Abstract
A novel form of acto-myosin regulation has been proposed in which polymerization of new actin filaments regulates motility of parasites of the apicomplexan class of protozoa. In vivo and in vitro parasite F-actin is very short and unstable, but the structural basis and details of filament dynamics remain unknown. Here, we show that long actin filaments can be obtained by polymerizing unlabeled rabbit skeletal actin (RS-actin) onto both ends of the short rhodamine-phalloidin-stabilized Plasmodium falciparum actin I (Pf-actin) filaments. Following annealing, hybrid filaments of micron length and “zebra-striped” appearance are observed by fluorescence microscopy that are stable enough to move over myosin class II motors in a gliding filament assay. Using negative stain electron microscopy we find that pure Pf-actin stabilized by jasplakinolide (JAS) also forms long filaments, indistinguishable in length from RS-actin filaments, and long enough to be characterized structurally. To compare structures in near physiological conditions in aqueous solution we imaged Pf-actin and RS-actin filaments by atomic force microscopy (AFM). We found the monomer stacking to be distinctly different for Pf-actin compared with RS-actin, such that the pitch of the double helix of Pf-actin filaments was 10% larger. Our results can be explained by a rotational angle between subunits that is larger in the parasite compared with RS-actin. Modeling of the AFM data using high-resolution actin filament models supports our interpretation of the data. The structural differences reported here may be a consequence of weaker inter- and intra-strand contacts, and may be critical for differences in filament dynamics and for regulation of parasite motility.
Keywords: Actin, Atomic Force Microscopy, Biophysics, Cytoskeleton, Parasitology
Introduction
Plasmodium falciparum (Pf)5 is a protozoan causing human malaria and belongs to the apicomplexan group of intracellular parasites. Apicomplexan motility and host cell invasion requires the parasite's own acto-myosin motor system (1–3). The organization of the acto-myosin machinery under the cell surface of Apicomplexa has been described by a linear model (4) (for review, see Refs. 3, 5–7). In this model, actin filaments are linked via receptor-ligand interactions to a substrate or the host cell surface, and the action of myosin motors moves the parasite forward. Interestingly, the treatment of the closely related apicomplexan parasite Toxoplasma gondii (Tg) with the actin filament-stabilizing toxin jasplakinolide (JAS) increased both actin polymerization and the speed of gliding (8–10). Based on these observations, a novel form of acto-myosin regulation has been proposed in which the availability of actin filaments regulates apicomplexan motility (8).
Many recent studies have since focused on actin filament dynamics in these parasites. In contrast to other cell types it appears that in Apicomplexa, actin exists primarily in a monomeric form (8, 10, 11). This is consistent with the fact that the apicomplexan actin, in vitro and in vivo, only forms very short and unstable filaments (8, 9, 12, 13). Unlike rabbit skeletal (RS)-actin, both Pf- and Tg-actins show a low sensitivity to phalloidin, while retaining high sensitivity to JAS (8–15). The differences between apicomplexan and other eukaryotic actins are also a feature of purified expressed parasite actins. Therefore the differences in sequence between apicomplexan and other eukaryotic actins are thought to be responsible for the differences in actin dynamics (9, 11–13, 16, 17). In addition, many protein families that are key actin regulators in other eukaryotes are absent in Apicomplexa. Those apicomplexan actin-binding proteins that are characterized to date can also not fully explain the unusual actin dynamics observed in these parasites (5, 7, 18–22). Differences also clearly exist within the Apicomplexa. It was found for example that both Pf- and Tg-actin only form very short filaments in vitro. However, while recombinant, expressed Pf-actin polymerized only in presence of gelsolin, expressed Tg-actin readily polymerized with a critical concentration (Cc) even lower than that of yeast and vertebrate actins (9, 13). Furthermore, while Pf-actin co-polymerized with vertebrate actin, Tg-actin did not (9, 11, 13).
Thus, the formation of short filaments appears to be an inherent property of apicomplexan actins, but in absence of high-resolution structures the microscopic mechanisms of their unusual filament dynamics remain unknown (9, 12, 13). In electron micrographs, parasite actins show some of the typical characteristics of RS-actin filaments. Tg- and Pf-actin filaments were described as double-stranded and helical with a width of about 9 nm, and for Pf-actin myosin binding was confirmed (9, 12). Almost all sequence differences between apicomplexan and other eukaryotic actins are likely to be located on the surface of the monomer. Therefore a similar structure for the parasite and other eukaryotic actin monomers has been predicted (9, 13). Interestingly, much of the variance in the sequence occurs in regions of the molecule thought to be involved in monomer-monomer contacts within the actin filament. This may lead to changed longitudinal and lateral contacts within F-actin, and the different polymerization properties observed.
When comparing apicomplexan and other eukaryotic actins, the dynamic nature and ATP-dependence of the filament structure has also to be taken into account (23). Following ATP hydrolysis, the release of inorganic phosphate from the filament is coupled to conformational changes that result in filament destabilization (24). Two conformations of the nucleotide-binding pocket of actin have been described; a closed conformation associated with bound ATP, and an open one with bound ADP. In Pf-actin the lack of methylation of histidine 73 may favor the open state and therefore a destabilized filament structure (12). The two states also show large differences in the DNase-I binding loop (25), a region highly divergent in both Pf-actin and Tg-actin (9, 12, 13). It is tempting to speculate that apicomplexan F-actin might prefer the ADP-bound open state and become more unstable after ATP hydrolysis than RS-actin. For RS-actin the critical concentration for ADP-actin is only about 3-fold higher than for ATP-actin (26) and ADP-actin still forms stable, long filaments (27), while Tg-actin also readily polymerizes in ATP and ADP but only forms short filaments (9). Examples with low polymer stability in ADP are Acanthamoeba actin, with a 50-fold higher critical concentration for ADP-actin compared with ATP-actin, or bacterial actin-like protein ParM the polymer stability of which seems to be regulated by ATP hydrolysis (28, 29).
Here we set out to characterize the structure of Pf-actin filaments and to investigate whether they co-polymerize with rabbit actin. Because preparations from merozoites yield only minute quantities we first needed to optimize preparation conditions. We extracted Pf-actin from merozoites and polymerized the protein in the presence of phalloidin and JAS. The use of JAS allowed us to investigate the structure of Pf-actin filaments using atomic force microscopy (AFM). We discuss our findings in the context of filament dynamics and parasite motility.
EXPERIMENTAL PROCEDURES
Protein Preparations
Rabbit skeletal actin was prepared as described by Pardee and Spudich (30). When rabbit F-actin was compared with parasite actin filaments, both proteins were used at the same concentration, with phalloidin or JAS added at the same molar excess in each case. Rabbit skeletal muscle heavy meromyosin (HMM) subfragment for the in vitro motility assay was prepared as described by Margossian and Lowey (31).
Purification of Pf-actin
P. falciparum line 3D7 parasites were cultured in vitro, and merozoites were prepared as described previously (32). Aliquots of 20 μl of packed merozoites were frozen in liquid nitrogen and stored at −80 °C. Merozoites were thawed and extracted with 100 μl of actin extraction buffer (2 mm Tris-HCl, 0.2 mm CaCl2, 0.2 mm ATP, 0.5 mm DTT, pH 8.0) for 30 min. The samples were then clarified by centrifugation at 500,000 × g for 20 min in a Beckman TLA 120.1 rotor (Pellet 1). An appropriate volume of 10× actin polymerization buffer (50 mm Tris-HCl, 500 mm KCl, 20 mm MgCl2, 10 mm ATP, pH 8.0) was added to the extracts. F-actin-stabilizing toxins, phalloidin or JAS, were added to concentrations up to 20 μm. To fluorescently label Pf-actin filaments, 20 μm of rhodamine-phalloidin was used. To test the effect of nucleotide on polymerization we prepared Pf-actin in an extraction buffer in which ATP was left out and replaced in the polymerization buffer by 5 mm of either AMP·PNP, ATPγS, ADP·BeFx, or ADP·AlF4. These experiments were all performed at 20 μm phalloidin. When polymerizations were performed at pH 6.0, Tris was replaced by 50 mm MES (pH 6.0). After 30 min of polymerization, the samples were spun again at 500,000 × g for 20 min (Pellet 2 and Supernatant).
Western Blotting
SDS-PAGE (Bio-Rad Mini-Protean 3 gel electrophoresis system) was performed with 10% (w/v) acrylamide gels (33). Gels were Western-blotted to detect the presence of actin using the anti-actin antibody C-11 (Santa Cruz Biotechnology sc-1615 HRP). Gels were pre-equilibrated in transfer buffer for 10 min (25 mm Tris, 192 mm glycine, pH 8.3), and the proteins were transferred to PVDF membranes at 30 V overnight using the Bio-Rad Mini Trans-Blot electrophoretic transfer cell. The membranes were soaked in 5% (w/v) MarvelTM milk powder in TBS (20 mm Tris-HCl, 0.5 m NaCl, 0.2% Tween-20, pH 7.5) for 1 h. For binding of the antibody, the membranes were incubated overnight in TBS/milk at an antibody dilution of 1:1000. Unbound antibody was washed off by five washes in TBS/milk for 5 min each. Before visualization, all membranes were washed five times in TBS without Tween-20 for 5 min each. The horseradish peroxidase-labeled antibody was visualized using an enhanced chemiluminescence kit (Amersham Biosciences) according to the manufacturer's instructions and recorded by exposure to autoradiograph film (Kodak BioMax MR) for between 5 s and 30 min. Protein concentrations were assessed by visual comparison with RS-actin standards quantified by spectrophotometry.
In Vitro Motility Assays
Malaria parasite F-actin pellets, obtained after polymerization in presence of 20 μm rhodamine-phalloidin, were resuspended in 100 μl of actin polymerization buffer. To investigate whether RS-actin can polymerize onto the ends of Pf-actin filaments, we further incubated these fluorescently labeled Pf-actin filaments with 1 μm unlabeled rabbit skeletal G-actin for 30 min. These hybrid filaments were studied in in vitro motility assays using skeletal muscle HMM deposited on nitrocellulose-coated surfaces following standard protocols (34, 35). We used buffer conditions and HMM concentrations (≈2 mg ml−1) as previously established for the in vitro motility assay with Pf-actin (12). Imaging was performed on a total internal reflection fluorescence microscopy set-up, built around a Zeiss Axiovert 200 inverted microscope, as described previously (36). A 532 nm laser was used for fluorescence excitation and images were collected with an EMCCD camera (iXon 897 BV, Andor, UK) at video rate. The gliding velocity of individual actin filaments was determined by centroid tracking using the imaging software GMimPro (37).
Electron Microscopy
F-actin pellets obtained after polymerization in the presence of 20 μm JAS were resuspended in 100-μl actin polymerization buffer containing the same concentration of JAS. The samples were applied to glow-discharged activated carbon-coated electron microscope grids and negatively stained with 1% (w/v) uranyl acetate. Images were recorded on an FEI Spirit electron microscope operated at 120 kV with an Eagle CCD camera or film. Films were scanned on a Z/I scanner densitometer.
Atomic Force Microscopy
Pf-actin polymerized in 20 μm JAS and RS-actin polmerised in either 20 μm JAS or phalloidin were resuspended in 100 μl actin polymerization buffer containing the same concentration of toxin. Prior to AFM-imaging, the proteins were diluted ∼200-fold in their own buffer. The actin filaments were bound to glass cover slips silanized with positively charged trimethoxysilylpropyl-diethylenetriamine (38). A sample volume of 20 μl was incubated on a coverslip for 10 min. Before imaging, another drop of 20 μl buffer was added resulting in a total sample volume of about 40 μl. The atomic force microscope (NanoWizard, JPK Instruments, Germany) was operated in buffer at room temperature in tapping mode (39). We used BL150 cantilevers (30 × 60 μm, 0.03 N.m−1, Olympus, Japan) oscillated with an amplitude of ∼5 nm at the resonance frequency of around 7 kHz. To reduce binding of the AFM tip to the sample, new cantilevers were irradiated for 20 min by a 254 nm, 5 watt mercury lamp (UVP, Cambridge, UK) prior to the experiment. The scans were performed at a resolution of 0.7–6.8 nm·pixel−1. Standard image processing for background subtraction was performed on all scans using WSxM software (Nanotec, Madrid, Spain) (40). The images of filaments shown in the figures are averages of trace and retrace scans obtained after aligning them by minimizing the cross-correlation function. A derivative filter was also applied to enhance the edges in these images. For frequency domain analysis the images (non-filtered and non-averaged) were saved as 8-bit greyscale bitmaps using the same software.
Image Processing
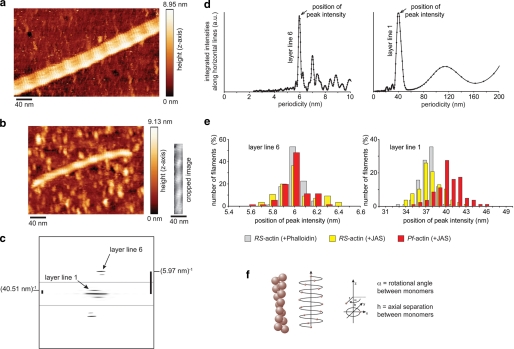
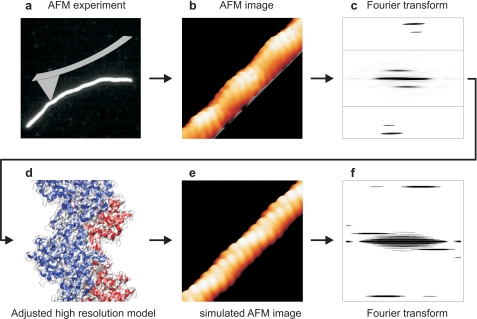
Straight segments of maximum possible length of the actin filaments were boxed out from the atomic force micrographs and rotated using the command “helixboxer” in EMAN software (41). Using SPIDER software, the background was cropped and the cropped image was padded to 2000 by 2000 nm to obtain a finer sampling of the calculated Fourier transform (42). As the two-dimensional Fourier transforms were asymmetric (Fig. 4c), we analyzed the top left and right quarters of these transforms independently. Intensities were integrated along horizontal lines to produce the projections shown in Fig. 4d. Average periodicities corresponding to the respective layer lines were determined by averaging results from all recorded images. As integrated intensities generally increase toward the equator of the transforms, it can become difficult to separate peaks associated with layer lines from background noise. We therefore only scored individual results when the layer line peaks were larger than noise peaks in the vicinity. Periodicity values were determined from each image as follows. For layer line 1 (half-pitch of the double helix), the position of the highest peak of integrated intensities in the range of 0–50 nm was taken, but only if there was no higher noise peak in the range of 50–100 nm. For layer line 6 (pitch of the genetic helix), the position of the highest peak of integrated intensities in the range of 0–8 nm was taken, but only if there was no higher noise peak in the range of 8–10 nm. The histograms of layer line 1 show the data from the top left quarter of the transforms, those of layer line 6 are from the top right quarter.
FIGURE 4.
Atomic force microscopy. Atomic force micrographs of (a) RS-actin (scan resolution = 1.43 nm·pixel−1) and (b) Pf-actin filaments (scan resolution = 1.17 nm·pixel−1), both polymerized in the presence of JAS. The cropped image shows the straight segment of the filament (172 nm) used for further analysis. c, Fourier transform of the straight segment of the Pf-actin filament shown in b after background cropping and padding to 2000 by 2000 nm (the area near the equator and the one further to the equator were individually optimized for contrast). d, intensities were integrated along horizontal lines to produce the projections shown here. For layer line 1 (half-pitch of the two-start helix) the position of the highest peak of integrated intensities (peak intensity) in the range of 0–50 nm was taken. For layer line 6 (pitch of the genetic helix) the highest peak of integrated intensities in the range of 0–8 nm was taken. The distributions of these peaks in d are shown in the histograms in e. For RS-actin +JAS the mean ± S.D. for layer line 1 was 37.7 ± 1.8 nm (n = 77), for layer line 6 we found 6.02 ± 0.17 nm (n = 55). For RS- actin +phalloidin, we measured 37.4 ± 1.3 nm (n = 76) for layer line 1 and 6.01 ± 0.08 nm (n = 62) for layer line 6. For Pf-actin, we found 40.4 ± 2.0 nm (n = 80) for layer line 1 and 6.00 ± 0.12 nm (n = 71) for layer line 6. f, model illustrating the genetic and the two-start helix.
Structure Modeling and Simulations of AFM Imaging
Graphic computer models of RS- and Pf-actin comprising 50 monomers were generated to simulate the way the conical AFM tip (∼20 nm tip radius) renders a spatially low-pass filtered image of the molecular surface. Details of the method will be described in a separate publication.6 In brief, the AFM imaging process was simulated using molecular models for monomeric and F-actin from the literature (Lorenz-model, 1993 (43)). A filament model was generated and the effect of the imaging AFM tip on surface profiling was modeled using the approach by Villarubia (44).
RESULTS
Purification of Pf-actin
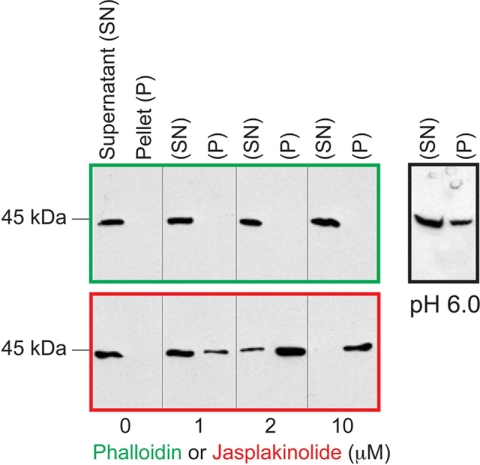
After extraction of actin from the merozoites with G-actin extraction buffer, no or only very little actin was left behind in the insoluble fraction (not shown). We found the total actin concentration in the extracts to be about 0.2 μm. Even after adding polymerization buffer to these extracts, actin could not be sedimented by centrifugation (Fig. 1). Likewise very little actin was sedimented in the presence of phalloidin (or rhodamine-phalloidin) at concentrations up to 20 μm. Replacing ATP with non-hydrolyzable analogues (AMP·PNP, ATPγS), mimicking the ADP·Pi state using ADP·BeFx or ADP·AlF4, using high concentrations of free Pi or an ATP-regenerating system (26, 45, 46) also showed no significant increase in the fraction of sedimented actin (not shown). However lowering the pH under high phalloidin conditions increased the yield of sedimented actin (Fig. 1), consistent with a lower Cc and higher rates of polymerization in more acidic conditions as described for RS-actin (47–51). In presence of JAS, in contrast to phalloidin, Pf-actin could be sedimented in a concentration-dependent manner (Fig. 1). At 20 μm JAS, most of the actin could be sedimented, with yields of about 1 μg of actin from 20 μl of merozoites. Nevertheless, although the yield with rhodamine-phalloidin was low, we could still sediment a sufficient amount of fluorescent Pf-actin filaments for studies in the in vitro motility assays.
FIGURE 1.
Pf-actin purification. Pf-actin was extracted from merozoites and then polymerized by adding actin polymerization buffer and 0–20 μm F-actin-stabilizing toxins phalloidin or jasplakinolide at pH 8.0. At pH 6.0 more actin could be polymerized in the presence of 20 μm phalloidin. Following centrifugation, soluble (monomeric actin) and the insoluble fractions (F-actin) were analyzed by SDS-PAGE and Western blotting to detect actin.
In Vitro Motility Assay
At pH 8.0, the rhodamine-phalloidin labeled Pf-actin filaments showed the same in vitro motility over rabbit skeletal HMM as reported earlier (12), and similar to that found for RS-actin filaments. The length of Pf-actin filaments was close to the diffraction limit (about 200 nm) of the microscope (12). In contrast, many of the filaments prepared at pH 6.0 were no longer spot-like, but clearly elongated, with lengths of around 1 μm (Fig. 2, b and c). When Pf-actin filaments were further incubated with unlabeled RS-G-actin at pH 8.0, we observed long filaments of “zebra-striped” appearance (Fig. 2a and supplemental movie S1). This shows that RS-actin can polymerize onto both ends of Pf-actin. The mechanical stability of the striped hybrid filaments in the in vitro motility assays appeared the same as that of pure RS-actin or Pf-actin filaments (12). This suggests that heterologous contact sites in the hybrid are stable enough to sustain forces imposed by the myosin motors during filament gliding.
FIGURE 2.
In vitro motility assay. a, Pf-actin filaments polymerized in the presence of 20 μm rhodamine-phalloidin at pH 8.0 were further incubated with unlabeled RS-actin. This produced long filaments of “zebra-striped” appearance with segments of fluorescent Pf-actin and unlabeled RS-actin. The hybrid filaments moved on a surface coated with skeletal muscle HMM (see supplemental movie S1). b, Pf-actin polymerized in the presence of 20 μm rhodamine-phalloidin at pH 6.0, and c, distribution of filament length of Pf-actin polymerized as in b.
Electron Microscopy
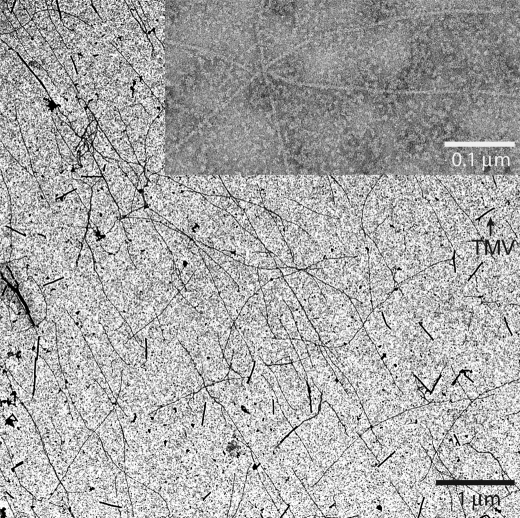
Negative stain electron micrographs of Pf-actin sedimented in the presence of JAS show individual filaments with a helical structure characteristic of rabbit F-actin (Fig. 3) (52). We found these filaments to be several micrometers long, and indistinguishable in average length from those formed by RS-actin.
FIGURE 3.
Electron microscopy. Electron micrographs of negatively stained Pf-actin filaments polymerized in the presence of jasplakinolide. Filaments are several μm in length. Tobacco mosaic virus (TMV) was used for calibration.
Atomic Force Microscopy and Image Processing
The number of straight segments analyzed for Pf-actin filaments (+JAS) was 151. The lengths of these segments varied between 50 and 508 nm, with an average length of 156 nm. The scan resolution varied between 0.7 and 6.8 nm·pixel−1, with an average of 1.9 nm·pixel−1. The respective values for the rabbit actin data sets were very similar. For RS-actin (+JAS), the number of segments was 113, their lengths 55 to 480 nm (average of 170 nm) and the scan resolution 1.1 to 3.1 nm·pixel−1 (average of 1.8 nm/pixel). For RS-actin (+phalloidin), the number of segments was 132, their lengths 48 to 590 nm (average of 202 nm) and the scan resolution 0.7 to 6.6 nm·pixel−1 (average of 1.9 nm·pixel−1). Filament heights as measured between background and filament ridge were close to 10 nm for both actins, consistent with the expected diameter of an individual RS-actin filament (Fig. 4, a and b) (43). While the measured height of the filaments is not affected by the AFM tip geometry, the measured filament width is broadened. As tip geometry varied, including double tips, the recorded filament width varied too (Fig. 4, a and b).
Searching for differences between Pf-actin and RS-actin filament structure we then focused on the comparison of the helix geometries. A (rigid) actin monomer has six degrees of freedom in space. In a helical assembly three rotational degrees of freedom and one translational degree of freedom are required to describe monomer location along the helix and monomer orientation (53). The rotational orientation defines the interaction surfaces. The helix itself is described by a pitch and diameter, assuming a circular cross-section. For the orientation of the monomers along the helix we assume helical symmetry. Monomer location on the helix is then described by only two parameters, the rotational angle α between successive monomers and their axial separation h. Each monomer is in contact with two lateral and two axial neighbors. Thus, alternatively to the genetic helix, which connects lateral neighbors, the actin filament can also be described by a two-start helix that connects axial neighbors (Fig. 4f). The periodicities or pitches of the one-start genetic and two-start helices of actin as well as their helical handedness (54) can be seen directly in the atomic force micrographs (Fig. 4, a–c). The Fourier transforms of the scanned filaments show strong layer lines for both periodicities, with the helical handedness reflected by the asymmetric distribution of these intensities (Fig. 4c). As for RS-actin, the genetic helix of Pf-actin is left-handed and the two-start helix right-handed. Although the monomers cannot be resolved directly in the AFM images, the parameters α and h can be calculated from the measured pitches P1 and P2. The pitch of the genetic helix is (55) shown in Equation 1,
 |
and the pitch of the two-start helix is shown in Equation 2.
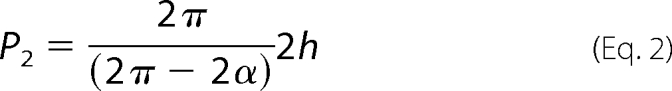 |
The factor of 2 in the term 2α comes from the fact that there are just over 2 monomers per turn in the one-start helix. In other words, the second lateral neighbor is the axial neighbor.
The Fourier transforms of RS actin filaments are dominated by intensities at 36 nm (corresponding to layer line 1, the half-pitch of the double helix) and 5.9 nm (corresponding to layer line 6, the pitch of the genetic helix) (56). The intensities of the Fourier transforms projected onto the filament axis showed strong peaks around these two values (Fig. 4d). The distributions of these peaks for all filaments analyzed are shown in the histograms in Fig. 4e. Where both pitches could be determined from the same stretch of filament, we calculated α and h from those pairs of values using equation 1 and 2. Measured from the same image, any calibration error will affect the measurement of both distances to the same degree. With α dimensionless and derived from the ratio of these two distances, the values for α will be free of calibration errors.
For RS-actin stabilized with JAS, P1 was 6.02 nm and P2 = 37.7 nm (n = 41), resulting in an average rotational angle between subunits α = 166.54 (±0.77) degrees, and an average axial separation between subunits h = 2.78 (±0.10) nm. From these values for α and h, we calculate 2.162 monomers per pitch for the genetic helix, and 13.37 monomers per half-pitch for the double helix. For RS-actin stabilized with phalloidin, similar values were measured for both pitches, P1 = 6.01 nm and P2 = 37.4 nm (n = 58). Here, we calculated α = 166.67 (±0.53) degrees and h = 2.78 (±0.04) nm. This results in 2.160 monomers per pitch for the genetic helix and 13.50 monomers per half-pitch for the double helix. Intriguingly for Pf-actin, the pitch of the genetic helix was again very similar to the values obtained for RS-actin, P1 = 6.02 nm (n = 37). However the half-pitch of the double helix was 10% larger with P2 = 40.4 nm. The resulting rotational angle between subunits here is slightly increased, α = 167.71 (±0.51) degrees. The axial separation between subunits was again unchanged compared with RS-actin, with h = 2.80 (±0.03) nm. Together these values for α and h correspond to 2.146 monomers per pitch of the genetic helix for Pf-actin, which is similar to the result for RS-actin. However the number of monomers (14.65) per half-pitch in the malaria actin double helix is increased by about one monomer (Table 1).
TABLE 1.
AFM image analysis
This table summarizes the results obtained from AFM image analysis.
| RS-actin + Phalloidin | RS-actin + JAS | Pf-actin + JAS | |
|---|---|---|---|
| P1 (nm) | 6.01 ± 0.08 | 6.02 ± 0.17 | 6.02 ± 0.12 |
| P2 (nm) | 37.4 ± 1.31 | 37.7 ± 1.83 | 40.4 ± 1.98 |
| α (degrees) | 166.67 ± 0.53 | 166.54 ± 0.77 | 167.71 ± 0.51 |
| h (nm) | 2.78 ± 0.04 | 2.78 ± 0.10 | 2.80 ± 0.03 |
| N | 58 | 41 | 37 |
| Monomers per pitch (genetic helix) | 2.16 | 2.16 | 2.15 |
| Monomers per half-pitch (double helix) | 13.50 | 13.37 | 14.65 |
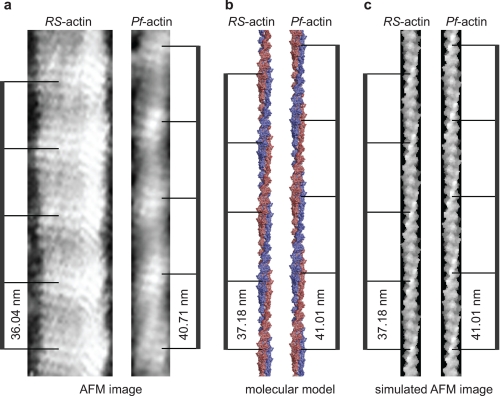
The simulations resulted in images strikingly similar to those obtained experimentally by AFM imaging (Figs. 5, a and c, and 6, b and e). As in the atomic force micrographs, both the genetic and two-start helices of actin are dominating features in the simulations. When the simulated images were analyzed in the same way as the experimental AFM images, the helical parameters measured for α and h were identical to those used to create the models. This consistency of experimental parameters and imaging simulation of atomistic models provides additional confidence in the interpretation of the structural differences between Pf-actin and RS-actin filaments. In summary, a small increase in the rotational angle between subunits can account for the 10% increase in half-pitch of the double helix we observe for the malaria parasite actin filament. This demonstrates that the half-pitch of the double helix is a very sensitive measure for the distance of monomers along the genetic actin helix (56).
FIGURE 5.
AFM image and molecular model. a, atomic force micrographs of RS-actin and Pf-actin filaments are shown. The AFM images were simulated using molecular models for monomeric and F-actin from the literature (43) and parameters α and h from the AFM experiments here. A filament model was produced and the surface of the model generated using standard software and a probe with a radius of 5 Å is shown in b. In c, the effect of the imaging tip on the surface profiling was modeled using an approach by Villarrubia (44), with a probe radius of 1.1 nm. The image matrix was generated with a resolution of 0.5 nm per pixel, similar to that of the experimental AFM images. The half-pitch of the double helix of the AFM experimental images (a), of the filament model (b), and the simulated AFM images (c) are indicated.
FIGURE 6.
Scheme of AFM imaging and modeling. a–c, AFM images were analyzed by Fourier transformation to determine the pitches of the genetic and the two-start helix for each image. From these pitches, the parameters α and h were calculated. d and e, molecular model was generated using α and h to simulate the AFM images analyzed using Fourier transformation (f).
DISCUSSION
It was already known that apicomplexan actins behave differently from other eukaryotic actins. Actin and myosin are required for motility but Apicomplexa appear to have an idiosyncratic way of motility regulation. One consistent observation on apicomplexan actins is the formation of short filaments. The structural details and what limits filament polymerization is unknown (9, 12, 13). The monomer structure of the parasite actin is predicted to be similar to known structures of other actins (9, 13) because almost all variant amino acids are located on the surface of the monomer. This fact however suggests that surface contacts between neighboring monomers are different. Our co-polymerization results show that these differences are not large enough to prevent RS-actin from polymerizing onto both ends of Pf-actin filaments. Hybrid filaments were moved by skeletal muscle myosin motors in an in vitro gliding filament assay. This demonstrates at least in a qualitative way the mechanical stability of the hybrid interface between heterologous monomers. We show that in the presence of the F-actin-stabilizing toxin JAS the yield and filament length of sedimented tissue-purified malaria parasite actin strongly increases, consistent with a high sensitivity of apicomplexan parasites toward JAS reported in the literature (8–11, 14, 15).
The use of JAS allowed us to visualize Pf-actin filaments by AFM in near physiological conditions in aqueous solution in order to investigate their structure. For comparison we also visualized rabbit skeletal actin filaments stabilized by JAS or phalloidin. Filament heights were close to 10 nm for parasite and rabbit actins, in line with the expected diameter of individual RS-actin filaments measured in EM (43). We find the same helical handedness for the Pf-actin as we do for RS-actin, consistent with the description of F-actin as a left-handed genetic helix or a right-handed double helix (54). Image processing of individual filaments often revealed the pitch of its genetic helix and/or the half-pitch of its double helix. Our analysis shows that the average value for the pitch of the genetic helix (6.0 nm – 6.02 nm) is the same for parasite and rabbit actins with less than 1% difference. Furthermore, for rabbit actin the average half-pitch of the double helix was insensitive to the presence of F-actin stabilizing phalloidin (P2 = 37.5 nm) or JAS (P2 = 37.2 nm). All these values are consistent with those reported elsewhere for other eukaryotic actins (54). In contrast, the average half-pitch of the double helix for Pf-actin was 10% larger (40.4 nm) than for rabbit actin. This increase in pitch of the double helix, with the pitch of the genetic helix unchanged, can be explained by a subtle geometrical difference in packing of the monomers, in particular the rotational angle α between subunits. This angle was very similar for the rabbit actins, with α = 166.67° (+phalloidin) and α = 166.54° (+JAS). However for the parasite actin we find the angle to be increased by more than 1° to α = 167.71°. Both helical pitches are directly proportional to h, the axial separation between subunits. However, the dependence on α differs for the two pitches. Therefore, a small increase in α by 1° from 166° to 167° reduces the pitch of the genetic helix by less than 1%, but increases the half-pitch of the double helix by more than 8%.
Modeling of the AFM data using high-resolution actin filament models confirms the validity of our image processing method and supports our interpretation of the data. Even though these first structural differences to be reported are small, monomer-monomer contacts within the filament could well be affected. An increased rotational angle between subunits possibly leads to weaker inter-strand contacts within the filament, while an increased axial separation between subunits possibly leads to weaker inter- and intra-strand contacts. These structural differences may be critical for the different filament dynamics observed and might be crucial for parasite motility. For RS-actin, phalloidin binds with high affinity at the interface of three monomers, two in the same strand with the other one in the other strand (43, 57). It is thought to stabilize F-actin by strengthening both inter- and intra-strand contacts. Our results indicate that phalloidin fails to significantly stabilize malaria parasite actin filaments in terms of length or yield. This is consistent with previous reports of a low sensitivity of apicomplexan parasites to phalloidin and a weaker binding of the toxin to apicomplexan F-actin (9, 13). The mapping of the phalloidin binding site on F-actin pointed to three aromatic residues on three different subunits, all candidates for important contacts to the aromatic ring of phalloidin (43). Interestingly, one of these, tyrosine 279, is absent in apicomplexan actins. Maybe phalloidin binds more weakly because this important contact cannot be made, or maybe, without this contact the toxin cannot overcome the instability of the parasite F-actin. Individual actin subunits in F-actin are rotated on average by >10° away from their ideal positions (58). This variability is the basis of F-actin's dynamic nature (59). The slow binding of phalloidin to conventional F-actin suggests that its binding site only becomes accessible due to the breathing motions of the filament (60). The idea that only certain filament conformations allow phalloidin to bind is strengthened by the observation that the F-actin-binding protein cofilin excludes phalloidin binding, even though their binding sites are different (61). Cofilin stabilizes a natural state of F-actin, which seems to be inaccessible to phalloidin, at a rotational angle between subunits of about 5° less than the average. As we observe an average rotational angle between subunits increased by more than 1° for the parasite actin, maybe the sensitivity of phalloidin binding to the twist of actin is the basis for its failure to stabilize the filaments. The binding site of JAS on F-actin has not been mapped, but is thought to be similar to that of phalloidin, with which it binds competitively (62, 63). These compounds are not structurally related, and differences have been described in their filament-stabilizing effects. We can therefore speculate that neither amino acid substitutions nor the changes in filament structure, which may prevent phalloidin from stabilizing the parasite filament, prevent JAS from binding. Like phalloidin, JAS is thought to bind three monomers, allowing it to strengthen both inter- and intra-strand contacts. Some actin-binding proteins are known to have such an effect. We can therefore speculate that the effect of JAS we observe is because the toxin fully compensates any weaker inter- and intra-strand contacts of the parasite actin filament structure, and that in vivo that role is taken up by an actin-binding protein in a regulatory fashion. Much higher rates for depolymerization and/or fragmentation for Pf-actin compared with RS-actin could therefore be the reasons for the observed instability, i.e. high critical concentration and short filament length, in the absence of JAS. Following ATP hydrolysis, the release of inorganic phosphate from the filaments of yeast actin was coupled to conformational changes that result in filament destabilization (24). Phalloidin also stabilizes RS-actin by inhibiting the dissociation of Pi from F-actin (64), and JAS might have similar effects. The idea that apicomplexan F-actin becomes more unstable after ATP hydrolysis than other eukaryotic actins is therefore intriguing. However, by stabilizing the ATP or ADP-Pi state, we did not find evidence for this, while lowering the pH, which is also known to stabilize conventional actin filaments, resulted in at least some stabilization (47–51).
In summary our findings show differences in the structure of Pf-actin from RS-actin, which we speculate could cause a reduced stability of Pf-actin filaments. This is a first step toward an understanding of the structural role of actin in apicomplexan motility. It will be critical to learn about the role of actin binding partners and their effect on the filament structure and dynamics.
Supplementary Material
Acknowledgments
We thank Christoph Schmidt and Justin E. Molloy for stimulating discussions and comments on the manuscript.
This work was funded by the UK Medical Research Council (Reference numbers: U117532067, U117570593, U117581331, and U117581334) and NWO and the EU through a Marie Curie Intra-European Fellowship (to I. A. T. S.).

The on-line version of this article (available at http://www.jbc.org) contains a supplemental movie.
C. Veigel, manuscript in preparation.
- Pf
- Plasmodium falciparum
- AFM
- atomic force microscopy
- Cc
- critical concentration
- HMM
- heavy meromyosin
- JAS
- jasplakinolide
- RS
- rabbit skeletal
- Tg
- Toxoplasma gondii
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- AMP·PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1.Dobrowolski J. M., Sibley L. D. (1996) Cell 84, 933–939 [DOI] [PubMed] [Google Scholar]
- 2.Dobrowolski J. M., Carruthers V. B., Sibley L. D. (1997) Mol. Microbiol. 26, 163–173 [DOI] [PubMed] [Google Scholar]
- 3.Fowler R. E., Margos G., Mitchell G. H. (2004) Adv. Parasitol. 56, 213–263 [DOI] [PubMed] [Google Scholar]
- 4.Bergman L. W., Kaiser K., Fujioka H., Coppens I., Daly T. M., Fox S., Matuschewski K., Nussenzweig V., Kappe S. H. I. (2003) J. Cell Sci. 116, 39–49 [DOI] [PubMed] [Google Scholar]
- 5.Baum J., Papenfuss A. T., Baum B., Speed T. P., Cowman A. F. (2006) Nat. Rev. Microbiol. 4, 621–628 [DOI] [PubMed] [Google Scholar]
- 6.Schüler H., Matuschewski K. (2006) Trends Parasitol. 22, 146–147 [DOI] [PubMed] [Google Scholar]
- 7.Schüler H., Matuschewski K. (2006) Traffic 7, 1433–1439 [DOI] [PubMed] [Google Scholar]
- 8.Wetzel D. M., Håkansson S., Hu K., Roos D., Sibley L. D. (2003) Mol. Biol. Cell 14, 396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahoo N., Beatty W., Heuser J., Sept D., Sibley L. D. (2006) Mol. Biol. Cell 17, 895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw M. K., Tilney L. G. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 9095–9099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrowolski J. M., Niesman I. R., Sibley L. D. (1997) Cell Motil. Cytoskeleton 37, 253–262 [DOI] [PubMed] [Google Scholar]
- 12.Schmitz S., Grainger M., Howell S., Calder L. J., Gaeb M., Pinder J. C., Holder A. A., Veigel C. (2005) J. Mol. Biol. 349, 113–125 [DOI] [PubMed] [Google Scholar]
- 13.Schüler H., Mueller A. K., Matuschewski K. (2005) FEBS Lett. 579, 655–660 [DOI] [PubMed] [Google Scholar]
- 14.Poupel O., Tardieux I. (1999) Microb. Infect. 1, 653–662 [DOI] [PubMed] [Google Scholar]
- 15.Mizuno Y., Makioka A., Kawazu S., Kano S., Kawai S., Akaki M., Aikawa M., Ohtomo H. (2002) Parasitol. Res. 88, 844–848 [DOI] [PubMed] [Google Scholar]
- 16.Wesseling J. G., de Ree J. M., Ponnudurai T., Smits M. A., Schoenmakers J. G. (1988) Mol. Biochem. Parasitol. 27, 313–320 [DOI] [PubMed] [Google Scholar]
- 17.Wesseling J. G., Smits M. A., Schoenmakers J. G. (1988) Mol. Biochem. Parasitol. 30, 143–153 [DOI] [PubMed] [Google Scholar]
- 18.Schüler H., Mueller A. K., Matuschewski K. (2005) Mol. Biol. Cell 16, 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum J., Tonkin C. J., Paul A., Smith B. J., Pollard T. D., Cowman A. F. (2008) Int. J. Parasitol. 38, S52 [Google Scholar]
- 20.Baum J., Tonkin C. J., Paul A. S., Rug M., Smith B. J., Gould S. B., Richard D., Pollard T. D., Cowman A. F. (2008) Cell Host Microbe 3, 188–198 [DOI] [PubMed] [Google Scholar]
- 21.Gordon J. L., Sibley L. D. (2005) BMC Genomics 6, 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poupel O., Boleti H., Axisa S., Couture-Tosi E., Tardieux I. (2000) Mol. Biol. Cell 11, 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galkin V. E., VanLoock M. S., Orlova A., Egelman E. H. (2002) Curr. Biol. 12, 570–575 [DOI] [PubMed] [Google Scholar]
- 24.Belmont L. D., Orlova A., Drubin D. G., Egelman E. H. (1999) Proc. Natl. Acad. Sci.U. S. A. 96, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuler H. (2001) Biochim. Biophys. Acta-Prot. Struct. Mol. Enzymol. 1549, 137–147 [DOI] [PubMed] [Google Scholar]
- 26.Cooke R. (1975) Biochemistry 14, 3250–3256 [DOI] [PubMed] [Google Scholar]
- 27.Pollard T. D., Goldberg I., Schwarz W. H. (1992) J. Biol. Chem. 267, 20339–20345 [PubMed] [Google Scholar]
- 28.Pollard T. D. (1984) J. Cell Biol. 99, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardee J. D., Spudich J. A. (1982) Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 31.Margossian S. S., Lowey S. (1982) Methods Enzymol. 85, 55–71 [DOI] [PubMed] [Google Scholar]
- 32.Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De la Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. (2003) Science 301, 1503–1508 [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34.Kron S. J., Spudich J. A. (1986) Proc. Natl. Acad. Sci. U. S. A. 83, 6272–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veigel C., Bartoo M. L., White D. C., Sparrow J. C., Molloy J. E. (1998) Biophys. J. 75, 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mashanov G. I., Tacon D., Knight A. E., Peckham M., Molloy J. E. (2003) Methods 29, 142–152 [DOI] [PubMed] [Google Scholar]
- 37.Mashanov G. I., Molloy J. E. (2007) Biophys. J. 92, 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaap I. A., de Pablo P. J., Schmidt C. F. (2004) Eur. Biophys. J. Biophys. Lett. 33, 462–467 [DOI] [PubMed] [Google Scholar]
- 39.Martin Y., Williams C. C., Wickramasinghe H. K. (1987) J. Appl. Phys. 61, 4723–4729 [Google Scholar]
- 40.Horcas I., Fernandez R., Gomez-Rodriguez J. M., Colchero J., Gomez-Herrero J., Baro A. M. (2007) Rev. Scientific Instr. 78, 013705_1– 013705_8 [DOI] [PubMed] [Google Scholar]
- 41.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 42.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 43.Lorenz M., Popp D., Holmes K. C. (1993) J. Mol. Biol. 234, 826–836 [DOI] [PubMed] [Google Scholar]
- 44.Villarrubia J. S. (1997) J. Res. Natl. Inst. Standards Technology 102, 425–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara I., Vavylonis D., Pollard T. D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Combeau C., Carlier M. F. (1988) J. Biol. Chem. 263, 17429–17436 [PubMed] [Google Scholar]
- 47.Wang F., Sampogna R., Ware B. R. (1988) Biophys. J. 53, A572–A572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukoyanova N., Orlova A., Egelman E. H. (2000) Biophys. J. 78, 1408Pos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerle C. T., Frieden C. (1988) Biochemistry 27, 7766–7772 [DOI] [PubMed] [Google Scholar]
- 50.Sampath P., Pollard T. D. (1991) Biochemistry 30, 1973–1980 [DOI] [PubMed] [Google Scholar]
- 51.Oda T., Makino K., Yamashita I., Namba K., Maéda Y. (2001) Biophys. J. 80, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanson J. (1967) Nature 213, 353–356 [Google Scholar]
- 53.André I., Bradley P., Wang C., Baker D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 17656–17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheterline P., Clayton J., Sparrow J. C. (1995) Protein Profile 2, 1–103 [PubMed] [Google Scholar]
- 55.Egelman E. H., Francis N., DeRosier D. J. (1982) Nature 298, 131–135 [DOI] [PubMed] [Google Scholar]
- 56.Stokes D. L., DeRosier D. J. (1987) J. Cell Biol. 104, 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieland T., Faulstich H. (1978) CRC Crit. Rev. Biochem. 5, 185–260 [DOI] [PubMed] [Google Scholar]
- 58.Schmid M. F., Sherman M. B., Matsudaira P., Chiu W. (2004) Nature 431, 104–107 [DOI] [PubMed] [Google Scholar]
- 59.Reisler E., Egelman E. H. (2007) J. Biol. Chem. 282, 36133–36137 [DOI] [PubMed] [Google Scholar]
- 60.DelaCruz E. M., Pollard T. D. (1996) Biochemistry 35, 14054–14061 [DOI] [PubMed] [Google Scholar]
- 61.McGough A., Pope B., Chiu W., Weeds A. (1997) J. Cell Biol. 138, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L., Korn E. D. (1994) J. Biol. Chem. 269, 14869–14871 [PubMed] [Google Scholar]
- 63.Visegrády B., Lorinczy D., Hild G., Somogyi B., Nyitrai M. (2005) FEBS Lett. 579, 6–10 [DOI] [PubMed] [Google Scholar]
- 64.Dancker P., Hess L. (1990) Biochim. Biophys. Acta 1035, 197–200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.