Abstract
Activin A, a member of the transforming growth factor-β family, plays important roles in hormonal homeostasis and embryogenesis. In this study, we produced recombinant human activin A and examined its abilities to bind to extracellular matrix proteins. Recombinant activin A expressed in 293-F cells was purified as complexes of mature dimeric activin A with its pro-region. Among a panel of extracellular matrix proteins tested, recombinant activin A bound to perlecan and agrin, but not to laminins, nidogens, collagens I and IV, fibronectin, and nephronectin. The binding of recombinant activin A to perlecan was inhibited by heparin and high concentrations of NaCl and abolished by heparitinase treatment of perlecan, suggesting that activin A binds to the heparan sulfate chains of perlecan. In support of this possibility, recombinant activin A was capable of directly binding to heparin and heparan sulfate chains. Site-directed mutagenesis of recombinant activin A revealed that clusters of basic amino acid residues, Lys259-Lys263 and Lys270-Lys272, in the pro-region were required for binding to perlecan. Interestingly, deletion of the peptide segment Lys259-Gly277 containing both basic amino acid clusters from the pro-region did not impair the activity of activin A to stimulate Smad-dependent gene expressions, although it completely ablated the perlecan-binding activity. The binding of activin A to basement membrane heparan sulfate proteoglycans through the basic residues in the pro-region was further confirmed by in situ activin A overlay assays using frozen tissue sections. Taken together, the present results indicate that activin A binds to heparan sulfate proteoglycans through its pro-region and thereby regulates its localization within tissues.
Keywords: Basement Membrane, Bone Morphogenetic Protein (BMP), Extracellular Matrix, Extracellular Matrix Proteins, Glycosaminoglycan, Growth Factors, Heparan Sulfate, Transforming Growth Factor β (TGFβ)
Introduction
Extracellular matrix (ECM)2 is a supramolecular assembly composed of proteins and glycosaminoglycans (GAGs) that is deposited outside cells. It serves as a scaffold for cell adhesion, and regulates cell survival, proliferation, migration, and gene expression through interactions with cell surface receptors including integrins and membrane-anchored proteoglycans (1). The ECM also binds to a variety of growth factors including fibroblast growth factor (FGF), hepatocyte growth factor (HGF), and vascular endothelial growth factor (VEGF) and thereby modulates the availability and biological activities of these growth factors (2). The binding of growth factors to ECM components, particularly heparan sulfate proteoglycans (HSPGs), is believed to protect them against proteolytic degradation, concentrate their activity in the vicinity of cells, and allow their rapid release during wound healing and tissue remodeling (2). The activities of such growth factors bound to HSPGs are considered to be regulated by heparanase, which cleaves heparan sulfate chains and consequently releases the growth factors from the ECM (3).
The interactions between growth factors and ECM components can be categorized into protein-GAG interactions and protein-protein interactions. For example, growth factors including FGF, HGF, and VEGF bind to HSPGs in the ECM via heparan sulfate chains (4–6) while transforming growth factor (TGF)-β1 interacts with fibronectin (7) and thrombospondin (8). Similarly, thrombospondin, fibronectin and collagen are binding partners of HGF in the ECM (9, 10). Type IV collagen binds to bone morphogenetic protein (BMP) and regulates its signaling events in Drosophila (11). The TGF-β family members play central roles in the regulation of multiple physiological processes such as cell differentiation, mitogenesis, embryogenesis, apoptosis, and inflammation (12). Many, if not all, TGF-β family members are produced as latent complexes of the mature dimeric growth factor and its pro-region (13). The pro-region of TGF-β1, which is known to be a latency-associated peptide, forms two disulfide bonds with latent TGF-β-binding proteins (LTBPs), thus targeting the latent TGF-β1 complexes to the ECM. The resulting latent TGF-β-LTBP complexes bind to fibrillins and fibronectins, and consequently facilitate the ECM targeting of TGF-β1. Fibrillin-1 has also been shown to bind to the pro-regions of BMP-2, -4, -7, and -10 at its N-terminal region (14), thereby sequestering a panel of TGF-β family proteins in the ECM. These observations indicate the importance of the pro-regions of TGF-β family proteins for their ECM deposition.
Activin A, a member of the TGF-β family, is a disulfide-linked homodimer of inhibin βA. The inhibin βA-subunit is synthesized as a precursor and processed by a furin protease to give an N-terminal pro-region and a C-terminal mature βA region (15). Activin A participates not only in the release of follicle-stimulating hormone from anterior pituitary cells (16) but also in the regulation of development, mitogenesis, apoptosis, and wound repair (17–20). Activin A has also been used to maintain the pluripotency of embryonic stem cells (21) and induce their differentiation into defined cell lineages (22). By analogy to TGF-β1 and some BMPs, the biological activity of activin A may also be modulated by the ECM, although the interactions of activin A with ECM components remain unexplored. In Xenopus, activin has been shown to act as a long-range dorsalizing signal to establish a concentration gradient (23), suggesting the potential associations of Xenopus activin with ECM components. In this study, we examined the interactions of activin A with a panel of ECM proteins as a first step toward addressing the roles of the ECM in the regulation of activin A function and tissue distribution. We found that recombinant activin A produced in human 293-F cells specifically bound to perlecan and agrin through its pro-region that was capable of binding to heparin/heparan sulfate chains. We mapped the regions responsible for the binding to perlecan to clusters of basic amino acid residues, Lys259-Lys263 and Lys270-Lys272, in the pro-region, although these residues were dispensable for the Smad-dependent signaling activity of activin A.
EXPERIMENTAL PROCEDURES
Chemical Reagents and Antibodies
KOD plus DNA polymerase was purchased from Toyobo (Osaka, Japan). Human follistatin was purchased from BioVision (Mountain View, CA). Bovine activin A was obtained from Wako (Osaka, Japan). Monoclonal antibodies (mAbs) against activin A (MAB3381) and human follistatin (MAB669) were obtained from R&D Systems (Minneapolis, MN). A mouse anti-perlecan mAb (7B5) was obtained from Zymed Laboratories Inc. (South San Francisco, CA). A horseradish peroxidase (HRP)-conjugated anti-pentaHis mAb and nickel-nitrilotriacetic (Ni-NTA) agarose were purchased from Qiagen (Piscataway, NJ). Heparin and hyaluronidase were obtained from Sigma. Chondroitinase ABC (Proteus vulgaris), heparitinase (Flavobacterium heparinum), and mAbs against heparan sulfate (3G10 and 10E4) were obtained from Seikagaku Kogyo (Tokyo, Japan). Heparin-Sepharose CL-6B dry resin was purchased from GE Healthcare (Piscataway, NJ).
ECM Proteins
Recombinant human laminins, agrin, nidogen 1, nidogen 2, and nephronectin were produced using a FreeStyleTM 293 Expression System (Invitrogen, Carlsbad, CA) and purified from conditioned medium by immunoaffinity chromatography or Ni-NTA agarose affinity chromatography. Perlecan was purified from conditioned medium of JAR human choriocarcinoma cells by immunoaffinity chromatography using an anti-perlecan mAb (kindly provided by Dr. Masahiko Katayama, Eisai Co. Ltd., Tsukuba, Japan). Recombinant perlecan was also expressed and purified as described below. Fibronectin was purified from outdated human plasma by gelatin affinity chromatography (24). Type I collagen was extracted from calf skin using 50 mm acetic acid and purified by salt fractionation under acidic and neutral conditions (25). Type IV collagen was extracted from bovine lens capsule using 0.5 m acetic acid (26).
Construction of Expression Vectors
pSecTag 2C (Invitrogen) was used to construct expression vectors for human activin A and its mutants. A full-length cDNA encoding the human inhibin βA subunit (GeneBankTM accession number: BC007858) was purchased from Open Biosystems (Huntsville, AL) and inserted into pSecTag 2C to produce the expression vector pSecTag-Act. Expression vectors for activin A with a His6 tag at either the N- or C-terminal end and a panel of activin A mutants with amino acid substitutions or deletions were prepared as follows: forward and reverse primers flanking the sites to be mutated were used to amplify pSecTag-Act by polymerase chain reaction (PCR), and the resulting linearized pSecTag-Act DNA fragments with added, deleted, or mutated nucleotides at their 3′- or 5′-ends were ligated again to obtain expression vectors for the individual mutants. A His6 tag was also added to the N terminus of the pro-region of the βA subunit (between Ser21 and Pro22) and the C terminus of the βA subunit. The nucleotide sequences of the resulting expression vectors were verified by DNA sequencing. A full-length cDNA encoding human perlecan was constructed from a human fetus cDNA library (Clontech, Palo Alto, CA) by PCR and inserted into pSecTag 2B (Invitrogen) between the HindIII and NotI sites with a His6 tag at its C terminus.
Expression and Purification of Recombinant Proteins
Recombinant activin A and its mutants were expressed and purified using the FreeStyleTM 293 Expression System according to the manufacturer's instructions. Briefly, 3 × 107 293-F cells were transfected with expression vectors and cultured for 3 days at 37 °C. The medium was initially centrifuged at 1,000 rpm for 5 min to remove the cells and then centrifuged at 10,000 rpm for 10 min. The supernatant was mixed with 1 ml of Ni-NTA agarose and incubated overnight at 4 °C with gentle agitation. Next, the Ni-NTA agarose was packed into a column and extensively washed with TBS (50 mm Tris-HCl, pH 7.4, 150 mm NaCl) containing 40 mm imidazole and 0.5 m NaCl. Activin A was eluted from the column with 0.2 m imidazole and dialyzed against phosphate-buffered saline (PBS). The purity of the purified activin A was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% gel, followed by silver staining. Purified activin A and its mutants were quantified by ELISA using bovine activin A as a standard. Expression and purification of recombinant perlecan were performed as described for activin A. Immunoblotting was carried out using specific mAbs and signals were detected with a chemiluminescence reagent (ECL; GE Healthcare) according to the manufacturer's protocol.
ECM Protein Binding Assays
96-well plates (MaxiSorpTM; Nunc, Roskilde, Denmark) were coated with individual ECM proteins at 10 nm overnight at 4 °C, followed by washing with TBS and blocking with 3% bovine serum albumin (BSA). The plates were then incubated with 50 nm recombinant activin A at room temperature for 1 h. After three washes with TBS containing 0.05% Tween-20, the bound recombinant activin A was detected with the HRP-conjugated anti-pentaHis mAb.
GAG Binding Assays
Phosphatidylethanolamine-conjugated chondroitin sulfate-A, chondroitin sulfate-C, chondroitin sulfate-D, chondroitin sulfate-E, dermatan sulfate, hyaluronic acid, heparan sulfate, and heparin were synthesized as described (27) and coated on 96-well MaxiSorpTM plates at 20 μg/ml overnight at 4 °C, followed by blocking with 3% BSA for 1 h at room temperature. The plates were then incubated with 50 nm recombinant activin A at room temperature for 1 h. After three washes with TBS containing 0.05% Tween-20, the bound recombinant activin A was detected with the HRP-conjugated anti-PentaHis mAb.
Chondroitinase and Heparitinase Treatments
Perlecan purified from JAR cells was coated on 96-well MaxiSorpTM plates at 10 nm overnight at 4 °C, followed by blocking with 3% BSA for 1 h. After washing with TBS, chondroitinase ABC (20 mU) or heparitinase (5 mU) was added, and the digestions were conducted in 50 mm Tris-HCl (pH 8.0) containing 25 mm sodium acetate and 5 mm CaCl2 at 37 °C for 3 h. The enzymes were removed by washing with TBS, and the binding activities of the enzyme-treated perlecan toward recombinant activin A were quantified by solid-phase binding assays as described above.
Pull-down Assay of Activin A Binding to Heparin
Fifteen microliters of recombinant activin A (300 ng) or bovine mature activin A (300 ng) was mixed with 10 μl of heparin-Sepharose beads and incubated on ice for 30 min. Unbound activin A was separated from the heparin-Sepharose beads by centrifugation at 10,000 rpm for 1 min. The heparin-Sepharose beads were washed three times with TBS, and the activin A bound to the beads was recovered in 15 μl of 2× SDS-PAGE sample loading buffer. The amounts of activin A in the supernatant and those recovered from the heparin-Sepharose beads were detected by SDS-PAGE using a 12% gel followed by silver staining.
Luciferase Assay of Activin Activity
The activin-responsive luciferase reporter plasmid p3TP-lux (Addgene, Cambridge, MA) and plasmid pSVβGal (Promega, Madison, WI) were used for activin A activity assays after transfection into Chinese hamster ovary (CHO) DG44 cells (28). The CHO cells were plated on 24-well plates (Falcon, San Jose, CA) at 150,000 cells/well and cultured in α-MEM containing 10% fetal bovine serum. After 24 h, the cells were transfected with p3TP-lux (600 ng) and pSVβGal (200 ng) using Lipofectamine LTX (Invitrogen). At 24 h after the transfection, the cells were incubated with increasing concentrations of recombinant activin A or bovine activin A for 20 h in α-MEM containing 0.1% fetal bovine serum. The activity of the induced luciferase was measured and normalized by the activity of β-galactosidase using a Dual-Light® System (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Immunohistochemistry
Serial sections of mouse embryos at embryonic day 16.5 (E16.5) were air-dried, fixed with acetone for 30 min and digested with chondroitinase ABC (5 U/ml) and hyaluronidase (1 U/ml) for 30 min at 37 °C. The sections were then incubated with recombinant activin A (5 μg/ml) in PBS containing 0.5% goat serum and HRP-conjugated anti-pentaHis mAb (1:100) at 4 °C overnight, followed by visualization of the bound HRP-conjugated mAbs with a DAKO Envision Kit (K3466; DAKO, Milan, Italy). For immunohistochemical detection of heparan sulfate chains, the sections were incubated with anti-heparan sulfate mAb 10E4 (1:500) in PBS containing 0.5% goat serum overnight at 4 °C. The sections were then incubated with HRP-conjugated goat anti-mouse IgG, followed by visualization of the bound antibodies with a DAKO Envision Kit. All sections were counterstained with hematoxylin.
RESULTS
Recombinant Activin A Binds to Perlecan
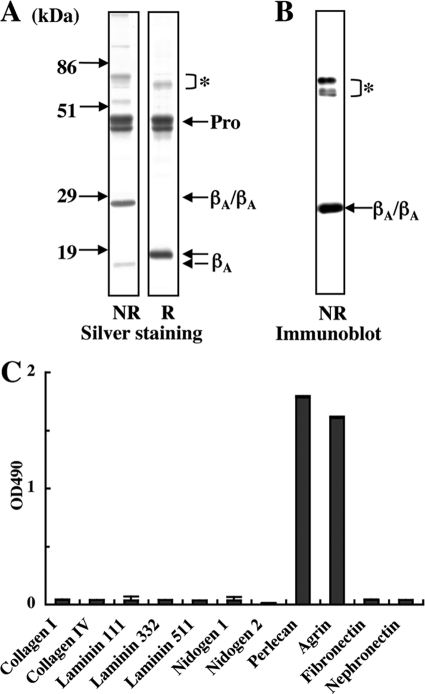
Activin A was expressed in 293-F cells with a C-terminal His6 tag and purified using Ni-NTA-agarose. After SDS-PAGE under non-reducing conditions, recombinant activin A produced a 25-kDa band that corresponded to the disulfide-linked mature activin A (“βA/βA”) and two closely spaced bands in the 44–46 kDa region that corresponded to the pro-region (Fig. 1A). The 25-kDa band was recognized by MAB3381, a function-blocking anti-activin A mAb that recognizes a conformation-dependent epitope on mature activin A (Fig. 1B). Under reducing conditions, the mature activin A produced a 15 kDa band, thus endorsing its disulfide-linked dimeric nature. Amino acid sequencing of the two major bands migrating in the 44–46-kDa region revealed that both had the same N-terminal sequence, SPTPG, which was identical to the deduced N-terminal sequence of the pro-region. The mass difference between these two bands could be caused by differential N-glycosylation and/or partial protease degradation. These results indicated that recombinant activin A expressed in 293-F cells was properly processed to yield a mature dimer that remained in a complex with its pro-region.
FIGURE 1.
Purification of recombinant activin A and its interactions with ECM proteins. A, SDS-PAGE of purified human recombinant activin A under non-reducing (NR) and reducing (R) conditions. Mature dimeric activin A (βA/βA) is purified as complexes with its pro-region (Pro). The precursor proteins that failed to be processed by a furin protease are indicated by the asterisk. B, immunoblot analysis of purified recombinant activin A with the conformational epitope-specific mAb MAB3381. C, solid-phase binding assays of recombinant activin A toward a panel of ECM proteins. ECM proteins were coated at 10 nm and incubated with 50 nm recombinant activin A. Bound activin A was quantified as described under “Experimental Procedures.” Each column and bar represent the mean and S.D. of triplicate assays, respectively.
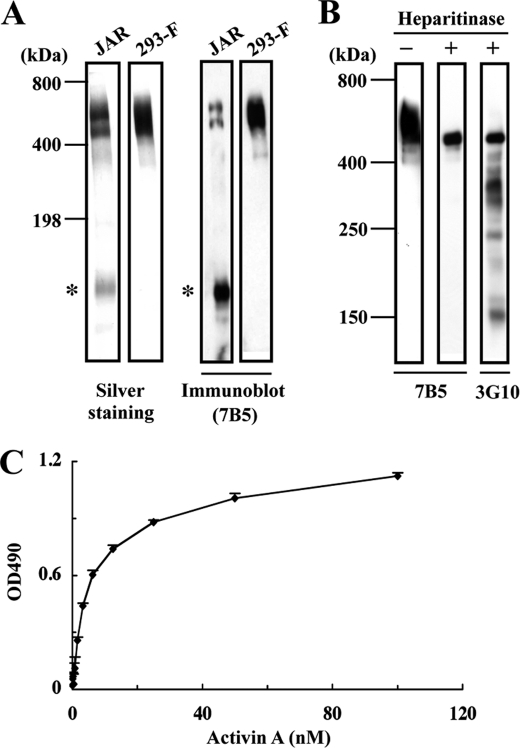
The ability of recombinant activin A to bind to the ECM was surveyed using a panel of ECM proteins including types I and IV collagens, laminin-111, -332, and -511, nidogens 1 and 2, perlecan, agrin, nephronectin, and fibronectin by solid-phase binding assays. Among these ECM proteins, recombinant activin A only bound to perlecan and agrin (Fig. 1C). None of the laminin isoforms examined, i.e. laminin-121, -211, -221, -411, -421, and -521, showed any affinities toward recombinant activin A (data not shown). The ECM proteins tested were recombinant proteins expressed in 293-F cells except for perlecan (purified from conditioned medium of JAR human choriocarcinoma cells), types I and IV collagens (extracted from calf skin and bovine lens, respectively) and fibronectin (purified from human plasma). To exclude the possibility that the binding of recombinant activin A to perlecan was caused by contaminants present in the JAR-derived perlecan, we produced recombinant human perlecan with a C-terminal His6 tag using 293-F cells and purified it with Ni-NTA agarose. The recombinant perlecan had a molecular weight similar to that of perlecan from JAR cells and was recognized by anti-perlecan mAb 7B5 (Fig. 2A). The attachment of heparan sulfate chains to the recombinant perlecan was verified by heparitinase treatment. Upon SDS-PAGE, the heparitinase-treated recombinant perlecan exhibited a thinner band with a decreased molecular mass (Fig. 2B). The presence of heparan sulfate chains was further corroborated by the reactivity of mAb 3G10, which specifically recognizes heparitinase-cleaved heparan sulfate chains (29), toward the heparitinase-treated recombinant perlecan (Fig. 2B). The recombinant perlecan bound to activin A in a dose-dependent manner (Fig. 2C), confirming the specific binding of recombinant activin A to perlecan.
FIGURE 2.
Binding of activin A to recombinant perlecan. A, SDS-PAGE and immunoblot analyses of human perlecan purified from the culture supernatants of JAR human choriocarcinoma cells and 293-F cells transfected with a full-length human perlecan cDNA. Both recombinant and JAR-derived perlecans produce bands migrating in the 500–600 kDa region that are recognized by an anti-perlecan mAb (7B5). A proteolytically degraded fragment (asterisks) is detected in JAR-derived perlecan but not recombinant perlecan. B, heparitinase treatment of recombinant perlecan. Recombinant perlecan was treated with heparitinase, followed by immunoblotting with anti-perlecan mAb 7B5 or with mAb 3G10, which recognizes a neo-epitope derived from heparan sulfate chains after heparitinase digestion. C, binding of activin A to recombinant perlecan. Recombinant perlecan was coated at 5 nm and incubated with increasing concentrations of recombinant activin A. Activin A bound to perlecan was quantified as described under “Experimental Procedures.” Each column and bar represent the mean and S.D. of triplicate assays, respectively.
Recombinant Activin A Binds to the Heparan Sulfate Chains of Perlecan
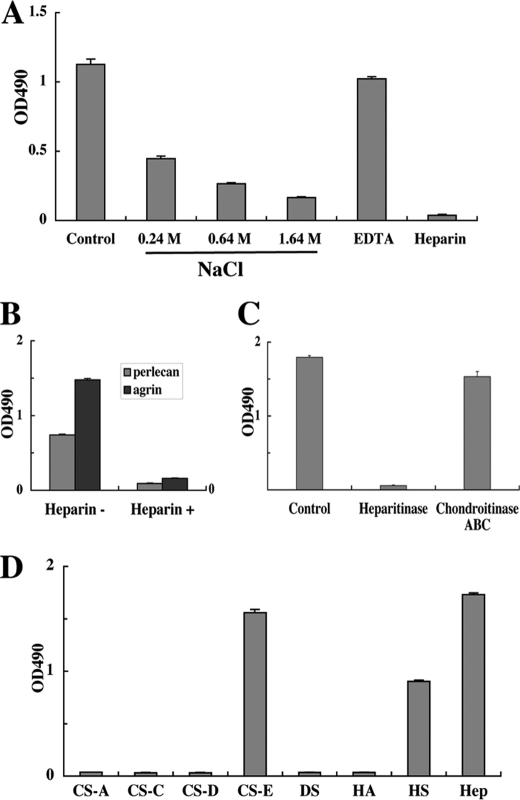
Because both perlecan and agrin are proteoglycans that contain heparan sulfate chains, which have been shown to bind many proteins including growth factors (30), we examined whether the heparan sulfate chains of perlecan were involved in the interaction with recombinant activin A. As shown in Fig. 3A, binding of recombinant activin A to perlecan was inhibited by increasing concentrations of NaCl, reaching >80% inhibition in the presence of 1.64 m NaCl. EDTA did not show any significant inhibitory effects. However, heparin at a concentration of 10 μg/ml completely abrogated the binding of recombinant activin A to perlecan. The strong inhibition by heparin was further confirmed by the binding of recombinant activin A to recombinant perlecan and agrin (Fig. 3B). These results indicated that the interactions of recombinant activin A with perlecan occur via the heparan sulfate chains on perlecan and are predominantly of an ionic nature.
FIGURE 3.
Involvement of heparan sulfate chains in activin A binding to perlecan. A, inhibitory effects of high salt concentrations, the chelating agent EDTA and heparin on the perlecan-binding activity of activin A. Recombinant activin A (50 nm) was incubated in 96-well microtiter plates coated with JAR-derived perlecan (10 nm) in the presence of increasing concentrations of NaCl (0.24–1.64 m), EDTA (5 mm), or heparin (10 μg/ml) for 1 h, followed by quantification of bound activin A as described under “Experimental Procedures.” B, heparin (10 μg/ml) inhibits the binding of activin A to recombinant perlecan and agrin, both of which were expressed in 293-F cells. C, abrogation of the perlecan binding activity of activin A after heparitinase treatment of perlecan. JAR-derived perlecan coated on 96-well microtiter plates at 10 nm was treated with heparitinase or chondroitinase ABC at 37 °C for 3 h, followed by incubation with recombinant activin A (50 nm) for 1 h. D, solid-phase binding assays of activin A with a panel of GAGs derivatized with phosphatidylethanolamine (PE-GAGs). PE-GAGs were coated at 20 μg/ml overnight and incubated with recombinant activin A (50 nm) for 1 h. Recombinant activin A bound to perlecan was quantified as described under “Experimental Procedures.” CS-A, chondroitin sulfate-A; CS-C, chondroitin sulfate-C; CS-D, chondroitin sulfate-D; CS-E, chondroitin sulfate-E; DS, dermatan sulfate; HA, hyaluronic acid; HS, heparan sulfate; Hep, heparin. Each column and bar represent the mean and S.D. of triplicate assays, respectively.
To corroborate this conclusion, perlecan was treated with heparitinase or chondroitinase ABC. Treatment with chondroitinase ABC barely compromised the interaction of activin A with perlecan, whereas treatment with heparitinase resulted in an almost complete loss of the binding activity (Fig. 3C). The interactions of recombinant activin A with heparan sulfate and other GAGs were further investigated using an array of GAGs. Recombinant activin A preferentially bound to heparin, heparan sulfate and chondroitin sulfate-E, but did not bind to chondroitin sulfate-A, -C, and -D, dermatan sulfate or hyaluronic acid (Fig. 3D). These results provided further evidence that recombinant activin A binds to the heparan sulfate chains of perlecan.
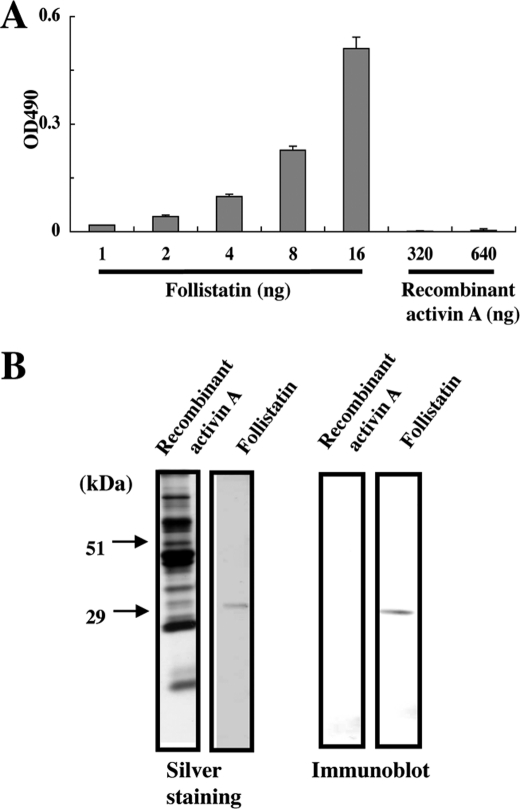
Follistatin is known to bind to activin A and other TGF-β family members. It also binds to heparan sulfate chains (31). It is therefore possible that the heparan sulfate-binding activity of recombinant activin A may be caused by follistatin co-purified with activin A. To address this possibility, we probed recombinant activin A coated on 96-well plates or electroblotted onto polyvinylidene fluoride membranes with an anti-follistatin mAb (Fig. 4, A and B). No follistatin signals were detected for recombinant activin A coated at 640 ng/well or loaded at 640 ng/lane, while control follistatin was detectable at concentrations as low as 1 ng/well, thus making it unlikely that the heparan sulfate-binding activity of recombinant activin A was caused by follistatin copurified with activin A.
FIGURE 4.
Detection of follistatin in recombinant activin A. A, detection of follistatin in recombinant activin A by ELISA. Recombinant activin A was coated on microtiter plates at 320 and 640 ng/well and probed with anti-follistatin mAb MAB669. Recombinant follistatin (1–16 ng/well) was analyzed by ELISA as a control. Each column and bar represent the mean and S.D. of triplicate assays, respectively. B, detection of follistatin in recombinant activin A by immunoblotting. Recombinant activin A and recombinant follistatin were loaded at 640 and 20 ng/lane, respectively, and detected with anti-follistatin mAb MAB669.
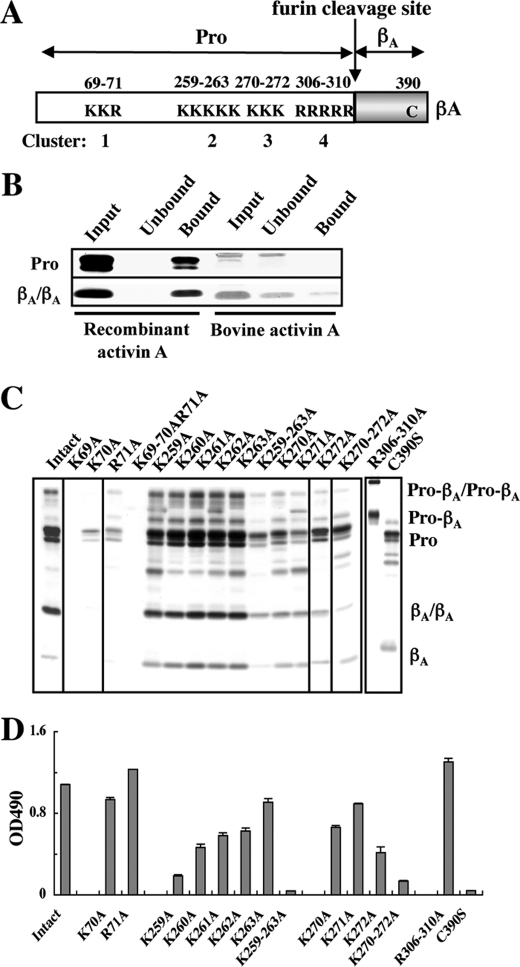
Clusters of Basic Amino Acid Residues in the Pro-region Are Responsible for Perlecan Binding
It has been reported that the binding sites for heparin/heparan sulfate chains are comprised of clusters of basic amino acid residues (32). There are four clusters of basic amino acid residues in the pro-region of the βA-subunit, comprising Lys69Lys70Arg71 (Cluster 1), Lys259Lys260Lys261Lys262Lys263 (Cluster 2), Lys270Lys271Lys272 (Cluster 3), and Arg306Arg307Arg308Arg309Arg310 (Cluster 4) (Fig. 5A), suggesting that recombinant activin A may bind to the heparan sulfate chains of perlecan through its pro-region. To explore the involvement of these basic amino acid clusters in the binding of recombinant activin A to perlecan, we first compared the heparin binding activity of recombinant activin A with that of bovine mature activin A without the pro-region by pull-down assays using heparin-Sepharose beads. SDS-PAGE analyses of the bound and unbound fractions from the pull-down assays revealed that the recombinant activin A was fully recovered in the bound fraction, while mature dimeric activin A was largely recovered in the unbound fraction (Fig. 5B). These results indicated that the pro-region harbors the major heparin-binding sites.
FIGURE 5.
Mapping of the perlecan-binding sites to basic amino acid clusters in the pro-region. A, schematic representation of the distributions of the clusters of basic amino acid residues in activin A. There are four clusters of basic amino acid residues, designated Clusters 1, 2, 3, and 4, in the pro-region (Pro). B, pull-down assays for the heparin-binding activity of activin A. Recombinant activin A and control bovine activin A that was free from the bound pro-region were incubated with heparin-Sepharose beads, followed by separation of the bound and unbound activin A by centrifugation and subsequent SDS-PAGE under non-reducing conditions. Proteins were visualized by silver staining. C, effects of amino acid substitutions within the basic amino acid clusters on the secretion of recombinant activin A. Equal volumes of the culture supernatants of 293-F cells that had been transfected with activin A mutants with amino acid substitutions were subjected to purification of the recombinant activin A mutants using Ni-NTA-agarose, followed by SDS-PAGE of equal volumes of the eluates. Proteins in the eluates were visualized by silver staining. D, solid-phase binding assays of the perlecan-binding activities of the activin A mutants. Activin A mutants with amino acid substitutions were incubated on microtiter plates coated with 10 nm JAR-derived perlecan for 1 h, followed by quantification of bound activin A as described under “Experimental Procedures.” Each column and bar represent the mean and S.D. of triplicate assays, respectively.
The involvement of the clusters of Lys/Arg residues in perlecan binding was investigated by alanine-scanning mutagenesis of these positively charged residues. Mutant proteins with individual or entire alanine substitutions at these basic amino acid clusters were expressed in 293-F cells and purified using Ni-NTA-agarose. Although individual alanine substitutions at Cluster 2 did not compromise the expressions and secretions of the mutant proteins, alanine substitutions at Cluster 1 resulted in severe reductions in the amounts of recombinant proteins recovered from Ni-NTA-agarose (Fig. 5C). Notably, we failed to obtain any detectable amount of the mutant with alanine substitution for Lys69. These results suggested that the basic amino acid residues in Cluster 1, particularly Lys69, play critical roles in the proper folding and/or subsequent secretion of activin A. On the other hand, amino acid substitutions at Clusters 3 and 4 exhibited only partial, if any, reductions in the amounts of the mutant proteins recovered from Ni-NTA-agarose, except that entire substitutions of all five arginine residues in Cluster 4 resulted in a failure of proteolytic processing by a furin protease into the pro-region and mature activin A.
The perlecan-binding activities of the mutant proteins were examined by solid phase binding assays (Fig. 5D). Entire substitutions of the basic amino acid residues in Clusters 2 and 3 caused ∼95 and ∼90% reductions in the perlecan binding activity, respectively. In contrast, entire substitutions of all five arginine residues in Cluster 4 did not compromise the activity. These results suggested that the basic residues in Clusters 2 and 3, but not those in Cluster 4, are critically involved in the perlecan-binding activity of recombinant activin A. Although the mutant with substitutions of all three basic residues in Cluster 1 failed to be expressed and its perlecan-binding activity could not be determined, individual substitutions at Lys70 and Lys71 showed no significant decreases in the perlecan binding activity, suggesting the possibility that Cluster 1 is not required for binding to perlecan.
Among the five mutant proteins with individual alanine substitutions at Cluster 2, the K259A mutant exhibited the most pronounced reduction in the perlecan-binding activity, while the K260A, K261A, and K262A mutants showed partial reductions and the K263A mutant showed only a moderate decrease in the activity. Among the three mutants with individual alanine substitutions in Cluster 3, the K272A mutant exhibited an ∼60% reduction in the perlecan-binding activity, while the K270A and K271A mutants showed ∼40 and ∼20% reductions, respectively. These results indicated that Lys259 in Cluster 2 and Lys272 in Cluster 3 are likely candidates that comprise the major heparan sulfate binding sites in the pro-region of activin A, although other residues in Clusters 2 and 3 also contribute to the heparan sulfate binding ability to variable extents.
Because activin A is expressed and secreted as a dimer, we next examined whether the dimer formation is required for perlecan binding by activin A. To this end, we produced a mutant protein in which Cys390 was substituted with serine to disrupt the disulfide bond required for dimer formation. The C390S mutant produced a 15-kDa band of the mature βA region upon SDS-PAGE under non-reducing conditions (Fig. 5C), confirming the requirement of Cys390 for the dimer formation. Solid-phase binding assays revealed that the C390S mutant was barely capable of binding to perlecan (Fig. 5D), thereby demonstrating the importance of the dimeric structure of activin A for binding to perlecan.
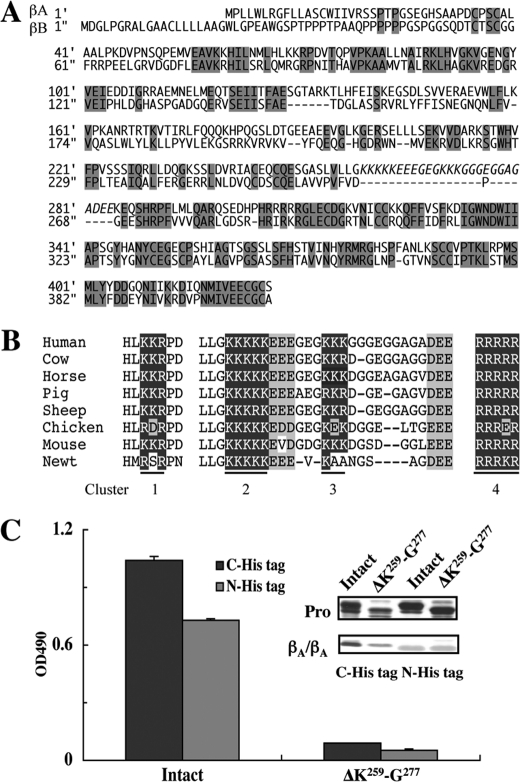
Deletion of the Heparan Sulfate Binding Sites Does Not Impair the Biological Activity of Activin A
The human inhibin βA- and βB-subunits share 39% amino acid sequence similarity. A comparison of the amino acid sequences of the βA- and βB-subunits revealed that the peptide segment Lys259-Glu284 containing Clusters 2 and 3 is absent from the βB-subunit (Fig. 6A). However, the amino acid sequence of this segment is highly conserved among different species (Fig. 6B). The significant conservation of the amino acid sequence of the Lys259-Glu284 segment together with its absence from the inhibin βB-subunit prompted us to hypothesize that this segment adopts an independent conformation and that the involvement of Clusters 2 and 3 in the heparan sulfate binding activity could be corroborated by deleting this segment from activin A. We produced a deletion mutant from which most of this segment including Clusters 2 and 3 (Lys259-Gly277) was deleted and examined its expression and perlecan-binding activity. As expected, this mutant was expressed and processed similar to the case for wild-type activin A, but was barely active in binding to perlecan (Fig. 6C).
FIGURE 6.
Abrogation of the perlecan binding activity of activin A by deletion of the inhibin βA subunit-specific region harboring Clusters 2 and 3. A, amino acid sequence alignment between the inhibin βA- and βB-subunits. The peptide segment Lys259-Glu284 containing Clusters 2 and 3 in the inhibin βA-subunit is absent from the inhibin βB-subunit (highlighted in italics). The conserved amino acid residues are shaded in gray. B, alignment of basic amino acid clusters in the amino acid sequences of the inhibin βA-subunits from different species. The amino acid sequence of the peptide segment Lys259-Glu284 harboring Clusters 2 and 3 is well conserved among the different species. The clusters of basic amino acid residues are shown in white letters on black whereas acidic amino acid residues are shaded in gray. C, the perlecan binding activities of recombinant activin A with a His6 tag at either the N terminus (N-His tag) or C terminus (C-His tag) and those with deletion of the Lys259-Gly277 segment (designated ΔK259-G277) were determined by solid-phase binding assays. The SDS-PAGE profiles of purified activin A with and without deletion of Clusters 2 and 3 are shown in the inset. Note that the pro-regions of intact and mutant activin A differ in their sizes because of the presence or absence of the Lys259-Gly277 segment.
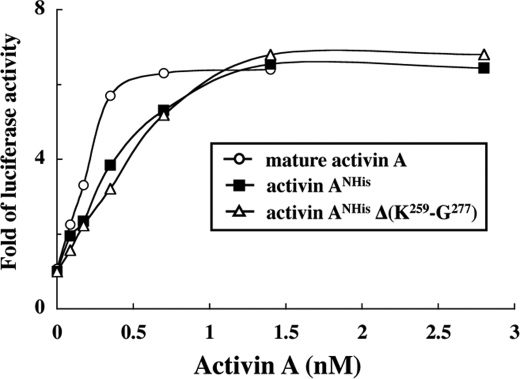
Next, we addressed whether the perlecan-binding activity of recombinant activin A is involved in the biological activities of activin A. To this end, we employed an activin-responsive luciferase reporter gene assay using CHO cells (28). As the recombinant activin A with a C-terminal His6 tag exhibited a low luciferase-inducing activity (data not shown), we produced another recombinant activin A with a His6 tag at the N terminus of the pro-region (designated activin ANHis). Activin ANHis was expressed well compared with the activin A with a C-terminal His6 tag, and retained the ability to bind to perlecan (Fig. 6C). The moderate decrease in its perlecan binding activity may be caused by differential effects of the position of the His6 tag on either the accessibility of the anti-His tag antibody or the binding affinity toward the heparan sulfate chains on perlecan. Deletion of the Lys259-Gly277 segment from activin ANHis abrogated its perlecan-binding activity (Fig. 6C), thus confirming the critical roles of Clusters 2 and 3 in the binding of activin A to perlecan.
Incubation of CHO cells transfected with the activin-responsive luciferase reporter gene with increasing concentrations of activin ANHis resulted in dose-dependent increases in the luciferase activity, reaching a maximum activity that was comparable to that of a commercially available mature activin A at >1.5 nm (Fig. 7). Activin ANHis exhibited lower luciferase activities than mature activin A at <1.5 nm, which mostly reflected the fact that only a fraction of activin ANHis gave rise to functionally active mature activin A upon dissociation, meaning that the effective concentration of mature activin A was significantly lower for recombinant activin A than for the mature activin A. Interestingly, the mutant activin ANHis lacking the Lys259-Gly277 segment was equally as potent as control activin ANHis, exhibiting the same dose-dependent luciferase activity (Fig. 7). These results indicated that the heparan sulfate-binding sites comprised of Clusters 2 and 3 in the pro-region are not required for receptor binding or subsequent signal transduction. Furthermore, neither the association with the pro-region nor the presence of the N-terminal His6 tag imposed deleterious effects on the biological activity of activin A.
FIGURE 7.
Smad-dependent signaling activities of recombinant activin A lacking Clusters 2 and 3. CHO cells cotransfected with an activin-responsive luciferase reporter gene and a β-galactosidase gene were stimulated with increasing concentrations of N-terminally His-tagged activin A with and without deletion of the Lys259-Gly277 segment (designated activin ANHis and activin ANHis(ΔK259-G277), respectively) or bovine activin A without the pro-region for 20 h in α-MEM containing 0.1% fetal bovine serum. The luciferase activities are expressed as the fold increases after normalization by the β-galactosidase activity. The results represent the means of duplicate determinations.
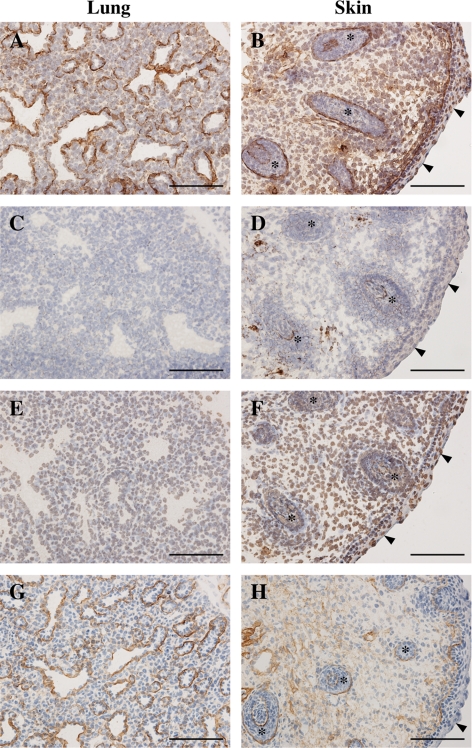
Activin A Binds to Heparan Sulfate in Tissues
Finally, we addressed the biological relevance of the heparan sulfate binding activity of the pro-region of activin A by in situ activin A overlay assays using frozen sections of mouse embryos, in which detection of bound activin A signals was facilitated by pretreatment of the tissue sections with chondroitinase ABC and hyaluronidase. A significant proportion of the signals for bound activin A were confined to the areas corresponding to the basement membranes of the lungs, hair follicles and epidermis (Fig. 8, A and B). The basement membrane regions of the retinas and kidneys were also positive for bound activin A signals (data not shown). Binding of activin A to the tissue sections was abrogated by pretreatment of the sections with heparitinase (Fig. 8, C and D), confirming that the binding of activin A to the tissues was heparan sulfate-dependent. The mutant activin A lacking the Lys259-Gly277 segment did not produce any significant signals for bound activin A (Fig. 8, E and F), consistent with the requirement of the basic amino acid clusters in the pro-region for the binding of activin A to the heparan sulfate chains of perlecan. Furthermore, the overall patterns of the activin A signals were reminiscent of those for mAb 10E4, which recognizes heparan sulfate chains (Fig. 8, G and H). Taken together, these results support the conclusion that activin A binds to heparan sulfate proteoglycans in basement membranes through its pro-region in a heparan sulfate-dependent manner.
FIGURE 8.
In situ activin A overlay assays on tissue sections. A–F, frozen sections of mouse E16.5 embryos were incubated with activin ANHis (A–D) or its mutant lacking the Lys259-Gly277 segment (E and F), followed by detection of the bound proteins by immunohistochemistry. Sections treated with heparitinase (C and D) were included in the overlay assays to confirm that the signals were heparan sulfate-dependent. G and H, additional sections were subjected to immunohistochemistry with anti-heparan sulfate mAb 10E4 to clarify the localizations of heparan sulfate chains. The signals for bound activin A are localized to the basement membranes of various organs including the lungs (A) and skin (B), where the epidermis and hair follicles are labeled with arrowheads and asterisks, respectively. Scale bar, 100 μm.
DISCUSSION
An important feature of activin A revealed in this study is that activin A specifically binds to perlecan and agrin among a variety of ECM proteins. Our data clearly show that activin A binds to perlecan through heparan sulfate chains. Consequently, the binding of activin A to perlecan was inhibited by high concentrations of NaCl and by heparin. Treatment with heparitinase, but not chondroitinase ABC, abolished the binding. Furthermore, activin A directly bound to heparin and heparan sulfate chains in solid-phase binding assays. To the best of our knowledge, this is the first report that a growth factor in the TGF-β family binds to HSPGs via its pro-region, although BMP-2 and BMP-4 have been reported to bind to HSPGs via the N-terminal regions of the mature growth factors (30, 33). Given that the pro-region of TGF-β1 binds to LTBPs (13) and those of BMP-2, -4, -7, and -10 bind to fibrillins (14), the pro-regions of TGF-β family proteins have distinctive specificities to localize these growth factors at target sites in the ECM and regulate their activities in a spatiotemporal manner. It should be noted that perlecan and agrin are major basement membrane components that are widely expressed in both embryonic and adult tissues (34–36). In embryogenesis, basement membranes serve as structural as well as functional interfaces of epithelial-mesenchymal interactions, through which many organs develop. Given the role of activin A as a morphogen, the associations of activin A with embryonic basement membranes through perlecan and agrin may be instrumental in regulating the morphogenetic activity of activin A during embryonic development. In support of this possibility, we demonstrated that activin A predominantly bound to the basement membrane regions of various tissues in a heparan sulfate-dependent manner.
The interactions of proteins with heparin/heparan sulfate chains are mainly contributed by ionic interactions between the negatively charged sulfate groups and positively charged amino and guanidinium groups of lysine and arginine residues, and in some cases are mediated by Ca2+ coordination (32). The strong inhibition of activin A binding to perlecan by high concentrations of NaCl and scarce inhibition by chelation of divalent cations with EDTA indicated that ionic interactions are more critical for the binding of recombinant activin A to perlecan. Because the binding of recombinant activin A to perlecan was not completely blocked at the highest concentration of NaCl examined (1.64 m), other types of chemical bonds such as hydrogen bonds and hydrophobic interactions may partially contribute to the interactions between recombinant activin A and perlecan (32). It should be noted that not only heparin and heparan sulfate but also chondroitin sulfate-E bound to activin A among the panel of GAGs tested in this study. The failures of chondroitin sulfate-A, -C, and -D, dermatan sulfate, hyaluronic acid, and highly sialylated nephronectin (37) to bind to activin A suggest that the interactions are not simply electrostatic and possibly require two closely positioned sulfate groups on repeating disaccharide units. Among the different forms of chondroitin sulfates, only chondroitin sulfate-E contains N-acetylgalactosamine with two sulfate groups at carbons 4 and 6. Consistent with these results, various heparin/heparan sulfate-binding growth factors including midkine, pleiotrophin, FGF-2, FGF-10, FGF-16, FGF-18, and heparin-binding epidermal growth factor are capable of binding to chondroitin sulfate-E (38). Although chondroitin sulfate-E is a minor component of mammalian chondroitin sulfate chains (39), it may contribute to the ECM deposition of activin A and the regulation of its biological functions.
Our data show that the basic amino acid clusters in the pro-region, which are well conserved among different species, function differently. Cluster 1 (Lys69-Lys70-Arg71) is critically required for secretion, possibly because of its involvement in the proper formation of the three-dimensional structure, while Clusters 2 (Lys259-Lys263) and 3 (Lys270-Lys272) contribute to perlecan binding. Cluster 4 serves as a recognition site for proteolytic processing by a furin protease. All five basic amino acid residues in Cluster 2 are fully conserved among different species, while Lys271 in Cluster 3 is replaced by Glu in the chicken and Lys271 and Lys272 are replaced by Ala in the newt (see Fig. 6B). The stringent conservation of the stretch of five Lys residues in Cluster 2 implies a critical role of Cluster 2 in heparan sulfate binding. The requirement for Clusters 2 and 3 in the heparan sulfate binding was further demonstrated by the deletion experiment in which the segment containing both Clusters 2 and 3 was deleted from the pro-region. There are five isoforms of inhibin β subunits, namely βA, βB, βC, βD, and βE (40). Among these, the βA- and βB-subunits share 39% amino acid sequence similarity. The Lys259-Glu284 segment that contains Clusters 2 and 3 is βA-subunit-specific and does not show any significant homology to other proteins including those with heparin binding activities (32). Because the deletion of a large part of this segment did not impair the growth factor activity of activin A, it is tempting to speculate that the Lys259-Glu284 segment adopts an independent conformation within the pro-region and that the βA-subunit could have acquired this extra segment during evolution to endow the binding activity toward heparan sulfate chains. Alternatively, this peptide segment could have been removed from the inhibin βB-subunit to attenuate the binding to heparan sulfate chains. It is interesting to note that there are two conserved clusters of negatively charged amino acid residues in the extra segment, which may regulate the heparan sulfate binding activity by antagonizing the positive electrostatic potential within the same segment.
The growth factors of the TGF-β family are synthesized as precursors and most of them are secreted as complexes of the N-terminal pro-region and the C-terminal mature growth factor (41). The pro-regions not only facilitate proper folding of the mature growth factors and their subsequent secretion from cells (15) but also confer latency on TGF-β1 (41) and growth differentiation factor-8 (42) and target TGF-β1, BMP-2, -4, -7, and -10 to the ECM (14). The latent TGF-β1 complexes are activated by binding to thrombospondin-1 (43) and integrin αvβ6 (44) and also by proteolytic cleavage of the pro-region (45, 46). The bioactivities of growth differentiation factor-8 and BMP-4 are also regulated by proteolytic cleavage of their pro-regions (45, 47–49). In contrast, the pro-regions of BMP-7 and BMP-9 form stable complexes with the mature growth factors but do not confer latency on the complexes (50, 51). Our results showed that activin A expressed in 293-F cells was purified as complexes with its pro-region but was functionally active in its complex form, as has been observed for BMP-7 and BMP-9 (50, 51). The failure of the pro-region to render activin A inactive may be caused by its relatively low binding affinity toward the mature growth factor (52), thus allowing activin A receptors on the cell surface to compete with the pro-region for the mature ligand. Consistent with this possibility, recombinant activin A exhibited lower biological activities at concentrations of <1.5 nm than bovine mature activin A that was free from a pro-region.
Dimerization of activin A is essential for its biological activity, since monomerization of activin A significantly decreases its receptor binding affinity as well as its activity to enhance the release of follicle-stimulating hormone (53). Our results showed that the activin A mutant C390S, which was unable to form a dimer, was devoid of the perlecan binding activity, thereby indicating that dimerization was also required for its heparan-sulfate binding activity. This finding is in striking contrast to many other heparin/heparan sulfate-binding growth factors including midkine, pleiotrophin, FGF-2, FGF-10, FGF-16, FGF-18, and heparin-binding epidermal growth factor, which can bind to heparin/heparan sulfates as monomers. The requirement for the dimeric structure of activin A may be caused by the relatively low heparan sulfate-binding activity of monomeric activin A, which necessitates bivalency for stable binding to heparan sulfate chains. Alternatively, the binding site for the heparan sulfate chains may be formed at the interface of the two pro-regions, thus requiring the juxtaposition of two monomers. The prerequisite of the dimeric structure for perlecan binding may also serve as a fail-safe quality control that rejects improperly folded monomeric activin A and prevents its deposition in the ECM near the target cells.
The negatively charged heparan sulfate chains that are covalently attached to perlecan, syndecan, agrin, and β-glycan have been shown to participate in the regulation of cell signaling and morphogenesis via binding to a variety of growth factors and/or their receptors (2, 54). Binding of heparan sulfate to FGF-2 stabilizes the complex formation with its receptor and prolongs its activity (55, 56). Release of the VEGF isoforms V165 and V189 from HSPGs by plasmin enables them to act on endothelial cells, leading to enhancement of vascular permeability (5). Regarding the TGF-β family, cell surface HSPGs mediate BMP-2 internalization and modulate BMP-2 osteogenic activity (57), while endogenous heparan sulfate chains modulate BMP-4 signaling and activity (58). The consequences of the sequestration of growth factors in the ECM and their subsequent regulated release are to prolong their action, facilitate their localization to the environment immediately adjacent to their target cells and tune their biological activity (56). Because deletion of the heparan sulfate-binding sites from recombinant activin A did not impair its biological activity, it is conceivable that the HSPG-binding activity of activin A facilitates the deposition of activin A in the ECM but does not regulate the growth factor activity per se. The physiological relevance of the heparan sulfate binding by activin A remains to be addressed in the context of histogenesis and organogenesis using animal models in which the heparan sulfate binding activity of activin A is ablated in particular cell lineages.
Acknowledgment
We thank Dr. Masahiko Katayama, Eisai Co. Ltd., for the generous gift of an anti-perlecan mAb.
This study was supported by Research Contract No. 06001294-0 with the New Energy and Industrial Technology Development Organization of Japan.
- ECM
- extracellular matrix
- GAG
- glycosaminoglycan
- HGF
- hepatocyte growth factor
- LTBP
- latent TGF-β-binding protein
- BMP
- bone morphogenetic protein
- HSPG
- heparan sulfate proteoglycan
- mAb
- monoclonal antibody
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1.Nelson C. M., Bissell M. J. (2006) Annu. Rev. Cell Dev. Biol. 22, 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macri L., Silverstein D., Clark R. A. (2007) Adv. Drug. Deliv. Rev. 59, 1366–1381 [DOI] [PubMed] [Google Scholar]
- 3.Vlodavsky I., Friedmann Y. (2001) J. Clin. Invest. 108, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asada M., Shinomiya M., Suzuki M., Honda E., Sugimoto R., Ikekita M., Imamura T. (2009) Biochim. Biophys. Acta 1790, 40–48 [DOI] [PubMed] [Google Scholar]
- 5.Houck K. A., Leung D. W., Rowland A. M., Winer J., Ferrara N. (1992) J. Biol. Chem. 267, 26031–26037 [PubMed] [Google Scholar]
- 6.Lyon M., Deakin J. A., Mizuno K., Nakamura T., Gallagher J. T. (1994) J. Biol. Chem. 269, 11216–11223 [PubMed] [Google Scholar]
- 7.Mooradian D. L., Lucas R. C., Weatherbee J. A., Furcht L. T. (1989) J. Cell. Biochem. 41, 189–200 [DOI] [PubMed] [Google Scholar]
- 8.Schultz-Cherry S., Murphy-Ullrich J. E. (1993) J. Cell Biol. 122, 923–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamszus K., Joseph A., Jin L., Yao Y., Chowdhury S., Fuchs A., Polverini P. J., Goldberg I. D., Rosen E. M. (1996) Am. J. Pathol. 149, 805–819 [PMC free article] [PubMed] [Google Scholar]
- 10.Schuppan D., Schmid M., Somasundaram R., Ackermann R., Ruehl M., Nakamura T., Riecken E. O. (1998) Gastroenterology 114, 139–152 [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Harris R. E., Bayston L. J., Ashe H. L. (2008) Nature 455, 72–77 [DOI] [PubMed] [Google Scholar]
- 12.Hogan B. L. (1996) Curr. Opin. Genet. Dev. 6, 432–438 [DOI] [PubMed] [Google Scholar]
- 13.Saharinen J., Hyytiäïnen M., Taipale J., Keski-Oja J. (1999) Cytokine Growth Factor Rev. 10, 99–117 [DOI] [PubMed] [Google Scholar]
- 14.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray A. M., Mason A. J. (1990) Science 247, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 16.Vale W., Rivier J., Vaughan J., McClintock R., Corrigan A., Woo W., Karr D., Spiess J. (1986) Nature 321, 776–779 [DOI] [PubMed] [Google Scholar]
- 17.Matzuk M. M., Kumar T. R., Shou W., Coerver K. A., Lau A. L., Behringer R. R., Finegold M. J. (1996) Recent Prog. Horm. Res. 51, 123–154, discussion 155–157 [PubMed] [Google Scholar]
- 18.Spencer S. J., Rabinovici J., Mesiano S., Goldsmith P. C., Jaffe R. B. (1992) J. Clin. Invest. 90, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishihara T., Okahashi N., Ueda N. (1993) Biochem. Biophys. Res. Commun. 197, 985–991 [DOI] [PubMed] [Google Scholar]
- 20.Munz B., Tretter Y. P., Hertel M., Engelhardt F., Alzheimer C., Werner S. (2001) Mol. Cell. Endocrinol. 180, 169–177 [DOI] [PubMed] [Google Scholar]
- 21.Beattie G. M., Lopez A. D., Bucay N., Hinton A., Firpo M. T., King C. C., Hayek A. (2005) Stem. Cells 23, 489–495 [DOI] [PubMed] [Google Scholar]
- 22.Asashima M., Michiue T., Kurisaki A. (2008) Dev. Growth Differ. 50, Suppl. 1, S35–S45 [DOI] [PubMed] [Google Scholar]
- 23.McDowell N., Zorn A. M., Crease D. J., Gurdon J. B. (1997) Curr. Biol. 7, 671–681 [DOI] [PubMed] [Google Scholar]
- 24.Sekiguchi K., Hakomori S., Funahashi M., Matsumoto I., Seno N. (1983) J. Biol. Chem. 258, 14359–14365 [PubMed] [Google Scholar]
- 25.Sato K., Ebihara T., Adachi E., Kawashima S., Hattori S., Irie S. (2000) J. Biol. Chem. 275, 25870–25875 [DOI] [PubMed] [Google Scholar]
- 26.Iwata M., Imamura Y., Sasaki T., Hayashi T. (1995) J. Biochem. 117, 1298–1304 [DOI] [PubMed] [Google Scholar]
- 27.Sugiura N., Sakurai K., Hori Y., Karasawa K., Suzuki S., Kimata K. (1993) J. Biol. Chem. 268, 15779–15787 [PubMed] [Google Scholar]
- 28.Arai K. Y., Tsuchida K., Li C., Watanabe G., Sugino H., Taya K., Nishiyama T. (2006) Protein Expr. Purif. 49, 78–82 [DOI] [PubMed] [Google Scholar]
- 29.David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitelock J. M., Melrose J., Iozzo R. V. (2008) Biochemistry 47, 11174–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Innis C. A., Hyvönen M. (2003) J. Biol. Chem. 278, 39969–39977 [DOI] [PubMed] [Google Scholar]
- 32.Gandhi N. S., Mancera R. L. (2008) Chem. Biol. Drug. Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 33.Ohkawara B., Iemura S., ten Dijke P., Ueno N. (2002) Curr. Biol. 12, 205–209 [DOI] [PubMed] [Google Scholar]
- 34.Manabe R., Tsutsui K., Yamada T., Kimura M., Nakano I., Shimono C., Sanzen N., Furutani Y., Fukuda T., Oguri Y., Shimamoto K., Kiyozumi D., Sato Y., Sado Y., Senoo H., Yamashina S., Fukuda S., Kawai J., Sugiura N., Kimata K., Hayashizaki Y., Sekiguchi K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12849–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murdoch A. D., Liu B., Schwarting R., Tuan R. S., Iozzo R. V. (1994) J. Histochem. Cytochem 42, 239–249 [DOI] [PubMed] [Google Scholar]
- 36.Groffen A. J., Buskens C. A., van Kuppevelt T. H., Veerkamp J. H., Monnens L. A., van den Heuvel L. P. (1998) Eur. J. Biochem. 254, 123–128 [DOI] [PubMed] [Google Scholar]
- 37.Brandenberger R., Schmidt A., Linton J., Wang D., Backus C., Denda S., Müller U., Reichardt L. F. (2001) J. Cell Biol. 154, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deepa S. S., Umehara Y., Higashiyama S., Itoh N., Sugahara K. (2002) J. Biol. Chem. 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- 39.Ueoka C., Kaneda N., Okazaki I., Nadanaka S., Muramatsu T., Sugahara K. (2000) J. Biol. Chem. 275, 37407–37413 [DOI] [PubMed] [Google Scholar]
- 40.Mellor S. L., Cranfield M., Ries R., Pedersen J., Cancilla B., de Kretser D., Groome N. P., Mason A. J., Risbridger G. P. (2000) J. Clin. Endocrinol. Metab. 85, 4851–4858 [DOI] [PubMed] [Google Scholar]
- 41.Rifkin D. B. (2005) J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
- 42.Lee S. J., McPherron A. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawford S. E., Stellmach V., Murphy-Ullrich J. E., Ribeiro S. M., Lawler J., Hynes R. O., Boivin G. P., Bouck N. (1998) Cell 93, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 44.Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 45.Lyons R. M., Keski-Oja J., Moses H. L. (1988) J. Cell Biol. 106, 1659–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge G., Greenspan D. S. (2006) J. Cell Biol. 175, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfman N. M., McPherron A. C., Pappano W. N., Davies M. V., Song K., Tomkinson K. N., Wright J. F., Zhao L., Sebald S. M., Greenspan D. S., Lee S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15842–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degnin C., Jean F., Thomas G., Christian J. L. (2004) Mol. Biol. Cell 15, 5012–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sopory S., Nelsen S. M., Degnin C., Wong C., Christian J. L. (2006) J. Biol. Chem. 281, 34021–34031 [DOI] [PubMed] [Google Scholar]
- 50.Sengle G., Ono R. N., Lyons K. M., Bächinger H. P., Sakai L. Y. (2008) J. Mol. Biol. 381, 1025–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown M. A., Zhao Q., Baker K. A., Naik C., Chen C., Pukac L., Singh M., Tsareva T., Parice Y., Mahoney A., Roschke V., Sanyal I., Choe S. (2005) J. Biol. Chem. 280, 25111–25118 [DOI] [PubMed] [Google Scholar]
- 52.Walton K. L., Makanji Y., Wilce M. C., Chan K. L., Robertson D. M., Harrison C. A. (2009) J. Biol. Chem. 284, 9311–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hüsken-Hindi P., Tsuchida K., Park M., Corrigan A. Z., Vaughan J. M., Vale W. W., Fischer W. H. (1994) J. Biol. Chem. 269, 19380–19384 [PubMed] [Google Scholar]
- 54.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 55.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 56.Flaumenhaft R., Rifkin D. B. (1991) Curr. Opin. Cell Biol. 3, 817–823 [DOI] [PubMed] [Google Scholar]
- 57.Jiao X., Billings P. C., O'Connell M. P., Kaplan F. S., Shore E. M., Glaser D. L. (2007) J. Biol. Chem. 282, 1080–1086 [DOI] [PubMed] [Google Scholar]
- 58.Khan S. A., Nelson M. S., Pan C., Gaffney P. M., Gupta P. (2008) Am. J. Physiol. Cell Physiol. 294, C1387–1397 [DOI] [PubMed] [Google Scholar]