Abstract
The structure of the dimeric ATP synthase from yeast mitochondria was analyzed by transmission electron microscopy and single particle image analysis. In addition to the previously reported side views of the dimer, top view and intermediate projections served to resolve the arrangement of the rotary c10 ring and the other stator subunits at the F0-F0 dimeric interface. A three-dimensional reconstruction of the complex was calculated from a data set of 9960 molecular images at a resolution of 27 Å. The structural model of the dimeric ATP synthase shows the two monomers arranged at an angle of ∼45°, consistent with our earlier analysis of the ATP synthase from bovine heart mitochondria (Minauro-Sanmiguel, F., Wilkens, S., and Garcia, J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12356–12358). In the ATP synthase dimer, the two peripheral stalks are located near the F1-F1 interface but are turned away from each other so that they are not in contact. Based on the three-dimensional reconstruction, a model of how dimeric ATP synthase assembles to form the higher order oligomeric structures that are required for mitochondrial cristae biogenesis is discussed.
Keywords: ATP Synthase, Electron Microscopy (EM), Membrane Structure, Mitochondria, Yeast
Introduction
The ATP synthase (F1F0-ATP synthase; F-ATPase) found in energy-transducing membranes from bacteria, chloroplasts, and mitochondria functions to harness the energy of a transmembrane proton-motive force for the synthesis of ATP from ADP and inorganic phosphate (1). It is now well documented that the energy-coupling mechanism involves a rotary motion of the central stalk of the F1F0 complex driven by proton conduction through F0 in the forward ATP synthesis direction and an opposite proton-pumping rotation backwards driven by the free energy change of ATP hydrolysis (2–6). The bacterial enzyme possesses a core rotor-stator structure containing eight essential subunits, five in F1 (α, β, γ, δ, and ϵ) and three in F0 (a, b, and c). This core structure is conserved in eukaryotic mitochondria, but it is complemented with numerous regulatory or supernumerary subunits, most of which are associated with the F0 proton channel, with the exception of the mitochondrial F1-ATPase inhibitor protein (IF1) (7) and a novel inhibitory ζ subunit in the ATP synthase of Paracoccus denitrificans and related α-proteobacteria (8). The role of some of the additional F0 subunits has been unveiled by native gel electrophoresis of the homodimeric mitochondrial ATP synthase (9), by proteomic identification of dimer-specific subunits (10), and by genetic deletion experiments in Saccharomyces cerevisiae, where removal of subunits e and g leads to a loss of the dimeric and oligomeric structures of the complex (11). Remarkably, these deletions (11) or genetic fusions and cross-linking (12) also lead to a loss of the cristae organization of the inner mitochondrial membrane. These observations thus indicate that the dimeric and higher oligomeric species of the ATP synthase exist constitutively in mitochondria promoting formation of mitochondrial cristae (11–20). In addition to the F0 dimerizing subunits e and g, subunit h in yeast (21) and bovine and rat IF1 (22) also promote dimerization of the whole mitochondrial F1F0. In consonance with these findings, it has been reported that overexpression or down-regulation of IF1 promotes or diminishes cristae formation in the mitochondria of cultured mammalian cells, respectively (23).
According to the mechanistic model for cristae formation originally introduced by Allen et al. (24), oligomerization of the dimeric ATP synthase promotes curvature and growth of tubular membrane structures (25). The formation of such tubular structures has been observed by electron tomography of intact mitochondria (26, 27). Previously, we resolved the first projection structure of dimeric ATP synthase by means of transmission electron microscopy and image analyses of the dimeric enzyme extracted from bovine heart mitochondria with the mild detergent digitonin (28). These images confirmed the predicted geometry of dimeric ATP synthase (25) with an angle of ∼40° between the two monomers. Subsequent studies confirmed the existence of a 40° angle in the dimer from S. cerevisiae, but structures with larger angles of about 70° in digitonin extracts of mitochondria from Polytomella sp. have also been observed (17, 29–31). The larger angle of the Polytomella enzyme is stabilized by additional subunits not present in yeast and animal ATP synthases, and this enzyme contains nine different proteins named ASA1 to ASA9 instead of the classical subunits that form the peripheral stalk as well as those involved in the dimerization of all other mitochondrial ATP synthases described (31, 32). ASA2, ASA4, ASA7, and ASA9 seem to be part of the peripheral stalk, although ASA1, ASA3, ASA5, ASA6, and ASA8 are likely contributing to the membranous dimerizing interface, with ASA6 being an integral membrane protein (31, 32). It is therefore unclear at this point what the significance of the different dimer angles in the different species is with respect to mitochondrial cristae formation. In this context, it is noted that the current structural information has been limited by the fact that only “side view” projections of the dimer have been reported. Furthermore, several proposals on the positioning of subunits on dimer and oligomer forming interfaces have been made (11, 30, 33–35); however, these lack the structural information of a dimeric three-dimensional model to accommodate properly these subunits in the quaternary structure of the ATP synthase dimer. It is noted, however, that a three-dimensional model of the monomeric yeast ATP synthase has been obtained by cryo-EM (36), and the position of dimerizing subunits e and g has been inferred from difference maps compared with the cryo-EM structure of the monomeric bovine mitochondrial enzyme (37). More recently, electron tomography has been used to visualize the three-dimensional structure of the ATP synthase dimer in sonicated mitochondrial fragments from bovine and rat (38) and in Polytomella mitochondria (39). Unfortunately, the current resolution of the tomographic reconstructions does not allow for a detailed description of the subunit arrangement in the dimeric enzyme.
To shed light on some of these issues, we have now analyzed the three-dimensional structure of the dimeric ATP synthase from the yeast S. cerevisiae by transmission electron microscopy and image reconstruction. Averaged two-dimensional projections displayed a variety of angles from ∼25° to about 160°, with the majority of molecules projected at an angle of about 40–45° as observed previously for the bovine enzyme (28). By including top and intermediate next to the previously reported side view orientations in the analysis, we were able to reconstruct a three-dimensional model of the dimeric ATP synthase complex at a resolution of 27 Å. The reconstruction shows that the peripheral stalks of each monomer are near the dimer interface but not in contact with each other. Based on the three-dimensional model of dimeric ATP synthase, we suggest that according to previous proposals (24, 25) oligomer formation occurs by packing dimers together with an offset so as to bring the peripheral stalks of two adjacent dimers into contact, consistent with available cross-linking data (11–15, 19, 40). The resulting diagonal polymer is in agreement with the model of a helical ATP synthase oligomer wrapping the tubular cristae as originally proposed by Allen et al. (24, 25).
EXPERIMENTAL PROCEDURES
Materials
Digitonin was obtained from Fluka (catalog number 370006) and was used without re-crystallization. Acrylamide was purchased from Bio-Rad, and Coomassie Blue G-250 was from Serva. All other chemicals were of analytical grade.
Yeast Strains and Growth Conditions
The yeast strain used in this study, W303-1A, was kindly provided by Prof. R. M. Stuart. Strains were grown in YPD medium supplemented with 80 mg/liter adenine and 20 mg/liter uracil at 30 °C using a 10-liter fermenter (Electrolab).
Isolation of Mitochondria
All steps were performed at 4 °C unless otherwise noted. Yeast cells were harvested during the second log phase of the diauxic growth by centrifugation at 2600 × g, washed in water, resuspended in isolation buffer (5 mm MES, pH 6.8, 600 mm mannitol, 1 mg/ml BSA), and disrupted using glass beads (0.25–0.5 mm) in a homemade bead beater at a temperature below 10 °C inside the chamber. Mitochondria were isolated as described previously (41) and stored at −80 °C until further use. Briefly, the initial cell homogenate was subjected to low speed centrifugation (750 × g, 5 min) to pellet cell debris, and the supernatant was then subjected to centrifugation (10,900 × g, 10 min) to pellet mitochondria. The pellet was resuspended in the extraction buffer and cleared of aggregated material by centrifugation at 3600 × g for 5 min, and mitochondria were collected from the supernatant by centrifugation at 17,000 × g.
Solubilization of Dimeric F1F0-ATP Synthase
Isolated mitochondria were diluted to 15 mg/ml (protein) in solubilization buffer (50 mm BisTris3-HCl, pH 7, 750 mm ϵ-amino caproic acid) and extracted with digitonin (2 mg of detergent per mg of protein). Afterward, the samples were centrifuged at 88,000 × g for 45 min, and the soluble fraction was recovered. Protein concentration determination was carried out by the method of Lowry et al. (42) as improved by TCA precipitation (43). Blue native (BN)-PAGE was carried out in gradient gels (4–11% acrylamide) according to Schägger and von Jagow (44) as described previously (28).
Enrichment of Dimeric F1F0-ATP Synthase
This was carried out as described previously for the bovine mitochondrial enzyme (28) with the following modifications: 4 mg (protein) of mitochondrial digitonin extract was loaded on a discontinuous glycerol gradient (15–40% (v/v), 2 mm MES, pH 7.0, 2 mm EDTA, 2 mm ADP, 2.5 mm digitonin) made by stepwise addition of glycerol solutions of different densities and centrifuged at 120,000 × g for 16 h. Afterward, 0.5-ml fractions were collected from the bottom of the gradient. 40 μl of glycerol gradient fractions were mixed with 10 μl of supplemental buffer (0.2 mg of Coomassie Blue/ml solubilization buffer) before running BN-PAGE.
In-gel ATPase Activity Staining
Solubilized mitochondrial inner membranes or glycerol gradient fractions were subjected to BN-PAGE. Lanes of interest were incubated overnight at room temperature with ATPase activity buffer (50 mm glycine, 10 mm MgATP, 0.15% (w/v) lead acetate, pH 8.5), and precipitated lead phosphate was observed by densitometry as white bands after scanning on a dark background.
Electron Microscopy
Fractions of dimeric F1F0-ATP synthase were diluted 10-fold (2 mm MES, pH 7, 2 mm EDTA, 2 mm ADP), and 5 μl of samples were applied to glow-discharged carbon-coated copper grids. Grids were incubated for 1 min at room temperature, washed once with water, and stained with 1% (w/v) uranyl acetate. Grids were examined in a JEM-2100 (JEOL) transmission electron microscope operating at 200 kV. Images were recorded in minimum dose mode using a charge-coupled device (F415MP, TVIPS Gauting, Germany) at ×40,000 and an underfocus of −1.5 μm. The pixel size on the specimen level was calibrated to be 2.82 Å using catalase as standard.
Image Analysis and Three-dimensional Reconstruction
CCD frames were displayed using Boxer in the EMAN package (45), and single molecule images were selected manually (192 × 192 pixels). A combined data set of 9960 particles was collected for the dimeric enzyme from yeast strain W303-1A. All subsequent image analysis was performed with the Imagic-5 software package (46) on a 12 CPU SGI Altix cluster running under SUSE Linux. The data sets were normalized and bandpass-filtered, and a circular mask was applied. The data sets were treated by reference-free alignment, and the best averages were used as references for multireference alignment as implemented in Imagic-5. After several rounds of alignment, the data set was sorted into 100 classes for measuring F1-F1 distances and angles as illustrated in Fig. 2.
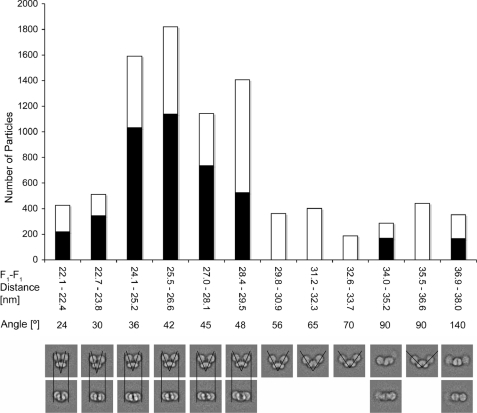
FIGURE 2.
Analysis of the dimer angles. The data set of 9960 ATP synthase dimer images was sorted into 100 classes, and the F1-F1 distances (edge to edge) and angles between monomers were measured. The white and black bars show the particle numbers of the “side” and “top” views shown at the bottom, respectively. For details, see text.
Three-dimensional Reconstruction
For starting up the three-dimensional reconstruction, two molecules of yeast F1c10 crystal structure (Protein Data Bank code 1qo1) (47) were arranged as a dimer so that projections of the model matched the projections of the enzyme obtained from the multireference alignment analysis (see Fig. 3 and supplemental Fig. S1). Peripheral stalks (Protein Data Bank code 2cly) (48), the in silico but not crystallographic or NMR model for E. coli a subunit (Protein Data Bank code 1c17) (49), and the N-terminal transmembrane domain of E. coli b subunits (Protein Data Bank code 1b9u) (50) together with the OSCP N-terminal domain (Protein Data Bank code 2bo5) (51) were docked onto the model guided by the cryo-EM reconstruction of bovine F-ATP synthase (see Fig. 3 and supplemental Fig. S1) (37). Model building was done in Chimera (52). In the first round of projection matching, the model was projected along 52 directions randomly distributed over the Euler sphere, and the projections then served as references to align the combined data set of 9960 images (“projection matching”). Averages were calculated based on cross-correlation between reference and raw images, and a first three-dimensional model of dimeric yeast ATP synthase was calculated. The model was projected along 52 directions, and projection matching was repeated. In later alignments, projection angles were limited to cover two-thirds of the Euler sphere (β-angle up to 136°) as only very few images aligned to projections with larger β-angles (probably due to preferred orientations of the complex on the carbon film used for electron microscopy). Subsequent refinement was performed by alignment of the data set with an increasing number of projections of the three-dimensional reconstruction from the preceding round of projection matching until stable results were obtained (9 iterations and 138 projections in the final iteration). At this stage of the analysis, the EM reconstruction was displayed in Chimera, and the reference crystal structure model was re-fitted to the EM density, resulting in a new reference atomic model (see supplemental Fig. S1). The refined atomic model was then used for a second cycle of projection matching. In this second cycle, the atomic models of Escherichia coli a and b subunits were omitted to avoid any bias in the alignment. The final three-dimensional reconstruction was obtained after eight iterations of projection matching. The three-dimensional models obtained after the first and second cycle of projection matching are very similar (compare supplemental Fig. S1 and Fig. 4, respectively), indicating that the structural features in the final reconstruction are not due to model bias. All three-dimensional reconstructions were performed assuming 2-fold symmetry of the ATP synthase dimer complex. About 37% of the data set was excluded from the final reconstruction. The excluded projections were from damaged molecules (e.g. missing one or both F1 domains) or had dimer angles larger than ∼48° (side view projections with smaller angles could be due to a rotation along the 2-fold axis of the dimer and were not excluded). For estimating the resolution in the final model, input averages were split in two groups, and the calculated three-dimensional models from both groups were compared via the FSC algorithm as implemented in Imagic 5 using 0.5 and the 3σ criterion as the cutoff value (see supplemental Fig. S3). The final three-dimensional model of the dimeric ATP synthase was filtered to a resolution of 20 Å (1st zero of contrast transfer function). Structure fitting and figures were generated with Chimera (52).
FIGURE 3.
A and D, top and side view projection of dimeric ATP synthase after multireference alignment and classification. C and F, atomic model of dimeric ATP synthase modeled after the projections shown in A and D. B and E, projections of the atomic model.
FIGURE 4.
Three-dimensional reconstruction of the yeast ATP synthase dimer. A, surface representation of the three-dimensional reconstruction of the yeast ATP synthase dimer. The model was calculated from 123 averages assuming 2-fold symmetry. B, “F1-F1” view of the same model as in A, showing a distance of ∼10 nm between each peripheral stalk and an angle of ∼30° in relation to the diameter perpendicular to the central 2-fold axis of the dimer. C and D, fitting of the atomic reference model into the three-dimensional EM reconstruction. As can be seen, the overall structure fits well with the EM reconstruction except for parts of the peripheral stalks, suggesting some rearrangement of the stator forming polypeptides as a result of the F1-F0 interaction and/or dimer formation. E and F, top and bottom view of the EM reconstruction fitted with the atomic reference model. Chains “s, t, u, and v” correspond to subunits OSCP, b, d, and F6, respectively (53). See text for further details.
RESULTS AND DISCUSSION
Purification of the ScF1F0 Dimer and Electron Microscopy Analysis
Previously, we provided the first electron microscopic images of the dimeric ATP synthase isolated from bovine heart mitochondria (28). The images showed the ATP synthase monomers arranged at an angle of ∼40°, and we suggested that this arrangement of the ATP synthase dimer may play a role in the formation of the curvature of mitochondrial inner membrane cristae. In this study, we obtained two-dimensional projections of the yeast mitochondrial ATP synthase dimer that allowed the calculation of a three-dimensional structural model of the complex at a resolution of 27 Å (0.5 FSC; 21 Å using the 3σ criterion, see supplemental Fig. S3). Fig. 1 summarizes the purification and electron microscopy of the dimeric ATP synthase from the yeast S. cerevisiae (ScF1F0). Fig. 1a depicts a typical in-gel ATPase assay developed after BN-PAGE of glycerol gradient fractions; the gel shows the effective separation of the dimeric ScF1F0 from its monomeric counterpart in fractions 5 and 6, which migrated at 30–35% glycerol in digitonin-containing buffer. Fig. 1b shows an electron microscopic image of purified dimeric ATP synthase, negatively stained with 1% uranyl acetate. Although the dimeric ATP synthase in side view orientation can be recognized by its typical double mushroom shape (Fig. 1b, circles), other orientations that show the molecule in top view or intermediate orientations were also observed (rectangles). Two data sets of ∼5000 particles each from two independent preparations were selected from electron micrographs and analyzed by alignment and classification protocols as described under “Experimental Procedures.” Because both data sets resulted in very similar averages (data not shown), the data sets were merged for subsequent analysis. A selection of averages is shown in Fig. 1c, images 1–7.
FIGURE 1.
Purification and transmission electron microscopy of the yeast F1F0-ATP synthase dimer. a, glycerol gradient fractionation of digitonin extracts of yeast mitochondrial inner membranes as analyzed by ATPase activity staining in native polyacrylamide gels. The 1st left lane was loaded with an aliquot of the digitonin mitochondrial extract that was subsequently loaded on the glycerol gradient; this shows the two white bands of the functional dimeric and monomeric forms of the S. cerevisiae ATP synthase. Fractions 1–9 collected after the glycerol gradient were loaded in a BN-PAGE gel run in parallel and are shown in the consecutive lanes as indicated. b, transmission electron microscopy of purified ATP synthase dimer. Fraction 7 of the gradient shown in A was diluted 1:10 in 2 mm MES, pH 7.0, 2 mm EDTA, 2 mm ADP to obtain a final concentration of 0.25 mm digitonin and negatively stained with 1% uranyl acetate on glow discharged carbon coated copper grids. Side and top views of the ATP synthase dimers are marked by circles and rectangles, respectively. c, alignment and classification of a data set of 9960 ATP synthase dimer images. The data set of 9960 images was sorted into 64 or 100 classes, and the averages 1–7 were calculated between 100 and 200 individual projections. For details, see the text.
In addition to the previously described side view of the dimeric enzyme (Fig. 1c, images 1–3) that show the central and peripheral stalks (see arrowheads and arrows, respectively, in image 2), we also obtained images of the ATP synthase dimer deposited in orientations other than the side view, namely top and intermediate orientations (Fig. 1c, images 4–7). Some of the top view averages clearly show the ring of c subunits of yeast F0 in projection (see arrows in Fig. 1c, image 4). In addition to the ring of c subunits, these averages show additional density at the dimer F0-F0 interface that in all likelihood corresponds to subunits a and b and the other subunits of the F0, including the polypeptides that are required for dimer formation, i.e. subunits e and g (see arrowheads in Fig. 1c, image 4).
As reported before for the enzyme isolated from S. cerevisiae (17, 30), projections with angles ranging from 35 to close to 180° are observed (see side views 1–3 in Figs. 1C and 2). In the side views, the different arrangement of the monomers can be expressed as differences in the angles between monomers or as differences in F1-F1 distances. In the top view and some of the intermediate orientations, the differences between F1-F1 distances can be measured reliably (compare views 4–7 in Fig. 1c). To determine the distribution of angles of the dimer, we measured the F1-F1 distances for the different averages. Fig. 2 shows the number of side and top/intermediate views as a function of F1-F1 distance with the white parts of the bars representing side and the black parts top/intermediate views, respectively. For the side view projection, monomer-monomer angles were also measured (see Fig. 2). Overall, the histogram showed that the dominant angle was close to 42°, similar to the angle reported previously for the bovine heart enzyme (28). As described before for the yeast enzyme (30), we also observed a continuum of larger dimer angles with a frequent loss of one or both of the F1 heads at angles larger than ∼65° (see bottom of Fig. 2). These data indicate that in our preparations the most stable form corresponds to a dimer with an angle of about 45° but also that the detergent-solubilized enzyme appears to exhibit some flexibility with respect to the dimer angles. Our data, however, do not seem to support the hypothesis that dimeric ATP synthase can exist as a “pseudo” dimer with ∼45° and a “true” dimer with ∼70° angle as proposed earlier (30).
Thomas et al. (17) recently reported a different distribution of angles toward larger values (83–93°) for the ATP synthase dimer extracted from yeast mitochondria with digitonin. At this point, we have no explanation for this discrepancy other than differences in the protocol used to purify the enzyme; for instance, unlike Thomas et al. (17), we included ADP in the buffers used for enzyme extraction and glycerol gradient centrifugation, both in this study and in our earlier investigation of the complex from bovine heart mitochondria, where we found a similar 40° angle (28). Therefore, ADP may stabilize the closed dimer conformer; however, this or other possibilities remain to be explored. For instance although sample deposition is similar in this and previous reports studying the ATP synthase dimer by EM, the possibility remains that adsorption of isolated ATP synthase dimers could affect differently the angle between monomers in our preparation. On the other hand, angles of between 55 and 95° have been found in averaged tomograms of frozen hydrated submitochondrial vesicles from rat liver and bovine heart (38), and the authors of that study concluded that the dimer interface must be flexible, consistent with our results summarized in Fig. 2.
Three-dimensional Reconstruction
Initial attempts to collect EM images of dimeric ATP synthase embedded in vitreous ice were not successful due to sample heterogeneity upon concentrating the glycerol gradient fractions to the range of protein concentration required for cryoelectron microscopy (several mg/ml, see Ref. 37). We therefore decided to reconstruct a first three-dimensional model of the dimeric complex from images of a negatively stained specimen. To determine a set of initial projection angles, we generated a reference model of the dimeric enzyme using the yeast F1-c10 crystal structure complemented with available atomic structures of the peripheral stalk, OSCP N-terminal domain, and the modeled structure for the E. coli a subunit. Docking of the x-ray models was guided by the cryo-EM model of monomeric bovine F-ATPase (37) as done in Dickson et al. (48). Two ATP synthase monomers were then arranged in such a way that the projections of the resulting model of the dimeric enzyme matched the most abundant EM projections observed with the yeast dimeric enzyme (see Figs. 1 and 2), both in terms of the angle between the two monomers (∼45°) in side view projections, as well as the distance of the two c10 oligomers in the top views (see Figs. 1C and 3). The model structure was projected along directions uniformly distributed over the Euler sphere, and the projections then served as references in a first round of multireference alignment. Subsequently, averages were calculated based on cross-correlation to the references (projection matching), and a first three-dimensional reconstruction of yeast ATP synthase dimer was computed from the averages using the angles of the initial reference projections. This first reconstruction was then used to generate a new set of reference projections for a second round of multireference alignment followed by averaging and computation of a new three-dimensional reconstruction. This procedure was iterated nine times with up to 139 reference projections until stable results were obtained. At this stage of the analysis, the resulting EM three-dimensional model was compared with the atomic model used for generating the initial references. The comparison is illustrated in supplemental Fig. S1, A and B. As can be seen, although there is an overall good match for the majority of the molecule, the densities of the peripheral stalks in the F1-F0 interface is displaced by about 2 nm (see arrows and arrowheads in supplemental Fig. S1B). In addition to the mismatch of the peripheral stalks, the EM reconstruction displays a slightly larger angle of the two ATP synthase monomers compared with the initial reference model as indicated by the unoccupied density at the left and right of the density corresponding to the F1 domains in the EM reconstruction (see arrowheads in supplemental Fig. S1A). Overall, this mismatch between the initial reference model and the final EM reconstruction indicates that the density of the peripheral stalks as well as the dimer angle in the EM reconstruction is not a result of bias introduced by the atomic reference model, but instead it is a genuine feature of the ATP synthase dimer.
However, to be sure that the position of the peripheral stalks is not influenced by the initial atomic reference model, we used three different model structures to start up the initial alignment of the EM images as follows: one with densities corresponding to α3β3γδOSCPntabntbctdf6c10, one with α3β3γδOSCPntabntc10, and one with α3β3γδc10 (see supplemental Fig. S2). As described above, projections of the reference models were used only in the first round of the projection matching (multireference alignment). Subsequent refinement of the three-dimensional reconstructions was again performed independently of the atomic model until stable results were obtained. It should be noted that irrespective of which crystal structure model was used to start up the alignment, the three final reconstructions were similar with respect to the overall arrangement of the dimer, including the F1-F1 distance and the positions of the peripheral stalks (see supplemental Fig. S2). This similarity provides again strong evidence that the atomic density representing the peripheral stalks in the three-dimensional reconstructions is a genuine feature of the dimeric ATP synthase and, as concluded above, not a result of a bias introduced by the crystal structure models.
The crystal structures for F1c10 and the peripheral stalks were then re-fitted to the EM reconstruction as shown in supplemental Fig. S1, C and D. As can be seen, a good match could be obtained for F1c10 as well as for the peripheral stalk complexes. The atomic model was subsequently “re-fitted,” guided by the EM density (for the lower portion of the peripheral stator) and by the recent crystallographic structure of the F1-stalk complex (Protein Data Bank code 2wss; for the upper portion of the stator). Note that deposited coordinates for the F1-stalk structure (Protein Data Bank code 2wss) only contain coordinates for parts of the peripheral stalk subunits (b, d, and F6 in the model), although the earlier crystal structure of the isolated peripheral stalk complex (Protein Data Bank code 2cly) is more complete. The re-fitted reference model (supplemental Fig. S1, C and D) was then used in a new cycle of projection matching and refinement. In this alignment cycle, the E. coli subunit a (in silico but not crystal or NMR model) and the subunit b membrane domains were omitted from the atomic reference model to rule out any bias that may originate from these densities in the final EM model. Stable results were obtained after eight iterations. About 37% of the data set was excluded from the final reconstruction. The excluded projections had dimer angles larger than ∼48° or were missing one or both of the F1 heads. However, most of the side view projections with smaller angles (see the left side of the top panel in Fig. 2) matched projections of the final model well and were therefore not excluded (projections with smaller angles could be due to a rotation along the 2-fold axis in the side view projection). Of the ∼6200 projections contained in the final model, about 3200 and 3000 were side view and top and intermediate view projections, respectively. A selection of input class sums and re-projections from the final model is shown in supplemental Fig. S4A.
Fig. 4 summarizes the final three-dimensional reconstruction of the ATP synthase dimer as obtained after the second cycle of projection matching and refinement. The model is shown as a surface representation as seen parallel to the membrane bilayer. Fig. 4A shows the model of the dimeric enzyme in the typical side view orientation with the 2-fold symmetry axis indicated by the vertical thin dashed line. Each ATP synthase monomer consists of an F1-ATPase (top) and a F0 proton channel domain (bottom). The F1 and F0 domains are connected by a central and by a peripheral stalk (see Fig. 4A, short arrow and arrowhead, respectively). The dimer interface is formed entirely by the two F0 domains (see large asterisk) despite their smaller size as compared with F1. Such an arrangement is possible because the long axes of the two monomers are arranged at an angle of ∼45°, resulting in a F1-F1 distance (center to center) of ∼15 nm. Fig. 4B shows the reconstruction as seen from the mitochondrial matrix toward the membrane. The peripheral stalks are projected at an angle of about 30° toward the periphery of the dimer, at a distance of ∼10 nm from each other.
Fitting of Available Crystal Structures
The re-fitted crystallographic dimer model used for the second cycle of projection matching was fitted into the negative stain EM map of the final three-dimensional EM reconstruction of the dimeric ATP synthase (Fig. 4, C–F). As can be seen, there is an overall good match, indicating that the refined reference model is a true representation of the three-dimensional structure of the yeast ATP synthase dimer. There are, however, some regions of the peripheral stalk that are not covered by negative stain EM map (see arrows in Fig. 4D), consistent with the idea that the peripheral stalks adopt a slightly different conformation in the intact ATP synthase monomer and/or dimer as seen in the F1-stalk complex (53). From the views shown in Fig. 4, D and F, it can be seen that there is considerable density next to the ring of c subunits, and we assume that this density is occupied by F0 subunits a, e, f, g, i, k, 8, and the N-terminal transmembrane α helices of subunit b. Again, we believe that this density is a true feature of the ATP synthase dimer as no atomic models were included in this region of the reference structure, except for models for subunit a and the N-terminal transmembrane helices of subunit b (see supplemental Fig. S2D). In support of this three-dimensional model of the ATP synthase dimer, this density at the F0-F0 interface is consistent with the position of dimerizing subunits e and g as determined by difference cryo-EM densities comparing the bovine and yeast F1F0 monomer (36). Taking as a reference the yeast monomeric structure determined by cryo-EM, the predicted position of e and g subunits is at or near the arrowhead of Fig. 4F, i.e. just at the dimerizing F0-F0 interface.
Monomer-Monomer Interface and Oligomer Formation
As mentioned above, the monomer-monomer interface is entirely formed by the two F0 domains of the dimer. Fig. 4F shows a view of the dimer from the intermembrane space perpendicular to the plane of the membrane bilayer. As can be seen, the subunit c rings have a center-to-center distance of about 10 nm. The two c subunit rings are in contact with two regions of protein density, one large density within each monomer and one smaller one that forms a bridge to the adjacent monomer (indicated by the hatched regions in Fig. 4F). A similar distance of the proteolipid rings was observed in atomic force microscopy images from the inter-membrane side of the inner mitochondrial membrane (33).
ATP synthase dimer and oligomer formation has been extensively characterized by genetic and chemical cross-linking analysis. According to the available data, the ATP synthase polymer exhibits contacts at cross-linking distance at two F0-F0 interfaces, one with subunits e and g (13–15) and the other with one subunit b from each monomer (19, 40). Because the dimer resolved here lacks contacts in the side stalks at its interface (see Fig. 4B), our working model of the ATP synthase polymer positions one peripheral stalk in close proximity to a contiguous dimer to account for the observed interdimeric cross-links (see Fig. 5). Such a diagonal model is consistent with the offset observed by Allen et al. (24, 25) in the first direct demonstration of the ATP synthase oligomer in tubular mitochondrial cristae and the images of the atomic force microscopy study mentioned above (33). In our model, the curvature of the membrane can then be induced either by the intrinsic angle of the dimer and/or by the curvature induced by interdimeric distances at interfaces forming the oligomer. Finally, further work will determine how other proteins such as IF1 may participate in the formation of the ATP synthase oligomer in mitochondria. This inhibitor stabilizes the ATP synthase dimer in bovine and rat mitochondria (22), and it also promotes mitochondrial cristae formation in cultured cells (23). The presence of endogenous IF1 in our dimer preparation is suggested by the 6–10-fold increase in the ATPase turnover by preincubation of the mitochondrial digitonin extract in IF1-release conditions (pH 8.0 and 100 mm KCl) as determined by different ATPase assays, including in gel-activity (Fig. 1a) or coupled enzyme assays (data not shown). This ATPase activation is accompanied by dissociation of the ATP synthase dimer into highly active F1F0-ATPase monomers (22). However, further structural and Western blotting analyses with yeast ΔINH1 deletion mutants are required to confirm the presence of endogenous IF1 in our yeast ATP synthase dimer preparation and to determine how IF1 participates in the stability and structure of the ATP synthase dimer and oligomer. Given that IF1 promotes ATP synthase oligomerization (22) and cristae formation (23), this protein may participate in interdimeric (Fig. 5A, red circles) if not intradimeric ATP synthase interactions. These studies are ongoing in our laboratories.
FIGURE 5.
Model of the ATP synthase oligomer. A diagonal polymer was built by taking the dimer model shown in Fig. 4A as the building block. The central 2-fold axis of each monomer was moved along a diagonal direction to get a close contact of inter-dimer peripheral stalks. Such an inter-dimer contact of the peripheral stalks would explain the cross-linking data observed for the in vivo oligomer (see text). The proposed arrangement may induce membrane curvature by slight separation of the (F1)2-(F1)2 interfaces, and/or by forming a slight angle of the central 2-fold axes with respect to one another as the polymer opens as a fan to form a helical oligomer that wraps tubular cristae. See text for further details.
In summary, the data presented in this study provide significant advances on the position at the ATP synthase dimer interface of the F0 c10 rings, the dimerizing subunits, and the side stalks as the first three-dimensional view of this dimer. This will be instrumental to understand how the ATP synthase oligomer contributes to give its cristae shape to the inner mitochondrial membrane. The model is consistent with the early models of the helical ATP synthase oligomer (24, 25) as well as the in situ atomic force microscopy data (33). Further studies, preferably by cryoelectron microscopy or tomography together with antibody labeling, for example, will be required to resolve the subunit architecture at the monomer-monomer interface.
Supplementary Material
Acknowledgments
The critical reading of this work is gratefully acknowledged to Professors Marietta Tuena de Gómez-Puyou and Armando Gómez-Puyou.
This work was supported in part by National Institutes of Health Grant GM58600. This work was also supported by a grant from Universidad Nacional Autónoma de México under the program “Dirección General de Asuntos del Personal Académico-Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica” 2009, Project IN213809-3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- OSCPnt
- the structure of the N-terminal domain of the oligomycin sensitivity conferring protein
- bnt and bct
- the structures of the N- and C-terminal domains of subunit b
- BN
- blue native.
REFERENCES
- 1.Mitchell P. (1961) Nature 191, 144–148 [DOI] [PubMed] [Google Scholar]
- 2.Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 3.Capaldi R. A., Aggeler R. (2002) Trends Biochem. Sci. 27, 154–160 [DOI] [PubMed] [Google Scholar]
- 4.Feniouk B. A., Yoshida M. (2008) Results Probl. Cell Differ. 45, 279–308 [DOI] [PubMed] [Google Scholar]
- 5.Wilkens S. (2005) Adv. Protein Chem. 71, 345–382 [DOI] [PubMed] [Google Scholar]
- 6.García-Trejo J. J., Morales-Ríos E. (2008) J. Biol. Phys. 39, 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pullman M. E., Monroy G. C. (1963) J. Biol. Chem. 238, 3762–3769 [PubMed] [Google Scholar]
- 8.Morales-Ríos E., de la Rosa-Morales F., Mendoza-Hernández G., Rodríguez-Zavala J. S., Celis H., Zarco-Zavala M., García-Trejo J. J. (2010) FASEB J. 24, 599–608 [DOI] [PubMed] [Google Scholar]
- 9.Schägger H. (2002) Biochim. Biophys. Acta 1555, 154–159 [DOI] [PubMed] [Google Scholar]
- 10.Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. (1998) EMBO J. 17, 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brèthes D., di Rago J. P., Velours J. (2002) EMBO J. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin P. D., Prescott M., Luff S. E., Devenish R. J. (2004) J. Cell Sci. 117, 2333–2343 [DOI] [PubMed] [Google Scholar]
- 13.Arselin G., Giraud M. F., Dautant A., Vaillier J., Brèthes D., Coulary-Salin B., Schaeffer J., Velours J. (2003) Eur. J. Biochem. 270, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 14.Bustos D. M., Velours J. (2005) J. Biol. Chem. 280, 29004–29010 [DOI] [PubMed] [Google Scholar]
- 15.Everard-Gigot V., Dunn C. D., Dolan B. M., Brunner S., Jensen R. E., Stuart R. A. (2005) Eukaryot. Cell 4, 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraud M. F., Paumard P., Soubannier V., Vaillier J., Arselin G., Salin B., Schaeffer J., Brèthes D., di Rago J. P., Velours J. (2002) Biochim. Biophys. Acta 1555, 174–180 [DOI] [PubMed] [Google Scholar]
- 17.Thomas D., Bron P., Weimann T., Dautant A., Giraud M. F., Paumard P., Salin B., Cavalier A., Velours J., Brèthes D. (2008) Biol. Cell 100, 591–601 [DOI] [PubMed] [Google Scholar]
- 18.Velours J., Dautant A., Salin B., Sagot I., Brèthes D. (2009) Int. J. Biochem. Cell Biol. 41, 1783–1789 [DOI] [PubMed] [Google Scholar]
- 19.Weimann T., Vaillier J., Salin B., Velours J. (2008) Biochemistry 47, 3556–3563 [DOI] [PubMed] [Google Scholar]
- 20.Yao H., Stuart R. A., Cai S., Sem D. S. (2008) Biochemistry 47, 1910–1917 [DOI] [PubMed] [Google Scholar]
- 21.Fronzes R., Weimann T., Vaillier J., Velours J., Brèthes D. (2006) Biochemistry 45, 6715–6723 [DOI] [PubMed] [Google Scholar]
- 22.García J. J., Morales-Ríos E., Cortés-Hernandez P., Rodríguez-Zavala J. S. (2006) Biochemistry 45, 12695–12703 [DOI] [PubMed] [Google Scholar]
- 23.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M. R., Abramov A. Y., Tinker A., Duchen M. R. (2008) Cell Metab. 8, 13–25 [DOI] [PubMed] [Google Scholar]
- 24.Allen R. D., Schroeder C. C., Fok A. K. (1989) J. Cell Biol. 108, 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen R. D. (1995) Protoplasma 189, 1–8 [Google Scholar]
- 26.Mannella C. A. (2006) Biochim. Biophys. Acta 1763, 542–548 [DOI] [PubMed] [Google Scholar]
- 27.Mannella C. A. (2006) Biochim. Biophys. Acta 1762, 140–147 [DOI] [PubMed] [Google Scholar]
- 28.Minauro-Sanmiguel F., Wilkens S., García J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12356–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J., Braun H. P. (2005) FEBS Lett. 579, 5769–5772 [DOI] [PubMed] [Google Scholar]
- 30.Dudkina N. V., Sunderhaus S., Braun H. P., Boekema E. J. (2006) FEBS Lett. 580, 3427–3432 [DOI] [PubMed] [Google Scholar]
- 31.Cano-Estrada A., Vazquez-Acevedo M., Villavicencio-Queijeiro A., Figueroa-Martinez F., Miranda-Astudillo H., Cordeiro Y., Mignaco J. A., Foguel D., Cardol P., Lapaille M., Remacle C., Wilkens S., Gonzalez-Halphen D. (2010) Biochim. Biophys. Acta 1797, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 32.Vázquez-Acevedo M., Cardol P., Cano-Estrada A., Lapaille M., Remacle C., González-Halphen D. (2006) J. Bioenerg. Biomembr. 38, 271–282 [DOI] [PubMed] [Google Scholar]
- 33.Buzhynskyy N., Sens P., Prima V., Sturgis J. N., Scheuring S. (2007) Biophys. J. 93, 2870–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vonck J., Schäfer E. (2009) Biochim. Biophys. Acta 1793, 117–124 [DOI] [PubMed] [Google Scholar]
- 35.Wittig I., Schägger H. (2009) Biochim. Biophys. Acta 1787, 672–680 [DOI] [PubMed] [Google Scholar]
- 36.Lau W. C., Baker L. A., Rubinstein J. L. (2008) J. Mol. Biol. 382, 1256–1264 [DOI] [PubMed] [Google Scholar]
- 37.Rubinstein J. L., Walker J. E., Henderson R. (2003) EMBO J. 22, 6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W. (2008) EMBO J. 27, 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudkina N. V., Oostergetel G. T., Lewejohann D., Braun H. P., Boekema E. J. (2010) Biochim. Biophys. Acta 1797, 272–277 [DOI] [PubMed] [Google Scholar]
- 40.Spannagel C., Vaillier J., Arselin G., Graves P. V., Grandier-Vazeille X., Velours J. (1998) Biochim. Biophys. Acta 1414, 260–264 [DOI] [PubMed] [Google Scholar]
- 41.Peña A., Piña M. Z., Escamilla E., Piña E. (1977) FEBS Lett. 80, 209–213 [DOI] [PubMed] [Google Scholar]
- 42.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 43.Peterson G. L. (1977) Anal. Biochem. 83, 346–356 [DOI] [PubMed] [Google Scholar]
- 44.Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 45.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 46.van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 47.Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 48.Dickson V. K., Silvester J. A., Fearnley I. M., Leslie A. G., Walker J. E. (2006) EMBO J. 25, 2911–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rastogi V. K., Girvin M. E. (1999) Nature 402, 263–268 [DOI] [PubMed] [Google Scholar]
- 50.Dmitriev O., Jones P. C., Jiang W., Fillingame R. H. (1999) J. Biol. Chem. 274, 15598–15604 [DOI] [PubMed] [Google Scholar]
- 51.Carbajo R. J., Kellas F. A., Runswick M. J., Montgomery M. G., Walker J. E., Neuhaus D. (2005) J. Mol. Biol. 351, 824–838 [DOI] [PubMed] [Google Scholar]
- 52.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 53.Rees D. M., Leslie A. G., Walker J. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21597–21601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.