Abstract
Hedgehog (Hh) signaling, via the key signal transducer Smoothened (SMO) and Gli transcription factors, is essential for embryonic development and carcinogenesis. At present, the molecular mechanism of Hh signaling-mediated carcinogenesis is not completely understood. Using a mouse model (K14cre/R26SmoM2) of SMO-mediated basal cell carcinoma development, we identified TGFβ2 as a major Hh-regulated gene. TGFβ2 expression was high in the keratinocytes, with activated TGFβ signaling (indicated by elevated expression of phosphorylated SMAD2/3) detected in both tumor and stroma. The significance of TGFβ signaling for SMO function was demonstrated in two assays. Down-regulation of TGFβ2 expression prevented Hh signaling-dependent osteoblast differentiation and motor neuron differentiation. Furthermore, inhibition of TGFβ signaling by TGFβ receptor I inhibitor SD208 significantly reduced tumor area in K14cre/R26SmoM2 mice. Tumor shrinkage in mice was associated with an increased number of lymphocytes, suggesting an immune suppression role of TGFβ signaling. The relevance of our results to human cancer is reflected by the fact that human basal cell carcinomas, which almost always harbor activated Hh signaling, have activated TGFβ signaling, as indicated by high levels of phosphorylated SMAD2 and SMAD3 in tumor and stroma. Together, our data indicate that TGFβ signaling is critical for Hh signaling-mediated carcinogenesis.
Keywords: Cancer Therapy, Signal Transduction, Skin, Transforming Growth Factor Beta (TGFbeta), Tumor, K14cre, Basal Cell Carcinoma, Hedgehog, Motor Neuron Differentiation, Smoothened
Introduction
The Hedgehog pathway plays an important role in cell differentiation, tissue polarity, cell proliferation, and carcinogenesis (1–4). The seven-transmembrane domain-containing protein Smoothened (SMO)4 serves as the key player for signal transduction of this pathway, whose function is inhibited by another transmembrane protein, Patched (PTC), in the absence of Hh ligands. Binding of Hh to its receptor PTC releases this inhibition, allowing SMO to signal downstream, leading to formation of active forms of Gli transcription factors. As transcription factors, Gli molecules can regulate target gene expression by direct association with a specific consensus sequence located at the promoter region of the target genes (5). In addition to the canonical pathways (ligand overexpression, altered expression of Hh signaling molecules, or gene mutations), recent studies indicate that Hh signaling can also be activated by other signaling pathways, such as K-Ras. Both canonical and non-canonical Hh signaling activation are found in many types of human cancer, including brain tumors, gastrointestinal, prostate, lung, and breast cancers (6–8).
Mounting evidence indicates that Hh signaling activation occurs frequently in a number of human cancers (9), but the underlying molecular basis remains largely elusive. To understand the molecular basis by which Hh signaling regulates carcinogenesis, we analyzed gene expression of a mouse model of basal cell carcinoma in which an activated form of SMO (SmoM2) replaces the wild type SMO allele and is expressed under the control of the keratin 14 promoter. Our results indicated that several signaling pathways associated with carcinogenesis are altered in this mouse model. In particular, TGFβ2 was significantly induced by SmoM2 expression. Elevated expression of phosphorylated SMAD2 and SMAD3, markers for TGFβ signaling activation, was seen in SmoM2-mediated tumors and in the stroma. TGFβ signaling is a known pathway important for tumor development. Depending on the stage of tumor development and cellular context, TGFβ signaling can function as tumor suppressor, tumor promoter, or modulator for immune surveillance and inflammation (10, 11). We investigated the significance of TGFβ signaling for SMO-mediated signaling. First, we examined the significance of TGFβ2 knockdown in SMO-mediated signaling in C3H10T1/2 cells and embryonic stem cells. Second, we tested the effect of inhibiting TGFβ receptor I function via topical application of small molecule kinase inhibitor SD208 in the K14cre/SmoM2 mouse model for BCCs.
MATERIALS AND METHODS
Mice
K14cre (Ref. 12, obtained from the Emice program) and R26-SmoM2YFP mice (Ref. 13), purchased from The Jackson Laboratory, Bar Harbor, ME) were maintained and mated under pathogen-free husbandry conditions. The offspring from the mating were screened using PCR to determine their transgenic status according to the instructions from the vendors. All animal studies were approved by the Institutional Animal Care and Use Committee at Indiana University.
Small Molecule Inhibitors and Animal Treatment Protocol
TGFβ receptor I-SD208 (14) was purchased from Sigma, dissolved in DMSO, and then diluted in 70% ethanol. Cyclopamine tartrate salt (hydroxy carboxylic acid) (15), kindly provided by Dr. Massoud Garrossian from Logan Natural Products Inc., was used as a control for Hh signaling inhibition and was dissolved in 70% ethanol. The final concentration of all the chemicals was 5 μm for in vivo study. Chemicals were applied topically on the skin surface in the upper and lower areas of abdomen, respectively. 70% ethanol was embrocated in the middle belly as a control. Topical application of chemicals was performed daily for 4 weeks. At the end of the study, skin biopsies were collected for hematoxylin and eosin (H&E) staining and gene expression analysis as described previously (16).
Histology and Microscopic BCC Analysis
Skin tissues were collected after each experiment. Half of the tissue was frozen on dry ice and immediately stored at −80 °C. The other half was fixed in formalin overnight, paraffin-embedded, sectioned at 5 μm, and stained with H&E. Eight epidermal areas were randomly chosen in each section (16). The viable tumor areas in selected tissue areas were quantified by manually demarcating the tumor boundary on an electronic image of an H&E-stained section. The proportion of tumor area to the total tissue area was quantified using MetaMorph imaging software (version 6.2r6, Downingtown, PA) or ImageJ.
Epidermis Separation
Newborn mouse skins were used to separate epidermis by emerging the skin in dispase solution (5 mg/ml) overnight at 4 °C. Epidermis of the skin was removed from dermis by forceps and used for total RNA extraction using the Ambion RNA extract kit. Expression of Hh target genes was examined by PCR as reported previously (17).
GeneChip Analysis
Purified RNAs from epidermis were used for labeling and hybridization to the Affymetrix GeneChip Mouse4302 according to the manufacturer's procedures. S-Plus Array Analyzer 2.1, a statistical program (TIBCO Software Inc., Palo Alto, CA), was used to analyze the microarray data. Probe level data analysis was performed on Affymetrix files using the robust multichip analysis method. Further, the probe sets absent across all the chips were filtered out before the differential expression testing. Differential expression testing was performed using Student's t test comparing epidermal gene expression between K14cre+/SmoM2+ and K14cre+/SmoM2− mice. The genes were filtered as significant from Student's t test at p value < = 0.05. In addition, the local pool error (LPE) (18) test was also done, taking into consideration the low number of replicates per experimental group. The LPE estimation pools error within genes and between arrays for genes with similar expression intensities, thus reducing the increased type I and II errors due to the low replicates. The hierarchical clustering on the significant genes from the Student's t test and LPE test with heat map was done using Spotfire (TIBCO Software Inc.). Further, IPA 8.6 (Ingenuity Systems, Redwood City, CA) was used for functional and network analysis to identify the pathways, biological functions that are involved due to the differential expression between the K14cre+/SmoM2+ and K14cre+/SmoM2− mice (17).
RT-PCR and Real-time PCR
Total RNA was isolated from the tissues using TRIzol reagent (Sigma) according to the manufacturer's instructions. One μg of total RNA was reverse-transcribed into cDNAs using the first-strand synthesis kit (Roche Applied Science). We performed RT-PCR of Hedgehog target genes with 32 cycles of 96 °C for 30 s, 57 °C for 45 s, and 72 °C for 45 s with the following primers: Hip, forward, 5′-CCTGTCGAGGCTACTTTTCG-3′, and reverse, 5′-GGGCAGGTTGAACTGTGACT-3′; Ptch1, forward, 5′-CTCAGGCAATACGAAGCACA-3′, and reverse, 5′-GACAAGGAGCCAGAGTCCAG-3′; Gli1, forward, 5′-GGTCCACCAACCAACTATGG-3′, and reverse, 5′-TGGCAGGGCTCTGACTAACT-3′. β-Actin was used as an internal control. PCR products were separated on a 1.5% agarose gel and visualized after ethidium bromide staining. Real-time PCR was performed as previously described (17).
Cell Culture and Lentivirus-mediated shRNA Expression
C3H10T1/2 cells, purchased from ATCC, were cultured according to ATCC instruction. Mouse embryonic stem (ES) cell line E14Tg2a derived from 129/O1a mice was purchased from the Mutant Mouse Regional Resource Centers (MMRRC) and cultured according to a procedure from the vendor (MMRRC Thawing and Culture Protocol for E14TG2a.4 feeder-independent ES cell clones). To obtain keratinocytes from mice, epidermis was first separated from the dermis as described earlier, digested in trypsin solution for 30 min at 37 °C, and passed through a cell strainer (BD Biosciences). Keratinocytes were cultured in keratinocyte-serum free medium (Invitrogen).
Lentivirus-mediated shRNA expression was used to silence gene expression of TGFβ2 or SMO according to a previously reported procedure (19). All shRNAs were purchased from Sigma. In brief, five clones of shRNAs specific for each target gene were used to generate lentiviruses. An shRNA clone to luciferase was used as a negative control. Viruses were purified by ultracentrifuge and used to infect either C3H10T1/2 or E14Tg2a cells. GFP-expressing lentiviruses were used to detect virus infection efficiency. Infected cells were selected with puromycin at 500–1000 μg/ml for 5 days. Gene silencing efficiency was assessed by RT-PCR of the target genes. In our experiments, three shRNA clones each for SMO and TGFβ2 were shown to effectively down-regulate target gene expression by 70%.
Osteoblast Cell Differentiation
Confluent C3H10T1/2 cells were treated with SMO agonist purmorphamine at 2.5 μm for 7 days. Expression of osteoblast cell marker alkaline phosphatase was achieved by incubating cells with alkaline phosphatase substrate, Fast Red TR/Naphthol AS-MX (Sigma), at 1 mg/ml for 30 min at 37 °C with continual agitation. The alkaline phosphatase activity was also measured by spectrophotometry using the Sensolyte p-nitrophenyl phosphate alkaline phosphatase assay kit (AnaSpec, San Jose, CA) following the vendor's instructions. The activity from each sample was determined and normalized to total protein content derived from the BCA protein assay (Pierce). The increase in absorbance at 570 nm was monitored every 5 min over 80 min, and the ALPase activity (nmol of p-nitrophenol produced/min) was calculated from a linear range. The specific activity of ALPase was given as nmol of p-nitrophenol/min/mg of protein.
Motor Neuron Differentiation from Mouse Embryonic Stem Cells
Motor neuron differentiation was performed according to Wichterle et al. (20). In short, embryonic bodies were formed from ES cells in ES cell medium without leukemia inhibitory factor in an ultra-low adhesive plate with 5 × 105 cells/ml for 2 days. Motor neurons were induced by the addition of retinoid acids (100 nm) and purmorphamine (2.5 μm) for 5–7 days (20, 21).
Statistical Analyses
Statistical analyses were performed using the Student's t test to compare the results, with p values of <0.05 indicating statistically significant differences.
RESULTS
Identification of TGFβ2 as an Hh Signaling-induced Gene
R26-SmoM2YFP mice (13) were crossed with K14cre mice (12) to generate mice expressing SmoM2YFP under the control of the keratin 14 promoter. Shortly after birth, the K14cre/R26SmoM2YFP mice exhibited hyperproliferation of basal cells in the epidermis on day 3, which became more severe on day 15 (Fig. 1A). The earliest visual phenotype in this model is the appearance of wrinkled ears due to tumor formation. These mice started to lose fur from week 3. By week 8, no fur was visible (Fig. 1A). Histology analysis revealed that BCC-like tumors appear in ear, penis, and skin all over the body. Most K14cre/R26-SmoM2YFP mice had reduced body weight (by 20–40%) in comparison with the wild type littermates. After dissection of the mice, no obvious tumor formation was seen in other organs examined. These phenotypes appeared in mice with B6, 129, CD1, or mixed genetic background, suggesting that genetic background does not affect the phenotype. These results indicate that keratin 14 promoter-driven expression of activated SMO, SmoM2, is sufficient to cause tumor formation in the skin.
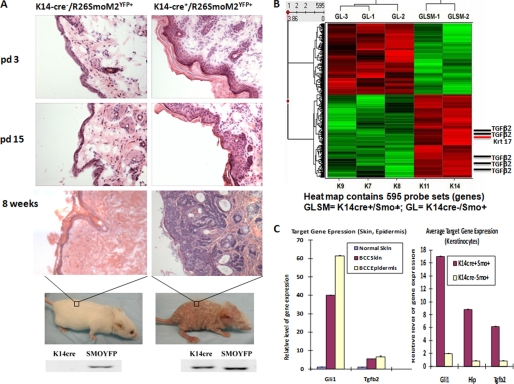
FIGURE 1.
Identification of TGFβ2 as an Hh signaling-regulated gene in mouse model of BCCs. A, using K14 promoter-driven expression of Cre recombinase, we established skin-specific expression of an active form of SMO, SmoM2, in mice. In this model, skin lesions were observed in newly born litters (day 3 (pd 3)). The phenotype gets worse over time. By week 8, BCC-like tumors appear all over the body, and little fur was seen in mice. Most mice were dead by 12 weeks. Genotyping results were shown at the bottom. pd 15, day 15. B, heat map with hierarchical clustering to show differentially expressed genes. TGFβ2 was shown to be an Hh signaling-regulated gene. Mouse epidermis was removed from mice with K14 promoter-driven expression of SmoM2, and the gene expression profile was compared with those from Cre negative mice following hybridization onto Affymetrix GeneChip microarrays. All five TGFβ2 probes from the chip showed elevated expression in epidermis of K14cre/R26SmoM2. C, analyses of gene expression in SmoM2-expressing skin, purified epidermis, or keratinocytes indicate that TGFβ2 was detected in keratinocytes, epidermis, as well as skin tissues.
To examine the molecular basis of Hedgehog signaling-mediated carcinogenesis in this mouse model, we isolated epidermis from different genotypes and performed GeneChip analysis. We found that many known Hedgehog target genes were up-regulated in K14cre/R26SmoM2YFP epidermis (in comparison with K14cre negative R26SmoM2YFP epidermis), such as krt 17, human homologue of hedgehog-interacting protein (Hip), and Ptch1 (supplemental Table S1 and Fig. 1C). In addition, we identified changes in a number of pathways involved in tumor microenvironment such as TGFβ, PDGF, IGF-1, and endothelin-1 from the LPE test (supplemental Fig. S1). As shown in Fig. 1B, TGFβ2 was one of the major genes highly expressed in SmoM2YFP-expressing epidermis. We further isolated keratinocytes from mouse litters with or without SmoM2 expression and detected gene expression of TGFβ2 and other known Hh target genes. As shown in Fig. 1C, SmoM2-expressing keratinocytes expressed a high level of TGFβ2 (over 5×) than those without SmoM2 expression. Consistent with this finding, we detected elevated levels of phosphorylated SMAD2 and SMAD3, markers for TGFβ signaling activation, in SmoM2YFP-mediated tumors and stroma (supplemental Fig. S2). The relevance of this finding to human cancer was our finding that human BCCs exhibit high levels of phosphorylated SMAD2 and SMAD3 in tumors as well as tumor-associated stroma (supplemental Fig. S3). Immunohistochemistry of SMAD molecules in both human BCCs and mouse tumors suggests that Hh signaling-induced TGFβ signaling activation occurs both in the tumor and in the stroma. Based on our data, we suggest that keratinocytes with activated Hh signaling (BCC cells) produce TGFβ2, which in turn acts via autocrine and paracrine signaling to activate the TGFβ pathway. We predict that TGFβ signaling mediates some functions of Hh signaling during development of BCCs.
The Role of TGFβ2 for Hh Signaling
To demonstrate the role of TGFβ2 for Hh signaling, we tested the effects of TGFβ2 depletion on Hh signaling-mediated motor neuron differentiation and osteoblast differentiation. The reason for choosing these two systems is the fact that we observed induced TGFβ signaling (as shown by elevated SMAD2 phosphorylation) (22) in the presence of SMO agonist purmorphamine (Fig. 2, A and B). Following Sonic Hh stimulation or the addition of purmorphamine, C3H101/2 cells can differentiate into osteoblast cell as distinguished by elevated expression of alkaline phosphatase (ALP) (Fig. 2C). Because we noticed that cells stimulated with Shh or purmorphamine had high levels of Hh target gene expression as well as a high level of phosphorylated SMAD2, we tested the effect of TGFβ2 down-regulation on Hh signaling-mediated osteoblast differentiation and found that knocking down the expression of TGFβ2 reduced the level of ALP. As shown in Fig. 2C and supplemental Fig. S4, cells with down-regulation of TGFβ2 had weak or non-detectable ALP activity, whereas the control cells (a non-functional shRNA or the parental cells) had a high level of ALP activity. This result indicates that Hh signaling-mediated osteoblast differentiation requires TGFβ2 signaling.
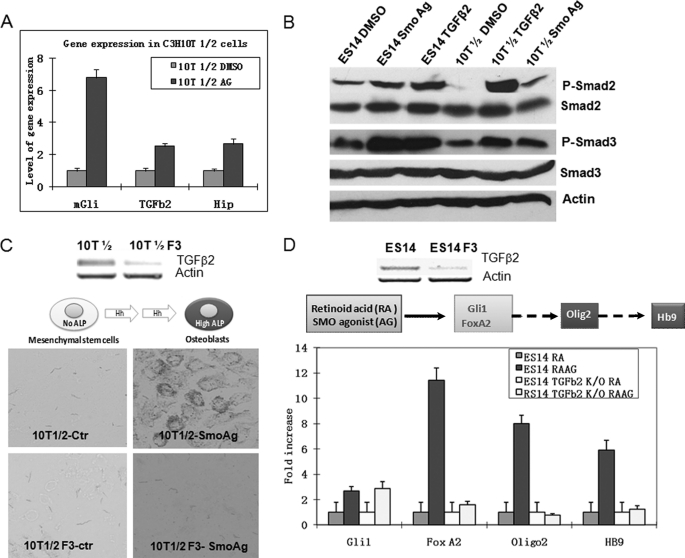
FIGURE 2.
Functional requirement of TGFβ2 for Hh signaling-mediated biological processes. A, real-time PCR analyses of gene expression of Gli1, Hip, and TGFβ2 in C3H10T1/2 cells following treatment with SMO agonist purmorphamine (indicated as Ag in the figure) for 24 h. Cells treated with purmorphamine were compared with those treated with DMSO for gene expression of mGli1, Hip, and TGFβ2 by real-time PCR. Error bars indicate S.D. B, detection of SMAD2 and SMAD3 phosphorylation in cells treated with purmorphamine. Mouse ES cells or C3H10T1/2 cells were incubated with purmorphamine or DMSO for 24 h, and levels of SMAD2/3 phosphorylation (P-Smad2/P-Smad3) were analyzed by Western blotting. The level of β-actin and the total proteins of SMAD2/3 were used as the controls. C, the requirement of TGFβ2 for Hh signaling-mediated osteoblast cell differentiation. Expression of TGFβ2 was down-regulated by shRNA-mediated gene silencing. Of the five shRNA clones, we found that three (F3, F5, and F7) were effective in silencing TGFβ2 expression, whereas two were not. We compared cells with down-regulation of TGFβ2 with either parental cells or cells with ineffective shRNAs for their abilities to express ALP. This figure shows TGFβ2 expression after F3 shRNA knockdown in C3H10T1/2 cells as compared with an ineffective clone (top) and their levels of alkaline phosphatase (bottom). F3 shRNA-derived cells were unable to express ALP, whereas the control (Ctr) cells had a high level. F5- and F7-derived cells were similar to F3-derived cells (data not shown). The level of ALP was also measured by optical density following incubation with the substrates for different time points (supplemental Fig. S4). D, the effect of TGFβ2 in Hh signaling-mediated motor neuron differentiation. Mouse ES cells were induced to differentiate into motor neurons after incubation with retinoic acid and purmorphamine. We examined TGFβ2 down-regulated cells with either parental cells or the control shRNA-derived cells for expression of motor neuron differentiation markers (Hb9, Olig2, and FoxA2) after induction. Cells with low expression of TGFβ2 (top) continued to express Gli1 in response to purmorphamine but failed to express Hb9, Olig2, and FoxA2 (bottom). All three shRNA-derived ES14 cells had similar results (not shown).
To substantiate our data in C3H10T1/2 cells, we examined the effects of TGFβ2 knockdown on Hh signaling-mediated motor neuron differentiation, a system known to be Hedgehog signaling-dependent (20). We showed that differentiation of motor neurons is associated with elevated levels of phosphorylated SMAD2 and SMAD3 (Fig. 2B). Using similar approaches to the one in C3H10T1/2 cells, we knocked down TGFβ2 expression by shRNAs and examined their capacities to differentiate into motor neurons according to previously established protocols (20, 21). We found that reduced expression of TGFβ2 prevented purmorphamine-induced motor neuron differentiation, as indicated by lack of HB9 expression and reduced expression of other markers (Fig. 2D). To determine whether TGFβ is sufficient to drive expression of some motor neuron markers in the absence of Hedgehog signaling, we knocked down SMO expression in ES cells and examined the effects of TGFβ2 on motor neuron differentiation. No Hb9 expression was induced by TGFβ2 plus retinoid acid in SMO knockdown cells (supplemental Fig. S5), suggesting that TGFβ signaling alone does not mediate all the functions of Hh signaling and thus is not sufficient to drive motor neuron differentiation. Based on our results, we conclude that TGFβ2 is required but may not be sufficient for Hh signaling-mediated osteoblast and motor neuron differentiation.
Effects of TGFβ Signaling Inhibitor SD208 on Tumor in K14cre/R26SmoM2YFP Mice
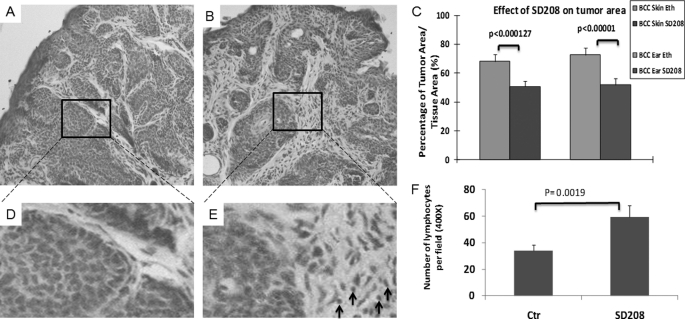
Because we already know that epidermal activation of Hh signaling is associated with elevated levels of phosphorylated SMAD2 and SMAD3 (supplemental Fig. S2), we tested the effect of SD208, a specific kinase inhibitor of TGFβRI, on SmoM2-mediated carcinogenesis. K14cre/R26SmoM2YFP mice were treated with SD208 by topical application daily for 4 weeks, and the tumor area in the skin of the SD208-treated group was compared with that of the control group. Following topical application of SD208, we did not see changes in animal behavior, body weight, or lifespan, suggesting that topical application of the compound is not very toxic. More importantly, we found that SD208 caused significant reduction of tumor area in the tissue in all mice (n = 6) in comparison with the control group. The reduction of tumor area in mice was about 20% (Fig. 3, A–C). Further analyses indicate that SD208 had no effects on Ki67 positivity (supplemental Fig. S6), the number of blood vessels (angiogenesis) or abnormal nuclear morphology resembling apoptosis in the skin tissues (not shown). Instead, SD208 treatment significantly increased the number of lymphocytes in the skin tissue, a phenotype often associated with reduced immune suppression (Fig. 3, D–F). It is known that TGFβ signaling can reduce the number of tumor-specific cytotoxic T and regulatory T cells (which are capable of eradicating tumor cells) but increase the number of myeloid-derived suppressor cells (which will exert systemic immune suppression leading to tumor tolerance), resulting in accelerated tumor development (23). We found that mice treated with SD208 exhibit decreased levels of phosphorylated SMAD2 and SMAD3, markers for TGFβ signaling activation (supplemental Fig. S7), indicating a specific effect of SD208. These results indicate that specific inhibition of TGFβ signaling results in tumor shrinkage derived from SmoM2, at least through regulation of immune surveillance.
FIGURE 3.
The effect of inhibiting TGFβ signaling by SD208 on BCC development of K14cre/R26 SmoM2 mice. Eight-week-old K14cre/R26-SmoM2 mice developed BCCs and were treated with TGFβRI inhibitor SD208 by topical application of the mouse skin for 4 weeks. Solvent 70% ethanol (Eth) was used as the control (Ctr). The effectiveness of SD208 was examined by comparing the tumor area of the two groups following H&E staining of the skin tissues (A and B) and ImageJ analyses of eight independent tissue areas (C). Although the skin in the control group contained 60–70% of tumor area in the tissue, SD208-treated skin had only 50% of tumor area in the tissue. SD208-induced change in lymphocytes (indicated by arrows) was detected after H&E staining (D and E). The number of lymphocytes per field (400×, ∼23.06 mm2) was calculated, and the average value from five mice was shown (F). Error bars in C and F indicate S.D.
DISCUSSION
Understanding the molecular basis by which Hh signaling regulates tumor development is critical for designing strategies for effective cancer therapeutics (24). Using a mouse model for SMO-mediated BCC development, we identified changes in several signaling pathways, particularly TGFβ signaling. TGFβ signaling is a known pathway important for tumor development (10, 11). Using two biological assays as well as one BCC mouse model, we demonstrated the critical role of TGFβ signaling in SMO-mediated signaling and cancer development. We also examined the relevance of our studies to human BCCs by examining levels of phosphorylated SMAD2 and SMAD3 (supplemental Fig. S3), which are markers for activated TGFβ signaling. Expression of phosphorylated SMAD2 and SMAD3 in tumor as well as stroma suggests that TGFβ signaling in BCCs functions via both autocrine and paracrine manners. Our data from the mouse model suggest that TGFβ molecules, which are produced in the tumor cells, affect immune surveillance system through paracrine signaling. Whether this finding can be applied to other types of cancer will require further examination. Supplemental Fig. S8 shows our proposed model of TGFβ functions in BCC development. Currently, it is unclear how the autocrine signaling of TGFβ affects BCC cells. Previous publications indicate that TGFβ signaling can regulate epithelial-mesenchymal transition, cell differentiation of cancer cells, and expression of matrix metalloproteinase molecules; the latter are known to regulate tumor invasiveness (28). Nevertheless, our data provide evidence to support that TGFβ signaling is required for Hh signaling-mediated BCC development.
Despite the critical role of TGFβ signaling in Hh-mediated biological processes, including carcinogenesis, our data in motor neuron differentiation showed that the addition of TGFβ2 alone was not sufficient to drive the Hh-mediated process in the absence of Hh signaling. Thus, it is predicted that the effect of SD208 (reduction of tumor area by 20%) will not be as effective as Hh signaling-specific inhibitors. Indeed, we found that SMO antagonists (such as cyclopamine derivative Cyc-T and the Genentech GDC-0449) can reduce tumor area by 40% in the same mouse model (data not shown here). Both C3H10T1/2 cells and E14Tg2a cells have the ability to differentiate into different cell types. The fact that TGFβ2 depletion in these cells decreases cell differentiation potentials does not necessarily support that autocrine TGFβ signaling (versus paracrine signaling) plays a major role. It is known that paracrine TGFβ signaling (among different progenitor cells or among the same types of cells) is critical in regulation of embryonic development.
Previously, several laboratories reported cross-talks between Hh signaling and the TGFβ pathway (25–27). In these reports, they showed direct up-regulation of Gli1/2 by TGFβ-SMAD signaling. Whether this mechanism occurs in our system is not known. Our major finding in this study is that TGFβ signaling is required for Hh signaling-mediated motor neuron differentiation, osteoblast differentiation, and BCC development. It is not known whether the above two signaling events occur in the same system to form a vicious cycle.
The canonical Hh signaling is through direct regulation of Hh target gene expression by transcriptional factor Gli molecules. Increasing data indicate that Hh signaling may also be activated by non-canonical pathways such as interactions between EGF and Hh and regulation of Gli molecules by Ras. Although we have shown in a number of biological systems that Hh signaling activation increases the expression of TGFβ signaling, the exact mechanisms remain to be identified. Analysis of TGFβ2 promoters did not reveal exact sequence matches to Gli binding sites, with three weak putative Gli binding sites (six of eight nucleotides matched).5 It is thus possible that TGFβ2 expression is regulated by interaction between Hh signaling and another pathway.
In summary, we provided biological evidence to indicate a critical role of TGFβ2 for Hh signaling-mediated biological processes, including tumor development. Despite the requirement of TGFβ signaling for Hh signaling-mediated biological processes, TGFβ alone is not sufficient to replace the roles of Hh signaling.
Supplementary Material
Acknowledgments
We thank Drs. Hailan Liu, Daohong Chen, and Dongsheng Gu for help with this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA94160 (to J. X.) and by a grant from the China Scholarship Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
Q. Fan, M. He, T. Sheng, X. Zhang, M. Sinha, B. Luxon, X. Zhao, and J. Xie, unpublished observations.
- SMO
- Smoothened
- Hh
- Hedgehog
- BCC
- basal cell carcinoma
- LPE
- local pool error
- ALP
- alkaline phosphatase
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Yang L., Xie G., Fan Q., Xie J. (2010) Oncogene 29, 469–481 [DOI] [PubMed] [Google Scholar]
- 2.Parkin C. A., Ingham P. W. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G363–G367 [DOI] [PubMed] [Google Scholar]
- 3.Jiang J., Hui C. C. (2008) Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahon A. P., Ingham P. W., Tabin C. J. (2003) Curr. Top. Dev. Biol. 53, 1–114 [DOI] [PubMed] [Google Scholar]
- 5.Kinzler K. W., Vogelstein B. (1990) Mol. Cell. Biol. 10, 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J. (2008) Acta Biochim. Biophys. Sin. 40, 670–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin L. L., de Sauvage F. J. (2006) Nat. Rev. Drug Discov. 5, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 8.Epstein E. H. (2008) Nat. Rev. Cancer 8, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theunissen J. W., de Sauvage F. J. (2009) Cancer Res. 69, 6007–6010 [DOI] [PubMed] [Google Scholar]
- 10.Owens P., Han G., Li A. G., Wang X. J. (2008) J. Invest. Derm. 128, 783–790 [DOI] [PubMed] [Google Scholar]
- 11.Stover D. G., Bierie B., Moses H. L. (2007) J. Cell. Biochem. 101, 851–861 [DOI] [PubMed] [Google Scholar]
- 12.Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. (2001) Nat. Genet. 29, 418–425 [DOI] [PubMed] [Google Scholar]
- 13.Mao J., Ligon K. L., Rakhlin E. Y., Thayer S. P., Bronson R. T., Rowitch D., McMahon A. P. (2006) Cancer Res. 66, 10171–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecanda J., Ganapathy V., D'Aquino-Ardalan C., Evans B., Cadacio C., Ayala A., Gold L. I. (2009) Cell Cycle 8, 724–756 [DOI] [PubMed] [Google Scholar]
- 15.Xie J., Garrossian M. (August13, 2009) World Intellectual Property Organization Patent PCT/US2009/000737
- 16.Athar M., Li C., Tang X., Chi S., Zhang X., Kim A. L., Tyring S. K., Kopelovich L., Hebert J., Epstein E. H., Jr., Bickers D. R., Xie J. (2004) Cancer Res. 64, 7545–7552 [DOI] [PubMed] [Google Scholar]
- 17.He J., Sheng T., Stelter A. A., Li C., Zhang X., Sinha M., Luxon B. A., Xie J. (2006) J. Biol. Chem. 281, 35598–35602 [DOI] [PubMed] [Google Scholar]
- 18.Jain N., Thatte J., Braciale T., Ley K., O'Connell M., Lee J. K. (2003) Bioinformatics 19, 1945–1951 [DOI] [PubMed] [Google Scholar]
- 19.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 20.Wichterle H., Lieberam I., Porter J. A., Jessell T. M. (2002) Cell 110, 385–397 [DOI] [PubMed] [Google Scholar]
- 21.Mao J., Barrow J., McMahon J., Vaughan J., McMahon A. P. (2005) Nucleic Acids Res. 33, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y. G., Wang X. F. (2009) Cell Res. 19, 1–2 [DOI] [PubMed] [Google Scholar]
- 23.Flavell R. A., Sanjabi S., Wrzesinski S. H., Licona-Limón P. (2010) Nat. Rev. Immunol. 10, 554–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X., Wang X. F. (2009) Cell Res. 19, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez J., Sohn P., Zeng X., Doetschman T., Robbins D. J., Serra R. (2002) Development 129, 1913–1924 [DOI] [PubMed] [Google Scholar]
- 26.Dennler S., André J., Alexaki I., Li A., Magnaldo T., ten Dijke P., Wang X. J., Verrecchia F., Mauviel A. (2007) Cancer Res. 67, 6981–6986 [DOI] [PubMed] [Google Scholar]
- 27.Yoo Y. A., Kang M. H., Kim J. S., Oh S. C. (2008) Carcinogenesis 29, 480–490 [DOI] [PubMed] [Google Scholar]
- 28.Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.