null Mice
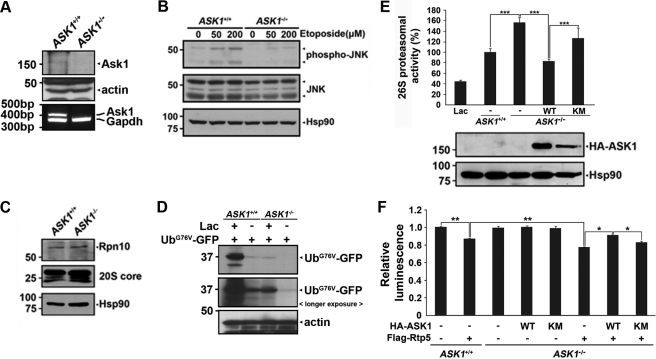
To understand the physiological significance of our cellular and in vitro findings, we compared 26 S proteasome activity in MEFs derived from ASK1−/− and control mice (ASK1+/+ MEFs). Immunoblot and RT-PCR analyses of cell lysates confirmed that intact ASK1 protein and mRNAs are absent in ASK1−/− MEFs (Fig. 8A). Knock-out of ASK1 in ASK1−/− MEFs was additionally verified by assaying the activation of JNK, a downstream target of ASK1. MEFs were exposed to etoposide, a genotoxic agent known to induce apoptosis via the ASK1-JNK signaling pathway. Western blot analysis showed that etoposide treatment led to phosphorylation of p46- and p54-JNK in ASK1+/+ MEFs, but not in ASK1−/− MEFs (Fig. 8B), confirming that ASK1 activation did not occur in ASK1−/− MEFs. Measurement of the steady-state levels of the 20 S and 19 S proteasome revealed that they were comparable between ASK1+/+ MEFs and ASK1−/− MEFs (Fig. 8C). The level of UbG76VGFP expression in ASK1+/+ MEFs was much higher than that in ASK1−/− MEFs (Fig. 8D), but the level of GFP was the same in both MEFs cells (supplemental Fig. S5). However, 26 S proteasome activity was ∼1.6-fold greater in ASK1−/− MEFs than in ASK1+/+ MEFs (Fig. 8E). Moreover, 26 S proteasome activity was reduced in ASK1+/+ MEFs treated with the proteasomal inhibitor lactacystin (Fig. 8E). To ensure the effect of ASK1 on 26 S proteasome, we reconstituted wild type or kinase-inactive mutant of ASK1 into ASK1−/− MEFs and examined its effect on the enhanced proteasomal activity. Immunoblot with anti-HA antibody confirmed that wild type ASK1 or its kinase-inactive mutant was well expressed in ASK1−/− MEFs (Fig. 8E). Moreover, increased 26 S proteasomal activity shown in ASK1−/− MEFs was lowered by reconstitution with wild type ASK1 to the similar degree of that shown in ASK1+/+ MEFs (Fig. 8E). However, overexpression of kinase-inactive ASK1 mutant into ASK1−/− MEFs failed to reduce 26 S proteasomal activity (Fig. 8E). Also, ATPase activity of Rpt5 was up-regulated in ASK1−/− MEFs and ASK1−/− MEFs reconstituted with kinase-inactive mutant of ASK1 when compared with ASK1+/+ MEFs and ASK1−/− MEFs reconstituted with wild type ASK1 (Fig. 8F). These data validate our culture finding that ASK1 negatively regulates 26 S proteasome activity in its kinase activity-dependent manner.
FIGURE 8.
Up-regulation of 26 S proteasomal activity in MEFs derived from ASK1−/− mice. A, total cell lysates of MEFs derived from ASK1+/+ or ASK1−/− mice were subjected to Western blot analysis using anti-ASK1 antibody (upper panel). Total RNAs of ASK1+/+ or ASK1−/− MEFs were subjected to RT-PCR analysis using mASK1 primers (lower panel). Hsp90 and mGAPDH served as loading controls. B, MEFs derived from ASK1+/+ or ASK1−/− mice were treated with 50 or 200 μm etoposide for 8 h, and total cell lysates were subjected to Western blot analysis using anti-phospho JNK or anti-JNK antibodies. Hsp90 served as a loading control. C, Western blot analysis of MEFs lysates using anti-Rpn10 or anti-20 S core antibodies. Hsp90 served as a loading control. D, MEFs derived from ASK1+/+ or ASK1−/− mice were transfected with UbG76V-GFP for 24 h and cultured in the presence or absence of 10 μm lactacystin (Lac) for 6 h. Total cell lysates were subjected to Western blot analysis using anti-GFP antibody. Actin served as a loading control. E, 26 S proteasomal activities in ASK1+/+ MEFs, ASK1−/− MEFs, or ASK1−/− MEFs reconstituted with wild type ASK1 (WT) or its kinase-inactive ASK1 K709R mutant (KM) were assessed by Suc-LLVY-AMC cleavage (***, p < 0.0001). Samples pretreated with 10 μm lactacystin (Lac) served as a positive control (upper panel). Data represent the mean ± S.E. of at least four experiments. ASK1−/− MEFs were transfected with HA-tagged wild type ASK1 or its kinase-inactive ASK1-K709R mutant. Total cell lysates were analyzed by Western blot using anti-HA antibody. Hsp90 served as a loading control (bottom panel). F, ASK1+/+ MEFs or ASK1−/− MEFs were transfected with FLAG-tagged Rpt5 alone or together with HA-ASK1-WT or HA-ASK1-KM. Anti-FLAG M2 affinity gel immunoprecipitates were used for in vitro ATPase activity assay. The luminescence of each sample was plotted (*, p < 0.05, **, p < 0.005).
Activated ASK1 by Stress Stimuli Decreased 26 S Proteasomal Activity
It has been well known that ASK1 is activated under stressful conditions such as oxidative stress and apoptosis (6–8). To examine whether ASK1 activation induced by stress stimuli mediates the down-regulation of 26 S proteasomal activity, we treated ASK1+/+ MEFs and ASK1−/− MEFs with H2O2 or etoposide. ASK1 activation by H2O2 or etoposide was verified by assaying the activation of JNK, a downstream target of ASK1 (Fig. 9, A and B). The addition of proteasomal inhibitor lactacystin increased the level of phospho-JNK (Fig. 9, A and B), which is consistent with the previous reports that proteasome inhibition can activate JNK (24–26). Treatment with H2O2 or etoposide all decreased 26 S proteasomal activity in ASK1+/+ MEFs, but not in ASK1−/− MEFs, which indicates that ASK1 is a key negative regulator of 26 S proteasome under stressful conditions (Fig. 9, A and B). Moreover, consistent with the previous report (27), we also found that H2O2 treatment in ASK1+/+ MEFs does not induce caspase-3 activation (data not shown), further confirming that the inhibition of 26 S proteasomal activity is mainly mediated by ASK1, not by caspase-3 activation.
FIGURE 9.
ASK1 mediates 26 S proteasomal dysfunction induced by H2O2 or etoposide. A and B, MEFs derived from ASK+/+ or ASK−/− mice were treated with 1 mm H2O2 for 1 h (A) or 200 μm etoposide (Eto) for 8 h (B), and total cell lysates were subjected to Western blot analysis using anti-phospho-JNK or anti-JNK antibodies (upper images). Hsp90 served as a loading control. Using these cell lysates, 26 S proteasomal activity was measured by Suc-LLVY-AMC cleavage (**, p < 0.005 versus control) (lower graphs). Samples pretreated with 10 μm lactacystin (Lac) served as a positive control. Data represent the mean ± S.E. of at least four experiments. No T., No treatment.
DISCUSSION
In this study, we have presented several lines of evidence that the activity of 26 S proteasome is negatively regulated by ASK1. ASK1 specifically interacts with ATPases of the 19 S regulatory particles and phosphorylates Rpt5 to inhibit its ATPase activity, an outcome that may underlie the negative regulation for the 26 S proteasome. Although the structure and function of the proteasome have been extensively investigated, the mechanisms responsible for regulation of its activity remain elusive. Recently, reports suggest that proteasome activity might be regulated by post-translational modification of the subunit components. Modification of Rpt2 by O-GlcNAc can inhibit its ATPase activity and subsequently inhibit proteasome function (28). Phosphorylation has been also detected in the core and regulatory subunits of the proteasome, and it has been shown to regulate the assembly of 26 S proteasome (29–32). For example, phosphorylation of the 20 S proteasome α subunit C8 substantially affects the stability of the 26 S proteasome (29). In addition, phosphorylation of Rpt6 promotes the association of the regulatory complex with the 20 S proteasome to form the 26 S proteasome (32). In contrast, the present study shows that ASK1 does not affect the assembly of the native 26 S proteasome. Instead, ASK1 phosphorylates Rpt5 and negatively affects the ATPase activity.
For a globular protein to be degraded, it must associate with the 19 S ATPases, undergo ATP-dependent unfolding, and be translocated into the 20 S core, an event that requires opening of the gate in the α ring (33). The 19 S regulatory particles contain six homologous AAA ATPases arranged as a ring that abuts the outer rings of the 20 S proteasome (34). This topology places them in a critical position to mediate key aspects of proteasome function that require ATP. The 19 S proteasomal ATPase subunits recognize the polyubiquitin degradation signal (35). However, the exact individual functions of the six 19 S proteasomal ATPases (Rpt1–Rpt6) remain unclear. Some reports suggest that Rpt2 inhibits the opening of the gating channel (33, 36). On the other hand, Rpt1 and Rpt6 have been shown to bind Ubr1 and Ufd4 E3s, suggesting that they play a role in recruitment of ubiquitination machineries (37). Rpt5 interacts with a substrate-attached polyubiquitin chain, suggesting that it functions in substrate recruitment (35). Our results show that the ATPase activity of Rpt5 can be regulated by ASK1, and they suggest that Rpt5 ATPase activity is critical for 26 S proteasomal activity. Therefore, in addition to binding to polyubiquitinated proteins, Rpt5 may provide the energy to unfold and translocate them into the 20 S core. Because a portion of the energy required to unfold proteins is used for the dissolution of intramolecular hydrophobic interactions, the loss of ATPase function due to ASK1 phosphorylation may affect this unfolding property. However, we cannot eliminate the possibility that the reduction of Rpt5 ATPase activity affects its ability to recognize polyubiquitinated proteins. Because several ATPase subunits and the 20 S proteasome are known to be phosphorylated within the cells (34, 38), further studies will be necessary to determine whether ASK1 could phosphorylate the other regulatory subunits and whether this phosphorylation may also be involved in the down-regulation of proteasome activity.
Although ATP binding has been reported to be necessary and sufficient for several aspects of 26 S proteasome function, including its assembly and activation (39), ATP hydrolysis is more likely to be required for ATP-dependent protein degradation of ubiquitinated substrates and for the 26 S proteasome assembly. Inactivation of any ATPase by mutation inhibits proteasome function (36). O-GluNAc transferase inhibition of Rpt2 ATPase activity induces proteasomal dysfunction (28). These reports lend support to the idea that ASK1-induced inhibition of Rpt5 ATPase activity can inhibit the proteolytic of the entire organelle.
The early decrease in 20 S and 26 S proteasome activities commonly observed during apoptosis occurs not due to a decrease in their protein concentration, but due to a down-regulation of their proteolytic activities (40). Oxidative stress was demonstrated to decrease the 26 S proteasome activity without affecting 20 S-mediated activity (41). Proteasome activity also declines with age (42, 43). The mechanisms responsible for the down-regulation of proteasome activity under these conditions have yet to be clarified. Although there were a few reports that caspase activated during apoptosis cleaves the regulatory subunits of proteasome and consequently inhibits proteasome function (44, 45), we proved that the dysfunction of proteasome in our system is independent of the caspase activation. Our findings claim that ASK1 mediates proteasomal dysfunction through Rpt5 phosphorylation and subsequently the reduction of Rpt5-ATPase activity. These findings may account for the reduction of 26 S proteasome activity under stressful conditions. In addition, we provided data to imply that the pro-apoptotic effect of ASK1 may be due to its inhibition of the proteasome. This idea is supported by the finding that ASK1−/− cells are resistant to proteasomal inhibitor-induced cell toxicity (11).
In summary, these findings provide an important expansion of the current understanding of the regulation of 26 S proteasome. They also suggest that ASK1 could be a new target for mechanistic approaches aimed at stabilizing proteasomal activity under various stresses.
Acknowledgments
We thank N. P. Dantuma for providing plasmids and Y. Park for technical assistance. We also thank H. Ichijo and E. J. Choi for providing plasmids and valuable comments.
This study was supported by the Brain Research Center of the 21st Century Frontier Research Program Technology (Grant 2009K-001251 to K. C. C.) funded by the Ministry of Education, Science and Technology (MEST), Republic of Korea. This work was also supported by the National Research Foundation of Korea (NRF) grant funded by MEST (Grant 2010-0018916 to K. C. C.) and by Basic Science Research Program through NRF (Grant 2010-0001668 to K. C. C.). This work was also partially supported by a grant from the Korea Healthcare technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (Grant A092004 to K. C. C.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- Rpn
- regulatory particle non-ATPase
- Rpt
- regulatory particle AAA ATPase
- ASK1
- apoptosis signal-regulating kinase 1
- GFP-ODC
- GFP-fused ornithine decarboxylase
- lactacystin
- clasto-lactacystin β-lactone
- MEF
- mouse embryonic fibroblast
- Z
- benzyloxycarbonyl
- Fmk
- fluoromethyl ketone
- AMC
- 7-amido-4-methylcoumarin
- Bz
- benzoyl
- Suc
- succinyl
- Ome
- Val-Ala-Asp
- TBST
- TBS with Tween
- Ub
- ubiquitin
- MKK
- MAPK kinase
- ex
- excitement
- em
- emission
- m
- mouse.
REFERENCES
- 1.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 2.Coux O., Tanaka K., Goldberg A. L. (1996) Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 3.Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A. (1998) EMBO J. 17, 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichijo H., Nishida E., Irie K., ten Dijke P., Saitoh M., Moriguchi T., Takagi M., Matsumoto K., Miyazono K., Gotoh Y. (1997) Science 275, 90–94 [DOI] [PubMed] [Google Scholar]
- 6.Chang H. Y., Nishitoh H., Yang X., Ichijo H., Baltimore D. (1998) Science 281, 1860–1863 [DOI] [PubMed] [Google Scholar]
- 7.Fujisawa T., Takeda K., Ichijo H. (2007) Mol. Biotechnol. 37, 13–18 [DOI] [PubMed] [Google Scholar]
- 8.Matsuzawa A., Ichijo H. (2008) Biochim. Biophys. Acta 1780, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 9.Matsuura H., Nishitoh H., Takeda K., Matsuzawa A., Amagasa T., Ito M., Yoshioka K., Ichijo H. (2002) J. Biol. Chem. 277, 40703–40709 [DOI] [PubMed] [Google Scholar]
- 10.He X., Liu Y., Sharma V., Dirksen R. T., Waugh R., Sheu S. S., Min W. (2003) Am. J. Pathol. 163, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. (2002) Genes Dev. 16, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G. (2000) Nat. Biotechnol. 18, 538–543 [DOI] [PubMed] [Google Scholar]
- 13.Park Y., Hwang Y. P., Lee J. S., Seo S. H., Yoon S. K., Yoon J. B. (2005) Mol. Cell. Biol. 25, 3842–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutuzov M. A., Andreeva A. V., Voyno-Yasenetskaya T. A. (2005) J. Biol. Chem. 280, 25388–25395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatai T., Matsuzawa A., Inoshita S., Mochida Y., Kuroda T., Sakamaki K., Kuida K., Yonehara S., Ichijo H., Takeda K. (2000) J. Biol. Chem. 275, 26576–26581 [DOI] [PubMed] [Google Scholar]
- 16.Hoyt M. A., Zhang M., Coffino P. (2003) J. Biol. Chem. 278, 12135–12143 [DOI] [PubMed] [Google Scholar]
- 17.Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., Kain S. R. (1998) J. Biol. Chem. 273, 34970–34975 [DOI] [PubMed] [Google Scholar]
- 18.Bence N. F., Bennett E. J., Kopito R. R., Raymond J. D. (2005) Methods Enzymol. 399, 481–490 [DOI] [PubMed] [Google Scholar]
- 19.Baeuerle P. A., Henkel T. (1994) Annu. Rev. Immunol. 12, 141–179 [DOI] [PubMed] [Google Scholar]
- 20.Baldwin A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683 [DOI] [PubMed] [Google Scholar]
- 21.Cho S. G., Lee Y. H., Park H. S., Ryoo K., Kang K. W., Park J., Eom S. J., Kim M. J., Chang T. S., Choi S. Y., Shim J., Kim Y., Dong M. S., Lee M. J., Kim S. G., Ichijo H., Choi E. J. (2001) J. Biol. Chem. 276, 12749–12755 [DOI] [PubMed] [Google Scholar]
- 22.Fujino G., Noguchi T., Matsuzawa A., Yamauchi S., Saitoh M., Takeda K., Ichijo H. (2007) Mol. Cell. Biol. 27, 8152–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsasser S., Schmidt M., Finley D., Raymond J. D. (2005) Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K., Furusu A., Xu Q., Konta T., Kitamura M. (2001) J. Immunol. 167, 1145–1150 [DOI] [PubMed] [Google Scholar]
- 25.Meriin A. B., Gabai V. L., Yaglom J., Shifrin V. I., Sherman M. Y. (1998) J. Biol. Chem. 273, 6373–6379 [DOI] [PubMed] [Google Scholar]
- 26.Starace D., Riccioli A., D'Alessio A., Giampietri C., Petrungaro S., Galli R., Filippini A., Ziparo E., De Cesaris P. (2005) FASEB J. 19, 473–475 [DOI] [PubMed] [Google Scholar]
- 27.Zhang S., Lin Y., Kim Y. S., Hande M. P., Liu Z. G., Shen H. M. (2007) Cell Death. Differ. 14, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 28.Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., Kudlow J. E. (2003) Cell 115, 715–725 [DOI] [PubMed] [Google Scholar]
- 29.Bose S., Stratford F. L., Broadfoot K. I., Mason G. G., Rivett A. J. (2004) Biochem. J. 378, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardag-Gorce F., Venkatesh R., Li J., French B. A., French S. W. (2004) Life Sciences 75, 585–597 [DOI] [PubMed] [Google Scholar]
- 31.Marambaud P., Wilk S., Checler F. (1996) J. Neurochem. 67, 2616–2619 [DOI] [PubMed] [Google Scholar]
- 32.Satoh K., Sasajima H., Nyoumura K. I., Yokosawa H., Sawada H. (2001) Biochemistry 40, 314–319 [DOI] [PubMed] [Google Scholar]
- 33.Köhler A., Bajorek M., Groll M., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2001) Biochimie. 83, 325–332 [DOI] [PubMed] [Google Scholar]
- 34.Ferrell K., Wilkinson C. R., Dubiel W., Gordon C. (2000) Trend Biochem. Sci. 25, 83–88 [DOI] [PubMed] [Google Scholar]
- 35.Lam Y. A., Lawson T. G., Velayutham M., Zweier J. L., Pickart C. M. (2002) Nature 416, 763–767 [DOI] [PubMed] [Google Scholar]
- 36.Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y., Varshavsky A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bose S., Mason G. G., Rivett A. J. (1999) Mol. Biol. Rep. 26, 11–14 [DOI] [PubMed] [Google Scholar]
- 39.Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beyette J., Mason G. G., Murray R. Z., Cohen G. M., Rivett A. J. (1998) Biochem. J. 332, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinheckel T., Grune T., Davies K. J. (2000) Methods Mol. Biol. 99, 49–60 [DOI] [PubMed] [Google Scholar]
- 42.Carrard G., Bulteau A. L., Petropoulos I., Friguet B. (2002) Int. J. Biochem. Cell Biol. 34, 1461–1474 [DOI] [PubMed] [Google Scholar]
- 43.Checler F., da Costa C. A., Ancolio K., Chevallier N., Lopez-Perez E., Marambaud P. (2000) Biochim. Biophys. Acta 1502, 133–138 [DOI] [PubMed] [Google Scholar]
- 44.Sun X. M., Butterworth M., MacFarlane M., Dubiel W., Ciechanover A., Cohen G. M. (2004) Mol. Cell 14, 81–93 [DOI] [PubMed] [Google Scholar]
- 45.Cohen G. M. (2005) Cell Death Differ. 12, 1218. [DOI] [PubMed] [Google Scholar]