Abstract
The envelope glycoproteins (Env) are the focus of HIV-1 vaccine development strategies based on the induction of humoral immunity, but the mechanisms the virus has evolved to limit the induction and binding of neutralizing antibodies (NAbs) constitute substantial obstacles. Conserved neutralization epitopes are shielded by variable regions and carbohydrates, so one strategy to increase their exposure and, it is hoped, their immunogenicity is to delete the overlying variable loops. However, deleting the variable regions from Env trimers can be problematic, because hydrophobic patches that are normally solvent-inaccessible now become exposed, causing protein misfolding or aggregation, for example. Here, we describe the construction and characterization of recombinant gp140 trimers lacking variable domains 1 and 2 (ΔV1V2). The design of the trimers was guided by HIV-1 evolution studies that identified compensatory changes in V1V2-deleted but functional Env proteins (Bontjer, I., Land, A., Eggink, D., Verkade, E., Tuin, K., Baldwin, C., Pollakis, G., Paxton, W. A., Braakman, I., Berkhout, B., and Sanders, R. W. (2009) J. Virol. 83, 368–383). We now show that specific compensatory changes improved the function of ΔV1V2 Env proteins and hence HIV-1 replication. The changes acted by reducing the exposure of a hydrophobic surface either by replacing a hydrophobic residue with a hydrophilic one or by covering the surface with a glycan. The compensatory changes allowed the efficient expression of well folded, soluble gp140 trimers derived from various HIV-1 isolates. The evolved ΔV1V2 Env viruses were extremely sensitive to NAbs, indicating that neutralization epitopes are well exposed, which was confirmed by studies of NAb binding to the soluble ΔV1V2 gp140 trimers. These evolved ΔV1V2 trimers could be useful reagents for immunogenicity and structural studies.
Keywords: Evolution, Glycoprotein, HIV, Protein Folding, Viral Immunology, Viral Replication, Virus Entry, Envelope Glycoprotein, Vaccine, Variable Loops
Introduction
HIV-1 entry into target cells is mediated by the envelope glycoprotein complex (Env).2 Env is the only external viral protein component and hence the only protein relevant for the induction of neutralizing antibodies (NAbs). Strategies to elicit an antiviral Env-specific humoral response are a central element of HIV-1 vaccine research. The identification of an increasing number of broadly reactive NAbs from HIV-infected individuals proves that Env can elicit such antibodies in humans (2–7). Unfortunately, raising NAbs of comparable specificity and potency with Env vaccination of animals or humans has proven challenging (reviewed in Ref. 8). Thus the design of Env-based vaccines requires improvement.
The functional HIV-1 Env complex consists of three gp120/gp41 heterodimers. The first stage of viral entry into target cells involves the binding of gp120 subunits to the CD4 receptor. Conformational changes that involve the rearrangement of the first, second, and third variable loops (V1, V2, and V3) then create and expose an additional binding site, one that interacts with the CCR5 or CXCR4 coreceptor (9–14). Further conformational changes in the trimeric complex lead to exposure of the hydrophobic fusion peptides of the gp41 subunits, which are inserted into the target membrane to link the cellular membranes prior to their fusion (15, 16).
The individual Env subunits, gp120 and gp41, have not been successful as vaccine antigens, probably because they mimic inadequately the native Env spike. Hence, gp120 and gp41 immunogens tend to induce antibodies that are nonreactive with the functional Env complex and hence devoid of neutralizing activity against relevant HIV-1 strains. The current generation of soluble Env trimers offer modest improvements over monomeric gp120 in respect to NAb induction but not to an extent that solves the problem (17–20). Considerable effort is therefore being applied to improve the design of such proteins. However, progress in this area has been hampered by the lack of atomic level structural data on the Env trimer.
We have previously developed a prototype recombinant trimer construct, SOSIP gp140, that is stabilized by an intermolecular disulfide bond between gp120 and gp41 and a substitution, I559P, in gp41 (21, 22). A unique feature of SOSIP gp140 trimers is that they are cleaved, which may make them better antigenic mimics of the native, cleaved spike than uncleaved trimers (21, 23–26). Single particle cryo-electron microscopy tomography studies show that SOSIP gp140 is indeed a reasonable mimic of the virion-associated Env spike, and it undergoes similar conformational changes upon CD4 and antibody binding to those occurring within the native spike under the same conditions (27). Hence, SOSIP gp140 is a useful lead molecule for vaccine development and structural studies.
HIV-1 has evolved many strategies to limit the induction and binding of NAbs, including the shielding of conserved and potentially vulnerable Env domains by both carbohydrates and flexible and sequence-variable loops (8, 28, 29). Thus, the sequences of variable domains can alter under Nab selection pressure without compromising Env function, whereas glycan residues are generally seen as self by the immune system and so only very rarely elicit NAbs (3, 6, 29–31). The variable domains are particularly heavily glycosylated, so these two defense mechanisms are interlinked. Deletion of the V1V2 domain from monomeric gp120 generally improves the exposure of underlying neutralization epitopes, particularly those around the CD4-binding site (CD4bs), and can increase immunogenicity to some extent (32–37). V1V2 deletion also facilitated the crystallization of gp120 monomers for atomic structure determination (14, 38). Indeed, there is still no gp120 structure that contains the V1V2 regions. However, deleting V1V2 from soluble trimers, with the same goals, has proven less straightforward. One particular problem with the loop-deleted trimers is that they can become misfolded and/or aggregated (1, 39–41).3 A possible explanation is that hydrophobic regions of Env that are normally occluded become solvent-accessible once the V1V2 loops are no longer present (1).
To address the adverse effects of V1V2 deletion, we conducted evolution experiments with ΔV1V2 mutants of the HIV-1 strain LAI. Similar gain-of-function studies using evolution have previously been performed with V3-deleted Env (42). The goal was to allow the virus to find a way to accommodate the loss of its V1V2 loops by evolving compensatory changes that increase trimer function and hence virus replication (1). We identified several such compensatory changes and proposed that they could be useful for designing improved Env trimer variants for immunogenicity and structural studies. However, the evolution experiments were, of necessity, conducted using an Env that lacked the SOS disulfide bond and the I559P substitutions we use to make stable, cleaved gp140 trimers, because these modifications are incompatible with virus replication (43, 44). Here, we address whether we can use the evolutionary changes that arose in ΔV1V2 HIV-1 LAI to improve the design of soluble Env trimers of heterologous sequences. We found that the forced evolution of ΔV1V2 LAI reduces the exposure of a hydrophobic protein surface, either via substitution of nonpolar amino acids by polar ones or by masking it with an added glycan. Moreover, these strategies can be used for expressing well folded, recombinant soluble ΔV1V2 Env trimers.
EXPERIMENTAL PROCEDURES
Plasmids
The full-length molecular clone of HIV-1 LAI (pLAI) was the source of WT and mutant viruses (45). The pRS1 plasmid was used to introduce mutations into env, as described previously (43). Mutant env genes were generated in pRS1 and cloned into pLAI as SalI-BamHI fragments. To facilitate subcloning of V1V2 deletion variants, we introduced two unique restriction sites into pRS1 that had silent mutations, HindIII and BmgBI, at the N- and C-terminal ends of V1V2, respectively. Mutations, deletions, and insertions were generated using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA), and the integrity of all plasmids was verified by sequencing. The pPPI4 vector for expressing gp140 proteins has been described elsewhere (21), as has the KNH1144 SOSIP.R6 construct (46). We have also previously described modifications to JR-FL SOSIP.R6 gp140 that include the addition of a GCN4-based trimerization domain (isoleucine zipper) and a C-terminal His8 tag (21, 22, 47, 48). The HindIII and BmgBI sites were also introduced into the pPPI4 plasmids used for expressing KNH1144 SOSIP.R6 gp140 and JR-FL SOSIP.R6-IZ-His gp140. The V1V2 deletion variants were cloned into pPPI4 from pRS1 using HindIII and BmgBI. Amino acid numbering is based on the HXB2 isolate.
Reagents
Monoclonal antibodies (mAbs) were obtained as follows: 2G12 (3), 4E10 (5), and 2F5 (7) were from Hermann Katinger through the National Institutes of Health AIDS Research and Reference Reagent Program; Z13e1 (49), b6, and b12 (50) were from Dennis Burton (The Scripps Research Institute, La Jolla, CA); 15e, 17b, 48d, 39F, and 19b (51) were from James Robinson (Tulane University, New Orleans, LA); 447-52D (52) was from Susan Zolla-Pazner through the National Institutes of Health AIDS Research and Reference Reagent Program; D50 (53) was from Patricia Earl through the National Institutes of Health AIDS Research and Reference Reagent Program; 412d (54), X5 (55), F105 (56), and F91 (51) were from Peter Kwong (Vaccine Research Center, Washington, D. C.). mAb PA-1 (12), recombinant gp120, soluble CD4 (sCD4), and CD4-IgG2 (57) were all gifts from William Olson (Progenics Pharmaceuticals Inc., Tarrytown, NY). HIVIg was acquired from Luis Barbosa through the National Institutes of Health AIDS Research and Reference Reagent Program. DC-SIGN-Fc was purchased from R & D Systems (Minneapolis, MN).
Cells and Transfections
SupT1 T cells, 293T cells, and 293S GnTI−/− were maintained in RPMI 1640 medium and Dulbecco's modified Eagle's medium (DMEM; Invitrogen), respectively, each supplemented with 10% fetal calf serum (FCS), penicillin (100 units/ml), and streptomycin (100 μg/ml) as described previously (58). TZM-bl cells were maintained in DMEM containing 10% FCS, minimal essential medium nonessential amino acids, and penicillin/streptomycin. SupT1 cells were transfected by electroporation as described elsewhere (59). 293T cells were transiently transfected with env genes using linear polyethyleneimine (PEI) (Mr 25,000), as described previously (48, 60). Briefly, Env-encoding DNA was diluted in DMEM to 0.1 of the final culture volume and mixed with PEI (0.15 mg/ml final concentration). After incubation for 20 min, the DNA/PEI mixture was added to the cells for 4 h before replacement with normal culture medium containing 0.01% BSA (Sigma) and minimal essential medium nonessential amino acids (0.1 mm, Invitrogen). Supernatants were harvested 48 h after transfection.
Viruses and Infections
Virus produced in SupT1 cells and virus from SupT1 evolution cultures were stored at −80 °C. The virus concentration was quantified by measuring its CA-p24 antigen content by ELISA, essentially as described previously (61). These values were used to normalize the amount of virus used in subsequent infection experiments, which were performed by adding 500 pg of CA-p24 to 4 × 105 SupT1 cells in each well of a 96-well plate. Virus production after 21 days was measured using the CA-p24 ELISA.
Single Cycle Infection and Neutralization
The TZM-bl reporter cell line stably expresses high levels of CD4 and HIV-1 coreceptors CCR5 and CXCR4 and contains the luciferase and β-galactosidase genes under control of the HIV-1 LTR promoter (62). The line was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc., Durham, NC. One day prior to infection, TZM-bl cells (17 × 103 cells) were added to a 96-well plate in DMEM containing 10% FCS, MEM nonessential amino acids, and penicillin/streptomycin (both at 100 units/ml) and incubated at 37 °C in an atmosphere containing 5% CO2. A fixed amount of virus (1.0 ng of CA-p24) was incubated for 30 min at room temperature with various concentrations of mAbs. The mAb/virus mixture was then added to the TZM-bl cells (70–80% confluency) in the presence of 400 nm saquinavir (Roche Applied Science) and 40 μg/ml DEAE, in a total volume of 200 μl. Two days post-infection, the medium was removed, then the cells were washed once with PBS and lysed in Reporter Lysis buffer (Promega, Madison, WI). Luciferase activity was measured using the luciferase assay kit (Promega) and a Glomax luminometer according to the manufacturer's instructions (Turner BioSystems, Sunnyvale, CA). All infections were performed in duplicate, and luciferase measurements were also made in duplicate. Uninfected cells were used to correct for background luciferase activity. The infectivity of each mutant without inhibitor was defined as 100%. Nonlinear regression curves were determined, and IC50 values were calculated using Prism software version 5.0c.
SDS-PAGE, Blue Native PAGE, and Western Blotting
SDS-PAGE and Western blotting with the anti-gp120 V3 loop mAb PA-1 (1:20,000 dilution to a final concentration of 0.2 μg/ml) and HRP-labeled goat anti-mouse IgG (1:5,000 dilution) were performed by following established protocols. Luminometric detection of envelope glycoproteins was performed using the Western Lightning ECL system (PerkinElmer Life Sciences). Blue native-PAGE was carried out as described previously (22, 40). In short, purified protein samples or cell culture supernatants were diluted with an equal volume of a buffer containing 100 mm MOPS, 100 mm Tris-HCl, pH 7.7, 40% glycerol, 0.1% Coomassie Blue, immediately prior to loading onto a 4–12% BisTris NuPAGE gel (Invitrogen). Typically, gel electrophoresis was performed for 2 h at 125 V (∼0.07 A) using 50 mm MOPS, 50 mm Tris, pH 7.7, as running buffer.
Immunoprecipitation Assays
Twenty-fold concentrated supernatants from transiently Env-transfected 293T cells were incubated overnight at 4 °C with mAbs or CD4-IgG2 (each at 150 ng/ml) in a 500-μl volume that included 100 μl of 5-fold concentrated RIPA buffer (250 mm Tris-HCl, pH 7.0, containing 750 mm NaCl and 5 mm EDTA). When appropriate, sCD4 (10 μg/ml) was also included to induce the exposure of CD4i epitopes. Next, 50 μl of protein G-coated agarose beads (Pierce) were added, with rotation mixing, for 2 h at 4 °C. The beads were then washed extensively with ice-cold RIPA buffer containing 0.01% Tween 20. Proteins were eluted by heating the beads at 100 °C for 5 min in 50 μl of SDS-PAGE loading buffer supplemented with 100 mm dithiothreitol (DTT). The immunoprecipitated proteins were analyzed on 8% SDS-polyacrylamide gels (125 V for 2 h).
Analytical Size Exclusion Chromatography
Twenty-fold concentrated, Env-containing supernatants (200 μl) from 293T cell cultures were fractionated using a Superose-6 10/300 GL column (GE Healthcare), equilibrated with phosphate-buffered saline, using an Äkta FPLC system (GE Healthcare). Fractions were analyzed by SDS-PAGE and Western blot.
Ni-NTA Trimer ELISA
Supernatants from HEK 293T cells transiently transfected with His-tagged Env proteins were diluted 1:3 in TBS, 10% FCS and added for 2 h to pre-blocked Ni-NTA His Sorb 96-well plates (Qiagen). After three washes using TSM (20 mm Tris, 150 mm NaCl, 1 mm CaCl2, 2 mm MgCl2), HIVIg, a mAb, or CD4-IgG2 in TSM, 5% BSA was then added at a defined concentration for 2 h, followed by three washes with TSM, 0.05% Tween 20. When appropriate, Env proteins were incubated with sCD4 for 30 min prior to addition of a mAb. HRP-labeled goat anti-human IgG (Jackson ImmunoResearch) was then added for 30 min at a 1:5000 dilution (final concentration 0.2 μg/ml) in TSM, 5% BSA, followed by five washes with TSM, 0.05% Tween 20. Colorimetric detection was performed using a solution containing 1% 3,3′,5,5′-tetramethylbenzidine (Sigma) and 0.01% H2O2 in a 100 mm sodium acetate, 100 mm citric acid buffer. The colorimetric reaction was stopped using 0.8 m H2SO4 when appropriate, and absorption was measured at 450 nm.
Molecular Models
The various mutant and evolved ΔV1V2 gp120 core variants were modeled using SWISS-MODEL (63) based on the coordinates of the gp120 core in complex with CD4 and 17b (Protein Data Bank code 1G9M) (64). Three gp120 molecules were aligned using a trimer model that was previously generated based on quantifiable parameters (65, 66). Glycans were modeled using GlyProt (67). Based on site-specific glycan profiling of gp120 (68–70), the most commonly found glycans were built onto gp120 as follows: Man6GlcNAc2 at position 241; Man7GlcNAc2 at positions 234, 262, 289, 356, and 448; Man8GlcNAc2 at position 230; Man9GlcNAc2 at positions 295, 332, 339, 386, and 392; the sialylated diantennary complex NeuAc2Gal2GlcNAc2Man3GlcNAc2Fuc at positions 197, 276, and 464; and the sialylated tetraantennary complex NeuAc4Gal4GlcNAc4Man3GlcNAc2Fuc at position 88. We realize that this assignment of N-glycans is imperfect and incomplete. Thus, the glycan composition can be heterogeneous at each gp120 site, it depends on the producer cell type, and it can vary between virus isolates. Furthermore, experimental determinations of the glycans on monomeric gp120 probably do not fully predict how a complete Env trimer is glycosylated (48).
Glycans were built onto gp120 using the default rotation angles computed by GlyProt. The resulting structures were inspected for clashes with neighboring glycans, other gp120 protomers within the trimer, and CD4. Except for the 197 glycan, no clashes were observed using the default parameters. The 197 glycans clashed with the other gp120 protomers at the trimer interface, so its conformation was adjusted to prevent both such clashes and ones with CD4. Electrostatic surface potentials were computed using PyMol and figures were prepared by PyMol (71).
RESULTS
Evolution-based Design of ΔV1V2 Viruses
Our aim was to identify amino acid substitutions that could be useful for improving the design of recombinant, soluble ΔV1V2 Env proteins. In previous studies, we selected several evolved HIV-1 LAI variants that replicated more efficiently than the input ΔV1V2 viruses. As the evolved viruses retained the V1V2 deletion, it seemed likely that compensatory sequence changes had helped overcome the adverse impact the loss of the V1V2 loops has on the structure and function of the native Env complex (1). Sequence analyses revealed several substitutions that could possibly account for the increase in HIV-1 replication, based on their frequency and the biochemical properties of the altered amino acids. However, because the evolution studies were carried out using a nonclonal virus (i.e. with quasi-species), the relevance of the observed changes needed to be investigated. We therefore introduced the most plausible changes into a molecular clone of HIV-1 LAI that also contained a V1V2 deletion, to allow us to directly assess the effect of specific compensatory changes on replication (Fig. 1).
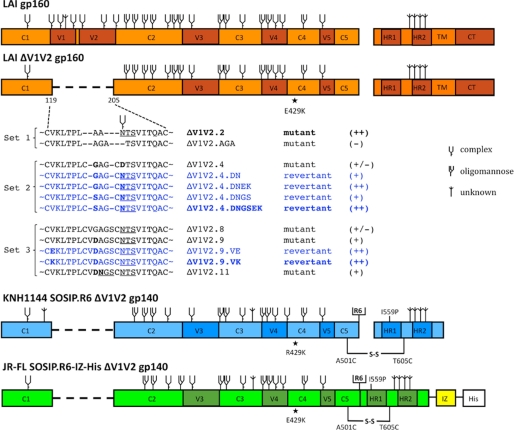
FIGURE 1.
Schematic of ΔV1V2 Env proteins. Constructs based on the sequences from LAI, JR-FL, and KNH1144 are indicated in orange, green, and blue, respectively. Three sets of ΔV1V2 Env proteins were used, including designed mutants (indicated in black) and variants derived from evolution studies (blue). A semi-quantitative assessment of Env function (see Fig. 2) is indicated in braces. Note that the E429K substitution is located far from the site of the V1V2 deletion. The three variants chosen for follow-up analyses are indicated in boldface. The KNH1144 SOSIP.R6 gp140 and JR-FL SOSIP.R6-IZ-His gp140s contain several modifications that have been described elsewhere, including the A501C and T605C substitutions to create the SOS disulfide bond (21), the I559P substitution to promote trimerization (22), and the hexa-arginine (R6) cleavage site to enhance cleavage (47). The SOSIP.R6-IZ-His gp140 also contains the GCN4-based isoleucine zipper (IZ) to promote trimerization (48) and a polyhistidine (His) tag. The N-linked glycan sites on Env are designated as oligomannose or complex, based on experimental determinations using IIIB gp120 (70). It is assumed that the glycans present at analogous sites are processed similarly in Env proteins from other isolates, but note that the carbohydrate composition at some sites has been reported to vary (68–70). Sites that are present on LAI, JR-FL, or KNH1144 gp120, but not on IIIB gp120, or sites that have not been characterized are designated as being of unknown glycan composition.
We studied three sets of viruses from which the V1V2 loops were deleted in slightly different ways, as well as their evolved mutants (Fig. 1). The first set included the designed mutant viruses ΔV1V2.2 and ΔV1V2.AGA. We did not analyze any evolved variants of ΔV1V2.2 because this particular mutant was already quite infectious and evolution yielded no improvement (1). The ΔV1V2.AGA virus is a new mutant that has not been subjected to evolution experiments. Both the ΔV1V2.2 and ΔV1V2.AGA viruses differ from traditional ΔV1V2 designs in that they lack the disulfide bond between residues 126 and 196; instead, residues 126–197 are replaced by an Ala-Ala-Asn or an Ala-Gly-Ala linker to create a continuous protein backbone (Fig. 1). The ΔV1V2.2 sequence contains the N-linked glycosylation site at position 197, although ΔV1V2.AGA does not.
In contrast, the second and third sets of viruses retained the above disulfide bond, with residues Cys-126 and Cys-196 connected by a small loop (Fig. 1). The second set included the original ΔV1V2.4 mutant in which residues 127–195 are replaced by Gly-Ala-Gly, as well as its evolved variants (Fig. 1) (1, 39). A D197N substitution was found in all four of the evolved viruses, suggesting that the N-linked glycan at position 197 is advantageous for replication of a ΔV1V2 virus. The ΔV1V2.4.DN variant was constructed to test this hypothesis directly. An additional E429K substitution arose during prolonged passage of ΔV1V2.4.DN, and a G127S substitution was identified in a subset of the evolved clones. The E429K change, a common polymorphism in LAI clones (72), is located in the bridging sheet immediately adjacent to the V1V2 stem, whereas G127S alters the linker region from Gly-Ala-Gly to Ser-Ala-Gly. Viruses containing both these individual changes were reconstructed (ΔV1V2.4.DNEK and ΔV1V2.4.DNGS), as well as one containing all three changes (ΔV1V2.4.DNGSEK) (Fig. 1).
The third set of mutants was based on the ΔV1V2 designs used in the first gp120 crystal structures (38, 73) and included viruses ΔV1V2.8, ΔV1V2.9, and ΔV1V2.11 (Fig. 1). In ΔV1V2.8, a Gly-Ala-Gly linker replaces residues 128–194. The ΔV1V2.9 design was based on a previous evolution experiment and contained an amino acid change in the linker (G128D: Gly-Ala-Gly to Asp-Ala-Gly (74)), whereas the ΔV1V2.11 variant has a different linker sequence that introduces an additional potential glycosylation site within the V1V2 stump (Asp-Asn-Gly). Two evolved variants of ΔV1V2.9 were included that contain either a V120E or a V120K substitution (ΔV1V2.9.VE and ΔV1V2.9.VK). Both variants emerged in evolution cultures of the ΔV1V2.9 virus (1).3
Compensatory Changes Improve the Replication and Infectivity of ΔV1V2 Viruses
We tested the abilities of the various viruses to replicate in SupT1 T cells in a multicycle assay and to infect TZM-bl reporter cells in a single cycle entry assay. The ΔV1V2.2 virus replicated efficiently in SupT1 cells, albeit with slightly delayed kinetics compared with the wild type (WT) virus (Fig. 2A). Furthermore, ΔV1V2.2 could infect TZM-bl cells, although less well than WT (Fig. 2B). In contrast, ΔV1V2.AGA could neither replicate in SupT1 cells nor infect TZM-bl cells (Fig. 2, A and B). The sequence differences between the ΔV1V2.2 and ΔV1V2.AGA viruses are only minor, but the latter lacks a glycosylation site at position 197 (Fig. 1).
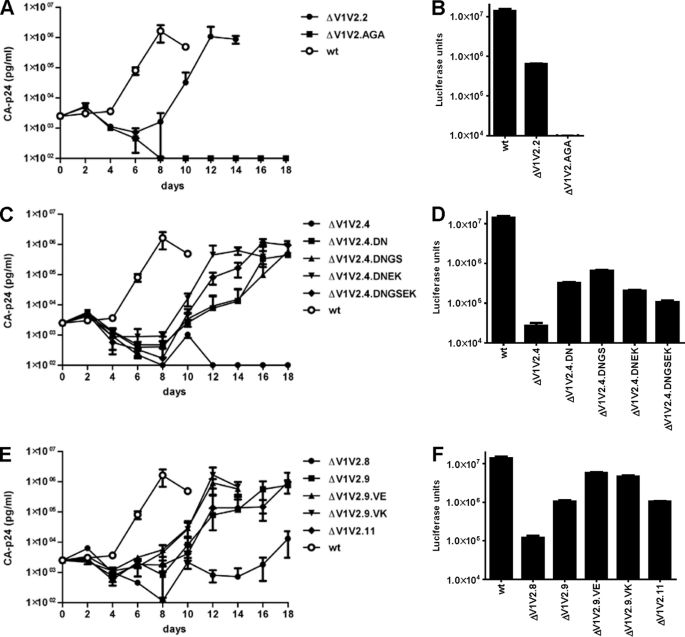
FIGURE 2.
Compensatory mutations improve ΔV1V2 virus replication and/or infectivity. A, C, and E, 4 × 105 SupT1 cells were infected with 500 pg of HIV-1, and replication was monitored for 21 days by CA-p24 ELISA. The curves are representative for at least two independent experiments. B, D, and F, TZM-bl reporter cells were infected with 1.0 ng of HIV-1 in the presence of SQV, and luciferase activity was measured after 48 h.
The ΔV1V2.4 virus was defective in replication, and the evolved ΔV1V2.4.DN and ΔV1V2.4.DNGS variants were also severely replication-impaired, with substantial delays in the production of virus in the SupT1 cell cultures (Fig. 2C). In contrast, the evolved variants ΔV1V2.4.DNEK and ΔV1V2.4.DNGSEK, which contain the E429K change, were both more replication-competent, although they were still delayed compared with WT (Fig. 2C). The ΔV1V2.4 virus was also inefficient at entering the TZM-bl reporter cells. In this assay, the D197N substitution improved infectivity, but the G127S and E429K changes did not (Fig. 2D). The replication and infectivity data were therefore not always concordant for this set of mutants. The varying outcomes may be explained by differing constraints on Env function that arise between single cycle infection assays and multicycle replication assays. For example, cell-free virus enters the TZM-bl cells, while spreading infections in T cell cultures are dominated by cell-to-cell virus transfer. Furthermore, any effects the various mutations have on Env synthesis will affect the single and multicycle assays differently.
The ΔV1V2.8 was severely impaired in replication, and the ΔV1V2.9 and ΔV1V2.11 viruses were quite delayed compared with WT, but the evolved variants ΔV1V2.9.VE and ΔV1V2.9.VK replicated efficiently in SupT1 cells (Fig. 2E). These results were corroborated in the single cycle assay. Thus, ΔV1V2.8 infected TZM-bl cells very inefficiently and ΔV1V2.9 and ΔV1V2.11 with intermediate efficiency, whereas the ΔV1V2.9.VE and ΔV1V2.9.VK variants were almost as infectious as the WT virus (Fig. 2F). Hence, the V120E/K substitutions improve the function of ΔV1V2 Env. Overall, we conclude that the reversions identified in evolution experiments (V120E, V120K, G127S, G128D, D197N, and E429K) do all help restore the function of Env proteins that lack the V1V2 loops.
Virus Evolution Counters the Adverse Effects of a Hydrophobic Patch on Trimeric ΔV1V2 Env
We were interested in how the substitutions identified in the evolution experiments acted to improve ΔV1V2 Env function. When the outer layers of a protein are deleted, hydrophobic domains that are normally hidden in the interior can become solvent-exposed, which could compromise protein folding, structure, or function. We had noted that some evolution substitutions in ΔV1V2 gp120 might act to counter this adverse effect (1). Two possible mechanisms are that the exposed hydrophobic residues can be deleted or replaced by hydrophilic ones, or they can be buried under a new, more hydrophilic surface.
We first used an Env trimer model to inspect the various original and evolved ΔV1V2 gp120 core variants. A ribbon diagram is shown for ΔV1V2.2, on which a red box highlights the V1V2 stem (Fig. 3A). We next computed and plotted the electrostatic surface potential of each variant. The surface of the original ΔV1V2 mutants all contain a relatively large, nonpolar patch located in the V1V2 stem that faces the target membrane, to which several hydrophobic residues contribute (Fig. 3B, shown for variant ΔV1V2.9). We anticipate that this patch would not exist on full-length gp120 because it would be occluded by the V1V2 loop and its associated N-glycans, although no structure exists to confirm this supposition. When we compared the electrostatic surface potentials of ΔV1V2.9 and ΔV1V2.9.VE, the V120E substitution clearly increases the polarity of the exposed patch (Fig. 3C). The acidic nature of the V120E substitution is probably not important per se because a V120K substitution also enhanced the surface polarity and improved ΔV1V2 Env function (Fig. 2, E and F). More likely is that any change from a nonpolar residue (Val) to any polar one (e.g. Glu or Lys) has a restorative effect. The G127S and G128D substitutions also directly increase the polarity of the ΔV1V2 trimer surface and are likely to improve Env folding and/or function via a similar mechanism (data not shown).
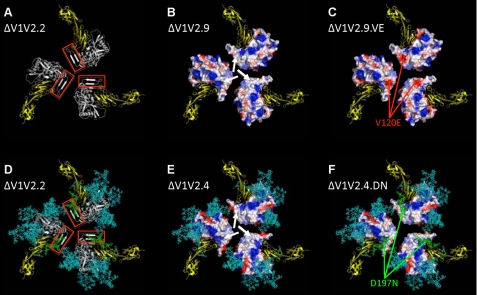
FIGURE 3.
Compensatory mutations affect a hydrophobic surface patch on ΔV1V2 Env trimers. A, ribbon diagram of a ΔV1V2.2 trimer model, depicted from the perspective of the target cell. The gp120 protomers are indicated in white, and the V1V2 stem, part of the bridging sheet, is highlighted by the red box. The two outer domains of CD4 are in yellow. B and C, rendering of the electrostatic surface potentials of the protein-associated, solvent-accessible surfaces of the ΔV1V2.9 and ΔV1V2.9.VE trimers, respectively. Acidic surfaces are in red; basic surfaces are in blue, and neutral surfaces are in white. The white arrows in B indicate a nonpolar surface patch, associated with hydrophobic residues, that is located at the stem of the V1V2 loop structure. The red arrows in C show the negative charge associated with the V120E substitution. D, ribbon diagram of a glycosylated ΔV1V2.2 trimer. The protein is depicted as in A but with the addition of N-glycans, which are represented as sticks in cyan except the one at position 197, which is in green. E and F, rendering of the electrostatic surface potentials of the protein-associated, solvent-accessible surfaces of the ΔV1V2.4 and ΔV1V2.4.DN trimers, respectively. The surface coloration is as in B and C, with the glycans represented as in D. The white arrows in E indicate a neutral surface similar to that shown in B. The N-glycan at residue 197, which partially covers the nonpolar protein-associated surface, is highlighted in green (F).
The D197N substitution does not increase the number of surface-associated polar amino acids, so this mechanism cannot explain why it improves ΔV1V2 Env function. However, the change creates a canonical site at position 197 that is N-glycosylated (see below; Fig. 5A). To assess the potential role of this N-glycan in shielding the hydrophobic surface, we generated models of mutant and revertant ΔV1V2 trimers containing all the N-glycans that are expected to be present at each canonical site, based on gp120 carbohydrate analyses (68–70). These glycans were built onto the gp120 subunits and aligned in the trimer (see “Experimental Procedures”). First, we added the glycans to the ΔV1V2.2 variant (Fig. 3D, cyan and green). As expected, much of the Env trimer protein surface is occluded by glycans (29, 65). However, a gap in the glycan shield of the ΔV1V2.4 trimer exposes the nonpolar surface patch associated with the V1V2 stem (Fig. 3E). Inspection of the glycosylated trimer model for the evolved variant ΔV1V2.4.DN shows that the D197N substitution introduces an N-glycan that partly occludes this nonpolar patch (Fig. 3F). We drew similar conclusions from the comparison of the trimer models based on the ΔV1V2.2 and ΔV1V2.AGA variants (data not shown). Thus the ΔV1V2.AGA virus, which lacks the N-glycan at position 197, is nonfunctional although ΔV1V2.2, which contains it, is infectious (Fig. 2, A and B).
FIGURE 5.
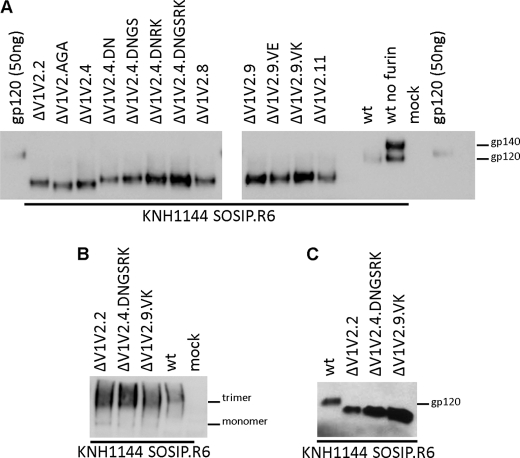
Compensatory mutations can facilitate the expression of soluble cleaved ΔV1V2 trimers. A, SDS-PAGE analysis of KNH1144 SOSIP.R6 gp140 variants expressed transiently in 293T cells, with co-transfection of furin to increase cleavage. Cleaved, full-length KNH1144 SOSIP.R6 and partially cleaved KNH1144 SOSIP.R6 (produced in the absence of furin) gp140s were included for comparison, as was purified KNH1144 gp120 (50 ng). B, blue native-PAGE analysis of WT and ΔV1V2 KNH1144 SOSIP.R6 gp140 variants expressed in 293T cells. C, SDS-PAGE analysis of WT and ΔV1V2 KNH1144 SOSIP.R6 gp140s expressed in 293S GnTI−/− cells.
Hence, HIV-1 uses at least two strategies to counter the adverse effect of a surface-exposed nonpolar patch on the ΔV1V2 Env trimer. One involves an amino acid substitution that introduces a polar residue within the patch, and the second is the addition of a glycan that masks it.
ΔV1V2 Viruses Are Extremely Neutralization-sensitive
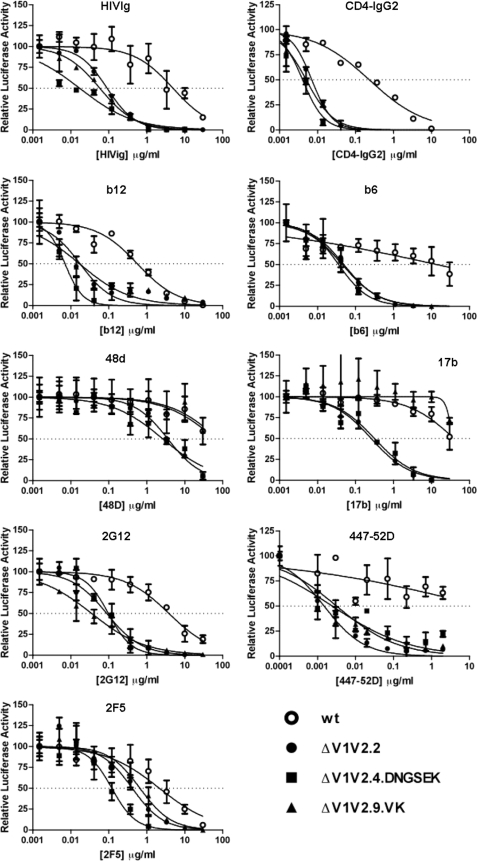
Deletion of the V1V2 domain is known to create neutralization-sensitive viruses on which important epitopes have become more exposed (1, 10, 75, 76). To study the accessibility of neutralization epitopes on the evolved ΔV1V2 viruses, we assessed the sensitivity of the most infectious members of each class (ΔV1V2.2; ΔV1V2.4.DNGSEK and ΔV1V2.9.VK) to polyclonal Ig from infected individuals (HIVIg), mAbs to various epitopes, and a CD4-mimetic protein, CD4-IgG2 (Fig. 4 and Table 1).
FIGURE 4.
ΔV1V2 viruses are extremely neutralization-sensitive. The various HIV-1 viruses (1.0 ng) were incubated with the indicated amount of mAb for 30 min at room temperature prior to addition to TZM-bl cells. Luciferase activity in the absence of antibody was defined as 100%.
TABLE 1.
Neutralization of ΔV1V2 LAI viruses
| mAb | WT IC50a | ΔV1V2.2 |
ΔV1V2.4.DNGSEK |
ΔV1V2.9.VK |
|||
|---|---|---|---|---|---|---|---|
| IC50a | n-foldb | IC50a | n-Foldb | IC50a | n-Foldb | ||
| HIVIg | 4.9 | 0.091 | 54 | 0.017 | 288 | 0.058 | 84 |
| CD4-IgG2 | 0.23 | 0.0071 | 32 | 0.0040 | 58 | 0.0050 | 46 |
| b12 | 0.57 | 0.018 | 32 | 0.0070 | 81 | 0.017 | 34 |
| b6 | 13.8 | 0.049 | 282 | 0.036 | 383 | 0.045 | 307 |
| 48d | 65.0 | 3.6 | 18 | 2.6 | 25 | 77.2 | 0.84 |
| 17b | 39.4 | 0.26 | 152 | 0.32 | 123 | 36.3 | 1.1 |
| 2G12 | 3.8 | 0.088 | 43 | 0.098 | 39 | 0.035 | 109 |
| 447–52D | 20.4 | 0.0015 | 13600 | 0.0025 | 8160 | 0.0032 | 6375 |
| 2F5 | 2.2 | 0.42 | 5.2 | 0.12 | 18 | 0.66 | 3.3 |
a IC50 values are given in μg/ml.
b The fold increase in sensitivity compared with WT is given.
Compared with WT virus, all three ΔV1V2 variants were much more sensitive to HIVIg, CD4-IgG2, and mAbs b6 and b12 against the CD4-binding site (CD4bs). mAbs 48d and 17b, which bind to a CD4-induced epitope formed when the V1V2 domain is rearranged, also neutralized the ΔV1V2 viruses more efficiently than WT. The ΔV1V2.9.VK virus was an exception, but for the trivial reason that it contains an inactivating V120K mutation in the epitopes for both 48d and 17b (38, 77). The glycan-dependent 2G12 mAb and the V3 mAb 447–52D also neutralized the ΔV1V2 variants more efficiently. Overall, the ΔV1V2 variants were 18–13,600-fold more sensitive to all the reagents directed against gp120, except when an amino acid substitution directly affected a mAb epitope. The variants were also more sensitive to mAb 2F5 against the gp41 MPER, but to a lesser extent (3–18-fold) than for the anti-gp120 mAbs. Overall, our ΔV1V2 variants have neutralization sensitivities that are consistent with previous reports on broadly comparable viruses (1, 10, 75, 76).
Compensatory Changes Facilitate the Expression of Soluble, Cleaved ΔV1V2 Env Trimers
The goal of the ΔV1V2 virus evolution studies was to identify whether compensatory changes, selected in the context of the native trimer, could be useful for improving the design of soluble, cleaved trimeric ΔV1V2 Env proteins. We therefore generated ΔV1V2 variants of SOSIP.R6 gp140, based on the KNH1144 genotype (Fig. 1). These subtype A trimers are relatively stable and are the basis of our collaborative efforts to determine an x-ray crystallography structure (46). Using a heterologous Env also allowed us to assess whether compensatory substitutions selected in one virus (LAI) might act more broadly. Thus, we first introduced the V1V2 deletions described above into KNH1144 SOSIP.R6 gp140, and we then added various compensatory substitutions (Fig. 1). The natural KNH1144 sequence contains an arginine at position 429; hence, the E429K substitution that arose in LAI translates to an R429K change in KNH1144 Env.
The various ΔV1V2 gp140 constructs were transiently expressed in 293T cells in the presence of furin to improve cleavage at the gp120-gp41 juncture (47). A full-length gp140 produced in the absence of furin, and hence ∼60% uncleaved, was included for comparison (Fig. 5A). There was considerable variation in the expression levels of the different ΔV1V2 gp140 constructs, but all the variants were cleaved efficiently (Fig. 5A). The original mutants ΔV1V2.2, ΔV1V2.AGA, ΔV1V2.4, ΔV1V2.8, and ΔV1V2.11, as well as the evolved variants ΔV1V2.4.DN, ΔV1V2.4.DNGS, and ΔV1V2.9.VE, were all expressed to similar extents. However, the ΔV1V2.4.DNRK and ΔV1V2.4.DNGSRK proteins were produced at higher levels, indicating that the G127S and R429K substitutions have beneficial effects on gp140 folding and/or secretion. The ΔV1V2.9 and ΔV1V2.9.VK gp140s were also expressed more efficiently, suggesting that the G128D substitution in the linker also improves production. The expression differences are unlikely to be attributable to differences in mRNA levels, because they match the improved functional data (Fig. 2), which were obtained using different expression cassettes. A native PAGE analysis showed that gp140 trimer formation was not affected by deletion of the V1V2 loops or the various compensatory changes (Fig. 5B and data not shown).
SDS-PAGE analyses revealed small size differences between the gp120 subunits of several ΔV1V2 gp140 variants. Because only a few amino acids differ among these variants, the size differences are likely to be attributable to changes in N-linked glycosylation. The ΔV1V2.AGA and ΔV1V2.4 proteins that lack the glycosylation site at position 197 were slightly smaller than the others, as expected. In contrast, the ΔV1V2.11 protein was indistinguishable in size from all except the ΔV1V2.AGA and ΔV1V2.4 proteins, suggesting that although its V1V2 stump contains two consensus N-linked glycosylation sites, only one of them is actually used.
The ΔV1V2 KNH1144 SOSIP.R6 gp140 proteins could also be produced in cells lacking N-acetylglucosaminyltransferase I (GnTI) (Fig. 5C). This procedure prevents the formation of complex N-glycans on Env, which reduces the size and heterogeneity of glycans (48, 78). Hence Env proteins produced in GnTI-negative cells might be useful for structural studies.
Soluble, Cleaved ΔV1V2 Trimers Expose Neutralization Epitopes
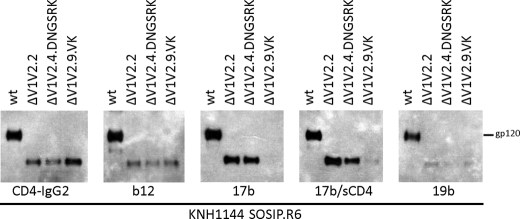
We next investigated the exposure of selected antibody epitopes on the best expressing ΔV1V2 trimer variant from each group. Both full-length and ΔV1V2 gp140 proteins interacted efficiently with CD4-IgG2, showing that their CD4bs were intact (Fig. 6). mAb b12 bound a little less well to the ΔV1V2 trimers than the full-length proteins, which is consistent with reports that components of the V1V2 stem contribute to the b12 epitope (Fig. 6) (79). A particular property of KNH1144 SOSIP.R6 gp140 is that it constitutively exposes CD4i epitopes and therefore binds mAb 17b both in the absence and presence of soluble CD4 (Fig. 6). The ΔV1V2 variants also interacted efficiently with 17b, with the exception of ΔV1V2.9.VK that contains the epitope-inactivating V120K change. Similar results were obtained with the CD4i mAbs 48d and 412d (data not shown). One concern with deleting the V1V2 loops from Env proteins is increased exposure of the immunodominant V3 domain, which may promote the induction of V3-directed antibodies with limited breadth of activity. However, we found that the V3-directed mAb 19b bound similarly to full-length and ΔV1V2 SOSIP.R6 gp140s (Fig. 6).
FIGURE 6.
Soluble, cleaved ΔV1V2 trimers expose neutralization epitopes. Immunoprecipitation analysis of cleaved KNH1144 SOSIP.R6 gp140 expressed in 293T cells with furin. The Env proteins were precipitated with CD4-IgG2, b12, or 17b and analyzed by SDS-PAGE and Western blotting.
Generation of Soluble, Uncleaved ΔV1V2 Trimers
Uncleaved Env trimers are not faithful mimics of the functional spike and expose a number of non-neutralizing antibody epitopes that are not accessible on cleaved trimers (21, 23–26). However, uncleaved trimers can be easier to produce. Moreover, it is possible to add protein domains such as stabilizing motifs, epitope and/or purification tags, or immunostimulatory sequences to their C termini (48).4 Such C-terminal extensions are problematic with cleaved gp140 trimers, as they interfere with the gp120/gp41 cleavage process (48),4 although we are currently studying the reasons for this interference and ways to overcome it.
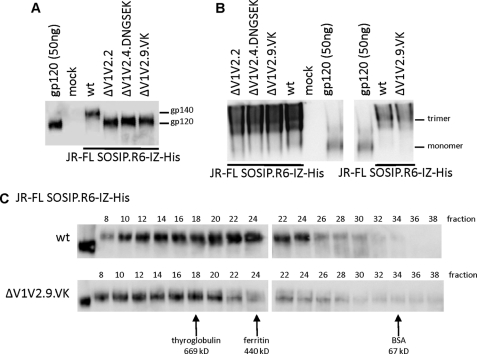
As a template for studying the effect of the ΔV1V2 compensatory changes on uncleaved trimers, we used the SOSIP.R6-IZ-His construct based on the subtype B JR-FL sequence. This protein contains a GCN4-based trimerization domain (isoleucine zipper) and a polyhistidine (His) tag at its C terminus (Fig. 1) (48).4 The three ΔV1V2 variants of this construct were well expressed in 293T cells and formed trimers as efficiently as the corresponding full-length protein, as judged by both Native PAGE and size exclusion chromatography (Fig. 7, A–C). The ΔV1V2.9.VK gp140s were eluted from the size exclusion column slightly earlier than their full-length counterparts (Fig. 7C), suggesting that the average ΔV1V2 gp140 protein component is larger than for full-length gp140. This difference is consistent with earlier findings that V1V2 deletion can improve trimerization at the expense of dimer formation (40).
FIGURE 7.
Soluble, uncleaved ΔV1V2 trimers can be expressed efficiently. A, SDS-PAGE analysis of uncleaved JR-FL SOSIP.R6-IZ-His gp140 variants expressed transiently in 293T cells. B, blue native-PAGE analysis of uncleaved JR-FL SOSIP.R6-IZ-His gp140 from two independent transfections (left and right panels). C, size exclusion analysis of gp140 trimers. Uncleaved JR-FL SOSIP.R6-IZ-His gp140 trimers produced in 293T cells were fractionated using a Superose-6 column. The fractions were analyzed by SDS-PAGE and Western blotting. The elution peaks of reference proteins are indicated.
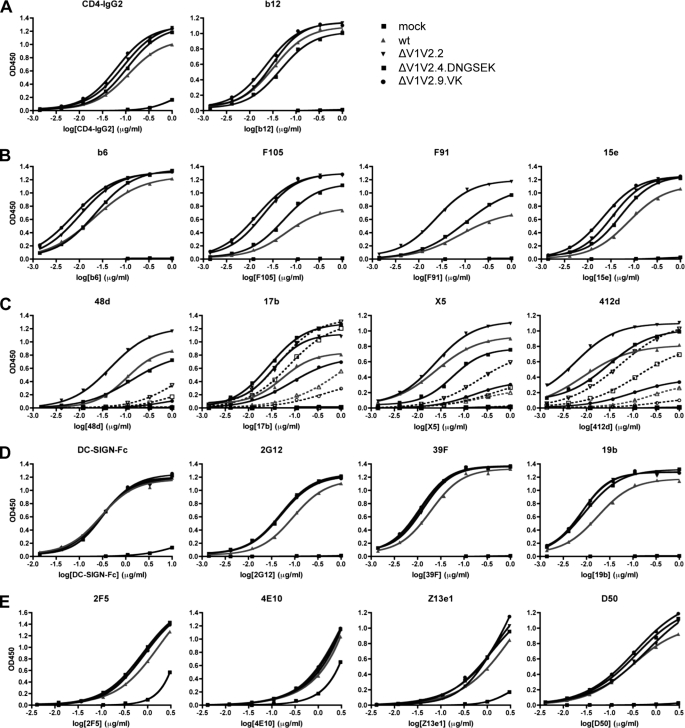
Uncleaved ΔV1V2 Trimers Expose Neutralization Epitopes
To study the exposure of neutralization epitopes on the uncleaved ΔV1V2 JR-FL gp140 constructs, we used an ELISA, in which trimers are captured onto a Ni-NTA-coated solid phase via a C-terminal His tag (Fig. 8 and Table 2). CD4-IgG2 bound slightly better to the three ΔV1V2 variant trimers than to the full-length trimer, but this was not the case with mAb b12 (Fig. 8 and Table 2). Several other CD4bs mAbs (b6, 15e, F91, and F105) did, however, bind better to the three ΔV1V2 trimers (Fig. 8 and Table 2). An exception was that F91 did not bind the ΔV1V2.9.VK trimers, which is probably attributable to the V120K change.
FIGURE 8.
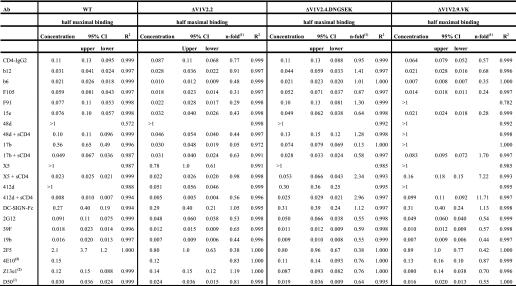
Soluble, uncleaved ΔV1V2 trimers expose neutralization epitopes. ELISA analysis of the binding of various mAbs and CD4-IgG2 to uncleaved JR-FL SOSIP.R6-IZ-His gp140. See Table 2 for half-maximal binding values and statistical analyses.
TABLE 2.
Binding data of soluble uncleaved ΔV1V2 trimers
1 The fold increase in half-maximal binding concentration compared with WT is given.
2 The end point concentrations are given instead of midpoint concentrations, because the binding curve did not reach a plateau.
Different mAbs to the CD4i epitope cluster varied in their reactivity with the uncleaved ΔV1V2 trimers (Fig. 8 and Table 2). mAbs 48d and X5 each bound inefficiently to the full-length proteins and the ΔV1V2.2 and ΔV1V2.4.DNGSEK variants in the absence of sCD4 but much more strongly when sCD4 was added. In contrast, the ΔV1V2.2 and ΔV1V2.4.DNGSEK variants, but not the full-length trimers, bound mAb 17b efficiently even when sCD4 was absent. Intermediate results were seen with a fourth CD4i-epitope mAb, 412d. Thus, without sCD4, the ΔV1V2.2 and ΔV1V2.4.DNGSEK trimers bound 412d more efficiently than the full-length trimers, but there was still an increase in reactivity when sCD4 was added. The variant ΔV1V2.9.VK trimers bound poorly to all the CD4i antibodies, which again probably relates to the epitope-impairing effect of the V120K substitution. Among the variants, ΔV1V2.2 was the one that bound all the CD4i antibodies most efficiently. Hence, the small disulfide-bonded V1V2 stump in ΔV1V2.4.DNGSEK and ΔV1V2.9.VK may interfere with the formation of CD4i epitopes.
DC-SIGN bound similarly to the ΔV1V2 variants and the full-length trimers, as expected because the oligomannose N-glycans on gp120 to which DC-SIGN binds are not located in the V1V2 domain (Fig. 8 and Table 2). In contrast, the ΔV1V2 variant trimers bound mAb 2G12 more efficiently than the full-length proteins (Fig. 8 and Table 2). The ΔV1V2 variants also interacted more efficiently with V3 mAbs 39F and 19b (Fig. 8 and Table 2), which differs slightly from observations made using cleaved KNH1144 trimers in immunoprecipitation assays (Fig. 6; see under “Discussion”). Finally, mAbs directed to the MPER region and neighboring cluster II epitopes in gp41 (2F5, 4E10, Z13e1, and D50) all interacted more efficiently with the ΔV1V2 variants than the full-length trimers, although the differences were minor (Fig. 8 and Table 2).
In summary, the global conformation of the various V1V2 loop-deleted trimers is comparable with that of the corresponding full-length proteins. However, deletion of the V1V2 domain increases the exposure of several antibody epitopes, to various extents.
DISCUSSION
Deleting the V1V2 loops from soluble monomeric HIV-1 Env proteins can be advantageous for both immunogenicity and structural studies (14, 32–38). However, the deletion interferes with the biosynthesis and integrity of soluble Env trimers, complicating the use of these proteins (1, 39–41).3 The principal defect caused by removing the variable loops may involve the exposure of hydrophobic regions of the protein that are normally occluded, leading to aggregation or misfolding (1). Our goal in this study was to devise ways to overcome these problems and thereby improve the design of soluble ΔV1V2 trimers. To achieve this, we applied lessons learned from analyzing the sequences of evolved ΔV1V2 viruses (1). The substitutions that compensate for the loss of the V1V2 loops, and thereby improve replication kinetics, involve changes from hydrophobic to hydrophilic amino acids, or the addition of a glycan site. They appear to act by reducing the extent to which a hydrophobic region of gp120, normally occluded by the V1V2 loops, is solvent-exposed. Our detailed findings are summarized as follows.
Frequently Used Replacement of V1V2 Sequences by a Gly-Ala-Gly Linker Is Not Optimal
Replacing the first glycine with aspartic acid improves HIV-1 LAI replication and/or infectivity (Fig. 2, E and F, compare ΔV1V2.9 with ΔV1V2.8). The G128D substitution also improves the expression of soluble trimers based on the subtype A virus, KNH1144 (Fig. 5A, compare ΔV1V2.9 with ΔV1V2.8). The serine substitution of the first glycine had an ambiguous effect on the function of LAI Env (Fig. 2, C and D, ΔV1V2.4.DNGS and ΔV1V2.4.DNGSEK), but it did improve the expression of soluble KNH1144 trimers (Fig. 5A, compare ΔV1V2.4.DNGS with ΔV1V2.4.DN and ΔV1V2.4.DNGSRK with ΔV1V2.4.DNRK).
Preserving the 126–196 Disulfide Bond Is Not Necessary
Replacing the natural disulfide-bond loop by an Ala-Ala linker (variant ΔV1V2.2) yielded a functional Env, and we could also generate soluble trimers based on this design. This strategy might be useful when designing trimers that contain new disulfide bonds by reducing the potential for their forming aberrant bonds with existing cysteine residues.5
Having an N-Linked Glycosylation Site at Position 197 Is Desirable
The ΔV1V2.4.DN variant was more infectious than ΔV1V2.4, confirming what we suspected from our earlier evolution experiments (1). Furthermore, ΔV1V2.2 was more infectious than ΔV1V2.AGA, and the principal difference is its possession of the Asn-197 glycan. The beneficial effect of this glycan is not necessarily linked to the presence of the neighboring 126–196 disulfide bond, despite the association between N-glycosylation and disulfide bond formation, particularly when the sites are proximal (81, 82). More likely is that the 197 glycan covers a hydrophobic patch that would otherwise cause aggregation and/or protein misfolding (Fig. 3F).
Hydrophobic Surface at the V1V2 Stem and Base Should Be Reduced by Substitutions in the V1V2 Stem and Bridging Sheet
We confirmed that replacement of valine 120 with glutamic acid (V120E) or lysine (V120K) substantially improved LAI infectivity and replication. Because either a positively or a negatively charged amino acid has this effect, the polar nature of the change, and not the charge per se, is important. The V120K substitution was particularly beneficial for enhancing the expression of soluble, V1V2 loop-deleted gp140 trimers (Fig. 5A).
Some Distant Site Changes May be Helpful
The E429K substitution improved the replication of ΔV1V2 LAI (Fig. 2C). Strikingly, we also found that the more subtle substitution R429K improved the expression of KNH1144 ΔV1V2 trimers (compare ΔV1V2.4.DNRK with ΔV1V2.4.DN and ΔV1V2.4.DNGSRK with ΔV1V2.4.DNGS in Fig. 5A).
The V1V2-deleted viruses were more sensitive to neutralization by the mannose-specific 2G12 mAb (Fig. 4), implying that the oligomannose N-glycans that constitute the 2G12 epitope are more accessible to 2G12 when the V1V2 domain is deleted. Paradoxically, V1V2 deletion apparently does not increase the exposure of the same N-glycans to the class I mannosidases in endoplasmic reticulum and/or Golgi that are responsible for trimming oligomannose N-glycans during their conversion to complex N-glycans (if this process did in fact occur, these N-glycans would be in complex form and hence 2G12 binding and neutralization would be lost). The explanation may lie in the different ways that 2G12 and mannosidases recognize oligomannose N-glycans. Thus, 2G12 interacts with the outer mannose moieties (30, 31, 83, 84), whereas class I mannosidases recognize the complete N-glycan, including components at the stem (85, 86). V1V2 deletion must therefore improve the accessibility of the outer carbohydrate residues but not the stems of the N-glycans that form the 2G12 epitope.
In our analyses of trimer antigenicity, we observed that V1V2 deletion affected the binding of mAbs to different (but similar) CD4i epitopes to varying extents. Thus, 17b interacted efficiently with ΔV1V2 Env in the absence of soluble CD4, although 48d and X5 did not. 48d and X5 binding to ΔV1V2 Env was enhanced very efficiently by soluble CD4, but less so for 17b because the binding of this CD4i mAb was already close to optimal in the absence of soluble CD4. The various CD4i epitopes may therefore be shielded to slightly different extents by the V1V2 domain. CD4 binding drives two conformational changes that contribute to the formation and exposure of these epitopes. First, the two 2-stranded β-sheets, β2-β3 and β20-β21, become united into the 4-stranded bridging sheet that is a critical element of all CD4i epitopes (14). Second, there is a repositioning of the V1V2 domain itself (9–14). An explanation for our observations may be that creating the 17b epitope is more dependent on the repositioning of V1V2, although creating the 48d and X5 epitopes is more dependent on the formation of the bridging sheet.
Will ΔV1V2 trimers be good immunogens? Variable loops have long been considered poor targets for broadly neutralizing antibodies, because their variability plays into the problem of HIV-1 sequence diversity. Furthermore, because they shield more conserved neutralization epitopes on the underlying protein domains, their removal can be beneficial. However, the recent isolation of the potent neutralizing antibodies PG9 and PG16 provides a counter-perspective. These mAbs appear to target conserved elements within the variable domains that may be important for immunogen design (6). Another argument against deleting the variable loops is that undesirable neo-epitopes, neutralization-irrelevant epitopes, or strain-restricted neutralization epitopes (for example in V3) may become more exposed, perhaps compromising the induction of broadly neutralizing antibodies. Along these lines, we observed that V1V2 deletion modestly increased Env binding and virus neutralization by some V3 mAbs and some weakly or non-neutralizing CD4bs mAbs. However, our understanding of the determinants of Env immunogenicity is incomplete. There may be a trade-off between sacrificing certain potential neutralization epitopes in favor of the optimal presentation of others, which could only be investigated by immunization studies. Moreover, an optimal Env-based vaccine might include more than one protein component with different properties, for example a protein that retains the V1V2 loops (and hence the PG9/PG16 epitopes) mixed with a loop-deleted protein that better exposes neutralization-relevant CD4bs epitopes.
Will ΔV1V2 trimers assist in structural analyses? The crystallization process is rarely predictable, but structural heterogeneity and flexibility are two major obstacles to obtaining Env trimer crystals. All the available gp120 structures lack the V1V2 domain and in most cases also V3, C1, and part of C5 (14, 38, 77, 80, 87). Furthermore, all the gp120s were deglycosylated. Trimeric gp140s are much more problematic for crystallography, even compared with gp120. The same techniques, and more, may therefore need to be used in trimer crystallization strategies. Hence, removal of the V1V2 domains and reduction or elimination of glycan may be necessary, if perhaps not sufficient, steps on the road to a trimer crystal structure. We have shown that glycan heterogeneity can be reduced dramatically by expressing Env proteins in a cell line lacking the GnTI enzyme (48, 78). We have since explored whether trimers expressed in GnTI knock-out cells can be deglycosylated, with encouraging results.6 We now show that ΔV1V2 trimers can also be produced efficiently in these cells (Fig. 5C), opening up the possibility of combining V1V2 deletion and deglycosylation strategies.
In conclusion, we have shown that virus evolution can guide the design of soluble, V1V2-deleted variant gp140 trimers with improved properties that might be valuable for immunogenicity and crystallography studies.
Acknowledgments
We are grateful to Stephan Heynen for technical assistance and to William Olson, Dennis Burton, James Robinson, and Peter Kwong for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants AI36082 and AI45463 (to J. P. M.). This work was also supported by AIDS Fund (Amsterdam, The Netherlands) Grants 2005021 (to B. B.) and 2008013 (to R. W. S.) and by the International AIDS Vaccine Initiative.
I. Bontjer, M. Melchers, D. Eggink, K. David, J. P. Moore, B. Berkhout, and R. W. Sanders, unpublished results.
M. Melchers, K. Matthews, R. P. de Vries, D. Eggink, T. van Montfort, I. Bontjer, C. E. Van de Sandt, K. B. David, B. Berkhout, J. P. Moore, and R. W. Sanders, submitted for publication.
R. W. Sanders, I. Braakman, and P. Kwong, unpublished observations.
R. W. Sanders, R. Pejchal, I. Wilson, and J. P. Moore, unpublished results.
- Env
- envelope glycoproteins complex
- NAb
- neutralizing antibody
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Ni-NTA
- nickel-nitrilotriacetic acid
- GnTI
- N-acetylglucosaminyltransferase I.
REFERENCES
- 1.Bontjer I., Land A., Eggink D., Verkade E., Tuin K., Baldwin C., Pollakis G., Paxton W. A., Braakman I., Berkhout B., Sanders R. W. (2009) J. Virol. 83, 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton D. R., Pyati J., Koduri R., Sharp S. J., Thornton G. B., Parren P. W., Sawyer L. S., Hendry R. M., Dunlop N., Nara P. L. (1994) Science 266, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 3.Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J. P., Katinger H. (1996) J. Virol. 70, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corti D., Langedijk J. P., Hinz A., Seaman M. S., Vanzetta F., Fernandez-Rodriguez B. M., Silacci C., Pinna D., Jarrossay D., Balla-Jhagjhoorsingh S., Willems B., Zekveld M. J., Dreja H., O'Sullivan E., Pade C., Orkin C., Jeffs S. A., Montefiori D. C., Davis D., Weissenhorn W., McKnight A., Heeney J. L., Sallusto F., Sattentau Q. J., Weiss R. A., Lanzavecchia A. (2010) PLoS ONE 5, e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., Katinger H. (2001) AIDS Res. Hum. Retroviruses 17, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 6.Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W., Kaminsky S., Zamb T., Moyle M., Koff W. C., Poignard P., Burton D. R. (2009) Science 326, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muster T., Guinea R., Trkola A., Purtscher M., Klima A., Steindl F., Palese P., Katinger H. (1994) J. Virol. 68, 4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascola J. R., Montefiori D. C. (2010) Annu. Rev. Immunol. 28, 413–444 [DOI] [PubMed] [Google Scholar]
- 9.Thali M., Moore J. P., Furman C., Charles M., Ho D. D., Robinson J., Sodroski J. (1993) J. Virol. 67, 3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyatt R., Moore J., Accola M., Desjardin E., Robinson J., Sodroski J. (1995) J. Virol. 69, 5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L., Gerard N. P., Wyatt R., Choe H., Parolin C., Ruffing N., Borsetti A., Cardoso A. A., Desjardin E., Newman W., Gerard C., Sodroski J. (1996) Nature 384, 179–183 [DOI] [PubMed] [Google Scholar]
- 12.Trkola A., Dragic T., Arthos J., Binley J. M., Olson W. C., Allaway G. P., Cheng-Mayer C., Robinson J., Maddon P. J., Moore J. P. (1996) Nature 384, 184–187 [DOI] [PubMed] [Google Scholar]
- 13.Sullivan N., Sun Y., Sattentau Q., Thali M., Wu D., Denisova G., Gershoni J., Robinson J., Moore J., Sodroski J. (1998) J. Virol. 72, 4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B., Vogan E. M., Gong H., Skehel J. J., Wiley D. C., Harrison S. C. (2005) Structure 13, 197–211 [DOI] [PubMed] [Google Scholar]
- 15.Jacobs A., Garg H., Viard M., Raviv Y., Puri A., Blumenthal R. (2008) Vaccine 26, 3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison J. E., Lynch J. B., Sierra L. J., Blackburn L. A., Ray N., Collman R. G., Doms R. W. (2008) J. Virol. 82, 11695–11704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earl P. L., Sugiura W., Montefiori D. C., Broder C. C., Lee S. A., Wild C., Lifson J., Moss B. (2001) J. Virol. 75, 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beddows S., Schülke N., Kirschner M., Barnes K., Franti M., Michael E., Ketas T., Sanders R. W., Maddon P. J., Olson W. C., Moore J. P. (2005) J. Virol. 79, 8812–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava I. K., Stamatatos L., Kan E., Vajdy M., Lian Y., Hilt S., Martin L., Vita C., Zhu P., Roux K. H., Vojtech L., C., Montefiori D., Donnelly J., Ulmer J. B., Barnett S. W. (2003) J. Virol. 77, 11244–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Wyatt R., Sodroski J. (2001) J. Virol. 75, 1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binley J. M., Sanders R. W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D. J., Maddon P. J., Olson W. C., Moore J. P. (2000) J. Virol. 74, 627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders R. W., Vesanen M., Schuelke N., Master A., Schiffner L., Kalyanaraman R., Paluch M., Berkhout B., Maddon P. J., Olson W. C., Lu M., Moore J. P. (2002) J. Virol. 76, 8875–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey A. K., David K. B., Lu M., Moore J. P. (2009) Virology 385, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Si Z., Phan N., Kiprilov E., Sodroski J. (2003) AIDS Res. Hum. Retroviruses 19, 217–226 [DOI] [PubMed] [Google Scholar]
- 25.Pancera M., Lebowitz J., Schön A., Zhu P., Freire E., Kwong P. D., Roux K. H., Sodroski J., Wyatt R. (2005) J. Virol. 79, 9954–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera C., Klasse P. J., Michael E., Kake S., Barnes K., Kibler C. W., Campbell-Gardener L., Si Z., Sodroski J., Moore J. P., Beddows S. (2005) Virology 338, 154–172 [DOI] [PubMed] [Google Scholar]
- 27.Subramaniam S. (2010) Keystone Symposia, HIV Vaccines (X5), Banff, Alberta, Canada, March 21–26, 2010, [Google Scholar]
- 28.Reitter J. N., Means R. E., Desrosiers R. C. (1998) Nat. Med. 4, 679–684 [DOI] [PubMed] [Google Scholar]
- 29.Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., Shaw G. M. (2003) Nature 422, 307–312 [DOI] [PubMed] [Google Scholar]
- 30.Sanders R. W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K. O., Kwong P. D., Moore J. P. (2002) J. Virol. 76, 7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann, Saphire E., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Rudd P. M., Burton D. R. (2002) J. Virol. 76, 7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnett S. W., Lu S., Srivastava I., Cherpelis S., Gettie A., Blanchard J., Wang S., Mboudjeka I., Leung L., Lian Y., Fong A., Buckner C., Ly A., Hilt S., Ulmer J., Wild C. T., Mascola J. R., Stamatatos L. (2001) J. Virol. 75, 5526–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gzyl J., Bolesta E., Wierzbicki A., Kmieciak D., Naito T., Honda M., Komuro K., Kaneko Y., Kozbor D. (2004) Virology 318, 493–506 [DOI] [PubMed] [Google Scholar]
- 34.Lu S., Wyatt R., Richmond J. F., Mustafa F., Wang S., Weng J., Montefiori D. C., Sodroski J., Robinson H. L. (1998) AIDS Res. Hum. Retroviruses 14, 151–155 [DOI] [PubMed] [Google Scholar]
- 35.Jeffs S. A., Shotton C., Balfe P., McKeating J. A. (2002) J. Gen. Virol. 83, 2723–2732 [DOI] [PubMed] [Google Scholar]
- 36.McKeating J. A., Shotton C., Jeffs S., Palmer C., Hammond A., Lewis J., Oliver K., May J., Balfe P. (1996) Immunol. Lett. 51, 101–105 [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. Y., Chakrabarti B. K., Xu L., Welcher B., Kong W. P., Leung K., Panet A., Mascola J. R., Nabel G. J. (2004) J. Virol. 78, 4029–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders R. W., Schiffner L., Master A., Kajumo F., Guo Y., Dragic T., Moore J. P., Binley J. M. (2000) J. Virol. 74, 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schülke N., Vesanen M. S., Sanders R. W., Zhu P., Lu M., Anselma D. J., Villa A. R., Parren P. W., Binley J. M., Roux K. H., Maddon P. J., Moore J. P., Olson W. C. (2002) J. Virol. 76, 7760–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Center R. J., Schuck P., Leapman R. D., Arthur L. O., Earl P. L., Moss B., Lebowitz J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laakso M. M., Lee F. H., Haggarty B., Agrawal C., Nolan K. M., Biscone M., Romano J., Jordan A. P., Leslie G. J., Meissner E. G., Su L., Hoxie J. A., Doms R. W. (2007) PLoS Pathog. 3, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders R. W., Dankers M. M., Busser E., Caffrey M., Moore J. P., Berkhout B. (2004) Retrovirology 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders R. W., Busser E., Moore J. P., Lu M., Berkhout B. (2004) AIDS Res. Hum. Retroviruses 20, 742–749 [DOI] [PubMed] [Google Scholar]
- 45.Peden K., Emerman M., Montagnier L. (1991) Virology 185, 661–672 [DOI] [PubMed] [Google Scholar]
- 46.Beddows S., Kirschner M., Campbell-Gardener L., Franti M., Dey A. K., Iyer S. P., Maddon P. J., Paluch M., Master A., Overbaugh J., VanCott T., Olson W. C., Moore J. P. (2006) AIDS Res. Hum. Retroviruses 22, 569–579 [DOI] [PubMed] [Google Scholar]
- 47.Binley J. M., Sanders R. W., Master A., Cayanan C. S., Wiley C. L., Schiffner L., Travis B., Kuhmann S., Burton D. R., Hu S. L., Olson W. C., Moore J. P. (2002) J. Virol. 76, 2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eggink D., Melchers M., Wuhrer M., van Montfort T., Dey A. K., Naaijkens B. A., David K. B., Le Douce V., Deelder A. M., Kang K., Olson W. C., Berkhout B., Hokke C. H., Moore J. P., Sanders R. W. (2010) Virology 401, 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson J. D., Brunel F. M., Jensen R., Crooks E. T., Cardoso R. M., Wang M., Hessell A., Wilson I. A., Binley J. M., Dawson P. E., Burton D. R., Zwick M. B. (2007) J. Virol. 81, 4033–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roben P., Moore J. P., Thali M., Sodroski J., Barbas C. F., 3rd., Burton D. R. (1994) J. Virol. 68, 4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson J. E., Yoshiyama H., Holton D., Elliot S., Ho D. D. (1992) J. Cell. Biochem. 50, Suppl.16E, 71 [Google Scholar]
- 52.Conley A. J., Gorny M. K., Kessler J. A., 2nd., Boots L. J., Ossorio-Castro M., Koenig S., Lineberger D. W., Emini E. A., Williams C., Zolla-Pazner S. (1994) J. Virol. 68, 6994–7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Earl P. L., Broder C. C., Doms R. W., Moss B. (1997) J. Virol. 71, 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang S. H., Farzan M., Si Z., Madani N., Wang L., Rosenberg E., Robinson J., Sodroski J. (2005) J. Virol. 79, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moulard M., Phogat S. K., Shu Y., Labrijn A. F., Xiao X., Binley J. M., Zhang M. Y., Sidorov I. A., Broder C. C., Robinson J., Parren P. W., Burton D. R., Dimitrov D. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6913–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Posner M. R., Hideshima T., Cannon T., Mukherjee M., Mayer K. H., Byrn R. A. (1991) J. Immunol. 146, 4325–4332 [PubMed] [Google Scholar]
- 57.Allaway G. P., Ryder A. M., Beaudry G. A., Maddon P. J. (1993) AIDS Res. Hum. Retroviruses 9, 581–587 [DOI] [PubMed] [Google Scholar]
- 58.Sanders R. W., de Jong E. C., Baldwin C. E., Schuitemaker J. H., Kapsenberg M. L., Berkhout B. (2002) J. Virol. 76, 7812–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das A. T., Land A., Braakman I., Klaver B., Berkhout B. (1999) Virology 263, 55–69 [DOI] [PubMed] [Google Scholar]
- 60.Kirschner M., Monrose V., Paluch M., Techodamrongsin N., Rethwilm A., Moore J. P. (2006) Protein Expr. Purif. 48, 61–68 [DOI] [PubMed] [Google Scholar]
- 61.Moore J. P., Jarrett R. F. (1988) AIDS Res. Hum. Retroviruses 4, 369–379 [DOI] [PubMed] [Google Scholar]
- 62.Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 64.Kwong P. D., Wyatt R., Majeed S., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (2000) Struct. Fold. Des. 8, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 65.Eggink D., Melchers M., Sanders R. W. (2007) Trends Microbiol. 15, 291–294 [DOI] [PubMed] [Google Scholar]
- 66.Kwong P. D., Wyatt R., Sattentau Q. J., Sodroski J., Hendrickson W. A. (2000) J. Virol. 74, 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bohne-Lang A., von der Lieth C. W. (2005) Nucleic Acids Res. 33, W214–W219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cutalo J. M., Deterding L. J., Tomer K. B. (2004) J. Am. Soc. Mass Spectrom. 15, 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu X., Borchers C., Bienstock R. J., Tomer K. B. (2000) Biochemistry 39, 11194–11204 [DOI] [PubMed] [Google Scholar]
- 70.Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. (1990) J. Biol. Chem. 265, 10373–10382 [PubMed] [Google Scholar]
- 71.Schrödinger L. L. (2010) The PyMOL Molecular Graphics System, Version 1.2r3pre Schrödinger, LLC, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 72.Moore J. P., Yoshiyama H., Ho D. D., Robinson J. E., Sodroski J. (1993) AIDS Res. Hum. Retroviruses 9, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 73.Kwong P. D., Wyatt R., Desjardins E., Robinson J., Culp J. S., Hellmig B. D., Sweet R. W., Sodroski J., Hendrickson W. A. (1999) J. Biol. Chem. 274, 4115–4123 [DOI] [PubMed] [Google Scholar]
- 74.Cao J., Sullivan N., Desjardin E., Parolin C., Robinson J., Wyatt R., Sodroski J. (1997) J. Virol. 71, 9808–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyatt R., Sullivan N., Thali M., Repke H., Ho D., Robinson J., Posner M., Sodroski J. (1993) J. Virol. 67, 4557–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stamatatos L., Wiskerchen M., Cheng-Mayer C. (1998) AIDS Res. Hum. Retroviruses 14, 1129–1139 [DOI] [PubMed] [Google Scholar]
- 77.Pancera M., Majeed S., Ban Y. E., Chen L., Huang C. C., Kong L., Kwon Y. D., Stuckey J., Zhou T., Robinson J. E., Schief W. R., Sodroski J., Wyatt R., Kwong P. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Binley J. M., Ban Y. E., Crooks E. T., Eggink D., Osawa K., Schief W. R., Sanders R. W. (2010) J. Virol. 84, 5637–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pantophlet R., Ollmann, Saphire E., Poignard P., Parren P. W., Wilson I. A., Burton D. R. (2003) J. Virol. 77, 642–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diskin R., Marcovecchio P. M., Bjorkman P. J. (2010) Nat. Struct. Mol. Biol. 17, 608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanders R. W., van Anken E., Nabatov A. A., Liscaljet I. M., Bontjer I., Eggink D., Melchers M., Busser E., Dankers M. M., Groot F., Braakman I., Berkhout B., Paxton W. A. (2008) Retrovirology 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daniels R., Kurowski B., Johnson A. E., Hebert D. N. (2003) Mol. Cell 11, 79–90 [DOI] [PubMed] [Google Scholar]
- 83.Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A. (2003) Science 300, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 84.Calarese D. A., Lee H. K., Huang C. Y., Best M. D., Astronomo R. D., Stanfield R. L., Katinger H., Burton D. R., Wong C. H., Wilson I. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vallée F., Lipari F., Yip P., Sleno B., Herscovics A., Howell P. L. (2000) EMBO J. 19, 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tempel W., Karaveg K., Liu Z. J., Rose J., Wang B. C., Moremen K. W. (2004) J. Biol. Chem. 279, 29774–29786 [DOI] [PubMed] [Google Scholar]
- 87.Zhou T., Xu L., Dey B., Hessell A. J., Van Ryk D., Xiang S. H., Yang X., Zhang M. Y., Zwick M. B., Arthos J., Burton D. R., Dimitrov D. S., Sodroski J., Wyatt R., Nabel G. J., Kwong P. D. (2007) Nature 445, 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]