Abstract
Human β-defensins (hBDs) are small, cationic antimicrobial peptides, secreted by mucosal epithelial cells that regulate adaptive immune functions. We previously reported that Fusobacterium nucleatum, a ubiquitous Gram-negative bacterium of the human oral cavity, induces human β-defensin 2 (hBD2) upon contact with primary oral epithelial cells. We now report the isolation and characterization of an F. nucleatum (ATCC 25586)-associated defensin inducer (FAD-I). Biochemical approaches revealed a cell wall fraction containing four proteins that stimulated the production of hBD2 in human oral epithelial cells (HOECs). Cross-referencing of the N-terminal sequences of these proteins with the F. nucleatum genome revealed that the genes encoding the proteins were FadA, FN1527, FN1529, and FN1792. Quantitative PCR of HOEC monolayers challenged with Escherichia coli clones expressing the respective cell wall proteins revealed that FN1527 was most active in the induction of hBD2 and hence was termed FAD-I. We tagged FN1527 with a c-myc epitope on the C-terminal end to identify and purify it from the E. coli clone. Purified FN1527 (FAD-I) induced hBD2 mRNA and protein expression in HOEC monolayers. F. nucleatum cell wall and FAD-I induced hBD2 via TLR2. Porphorymonas gingivalis, an oral pathogen that does not induce hBD2 in HOECs, was able to significantly induce expression of hBD2 in HOECs only when transformed to express FAD-I. FAD-I or its derivates offer a potentially new paradigm in immunoregulatory therapeutics because they may one day be used to bolster the innate defenses of vulnerable mucosae.
Keywords: Bacteria, Cell Wall, Defensins, Epithelial cell, Transformation, Beta-Defensins, FAD-I, Fusobacterium nucleatum, Porphyromonas gingivalis, Human Oral Epithelial Cells
Introduction
Oral mucosal epithelium represents the first line of defense against invading microbes. This tissue expresses antimicrobial peptides, including human β-defensins (hBDs),2 that play an important role in immune defense against invading pathogens. Recent studies have shown that in addition to their antibacterial, antifungal, and antiviral properties (1–8), hBDs also contribute to immunoregulatory functions of mucosal barriers. hBDs engage the CCR6 receptor on selected immune effector cells, such as immature dendritic cells and T cells, and evoke a chemokine response, thereby recruiting these cells to the site of interest (9). In addition, mouse β-defensin 2 has been shown to induce dendritic cell maturation (10) through TLR4 (Toll-like receptor 4), and mouse β-defensin 2-based vaccines elicit potent cell-mediated responses and antitumor immunity (11, 12). Our group's recent discoveries demonstrate that hBD3 induces maturation of immature dendritic cells and monocytes through interaction with TLR1/2 (13) and interacts with the HIV co-receptor CXCR4, promoting receptor internalization and antagonism (14). Recently, we discovered that hBD3 demonstrates an affinity for CCR2 and can chemoattract monocytes/macrophages into the oral mucosae (15). Interestingly, hBD2 can also chemoattract myeloid cells via CCR2 (16).
We previously showed that the cell wall of Fusobacterium nucleatum, an indigenous Gram-negative bacterium of the human oral cavity present in periodontal health and disease, can induce hBD2 mRNA expression in normal human oral epithelial cells (HOECs) (17). Herein, we present novel information regarding the isolation and characterization of a Fusobacterium-associated defensin inducer (FAD-I). A systematic biochemical fractionation of the F. nucleatum cell wall (ATCC 25586), followed by isoelectric focusing, led to an active fraction containing four candidate proteins, whose genes were identified using the F. nucleatum genome (18). Expressing each of the candidate proteins in Escherichia coli and challenging HOECs with them demonstrated their respective capacity to induce hBD2. FN1527 (annotation based on a gene from the F. nucleatum genome (18)) consistently induced hBD2 to the highest levels. Finally, by expressing FN1527 in Porphyromonas gingivalis, an opportunistic Gram-negative bacterium strongly implicated in periodontal disease and an organism that does not induce appreciable levels of hBD mRNA in HOECs when compared with F. nucleatum (17), we were able to show that the transformed bacterium could induce hBD2 significantly more than the parent strain. FAD-I or its derivates offer a potentially new paradigm in immunoregulatory therapeutics because they may one day be used as novel agents to bolster the innate defenses of vulnerable mucosae.
EXPERIMENTAL PROCEDURES
Oral Epithelial Cell Culture
Our studies were performed according to the policies of the Institutional Review Board at Case Western Reserve University. After obtaining informed consent, healthy oral tissue overlying impacted third molars of normal adults were extracted and used to isolate HOECs, as described previously (19). Cells were cultured in EpiLife growth medium (Cascade Biologists, Portland, OR) and maintained at 37 °C in 5% CO2. Primary cells were grown in serum-free conditions as a monolayer (19). At confluence, cells from at least three donors were trypsinized, detached, pooled, and reseeded at 4 × 104 cells/well in 6-well culture dishes in EpiLife medium. The cells were cultured until they were ∼80% confluent (∼3 × 105 cells/well) prior to challenge with different E. coli constructs.
Preparation of F. nucleatum Cell Wall
Cell wall from F. nucleatum was prepared as described previously (20). Briefly, F. nucleatum (ATCC 25586) was grown anaerobically in Columbia broth (BD Biosciences) overnight. Crude cell wall preparations were prepared by French pressure cell disruption of freshly harvested whole cells (7.1 g wet biomass) in 15 ml of phosphate-buffered saline PBS (pH 7.2) at 15,000 pounds/inch2. The cell walls were recovered after low speed centrifugation (1,000 × g, 15 min), followed by high speed centrifugation (138,000 × g, 30 min) of the supernatant (20). The cell wall pellet obtained from the high speed centrifugation was lyophilized and subjected to purification methods to isolate the active fraction as described below.
Isolation of the “Active Fraction”
Isoelectric focusing was performed with a Rotofor cell (Bio-Rad) with ampholytes in the pH range of 3–10 for 3–4 h. Fractions were collected, and ampholytes were removed by Centricon Plus filters (3000 molecular weight cut-off membranes; Amicon (Bedford, MA)). HPLC was performed on a BREEZE chromatography system (Waters, Milford, MA) using a Symmetry C4, 5-μm, 4.6 × 150-mm column with an acetonitrile gradient. Fractions collected in the pI range of 4–5 were found to induce hBD2 mRNA in HOECs. These fractions were charged onto a C4 HPLC column and eluted at various time points in an acetonitrile gradient (Fig. 2C, blue line). Candidate peaks from 30–40 min elutions were found to induce hBD2. These were collected into a fraction that was subsequently termed the “active fraction.”
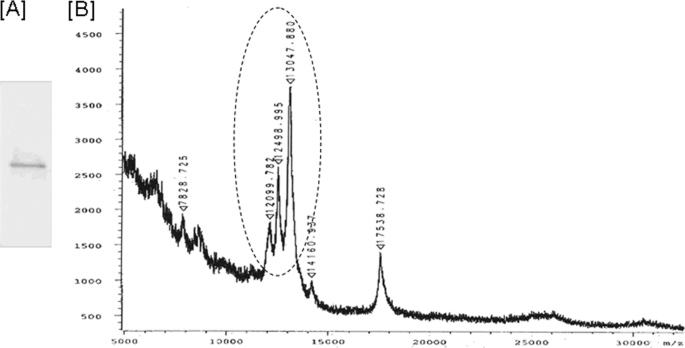
FIGURE 2.
SDS-PAGE and MALDI-MS analysis of the active fraction. A, SDS-PAGE of active fraction obtained from HPLC purification in Fig. 1. The only band found in the active fraction near 12–14 kDa was excised for trypsin digest and amino acid sequencing. B, MALDI-MS was performed in a solvent of 1:1 mixture of acetonitrile and water with 0.1% TFA. The sample was mixed 1:1 with the matrix sinapinic acid, and 1 μl was spotted onto the target. The samples were run on a Bucher Reflex II MALDI-TOF instrument operating in linear and positive ion modes. The candidate peaks are indicated with a circle.
Identification of the Components from the hBD2-inducing Active Fraction by MALDI-MS
SDS-PAGE analysis of the active fraction was performed with gradient gels (4–20%) and stained with Coomassie Blue. The only protein band observed in the active fraction near 12–14 kDa (Fig. 3A) was excised for trypsin digest and amino acid sequencing (Mass Spectrometry Laboratory for Protein sequencing, Lerner Research Institute, Cleveland Clinic Foundation). To perform MALDI-MS analysis on the active fraction, the solvent used was a 1:1 mixture of acetonitrile and water with 0.1% TFA. The sample was mixed 1:1 with the matrix sinapinic acid, and 1 μl was spotted onto the target. The samples were run on a Bucher Reflex II MALDI TOF instrument operating in linear and positive ion modes. The data were analyzed by using collision-induced dissociation spectra to search the NCBI non-redundant data base, including the entire genome sequence of F. nucleatum 25586 (18), with the search program TurboSEQUEST® (Thermo Electron Corp.).
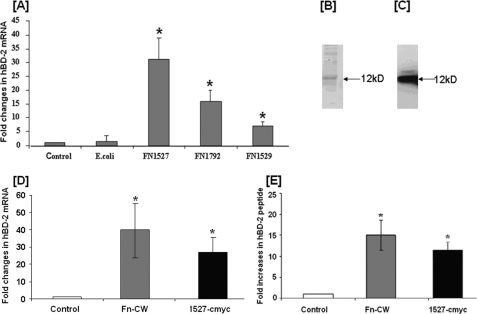
FIGURE 3.
HOEC expression of hBD2 following challenge by cell extracts from E. coli expressing different F. nucleatum recombinant proteins and induction of hBD2 by affinity-purified FN1527 c-Myc. A, HOECs were treated with extracts (100 μg/ml total protein) of lysed E. coli expressing protein FN1527, FN1529, or FN1792. Total RNA was extracted, followed by real-time PCR using primers specific for hBD2 (Table 2). -Fold change in mRNA levels represents mean ± S.D. (error bars) of triplicate experiments. p values were calculated by Student's t test (*, p < 0.05). B, analysis of affinity-purified recombinant FN1527 c-Myc by Coomassie staining of an SDS-polyacrylamide gel. C, Western blot of affinity-purified recombinant FN1527 c-Myc using c-Myc antibody. D, HOECs were stimulated with either 10 μg/ml F. nucleatum cell wall (Fn-CW) or 10 μg/ml of 1527 c-Myc for 18 h. Total RNA extracted from cells of experimental and control cultures were screened by real-time PCR for hBD2 mRNA using specific primers. -Fold change in mRNA levels represents mean ± S.D. of triplicate experiments. p values were calculated by Student's t test (*, p < 0.05). E, HOECs were stimulated with either 10 μg/ml F. nucleatum cell wall or 10 μg/ml 1527 c-Myc for 18 h. Cell-free supernatants from experimental and control cultures were screened by ELISA for hBD2 peptide. -Fold increase represents mean ± S.D. of triplicate experiments. p values were calculated by Student's t test. *, p < 0.05. C, HOECs were challenged with heat-treated (100 °C, 10 min) F. nucleatum cell wall, FN1527. After 18 h, cell supernatants were collected and assayed for hBD2 by ELISA. -Fold increase represents mean ± S.D. of triplicate experiments. p values were calculated by Student's t test. *, p < 0.05.
Preparation of Genomic DNA of F. nucleatum
Genomic DNA was isolated from F. nucleatum (ATCC 25586). Briefly, the cell pellet was resuspended in buffer containing 10 mm Tris, pH 8.0, and 1 mm EDTA. 0.5% SDS, and 200 μg/ml proteinase K were added to the cell suspension. The mix was then incubated at 55 °C for 1 h and extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1) mix, followed by precipitation of the DNA with ethanol at −20 °C for 2 h. The DNA pellet was collected by centrifugation; the supernatant was discarded; and the pellet was washed with 70% ethanol, dried at room temperature for 10 min, and resuspended in nuclease-free water.
Cloning and Expression of the Recombinant Peptides
Except for FN0264 (FadA), which was kindly provided by Dr. Y. W. Han (Department of Periodontics, Case Western Reserve University School of Dental Medicine), DNAs encoding the other three candidate peptides (Table 1) were amplified by PCR using specific primers from genomic DNA of F. nucleatum and inserted in pET17b vector (EMD Biosciences, San Diego, CA) under the T7 promoter for expression in E. coli. E. coli BL21 DE3 (Invitrogen) transformed with the pET17b vector containing the DNAs encoding the peptides was cultured at 37 °C to A600 of 0.6–1, treated with 0.1 mm IPTG, and cultured for an additional 3 h. Organisms from the respective E. coli clones were harvested by centrifugation, the cell pellets were washed with PBS, and extracts were made, which were used first to determine hBD2 induction in HOECs, and subsequently the clone expressing FN1527 with the c-myc tag was used for isolating the inducing protein.
TABLE 1.
Proteins identified from the active fraction
NA, not available; OM, outer membrane; PP, periplasmic protein; CM, cytoplasmic membrane proteins.
| Gene name | Preprotein |
Mature |
Function | Identity | Location | ||
|---|---|---|---|---|---|---|---|
| Mass | pI | Mass | pI | ||||
| kDa | kDa | % | |||||
| FN0264 | 14.5 | 4.8 | 12.6 | 4.6 | FadA | 75 | OM-PP |
| FN1529 | 14.2 | 5.4 | 12.2 | 5.1 | Hypothetical | 67 | CM |
| FN1792 | NA | NA | 12.5 | 4.3 | Hypothetical | 65 and 38 | Cytoplasm |
| FN1527 | 14.8 | NA | 13.1 | 4.8 | Hypothetical | 33 | OM-CM |
Isolation and Purification of FN1527 from E. coli
To facilitate identification and purification of FN1527, a c-Myc epitope (EQKLISEEDL) was introduced at its C terminus using standard recombinant DNA techniques. The cell pellet from a 250-ml culture of the E. coli expressing FN1527 was resuspended in 15 ml of PBS containing 1% Triton X-100. The resuspended pellet was sonicated and centrifuged, after which the pellet containing the cell debris was discarded, and recombinant FN1527 was enriched from the supernatant using the c-Myc tag for immunoprecipitation. Briefly, 5 μg of polyclonal c-Myc antibody (GenScript, Piscataway, NJ) was added to the supernatant and incubated with rotary shaking at 4 °C for 1 h. 100 μl of Protein A/G-Sepharose (GE Healthcare) was added, and the lysate was incubated at 4 °C for 2 h with rotary shaking. The beads were precipitated by low speed centrifugation, and the supernatant was discarded. The beads were washed three times with PBS containing 1% Triton X-100.
Western Blot Analysis
Western blot analysis was used to identify c-Myc-tagged FN1527 using standard procedures and c-Myc monoclonal antibody 9E-10 (Covance, Harrisburg, PA). Additionally, a polyclonal antibody to FN1527 was generated (GenScript) for experiments involving the identification of FN1527 in F. nucleatum and P. gingivalis.
Insertion of c-Myc-tagged FN1527 Gene in the Genome of P. gingivalis
To facilitate expression in P. gingivalis, the FN1527 gene was fused to the promoter region of the P. gingivalis fimA gene (21) by fusion PCR (22). Briefly, FN1527 was amplified by PCR from plasmid (pET17b with FN1527) using primers BB28 and BB11 (Table 2), and the promoter-containing region upstream of the P. gingivalis fimA was amplified from P. gingivalis genomic DNA using primers BB25 and BB27. The primers were designed to generate products that have 30 base pairs of overlap to allow fusion in the next reaction. Purified products were combined and cycled for 30 s at 95 °C, 30 s at 58 °C, and 2 min at 68 °C for 30 cycles. The resulting product was then used as a template for amplification of the fused product using primers BB11 and BB26. These primers included additional sequence to add BamHI and SphI sites as well as a c-Myc epitope on the C terminus. The fusion product was gel-purified, digested with SphI and BamHI, ligated into similarly digested pT-COW plasmid (23), and cloned into TOP10F competent E. coli (Invitrogen). Plasmid was electroporated into P. gingivalis strain 33277 as described previously by Smith (24), and transformants were selected by plating on blood agar plates containing 1 μg/ml tetracycline.
TABLE 2.
List of primers
| Primer name | Primer sequence |
|---|---|
| HBD-2 | 5′-ATC AGC CAT GAG GGT CTT GT |
| 3′-GAG ACC ACA GGT GCC AAT TT | |
| HK-5 | 5′-GTC CTC TCC ATG GAC AAC AAC |
| 3′-TGT CAA TCT CGG CTC TCA GCC | |
| FN1527 | 5′-CTA GTC TAG AAT GAA AAA AAT ATT ATT ACT A |
| 3′-CGC GGA TCC TTA TTT TAT TCC TGC ATT ATT | |
| FN1527 reverse c myc | 3′-CGC GGA TCC TTA CAG ATC TTC TTC AGA AAT AAG TTT TTG TTC TTT TAT TCC TGC ATT ATT TAA |
| FN1792 | 5′-CTA GTC TAG AAT GAG TTT ATT CTT AGT AGC T |
| 3′-CGC GGA TCC CTA TTT AGC TTC AAC AGT TAC | |
| FN1529 | 5′-CTA GTC TAG AAT GAA AAA AGT TAT TTT AAC A |
| 3′-CGC GGA TCC CTA TCT TAT TTT TTG AAT TTT | |
| BB25 | 5′-CTC GTC TGA GTC TGG CAG AGG TTT |
| BB27 | 3′-ACT TTG TTT TTT TCA TCT CGT TTT |
| BB11 | 5′-CGC GGA TCC TTA CAG ATC TTC TTC AGA AAT AAG TTT TTG TTC TTT TAT TCC TGC ATT ATT TAA |
| BB26 | 3′-ACA TGA GCA TGC GGG AGC CAC GAA CGC TAC GAA ACG |
Treatment of HOEC Monolayers with Transformed E. coli, P. gingivalis Recombinant Proteins, and Specific Antibodies
HOEC monolayers (70–85% confluent) were incubated with respective recombinant peptide expressing E. coli cell extract (100 μg/ml); 10 μg/ml (unless otherwise mentioned) purified FN1527 c-Myc protein; or 10 μg/ml cell wall preparations of the parent P. gingivalis strain, the P. gingivalis vector control strain, and/or the transformed FN1527-expressing strain, respectively. After an 18-h incubation at 37 °C, 5% CO2, HOECs were lysed for RNA isolation. Negative (unchallenged cells) and positive (10 μg/ml of F. nucleatum cell wall preparation) controls were included in each study. For blocking experiments, anti-TLR2 antibody (eBioscience, San Diego, CA) or isotype control (Invitrogen) was added at 5 μg/ml and incubated for 30 min prior to treatment with F. nucleatum cell wall or FN1527 c-Myc protein.
RNA Preparation and Analysis
Cells were lysed using TRIzol reagent (Invitrogen), and total RNA was isolated in accordance with the manufacturer's instruction. RNA concentration was measured by UV absorbance at 260/280 nm using a Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE). RT-PCR of the samples was done using the procedure we described previously (25). Real-time PCR was done using the Bio-Rad MyIQ system with the iScript cDNA Synthesis Kit and iQ SYBR Green Supermix according to the manufacturer's protocol. The sequences of the specific primers used are found in Table 2.
Detection of hBD2 in Medium Supernatants
Detection of hBD2 in medium supernatant was done using an enzyme-linked immunosorbent assay (ELISA) method as described previously (26, 27).
RESULTS
Isolation of a Cell Wall Fraction of F. nucleatum Responsible for Defensin Production in HOECs and Identification of Its Components
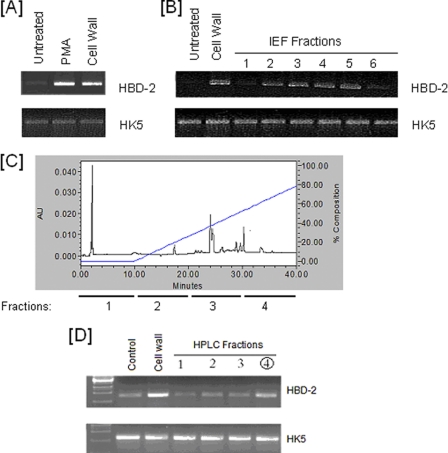
The cell wall preparation of F. nucleatum that induces hBD2 mRNA in HOECs (Fig. 1A) (17) was subjected to isoelectric focusing (pI ∼3–10), and fractions were analyzed for their ability to induce hBD2 mRNA. pI fractions ranging from 4 to 6 were found to induce hBD2 mRNA production in HOECs (Fig. 1B), whereas pI fractions above 6 did not induce hBD2. The pooled active fractions were then subjected to HPLC using a C4 column (Fig. 1C), and fractions eluting at different times (0–10 min, 10–20 min, 20–30 min, and 30–40 min) were tested for hBD2 induction. Fig. 2D shows that fraction 4 (elution time 30–40 min at 52–66% acetonitrile concentration) induced the highest level of hBD2 mRNA expression and was thus termed the “active fraction.” This fraction was subsequently run on PAGE to identify its component proteins. The resulting fraction revealed a single 12–14 kDa protein band (Fig. 2A, a representative SDS-PAGE of multiple runs), which was excised for amino acid sequencing and MALDI-MS analysis (Fig. 2B). By associating fragments of sequences with genes from the F. nucleatum genome data base (18), we were able to identify four candidate proteins (Table 1). One of the four proteins identified was FN0264, which had been previously identified as an F. nucleatum adhesin (FadA; 12.6 kDa, pI 4.6) (28). Recombinant FadA did not stimulate hBD2 mRNA production by HOECs (data not shown). The other three (hypothetical) proteins identified were FN1527 (covering 33% of the sequence, 13.1 kDa, pI 4.8), FN1529 (covering 67% of the protein sequence; 12.2 kDa, pI 5.1), and FN1792 (covering 33% of the sequence; 12.5 kDa and pI 4.8). The molecular mass of these three hypothetical proteins corresponded well with the major protein peaks found by MS analysis (Fig. 2B).
FIGURE 1.
RT-PCR analysis of hBD2 mRNA induction in HOECs at different steps of purification. A, RT-PCR analysis was performed with RNA from HOEC monolayers treated with phorbol 12-myristate 13-acetate (PMA) as positive control (lane 2) or 10 μg of F. nucleatum cell wall preparation (lane 3). Lane 1 shows untreated cells. B, RT-PCR analysis of RNA prepared from HOECs treated with isoelectric focusing-isolated fractions of mean pI 3.7 (lane 1), pI 4.0 (lane 2), pI 4.1 (lane 3), pI 4.5 (lane 4), pI 5.0 (lane 5), or pI 5.8 (lane 6). RT-PCR of RNA from untreated cells and cells treated with 10 μg of F. nucleatum cell wall are also shown. The bottom panel shows RT-PCR for HK-5 in the above samples. Fractions used in lanes 2–5 were pooled and used for HPLC separation using a C4 column. C, isoelectric focusing fraction of pI 4–5 was charged onto a C4 HPLC column and eluted at various time points in an acetonitrile gradient (blue line). Fractions were assayed for their ability to induce hBD2. D, HPLC fractions were incubated with HOEC monolayers, and RT-PCR analysis was performed. Lane 1, HPLC fraction at 0–10 min of elution (fraction 1 in C); lane 2, HPLC fraction at 10–20 min of elution (fraction 2 in C); lane 3, HPLC fraction at 20–30 min of elution (fraction 3 in C); lane 4, HPLC fraction at 30–40 min of elution (fraction 4 in C). Lane 4 represents the active fraction and was analyzed further by MALDI-MS.
FN1527 Induces hBD2 mRNA in HOECs
E. coli BL21 DE3 was transformed to express recombinant forms of the FN1527, FN1529, and FN1792 proteins, respectively. Cell lysate prepared from E. coli expressing the recombinant proteins was applied to HOEC monolayers. Fig. 3A shows that HOECs challenged with cell lysate from FN1527-expressing E. coli induced the highest level of hBD2 mRNA (>30-fold above base line), followed by FN1792-expressing E. coli (15 fold above base line), with FN1529-expressing E. coli inducing the lowest amount (5-fold above base line). The parent E. coli cell lysate did not induce hBD2 mRNA synthesis to any significant degree.
Expression and Purification of c-Myc FN1527 from E. coli
The c-Myc-tagged FN1527 was expressed in E. coli as described under “Experimental Procedures,” and the recombinant protein was purified by immunoprecipitation using an antibody to c-Myc. The protein was separated by SDS-PAGE followed by either Coomassie staining (Fig. 3B) or Western blot using anti-c-Myc antibody (Fig. 3C). The major protein band on the gel migrating at 12 kDa that reacts with the c-Myc antibody on the Western blot is FN1527. A minor band of higher molecular weight that appears in both Western blot and Coomassie-stained gel may represent the unprocessed preprotein because a signal peptide sequence at the N terminus of FN1527 is predicted by the PROSITE program (available on the World Wide Web).
Recombinant FN1527 Induces hBD2 Transcript and Protein in HOEC
Affinity-purified recombinant FN1527 was applied to HOEC monolayers to assay for induction of hBD2 mRNA and secreted protein. Fig. 3D shows that purified FN1527 induced hBD2 message comparably with that determined for the F. nucleatum cell wall. hBD2 protein expression in the supernatant was also assessed from challenged HOEC monolayers. Fig. 3E shows that FN1527 induced hBD2 peptide ∼13-fold above base line, comparable to induction by F. nucleatum cell wall.
FN1527 Induces hBD2 in HOECs through TLR2
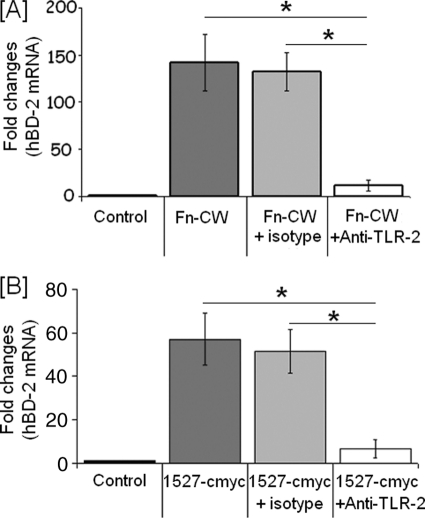
It was previously reported (29, 30) that F. nucleatum induces hBD2 via TLR2 (29). To determine if FN1527 also stimulates hBD2 induction in HOECs through TLR2, we treated the cells with anti-TLR2 antibody for 30 min before the addition of FN1527. As shown in Fig. 4, treatment with anti-TLR2 antibody significantly inhibits hBD2 induction in HOECs both by F. nucleatum cell wall (Fig. 4A) and FN1527 (Fig. 4B).
FIGURE 4.
F. nucleatum cell wall and FAD-I induce hBD2 via TLR2. HOECs were treated with 5 μg/ml anti-TLR2 antibody or isotype control for 30 min and then stimulated with either 10 μg/ml F. nucleatum cell wall (Fn-CW) (A) or 10 μg/ml 1527 c-Myc for 18 h (B). Total RNA extracted from cells of experimental and control cultures were screened by real time PCR for hBD2 mRNA using specific primers. -Fold change in mRNA levels represents mean ± S.D. (error bars) of triplicate experiments. p values were calculated by Student's t test (*, p < 0.05).
Confirmation of FN1527 as FAD-I Using a P. gingivalis Expression System
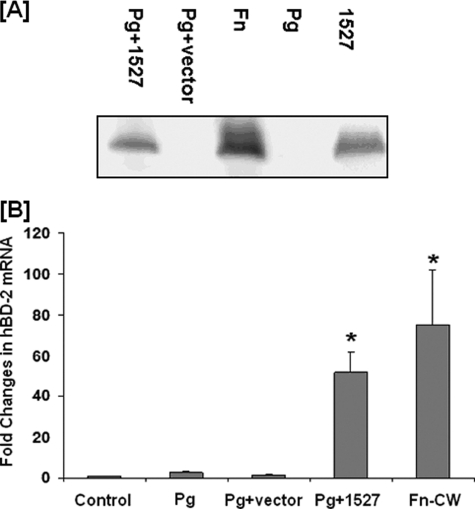
We previously published (17) and continuously observed that, when compared with F. nucleatum, P. gingivalis does not induce hBD2 expression in HOECs. We therefore expressed FN1527 in P. gingivalis 33277 (see “Experimental Procedures” for details) and determined if the transformed strain could induce hBD2 transcript and protein in HOECs. Fig. 5A is representative of numerous Western blots showing that the antibody to FN1527 exposed a 12 kDa band appearing in the lanes run with cell wall fractions from the Pg1527-transformed strain, cell wall fractions from F. nucleatum (Fn), and the recombinant 1527 against which the antibody was made but not in cell walls from wild type P. gingivalis (Pg) or from the P. gingivalis plus vector (Pg+vector) control lanes. Moreover, Pg1527 cell wall fractions induced the hBD2 transcript in HOECs to levels comparable with that seen with F. nucleatum cell wall (Fig. 5B).
FIGURE 5.
Induction of hBD2 mRNA and protein by Pg1527. A, Western blot analysis of cell wall preparations from P. gingivalis transformed with FN1527 (Pg + 1527), P. gingivalis transformed with vector only (Pg + vector), F. nucleatum 25586 cell wall (Fn), or P. gingivalis 33277 cell wall (Pg) using anti-FN1527 antibody (GenScript). A representative image of numerous repeated experiments is shown. B, HOECs were treated with 10 μg/ml P. gingivalis 33277 cell wall, P. gingivalis transformed with vector only, P. gingivalis transformed with FN1527, or F. nucleatum cell wall (Fn-CW) for 18 h. Total RNA extracted from cells of experimental and control cultures were screened by real-time PCR for hBD2 mRNA using specific primers. -Fold change in mRNA levels represents mean ± S.D. (error bars) of triplicate experiments (*, p < 0.05).
Pg1527 Induces hBD2 Peptide Production/Release by HOECs
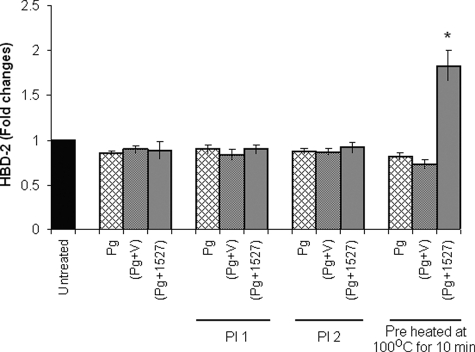
Earlier reports indicated that P. gingivalis-associated proteases can degrade hBD2 produced by HOECs (31). We therefore pretreated respective fractions with a mixture of protease inhibitors (Halt protease inhibitor mixture (Thermo Fisher Scientific) and protease inhibitor mixture (Sigma)) before applying them to HOEC monolayers. Because we did not detect any hBD2 in the cell supernatants (Fig. 6), although Pg1527 induced hBD2 transcript significantly above base line (Fig. 5B), we speculated that P. gingivalis-related proteases were still active. Because we knew that the F. nucleatum cell wall-associated FAD-I was heat-stable (see “Discussion”), we boiled cell wall preparations of Pg, Pg+ vector, and Pg1527, respectively, for 10 min before applying them to HOEC monolayers. Fig. 6 shows that, after boiling, only Pg1527 was able to induce hBD2 peptide release/production by HOECs.
FIGURE 6.
Pg1527 induces release of hBD2 by HOECs. HOECs were challenged with cell wall preparations of P. gingivalis 33277 (Pg), P. gingivalis transformed with vector only (Pg+V), or P. gingivalis transformed with FN1527 (Pg + 1527) and with PI 1-treated (Halt protease inhibitor mixture, Thermo Fisher Scientific), PI 2-treated (protease inhibitor mixture, Sigma), or boiled (10 min) cell wall preparations of P. gingivalis 33277, P. gingivalis transformed with vector only, or P. gingivalis transformed with FN1527. After 18 h, cell supernatants were collected and assayed for hBD2 by ELISA. The results represent means of three independent experiments. -Fold change in peptide levels represents mean ± S.D. (error bars) of triplicate experiments. p values were calculated by Student's t test. *, p < 0.05.
DISCUSSION
Using a combination of biochemical and molecular biological techniques, we were able to identify and isolate an F. nucleatum cell wall-associated peptide that induced hBD2. We identified this agent as FN1527. It is worth mentioning that in addition to inducing hBD2, FN1527 also induces hBD3 in HOECs, albeit to a lesser degree than hBD2 (data not shown). Interestingly, the chemokine IL-8 was not induced by FAD-I in these cells (data not shown), contrary to what we and others have reported for F. nucleatum whole bacteria and its cell wall fraction (17, 29, 30, 32–34).
It has been reported that F. nucleatum induces hBD2 via Toll-like receptor 2 (TLR2) (29, 30), and we have confirmed this finding, both for cell wall of F. nucleatum and recombinant FN1527 (Fig. 4). The fact that recombinant FN1527 promotes innate response activation via TLR2 is not unusual because other pure proteins, such as heat shock proteins and hBD3, have been reported to interact with TLR2 (13, 35–37). Because it is known that certain bacterial lipopeptides interact with TLR2 via either heterodimerization with TLR1 (Pam3-Cys-Ser-Lys4) or TLR6 (macrophage-activating lipopeptide-2 (MALP-2)) (38–41), our ongoing studies are focused on determining if native FN1527 is also a lipopeptide and whether TLR1 or TLR6 is involved together with TLR2 in hBD2 induction by native FN1527 (see further discussion below related to a post-translational modification in FN1527). However, the fact that IL-8 is not induced by recombinant FN1527, whereas hBD2 is, may suggest that the recombinant version of FN1527 (i.e. not expressing a post-translational lipid moiety) may promote the expression of certain innate response elements but not others. Moreover, the role of the canonical versus the noncanonical NF-κB-dependent pathways (42–45) as well as the role of co-receptors in TLR2 activation processes (46–48) may add further complexity. These signaling pathway related issues are currently under active investigation by our group.
Due to the fact that we tested three different sources for FAD-I activity; 1) native components in the active fraction, 2) the E. coli clones expressing FAD-I candidates, and 3) the recombinant FN1527) and that the outcomes were determined using different donor HOECs, with inherent interpersonal variability, we could not establish the specific activity of FAD-I, as would be expected when purifying an active biological agent. Instead, we resorted to expressing the functionality of FN1527 as inducing relative -fold changes of hBD2 transcript and peptide by HOECs above base line.
During our study, we repeatedly noted that the active fraction was a better inducer of hBD2 than the recombinant FN1527. The answer may lie in there being more than one cell wall-associated inducer acting additively or synergistically to induce hBD2, and/or there may be a post-translational modification in FN1527 that is absent in the recombinant generated in E. coli that contributes to enhanced induction of hBD2. Based on our initial observations, the cell wall-associated FAD-I and the active fraction are heat-stable (i.e. retain their ability to induce hBD2 following boiling), whereas the recombinant FN1527 is heat-labile, suggesting a post-translational lipid moiety bound to the protein backbone of FAD-I. This observation is not unique to F. nucleatum because it has been shown that the hBD2-inducing factors from Salmonella enteritidis and E. coli Nissle 1917 are also heat-resistant (49, 50). By searching for prokaryotic membrane lipoprotein lipid attachment site profiles using PROSITE (available on the World Wide Web), we found a potential diacylglycerol moiety bound to cysteine at position 16 in the FN1527 molecule. Additionally, when a model for FN1527 was built using I-TASSER (51–53), cysteine at position 16 was found to be one of the potential binding sites. The fact that Pg1527 cell wall induced hBD2 in HOECs to levels achieved with F. nucleatum cell wall suggests to us that a post-translational modification may have occurred in P. gingivalis (which is lacking in recombinant FN1527) (i.e. mimicking what occurs in the wild type F. nucleatum strain).
What is the function of FN1527 in F. nucleatum, and is it the only source of hBD2 induction by this organism? Clearly, deleting this gene from F. nucleatum could be very informative. The transformation of F. nucleatum is strain-specific, due to the existence of native restriction-modification systems (54). Because we conducted the present studies in F. nucleatum 25586, a strain that to date has proved intractable to molecular manipulation, we will need to identify a potentially useful alternative strain that is readily transformable and useful in systematic approaches to mutagenesis.
Our strategy to decisively demonstrate that FN1527 has the capacity to induce hBD2 in HOECs was to introduce the recombinant DNA encoding FN1527 into P. gingivalis, an organism that we previously showed does not induce hBD2 to any appreciable degree (17). Upon introduction of FN1527 into the genome of P. gingivalis, Western blot analysis confirmed that the transformed strain (Pg1527) expressed FN1527 and that Pg1527 induced hBD2 not only to a much higher level than the original parent strain of P. gingivalis but to levels comparable to those induced by cell wall preparations from F. nucleatum. Although reporter enzymes such as lacZ (55) have been expressed in P. gingivalis, this represents the first time that a functional cell wall protein of a heterologous oral organism has been expressed in the membrane of P. gingivalis.
Interestingly, although we noticed induction of hBD2 transcript by Pg1527, we were unable to detect the production of hBD2 peptide (Fig. 6). This result is not surprising because mass spectrometry has shown that hBD peptides are degraded in the presence of P. gingivalis (31). It has also been shown that P. gingivalis antagonizes F. nucleatum-induced epithelial cell secretion of IL-8 (33, 56–58). Moreover, it has been reported that P. gingivalis induces IL-8 mRNA but not the peptide in HUVEC cells (58). These reports are in agreement with findings demonstrating that by boiling the Pg1527 cell wall, we were able to neutralize the associated proteases and detect hBD2 peptide release/production in HOECs by ELISA (Fig. 6).
Although others have reported hBD2 mRNA induction by P. gingivalis-challenged HOECs (59), this pales in comparison to that elicited by F. nucleatum (i.e. about 8–10-fold for P. gingivalis (60) versus 30- to >300-fold for F. nucleatum), probably due to interpersonal variability in HOECs. In addition, our ELISA data suggest that hBD2 is degraded upon release by P. gingivalis-associated proteases, whereas it is produced significantly above base line by F. nucleatum. Dommisch et al. (60) indicated that the low level induction of hBD2 mRNA is due to P. gingivalis activation of protease-activated receptor 2 (PAR-2). Interestingly, PAR-2 was recently shown to activate stress-activated protein kinase (61), suggesting that severe stress may be related to the low level induction of hBD2 by P. gingivalis. The authors observed extensive HOEC lysis at high P. gingivalis multiplicities of infection, something that we never observed with comparable multiplicities of infection of F. nucleatum. The possibility that HOECs are stressed after P. gingivalis challenge, with appreciably lower total RNA yields when compared with F. nucleatum-challenged cells, and that the mechanism for hBD2 activation by P. gingivalis is NF-κB-dependent (59), although independent with F. nucleatum (62), indicates to us a basic difference between an opportunistic organism that may have evolved to evade innate immune effector activity, such as hBDs (56, 63), and a potentially beneficial oral commensal bacterium that promotes the active expression of these agents.
A notable difference between normal oral and most other normal epithelia is the expression of hBD2. This defensin is induced only in the presence of infection or inflammation in most tissues, including skin (64, 65), trachea (66), and gut epithelium (67, 68). However, it is expressed in normal uninflamed oral tissue (69–72). Our hypothesis is that this base-line level of hBD2 expression is in part due to the exposure of the tissue to specific oral commensal bacteria, such as F. nucleatum expressing FN1527. In contrast, opportunistic bacteria strongly implicated in the etiology of periodontal disease, such as P. gingivalis, display stealthlike qualities when in contact with the epithelium (73, 74), including the lack of induction of β-defensins, as demonstrated herein and reported previously (17).
By better understanding possible inherent strategies of symbiosis between certain commensal bacteria and humans, we may be able to exploit these strategies and apply them as prophylactic agents to prevent unwanted opportunistic biofilms from forming in vulnerable mucosal sites of the body. FAD-I or its derivates may one day offer a new paradigm in immunoregulatory therapeutics by bolstering expression of innate response elements with both antimicrobial and adjuvant capabilities.
Acknowledgments
We thank Dr. J. R. Blakemore, Dr. E. K. Schneider, Dr. W. S. Blood, and Dr. F. Faddoul for providing normal human oral tissue.
This work was supported, in whole or in part, by National Institutes of Health Grants DE018276 (to A. W.) and DE11111 (to R. J. L.).
- hBD
- human β-defensin
- HOEC
- human oral epithelial cell.
REFERENCES
- 1.De Smet K., Contreras R. (2005) Biotechnol. Lett. 27, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. (2002) N. Engl. J. Med. 347, 1199–1200 [DOI] [PubMed] [Google Scholar]
- 3.Ganz T., Lehrer R. I. (1998) Curr. Opin Immunol. 10, 41–44 [DOI] [PubMed] [Google Scholar]
- 4.Ganz T., Oren A., Lehrer R. I. (1992) Med. Microbiol. Immunol. 181, 99–105 [DOI] [PubMed] [Google Scholar]
- 5.Quiñones-Mateu M. E., Lederman M. M., Feng Z., Chakraborty B., Weber J., Rangel H. R., Marotta M. L., Mirza M., Jiang B., Kiser P., Medvik K., Sieg S. F., Weinberg A. (2003) AIDS 17, F39–F48 [DOI] [PubMed] [Google Scholar]
- 6.Feng Z., Jiang B., Chandra J., Ghannoum M., Nelson S., Weinberg A. (2005) J. Dent Res. 84, 445–450 [DOI] [PubMed] [Google Scholar]
- 7.Yadava P., Zhang C., Sun J., Hughes J. A. (2006) Int. J. Antimicrob. Agents 28, 132–137 [DOI] [PubMed] [Google Scholar]
- 8.Selsted M. E., Tang Y. Q., Morris W. L., McGuire P. A., Novotny M. J., Smith W., Henschen A. H., Cullor J. S. (1993) J. Biol. Chem. 268, 6641–6648 [PubMed] [Google Scholar]
- 9.Aliprantis A. O., Yang R. B., Mark M. R., Suggett S., Devaux B., Radolf J. D., Klimpel G. R., Godowski P., Zychlinsky A. (1999) Science 285, 736–739 [DOI] [PubMed] [Google Scholar]
- 10.Biragyn A., Ruffini P. A., Leifer C. A., Klyushnenkova E., Shakhov A., Chertov O., Shirakawa A. K., Farber J. M., Segal D. M., Oppenheim J. J., Kwak L. W. (2002) Science 298, 1025–1029 [DOI] [PubMed] [Google Scholar]
- 11.Biragyn A., Surenhu M., Yang D., Ruffini P. A., Haines B. A., Klyushnenkova E., Oppenheim J. J., Kwak L. W. (2001) J. Immunol. 167, 6644–6653 [DOI] [PubMed] [Google Scholar]
- 12.Biragyn A., Belyakov I. M., Chow Y. H., Dimitrov D. S., Berzofsky J. A., Kwak L. W. (2002) Blood 100, 1153–1159 [DOI] [PubMed] [Google Scholar]
- 13.Funderburg N., Lederman M. M., Feng Z., Drage M. G., Jadlowsky J., Harding C. V., Weinberg A., Sieg S. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18631–18635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Z., Dubyak G. R., Lederman M. M., Weinberg A. (2006) J. Immunol. 177, 782–786 [DOI] [PubMed] [Google Scholar]
- 15.Jin G., Kawsar H. I., Hirsch S. A., Zeng C., Jia X., Feng Z., Ghosh S. K., Zheng Q. Y., Zhou A., McIntyre T. M., Weinberg A. (2010) PLoS One 5, e10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Röhrl J., Yang D., Oppenheim J. J., Hehlgans T. (2010) J. Immunol. 184, 6688–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krisanaprakornkit S., Kimball J. R., Weinberg A., Darveau R. P., Bainbridge B. W., Dale B. A. (2000) Infect. Immun. 68, 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapatral V., Anderson I., Ivanova N., Reznik G., Los T., Lykidis A., Bhattacharyya A., Bartman A., Gardner W., Grechkin G., Zhu L., Vasieva O., Chu L., Kogan Y., Chaga O., Goltsman E., Bernal A., Larsen N., D'Souza M., Walunas T., Pusch G., Haselkorn R., Fonstein M., Kyrpides N., Overbeek R. (2002) J. Bacteriol. 184, 2005–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oda D., Watson E. (1990) In Vitro Cell Dev. Biol. 26, 589–595 [DOI] [PubMed] [Google Scholar]
- 20.Kennell W., Holt S. C. (1990) Oral Microbiol. Immunol. 5, 121–130 [DOI] [PubMed] [Google Scholar]
- 21.Park Y., Xie H., Lamont R. J. (2007) FEMS Microbiol. Lett. 273, 103–108 [DOI] [PubMed] [Google Scholar]
- 22.Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., Tanaka Y. (2002) Nucleic Acids Res. 30, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner R. G., Russell J. B., Wilson D. B., Wang G. R., Shoemaker N. B. (1996) Appl. Environ. Microbiol. 62, 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith C. J. (1995) Methods Mol. Biol. 47, 161–169 [DOI] [PubMed] [Google Scholar]
- 25.Krisanaprakornkit S., Weinberg A., Perez C. N., Dale B. A. (1998) Infect. Immun. 66, 4222–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawsar H. I., Ghosh S. K., Hirsch S. A., Koon H. B., Weinberg A., Jin G. (2010) Peptides 31, 195–201 [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S. K., Gerken T. A., Schneider K. M., Feng Z., McCormick T. S., Weinberg A. (2007) Clin. Chem. 53, 757–765 [DOI] [PubMed] [Google Scholar]
- 28.Han Y. W., Ikegami A., Rajanna C., Kawsar H. I., Zhou Y., Li M., Sojar H. T., Genco R. J., Kuramitsu H. K., Deng C. X. (2005) J. Bacteriol. 187, 5330–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peyret-Lacombe A., Brunel G., Watts M., Charveron M., Duplan H. (2009) Cytokine 46, 201–210 [DOI] [PubMed] [Google Scholar]
- 30.Ji S., Shin J. E., Kim Y. S., Oh J. E., Min B. M., Choi Y. (2009) Infect. Immun. 77, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlisle M. D., Srikantha R. N., Brogden K. A. (2009) J. Innate Immun. 1, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y. W., Shi W., Huang G. T., Kinder Haake S., Park N. H., Kuramitsu H., Genco R. J. (2000) Infect. Immun. 68, 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang G. T., Kim D., Lee J. K., Kuramitsu H. K., Haake S. K. (2001) Infect. Immun. 69, 1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gursoy U. K., Könönen E., Uitto V. J. (2008) Oral Microbiol. Immunol. 23, 432–434 [DOI] [PubMed] [Google Scholar]
- 35.Gong J., Zhu B., Murshid A., Adachi H., Song B., Lee A., Liu C., Calderwood S. K. (2009) J. Immunol. 183, 3092–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Q. Q., Sobkoviak R., Jockheck-Clark A. R., Shi B., Mandelin A. M., 2nd, Tak P. P., Haines G. K., 3rd, Nicchitta C. V., Pope R. M. (2009) J. Immunol. 182, 4965–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanin-Zhorov A., Nussbaum G., Franitza S., Cohen I. R., Lider O. (2003) FASEB J. 17, 1567–1569 [DOI] [PubMed] [Google Scholar]
- 38.Hajjar A. M., O'Mahony D. S., Ozinsky A., Underhill D. M., Aderem A., Klebanoff S. J., Wilson C. B. (2001) J. Immunol. 166, 15–19 [DOI] [PubMed] [Google Scholar]
- 39.Omueti K. O., Beyer J. M., Johnson C. M., Lyle E. A., Tapping R. I. (2005) J. Biol. Chem. 280, 36616–36625 [DOI] [PubMed] [Google Scholar]
- 40.Sandor F., Latz E., Re F., Mandell L., Repik G., Golenbock D. T., Espevik T., Kurt-Jones E. A., Finberg R. W. (2003) J. Cell Biol. 162, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., Aderem A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayden M. S., West A. P., Ghosh S. (2006) Oncogene 25, 6758–6780 [DOI] [PubMed] [Google Scholar]
- 43.Hiscott J., Marois J., Garoufalis J., D'Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., Bensi G. (1993) Mol. Cell. Biol. 13, 6231–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarnicki A. G., Conroy H., Brereton C., Donnelly G., Toomey D., Walsh K., Sweeney C., Leavy O., Fletcher J., Lavelle E. C., Dunne P., Mills K. H. (2008) J. Immunol. 180, 3797–3806 [DOI] [PubMed] [Google Scholar]
- 45.Caamaño J., Tato C., Cai G., Villegas E. N., Speirs K., Craig L., Alexander J., Hunter C. A. (2000) J. Immunol. 165, 5720–5728 [DOI] [PubMed] [Google Scholar]
- 46.Drage M. G., Pecora N. D., Hise A. G., Febbraio M., Silverstein R. L., Golenbock D. T., Boom W. H., Harding C. V. (2009) Cell. Immunol. 258, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang S., Wang M., Tapping R. I., Stepensky V., Nawar H. F., Triantafilou M., Triantafilou K., Connell T. D., Hajishengallis G. (2007) J. Biol. Chem. 282, 7532–7542 [DOI] [PubMed] [Google Scholar]
- 48.Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., Silverstein R. L. (2006) Cell Metab. 4, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogushi K., Wada A., Niidome T., Mori N., Oishi K., Nagatake T., Takahashi A., Asakura H., Makino S., Hojo H., Nakahara Y., Ohsaki M., Hatakeyama T., Aoyagi H., Kurazono H., Moss J., Hirayama T. (2001) J. Biol. Chem. 276, 30521–30526 [DOI] [PubMed] [Google Scholar]
- 50.Takahashi A., Wada A., Ogushi K., Maeda K., Kawahara T., Mawatari K., Kurazono H., Moss J., Hirayama T., Nakaya Y. (2001) FEBS Lett. 508, 484–488 [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y. (2007) Proteins 69, Suppl. 8, 108–117 [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y. (2008) BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S., Skolnick J., Zhang Y. (2007) BMC Biol. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haake S. K., Yoder S. C., Attarian G., Podkaminer K. (2000) J. Bacteriol. 182, 1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie H., Chung W. O., Park Y., Lamont R. J. (2000) Infect. Immun. 68, 6574–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasegawa Y., Tribble G. D., Baker H. V., Mans J. J., Handfield M., Lamont R. J. (2008) Infect. Immun. 76, 2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madianos P. N., Papapanou P. N., Sandros J. (1997) Infect. Immun. 65, 3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi-Sakamoto M., Isogai E., Hirose K. (2003) Curr. Microbiol. 46, 109–114 [DOI] [PubMed] [Google Scholar]
- 59.Chung W. O., Hansen S. R., Rao D., Dale B. A. (2004) J. Immunol. 173, 5165–5170 [DOI] [PubMed] [Google Scholar]
- 60.Dommisch H., Chung W. O., Rohani M. G., Williams D., Rangarajan M., Curtis M. A., Dale B. A. (2007) Infect. Immun. 75, 4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanke T., Macfarlane S. R., Seatter M. J., Davenport E., Paul A., McKenzie R. C., Plevin R. (2001) J. Biol. Chem. 276, 31657–31666 [DOI] [PubMed] [Google Scholar]
- 62.Krisanaprakornkit S., Kimball J. R., Dale B. A. (2002) J. Immunol. 168, 316–324 [DOI] [PubMed] [Google Scholar]
- 63.Lu Q., Darveau R. P., Samaranayake L. P., Wang C. Y., Jin L. (2009) Innate Immun. 15, 325–335 [DOI] [PubMed] [Google Scholar]
- 64.Ong P. Y., Ohtake T., Brandt C., Strickland I., Boguniewicz M., Ganz T., Gallo R. L., Leung D. Y. (2002) N. Engl. J. Med. 347, 1151–1160 [DOI] [PubMed] [Google Scholar]
- 65.Liu L., Wang L., Jia H. P., Zhao C., Heng H. H., Schutte B. C., McCray P. B., Jr., Ganz T. (1998) Gene 222, 237–244 [DOI] [PubMed] [Google Scholar]
- 66.Starner T. D., Agerberth B., Gudmundsson G. H., McCray P. B., Jr. (2005) J. Immunol. 174, 1608–1615 [DOI] [PubMed] [Google Scholar]
- 67.O'Neil D. A., Porter E. M., Elewaut D., Anderson G. M., Eckmann L., Ganz T., Kagnoff M. F. (1999) J. Immunol. 163, 6718–6724 [PubMed] [Google Scholar]
- 68.Wehkamp J., Fellermann K., Herrlinger K. R., Baxmann S., Schmidt K., Schwind B., Duchrow M., Wohlschläger C., Feller A. C., Stange E. F. (2002) Eur. J. Gastroenterol. Hepatol. 14, 745–752 [DOI] [PubMed] [Google Scholar]
- 69.Dale B. A., Kimball J. R., Krisanaprakornkit S., Roberts F., Robinovitch M., O'Neal R., Valore E. V., Ganz T., Anderson G. M., Weinberg A. (2001) J. Periodontal Res. 36, 285–294 [DOI] [PubMed] [Google Scholar]
- 70.Jurevic R. J., Bai M., Chadwick R. B., White T. C., Dale B. A. (2003) J. Clin. Microbiol. 41, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pazgier M., Hoover D. M., Yang D., Lu W., Lubkowski J. (2006) Cell Mol. Life Sci. 63, 1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang D., Liu Z. H., Tewary P., Chen Q., de la Rosa G., Oppenheim J. J. (2007) Curr. Pharm. Des. 13, 3131–3139 [DOI] [PubMed] [Google Scholar]
- 73.Darveau R. P., Belton C. M., Reife R. A., Lamont R. J. (1998) Infect. Immun. 66, 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamont R. J., Jenkinson H. F. (1998) Microbiol. Mol. Biol. Rev. 62, 1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]