Abstract
The Na+/H+ exchanger isoform 1 is a ubiquitously expressed integral membrane protein. It resides on the plasma membrane of cells and regulates intracellular pH in mammals by extruding an intracellular H+ in exchange for one extracellular Na+. We characterized structural and functional aspects of the transmembrane segment (TM) VI (residues 227–249) by using cysteine scanning mutagenesis and high resolution NMR. Each residue of TM VI was mutated to cysteine in the background of the cysteineless NHE1 protein, and the sensitivity to water-soluble sulfhydryl-reactive compounds (2-(trimethylammonium)ethyl)methanethiosulfonate (MTSET) and (2-sulfonatoethyl)methanethiosulfonate (MTSES) was determined for those residues with significant activity remaining. Three residues were essentially inactive when mutated to Cys: Asp238, Pro239, and Glu247. Of the remaining residues, proteins with the mutations N227C, I233C, and L243C were strongly inhibited by MTSET, whereas amino acids Phe230, Gly231, Ala236, Val237, Ala244, Val245, and Glu248 were partially inhibited by MTSET. MTSES did not affect the activity of the mutant NHE1 proteins. The structure of a peptide representing TM VI was determined using high resolution NMR spectroscopy in dodecylphosphocholine micelles. TM VI contains two helical regions oriented at an approximate right angle to each other (residues 229–236 and 239–250) surrounding a central unwound region. This structure bears a resemblance to TM IV of the Escherichia coli protein NhaA. The results demonstrate that TM VI of NHE1 is a discontinuous pore-lining helix with residues Asn227, Ile233, and Leu243 lining the translocation pore.
Keywords: Cell pH, Exchangers, Membrane, Membrane Proteins, NMR, Proton Transport, Sodium Transport, Sodium Proton Exchange
Introduction
Mammalian NHE1 (Na+/H+ exchanger isoform 1) is a ubiquitous integral plasma membrane protein. It regulates intracellular pH by mediating removal of one intracellular proton in exchange for one extracellular sodium ion (1, 2). NHE1 also has several other additional functions (3). These include promoting cell growth and differentiation (4), promoting inward sodium flux in response to osmotic shrinkage (5), and facilitating cell motility (6). NHE1 also has several pathological roles, including promoting invasiveness of neoplastic breast cancer cells (7). Additionally, its activity promotes pathological heart hypertrophy and is involved in the damage that occurs during ischemia and reperfusion. Inhibition of NHE1 with Na+/H+ exchanger inhibitors protects the myocardium during various disease states (8–11).
NHE1 consists of an N-terminal membrane domain of ∼500 amino acids and a C-terminal regulatory domain of about 315 amino acids (1, 9). The membrane domain is responsible for ion movement. Two models of the topology of NHE1 exist. An analysis of topology by cysteine scanning accessibility suggested that NHE1 has three membrane-associated re-entrant segments and 12 integral transmembrane segments (12). A different model of the topology of NHE1 was more recently suggested (13). It was based on computational methods, including evolutionary conservation analyses and fold alignment methods with NhaA, a partially homologous bacterial Na+/H+ antiporter whose crystal structure has been determined (14). A comparison of the models shows some similarities and some differences (15). Amino acids 227–249 are predicted to form a transmembrane segment with the same orientation in both models and were initially referred to as TM5 VI in the model of Wakabayashi et al. (12).
The mechanism of transport of the membrane domain is both of great interest from a scientific viewpoint and potentially helpful in the design of improved NHE1 inhibitors for clinical use (15). In this regard, we have recently characterized the functionally important residues and the structure of TM IV (residues 155–180), TM VII (residues 250–275), TM IX (residues 338–365), and TM IX (residues 447–472). We have used a combination of cysteine scanning mutagenesis, site-specific mutagenesis, and NMR analysis to analyze the details of the NHE1 membrane domain. Cysteine scanning mutagenesis showed that Phe161 of TM IV is a pore-lining residue critical to transport. Analysis of the structure of TM IV showed that it is composed of one region of β-turns, an extended middle region including Pro167–Pro168, and a helical region (16). TM VII was a more typical transmembrane helix, although it was interrupted with a break in the helix at the functionally critical residues Gly261–Glu262 (17). TM IX contains two structurally conserved regions containing α-helix structure at residues 340–344 and 353–359, with a 90° kink at Ser351 between the two regions that could potentially provide flexibility. Cysteine scanning mutagenesis showed that both residues Ser351 and Glu346, between the helical regions, are pore-lining (18). TM XI contains two helical regions, residues 447–454 and 460–471, and Leu465 was identified as pore-accessible through cysteine scanning mutagenesis and reaction with MTSET (19). Extracellular loop 2 (amino acids 153–155) was also recently characterized in this fashion, and the loop had a specific structure, disruption of which affected protein function (20).
TM VI of NHE1 has not been well studied. In the proposed new topology of NHE1 suggested by Landau et al. (13), TM VI (amino acids 227–249) is suggested to be TM IV. This region corresponds to TM IV of the bacterial antiporter NhaA. In the high resolution structure of NhaA, TM IV was an important part of a novel fold of two TMs that crossed each other in the center of the membrane and played a key role in ion binding and transport (14). It was proposed that amino acids 227–249 of NHE1 provide an equivalent role for this protein, forming part of the TM4-TM11 assembly (13). We therefore decided to investigate TM VI (amino acids 227–249) of NHE1 in detail. We used cysteine scanning mutagenesis to identify and characterize critical pore-lining residues of the protein. We also used nuclear magnetic resonance (NMR) spectroscopy to characterize the structure of a synthetic peptide representing TM VI in dodecylphosphocholine (DPC) micelles, including paramagnetic relaxation NMR experiments (21, 22) to examine the location of the peptide in the micelle and its interaction with a cation. Studies have shown that TMs of membrane proteins possess all of the structural information required to form their higher order structures in their amino acid sequence (23). This has been demonstrated in earlier studies on membrane protein segments, such as the cystic fibrosis transmembrane conductance regulator (24), bacteriorhodopsin (25, 26), a fungal G-protein-coupled receptor (27), and rhodopsin (28). The use of DPC micelles has also been shown to be an excellent membrane mimetic environment for these studies (29, 30).
Our results identify TM VI as a critical pore-lining residue of NHE1. Three residues were identified as lining the pore, and several others were affected by changing them to Cys and reacting with MTSET. We also demonstrate that TM VI is an interrupted helix with a similarity to TM IV of NhaA.
EXPERIMENTAL PROCEDURES
Materials
MTSET and MTSES were purchased from Toronto Research Chemicals, (Toronto, Canada), and LipofectamineTM 2000 reagent was purchased from Invitrogen. PWO DNA polymerase was obtained from Roche Applied Science, and sulfo-NHS-SS-biotin was from Pierce. Immobilized streptavidin resin was from Sigma-Aldrich, and deuterated DPC-d38 was purchased from Cambridge Isotope Laboratories (Andover, MA).
Site-directed Mutagenesis
The expression plasmid pYN4+ contains a hemagglutinin (HA)-tagged human NHE1 isoform of the Na+/H+ exchanger. The plasmid contains the cDNA for the entire coding region of the Na+/H+ exchanger. We previously demonstrated that the tagged expressed protein functions normally (31). Mutations to the NHE1 cDNA in pYN4+ changed the indicated amino acids to cysteine (supplemental Table 1), using the functional cysteineless NHE1 protein that we described earlier (16). Site-directed mutagenesis was done using amplification by PWO DNA polymerase and using the Stratagene (La Jolla, CA) QuikChangeTM site-directed mutagenesis kit. Mutations created or deleted a restriction enzyme site that was used in plasmid screening. DNA sequencing confirmed the fidelity of DNA amplification.
Cell Culture and Transfections
Using control and mutant pYN4+ DNA, stable cell lines were made using AP-1 cells that lack an endogenous Na+/H+ exchanger as described earlier (32). Transfection was with LipofectamineTM 2000 reagent (Invitrogen) (31). Stable cell lines for experiments were regularly reestablished from frozen stocks at passage numbers between 5 and 9. At least two independently made clones of each mutant were made, and results are shown from one mutant. Independently made clones all had very similar functional characteristics.
SDS-PAGE and Immunoblotting
Cell lysates made as described earlier (31) were used for Western blot analysis on 10% SDS-polyacrylamide gels. Nitrocellulose transfers were immunostained using anti-HA monoclonal antibody that recognizes the HA tag on the expressed Na+/H+ exchanger protein. The second antibody was peroxidase-conjugated goat anti-mouse antibody. Chemiluminescence was used to visualize immunoreactive proteins. ImageJ 1.35 software (National Institutes of Health, Bethesda, MD) was used for analysis of protein expression on x-ray films.
Cell Surface Expression
The relative levels of NHE1 cell surface expression were measured essentially as described earlier (31). Briefly, cell surfaces were labeled with sulfo-NHS-SS-biotin (Pierce), and immobilized streptavidin resin was used to remove plasma membrane NHE1 protein. Equal amounts of total and unbound proteins were analyzed by Western blotting and densitometry measuring immunoreactive (HA-tagged) NHE1 protein. It was not possible to efficiently and reproducibly elute proteins bound to immobilized streptavidin resin. The relative amount of NHE1 on the cell surface was calculated for both the 110-kDa and the 95-kDa (partial or deglycosylated) HA-immunoreactive species in Western blots. Calculations of surface targeting and corrections for activity were for the fully glycosylated protein, as indicated in the figure legends.
Na+/H+ Exchange Activity
Na+/H+ exchange activity was measured as described earlier (17) using a spectrofluorometer. Briefly, after ammonium chloride-induced acute acid load, the initial rate of Na+ (135 mm)-induced recovery of cytosolic pH (pHi) was measured using 2′,7-bis(2-carboxyethyl)-5(6)-carboxyfluorescein-AM (Molecular Probes, Inc., Eugene, OR). Buffering capacities of stable cell lines did not vary, and the pHi recovery was from equivalent initial pH values. Where indicated, Na+/H+ exchanger activity was corrected for the targeting of the protein to the cell surface and for the level of protein expression. In some experiments, cells were treated with MTSET or MTSES using a two-pulse acidification assay. Cells were treated with ammonium chloride two times and allowed to recover twice in NaCl-containing medium (16). One pulse was in the presence of inhibitor, and one was in the absence of either 10 mm MTSET or MTSES for 10 min. The starting pH of recovery was equivalent in the two pulses. The rate of recovery from the acid load in the presence of inhibitor was compared with the rate in the absence of inhibitor. A two-pulse assay in the absence of inhibitors was used to correct for any time-dependent changes in NHE1 activity. The calculation used for residual activity was as follows.
 |
Results are shown as mean ± S.E., and statistical significance was determined using a Mann-Whitney U test.
Peptide Synthesis, Purification, and Preparation
Peptide representing TM VI (sequence acetyl-KKKDNLLFGSIISAVDPVAVLAVFEEIHKKK-amide) was purchased from GI Biochem (Shanghai) Ltd. The peptide was purified by HPLC and the identity of the peptide was confirmed by using matrix-assisted laser desorption ionization mass spectrometry and by sequential assignment of the NMR spectra. 2 mm purified peptide and 150 mm DPC-d38 were dissolved in double-distilled H2O containing 5% D2O and 0.25 mm DSS-d6 (Chenomx) as an NMR internal standard. The pH was adjusted to 4.9 without consideration of the deuterium effect on the glass electrode.
NMR Spectroscopy
One-dimensional 1H and two-dimensional 1H-1H DQFCOSY, TOCSY (60-ms mixing time) and NOESY (225-ms mixing time) spectra were acquired on a Varian Inova 600-MHz NMR spectrometer, and a two-dimensional NOESY (225-ms mixing time) spectrum was also acquired on an Varian Inova 800-MHz spectrometer. All spectra were acquired at 30 °C. The NMR spectra were processed using NMRPipe (33) and analyzed in NMRView (34). Sequential assignment of spectra followed standard procedures (35). Peaks in the spectra were manually assigned in NMRView. Distance restraints for structure calculation were obtained from the peaks in the NOESY spectrum acquired at 800 MHz. The restraints were sorted by peak intensity into strong (1.8–2.8 Å), medium (1.8–3.4 Å), and weak (1.8–5.0 Å) distance ranges. Restraints were extended to an additional very weak (1.8–6.0 Å) category as necessary during structure refinement. Structures were calculated using a torsion angle dynamics simulated annealing protocol in Xplor-NIH (36) and following the peptide structure calculation protocol we have used previously (37). In each round of structure calculation, an extended polypeptide was generated and subjected to simulated annealing with restraints, with 50 structures generated in each round. A soft square well potential was used for the NOE restraining function in the calculations. After each round, violated restraints were examined and either lengthened or removed. Initially, violations of >0.5 Å in >50% of the structures calculated were modified. The cut-off for violations that were modified increased in stringency in subsequent rounds until violations of >0.1 Å and >10% were considered. Several further rounds of structure calculation were performed with the inclusion of dihedral angle restraints, with the scaling factors of 10, 50, 100, 100, 50, and 25 in each round.
Paramagnetic Relaxation Experiments
Paramagnetic relaxation enhancement (PRE) rates were measured by titrating MnCl2 stock solution into a sample of TM VI in DPC to concentrations of 0, 0.1, 0.2, 0.5, and 1.0 mm Mn2+ and measuring proton T1 relaxation rates at each titration point using a series of two-dimensional 1H-1H NOESY spectra with a saturation recovery sequence at the beginning (22). The saturation recovery sequence at the beginning of each NOESY utilized the gradient homospoil option built into the Varian tnnoesy pulse sequence. The relaxation delays were 200, 314, 493, 775, 1216, 1910, and 3000 ms for the series of NOESY experiments at each titration point. Nmrview (34) was used for peak assignment and intensity measurement. Curve fitting was performed using the program Xcrvfit (Version 4.0.12; R. Boyko and B. D. Sykes) for determining T1 and PRE values.
RESULTS
Fig. 1 shows models of the NHE1 isoform of the Na+/H+ exchanger. Fig. 1A is a schematic based on the topology deduced by cysteine scanning accessibility studies (12), whereas 1B is a model of NHE1 based on homology modeling with bacterial NhaA (13). Residues corresponding to TM VI in Fig. 1A are predicted to be in the same orientation and relative position in Fig. 1B. The first two TMs are believed to be cleaved and removed in this model; therefore, TM VI is relabeled TM IV. Fig. 1C shows a schematic model illustrating the amino acids of this TM (herein referred to as TM VI based on (12). To examine which amino acids of TM VI are lining the protein cation pore, we performed a cysteine scanning accessibility study of this TM. Each residue in TM VI of cysteineless Na+/H+ exchanger (cNHE1) was mutated to a cysteine residue. Initially, we determined whether these mutant forms of the Na+/H+ exchanger were active enough to permit functional analysis. Fig. 2, A–C, illustrates the results of experiments determining expression, targeting, and activity levels of the wild type and TM VI mutant NHE1 proteins. Expression levels (Fig. 2A) of some of the TM VI Cys scanning mutants decreased greatly. Changing the amino acids Asn227 and Ala236 to Cys decreased expression to less than one-third of control. The mutants I233C and P239C had expression reduced to 60 and 40% of the cNHE1 protein, respectively. In all cases in the mutants with reduced NHE1 expression, there was a great reduction in the level of mature, fully glycosylated protein. It was also noticeable that in the mutants with P239C and E247C mutations, expression was almost exclusively of the lower molecular weight, partial or deglycosylated form of the NHE1 protein.
FIGURE 1.
Models of NHE1 isoform of the Na+/H+ exchanger. A, topological model of the transmembrane domain of the NHE1 isoform of the Na+/H+ exchanger based on cysteine accessibility studies (12). B, topological model of NHE1 based on comparison with crystal structure of bacterial homologue NhaA (13). C, schematic diagram of amino acids present in TM VI.
FIGURE 2.
Analysis of wild type and mutant NHE1 proteins. A, Western blot of whole cell extracts of stable transfectants expressing Na+/H+ exchanger TM VI mutants or control proteins. All mutations were to cysteine. 75 μg of total protein was loaded in each lane. The numbers below the lanes indicate the mean values (n = 3–4) obtained from densitometric scans of both the 110 and 95 kDa bands relative to wild type NHE. AP-1, mock-transfected AP-1 cells. Wt and cNHE1, cells stably expressing wild type Na+/H+ exchanger protein and the cysteineless NHE1, respectively. *, significantly different from cNHE1 at p < 0.05. B, surface localization of cells expressing control and TM VI mutants, as described under “Experimental Procedures.” Equal amounts of total cell lysate (left lane) and unbound intracellular lysate (right lane) were examined by Western blotting with anti-HA antibody to identify NHE1 protein. cNHE1 Ct, a control experiment in which nonspecific binding to streptavidin-agarose beads was carried out following the standard procedure but without labeling cells with biotin. The percentage of the total NHE1 protein found on the plasma membrane is indicated for each mutant; calculations were based on the fully glycosylated protein only. For the control experiment, this indicates the amount of nonspecific binding to streptavidin-agarose beads. Results are the means ± S.E. (n ≥ 6 determinations. Autoradiography exposure times were increased for mutants expressing lower levels of protein. +, significantly different from that of cNHE1 at p < 0.01. C, summary of the rate of recovery from an acute acid load of AP-1 cells transfected with cNHE and TM VI Na+/H+ exchanger mutants. The mean activity of cNHE1 stably transfected with NHE1 was 0.01 ΔpH/s, and this value was set to 100%. Activities are a percentage of those of cNHE. Values are the mean ± S.E. (error bars) of 6–10 determinations. Results are shown for mean activity, both uncorrected (black) and normalized for surface processing (of glycosylated protein) and expression levels (gray). Mutants P239C and E247C were not corrected for surface targeting. *, mutants with uncorrected activity that is significantly different from that of cNHE1 at p < 0.001.
Mutation of amino acids of TMs of NHE1 can cause intracellular retention of the NHE1 protein (16). We wanted to determine if changes in NHE1 activity that we observed were due to effects on the protein itself or were due in whole or in part to intracellular retention of the protein. We therefore examined intracellular targeting of the NHE1 TM VI-expressing mutant cell lines as described under “Experimental Procedures.” Sulfo-NHS-SS-biotin cell surface-labeled proteins were bound to streptavidin-agarose beads, and equal amounts of total cell lysates and unbound lysates (representing intracellular protein) were examined by using SDS-PAGE. Western blotting against the anti-HA tag identified the relative amount of NHE1 protein in the total and intracellular fractions (Fig. 2B). Supplemental Table 2 shows a summary of the plasma membrane localization of the glycosylated and unglycosylated proteins. Although some of this unglycosylated protein may be functional (39) and may still target to the cell surface (19), a majority of the surface protein is of the glycosylated protein (40), so the activity was corrected for surface-localized glycosylated NHE1 protein. The results are shown in Fig. 2B and supplemental Table 2. The fully glycosylated protein is more highly targeted to the cell surface, relative to the partial or deglycosylated protein. The relative amounts of the partial or deglycosylated protein targeted to the plasma membrane varied from ∼30 to 15% of the fully glycosylated levels. This was noticeably decreased for mutants F230C and D238C, although the significance of this observation is not yet clear. A number of the mutants (L229C, I234C, S235C, P239C, A241C, V242C, V245C, and E247C) had significantly less targeting of the fully glycosylated protein to the cell surface. For some of the mutants, the difference between the cNHE1 targeting and the mutants was not very large (L229C, I234C, S235C, A241C, V242C, and V245C); however, for other mutants, the amount of glycosylated protein targeted to the surface was negligible (P239C and E247C). For mutants I233C and L243C, surface targeting of the glycosylated band was slightly increased, although the significance of this observation is not yet clear.
We also determined the Na+/H+ exchanger activity of the NHE1 TM VI mutants. An acute acid load was induced as described earlier (40), and the rate of recovery from the acute acid load was determined in stable cell lines transfected with either wild type Na+/H+ exchanger or mutants of TM VI. To determine if decreased activity was due to effects on the protein itself or due to changes in surface targeting or expression, the rate of recovery was also corrected for both of these values, relative to cNHE1 (Fig. 2C and supplemental Table 3). Several of the mutants (D238C, P239C, and E247C) had greatly reduced activity that was less than 15% of the control levels. It was not possible to accurately work with them for further experiments. Several other mutants (N227C, I233C, A236C, L243C, and E248C) had activity from 15 to 40% that of cNHE1. Their activity, although reduced, was still substantial enough for further experimentation. It was notable that for the mutants N227C, A236C, A241C, and V242C, the NHE1 activity was essentially equivalent to controls when corrected for effects on surface processing and expression levels. This indicated that the protein functioned normally but that the reduced activity we observed was due to decreased expression or targeting of the protein. The N227C and A236C mutants had large decreases in expression levels but normal surface targeting. (Both the A241C and V242C mutants had significant decreases in surface targeting and minor decreases in expression levels.) Two mutants, S235C and I249C, had somewhat elevated levels of NHE1 activity. The significance of this observation is not yet clear, although for I249C it may reflect a higher level of surface targeting.
We next determined the sensitivity of the Cys mutants to MTSET or MTSES. Fig. 3A illustrates one example of the findings with a mutant that is sensitive to MTSET, and Fig. 3B shows a summary of the results of the various mutants tested. Of the active Na+/H+ exchangers, Asn227, Ile233, and Leu243 were very sensitive to MTSET treatment. This resulted in elimination of most of the activity of the protein. Seven other mutants, F230C, G231C, A236C, V237C, A244C, V245C, and E248C, were partially inhibited by MTSET although to a more limited extent. Negatively charged MTSES had no effect on any of the mutant proteins. As a control, we examined the sensitivity of an F161C mutant that we have previously determined was sensitive to both MTSET and MTSES (16). This mutant was again inhibited by both MTSET and MTSES.
FIGURE 3.
Effect of sulfhydryl-reactive compounds, MTSET and MTSES, on activity of cNHE1 and single cysteine NHE1 mutant-containing cell lines. A, example of results of the effect of MTSET or MTSES on activity of cNHE1 and I233C mutant. cNHE and I233C NHE1 protein activity was assayed in stably transfected AP-1 cells as described under “Experimental Procedures.” Activity was measured after two acid pulses. The first pulse in the absence of MTSET is shown. For ease of viewing, only the recovery from acidosis is shown for the second pulse, in which cells were treated with MTSES or MTSET. NH4Cl, treatment with ammonium chloride; Na Free, treatment with Na+-free buffer to induce acidosis; NaCl, recovery from acidosis in NaCl-containing buffer (for the second pulse, this contained MTSET/MTSES, and cells were pretreated with MTSET/MTSES for 10 min prior to NH4Cl-induced acid load). B, summary of results of mutant and control activities of TM VI mutants. Activity was measured after two ammonium chloride pulses as described under “Experimental Procedures.” The second acidification was after cells were treated with 10 mm reagent. Results are presented as the percentage of activity of the second acid load relative to the first. * or †, the second recovery from acid load was significantly lower than the first at p < 0.01 or p < 0.05, respectively. Solid filled bars, MTSET treatments; lightly shaded bars, MTSES treatments.
NMR spectroscopy was used to determine the structure of a peptide containing the TM VI sequence. The peptide included residues Asp226–His250, which encompasses the predicted transmembrane helix in both models of the NHE1 topology (Fig. 1, A–C). We added cationic lysine residues at the termini of this peptide as we have earlier (19) because this aids in their purification and in the maintenance of correct transbilayer orientation (41). N- and C-terminal acetyl and amide caps, respectively, were added to remove charges at the peptide termini similar to our earlier procedure (19) to better represent the full-length protein. We have previously used DPC to act as a membrane mimetic for transmembrane segments of NHE1 (18, 19). TMVI peptide was solubilized by a similar procedure and provided a stable, well behaved sample suitable for structure determination by NMR. A one-dimensional 1H NMR spectrum of the peptide in DPC showed relatively narrow lines and reasonable spectral dispersion, indicative of structure formation in the peptide (results not shown).
Sequential assignment of the peptide was accomplished using the two-dimensional 1H-1H Double-quantum filtered correlation spectroscopy (DQFCOSY), total correlation spectroscopy (TOCSY), and NOESY spectra using standard procedures (35). The assignment of the peptide was complete except for some Lys Hϵ atoms. Strong NOE peaks observed between Asp238 Hα and Pro239 Hδ indicate a trans conformation for the peptide bond between Asp238 and Pro239. Additional resonances that would correspond to a cis peptide bond conformation were not detected, suggesting that the dominant conformation is the trans-proline conformation. Distance restraints for structure calculation were obtained from the two-dimensional NOESY spectrum acquired at 800 MHz. Distances were calibrated using peak intensity rather than peak volume to compensate for peak overlap. A soft square well potential was used for the NOE potential to compensate for possible erroneous assignments or overrestrained distances. Structures were initially calculated without dihedral angle restraints, so the structure would be influenced primarily by the NOE distance restraints. Dihedral angle restrains were added in at a later stage of refinement. NOE restraints and chemical shift index prediction of secondary structure (42) (supplemental Fig. S1) suggested that the peptide was helical at approximately residues Leu229–Ala236 and Val240–Lys251. Helical dihedral angle restraints for φ (−60 ± 30°) and ψ (−40 ± 40°) bonds were applied to these residues for several further rounds of calculation. The 40 lowest energy structures of the 50 structures of the final structure calculation were kept for analysis, and the structure calculation statistics are summarized in supplemental Table 4.
The ensemble of peptide structures do not superimpose particularly well with each other over the entire length of the peptide. Attempts to superimpose small regions of the ensemble of structures show two overlapping segments of the peptide at residues Leu228–Asp238 and Val237–Lys253, which superimpose well, where the average root mean square deviation per residue over the ensemble of structures is less than 1 Å (Fig. 4) (supplemental Fig. S2). Superimposition was performed using the method of Kabasch (43), as implemented in the CCP4 suite (44). The lowest energy structure is shown in Fig. 4C. The two structurally conserved regions contain two α-helices at residues Leu229–Ala236 and Pro239–Lys252, based on the dihedral angles in that region, and are maintained at approximately right angles to each other primarily due to restraints from Val237–Asp238 in the extended region to residues in the helices on either side (Fig. 5). This extended segment allows for some variation in the relative orientations of the two helices among the ensemble members (supplemental Fig. S2). A plot of the dihedral angle order parameters (37) (supplemental Fig. 3) also shows the two structured regions of the peptide, corresponding to the helices, and a short, more structurally variable region, corresponding to the extended region. The N-terminal end of the peptide, Lys223–Leu228, appears to be relatively unstructured, as shown by the lack of NOE contacts in that region and by chemical shift index values suggesting random coil conformations. The C-terminal helix is slightly curved, which could reflect the peptide's conformation in the micelle or be a result of the smaller number of NOE contacts between the residues on the outside of the curve.
FIGURE 4.
NMR structure of TMXI in DPC micelles. Shown is superimposition of structurally conserved regions of the TMVI peptide structure. Superimposition of the backbone atoms of the structurally conserved regions 228–238 (A) and 237–253 (B) is shown. The backbone is shown in black, and side chains are shown in color. C, views of a single ensemble member with MTSET-sensitive residues labeled shown from two sides.
FIGURE 5.
Distance restraints maintaining the kinked structure of TM VI. The lowest energy ensemble member is shown in both a schematic diagram and a stick representation. Distance restraints to and from atoms in residues Val237 and Asp238 to other atoms are shown in black dotted lines.
A crystal structure of an E. coli Na+/H+ antiporter, NhaA, has been determined (14). NHE1 and NhaA share similar activities and are distantly related; therefore, it is possible that they have similar structures, as has been suggested earlier (13). Pairwise alignment of TM VI of NHE1 onto the sequence of NhaA suggests that TM VI aligns with TM VI of NhaA (residues 180–200); this alignment is shown in Fig. 6a. TM VI in NhaA is a straight helix and primarily hydrophobic, located on the periphery of the protein in the NhaA crystal structure. This alignment does not support the critical role of NHE1 TM VI found in this paper. Using more advanced alignment and structure prediction techniques, Landau et al. (13) suggest that the sequence of TM VI of NHE1 is equivalent to TM IV of NhaA. A SEQSEE (45) alignment of these two helices (not shown) suggests that the Pro239 in NHE1 lines up with the Asp133 in NhaA. This is close to the alignment proposed in the Landau model that aligns NHE1 Asp238 and NhaA Asp133. The latter alignment is shown in Fig. 6B and is used for superimposition of the NMR structure of TM VI with the structure of TM IV from NhaA (Fig. 6C).
FIGURE 6.
Comparison of TM VI of NHE1 with TM IV of NhaA. A, region identified using the SEQSEE program (45) for the optimal pairwise alignment of the sequence of NHE1 TM VI against the entire NhaA sequence. B, alignment of the sequences of NHE1 TM VI and NhaA TM IV, as suggested by Landau et al. (13), with the extended regions highlighted. A vertical bar indicates amino acid identity. *, similar amino acids. MTSET-sensitive residues and the corresponding residues in NhaA are shown in boldface type. C, comparison of a representative NMR structure of TM VI of NHE1 (right) with that of TM IV of NhaA (left). Amino acids 223–253 of NHE1 and amino acids 121–143 of NhaA are shown. The conserved Asp residue and MSTET-sensitive residues in NHE1 and the corresponding residues in NhaA are labeled.
No member of the ensemble of structures of the entire length of TM VI, determined by NMR, superimposed well onto the entire structure of TM IV from NhaA. The L-shaped conformation of the NMR structure inhibits superimposition of the entire structure onto NhaA TM IV, which has a more linear conformation. Differences in the conformations of the helical regions also prevent a reasonable superimposition (root mean square deviation <1 Å) of the helices between NHE1 TM VI and NhaA TM IV. However, smaller, overlapping segments of the NMR structure of TM VI of NHE1 can superimpose well onto the crystal structure of TM IV of NhaA, covering the two helical regions in the structures. The central extended region in the NMR structure does not superimpose particularly well with the extended region in the crystal structure of TM IV of NhaA. At best, Ala130–Asp133 of NhaA and Ser235–Asp238 of NHE1 align structurally, with root mean square deviations of about 1–2 Å. Despite the poor superimposition of the NMR and crystal structures, there is nevertheless a qualitative resemblance between them. The lowest energy ensemble member of the TM VI NMR structure and the structure of TM IV from NhaA are shown in Fig. 6c. Both contain two α-helices at each end separated by an extended region of two or three residues in the middle. Both the NMR structure of NHE1 TM VI and the crystal structure of NhaA TM IV contain an evolutionarily conserved Asp residue at the beginning of the C-terminal helix. This seems to support the sequence alignment and three-dimensional structural model of NHE1 proposed by Landau et al. (13). The residues identified in NHE1 TM VI that are most sensitive to MTSET, Asn227, Ile233, and Leu243, correspond to Thr122, Ile128, and Leu138, respectively, based on this alignment.
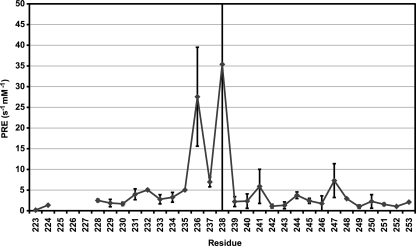
To determine the location of the TM IV peptide with respect to the DPC micelles, we measured the paramagnetic relaxation enhancement rates for the peptide in the presence of Mn2+. Broadening and overlap of peaks in the one-dimensional NMR spectra prevent accurate measurement of relaxation times. In the absence of isotopic labeling, such as for synthetic peptides, homonuclear two-dimensional spectra have been used to resolve peaks and measure the relaxation rates of samples (22, 46). We used a two-dimensional NOESY with a saturation recovery pulse sequence to measure the T1 relaxation times of the peptide. Only resolved peaks were used for curve fitting to determine T1 times. PRE values are determined by linear fitting of 1/T1 versus Mn2+ concentrations. PRE values were averaged together in the case of multiple measurable peaks representing the same proton. PRE values for backbone Hα atoms are shown in Fig. 7. The regions of the peptide that were determined to be helical in the NMR structure show a periodic variation in PRE values, with maxima at Gly231-Ser232, Ser235-Ala236, Ala241, Ala244, and Glu247. PRE values increase dramatically at residues within or near the extended region of the NMR structure, Ala236–Asp238. Due to overlap of peaks even in the two-dimensional spectra, only a few peaks from the terminal lysines could be used for measurement. The terminal Lys Hα appear to have relatively low PRE values compared with the rest of the peptide. The paramagnetic relaxation experiments (Fig. 7) provided additional details about the structure of the peptide and its interactions with the micelle (see “Discussion”).
FIGURE 7.
Paramagnetic relaxation enhancement rates for TM VI in DPC micelles. Backbone Hα PRE values are shown for TM VI. Higher PRE values represent a greater change in the measured T1 relaxation rates with respect to Mn2+ concentration and consequently represent regions of the peptide that are closer to the surface of the detergent micelle that are more accessible to the water-soluble Mn2+ ions.
DISCUSSION
In this work, we characterized both the structural and functional characteristics of amino acids 228–249 of the NHE1 isoform of the Na+/H+ exchanger. These residues are purported to be either TM VI (12) or TM IV (13) of the protein. This TM has not been well studied earlier, but molecular modeling by Landau et al. (13) and our own (not shown) suggested that it may be an important pore-lining segment, critical in activity of the protein. Initial experiments therefore used cysteine scanning mutagenesis to examine if this segment was pore-lining. All of the amino acids of the segment from Asn227 to Ile249 were mutated to Cys in the background of the cysteineless protein that we have earlier shown is fully functional (16, 19). With mutation to Cys, the activity of many residues was reduced. For the amino acids Asp238, Pro239, and Glu247, the activity was reduced so greatly that it was not possible to assay these mutants further. For 12 other mutants, the activity was reduced but was sufficient for measurement. We have found that some TMs (TM IV (47) and TM IX (18)) are more tolerant of mutations to Cys. Others, such as TM XI (19), appear to be intolerant of changes, and this differential sensitivity has been noted in other proteins (48). In several cases (Asn227, Ala236, Ala241, and Val242), partially decreased activity of the protein was due to decreased level of expression or targeting of the protein. This indicated that these proteins were functional but were not processed properly.
When mutated to Cys, amino acids of TM VI (amino acids 228–249) were exquisitely sensitive to reaction with MTSET. Of the 20 mutant locations that were active and were changed to Cys residues, treatment with MTSET resulted in inhibition in 10 amino acid positions. This made TM VI more susceptible than any of TM IV, IX, or XI. Three of the mutants (at amino acids Asn227, Ile233, and Leu243) were greatly inhibited by MTSET, whereas seven others were inhibited to a lesser degree. In no cases did reactivity with MTSES cause any inhibition with the amino acids of TM VI. A positive control, Phe161 of TM IV, was inhibited by both MTSES and MTSET. Positively charged MTSET may be inhibitory because of direct electrostatic repulsion of a Na+ or H+. These results suggested that many residues of TM VI were pore-lining and that this segment plays an important role in forming the pore of the NHE1 protein. Although the residues strongly inhibited by MTSET (Asn227, Ile233, and Leu243) are strong candidates for pore-lining residues, those partially inhibited (Phe230, Gly231, Ala236, Val237, Ala244, Val245, and Glu248) are less clear cases. Residues Ala244 and Val245 are next to Leu243 and may have partial access to the pore. We have previously found a similar phenomenon; in TM IX, residues adjacent to strongly inhibited residues also tended to be partially inhibited (18). Glu248 is on a similar face of TM VI as Ala244 (Fig. 4), which may explain its partial accessibility. Residues Asn227, Phe230, Ala236, and Val237 also are on the same face of the peptide as Asn227 (Fig. 4), which may again account for their partial accessibility.
The structure of TM VI was that of two α-helices between residues Leu229 and Ala236 and residues Pro239 and Lys252, separated by an extended region at Val237-Asp238. We have also found that in the cases of TM XI of NHE1 (19) and TM IV of NHE1 (16), the TM is not a continuous unbroken helix. Recent structures of membrane proteins have suggested that this is not an uncommon occurrence. For example, TMs of rhodopsin are interrupted or kinked at multiple sites (49). Up to 40% of TM helices contain kinks or other distortions from an ideal helix conformation, and these residues tend to be highly conserved and often directly involved in the function of transporters and channels (50). These regions also tend to have an increased incidence of glycine or prolines (50), and we found that the extended region of Val237-Asp238 was immediately upstream of Pro239. Proline residues are considered to be helix breakers because they lack an amide hydrogen and cause a kink of ∼26° in the α-helix (51, 52). This causes the i − 4 backbone carbonyl to not have its normal hydrogen bond donor and prevents formation of the (i − 3)-carbonyl-(i + 1)-amide backbone hydrogen bond (51). With each proline, the α-helix also has two free backbone carbonyls that could coordinate cations or interact with inhibitors (52). All three residues upstream of Pro239 (Ala236, Val237, and Asp238) were important in activity or expression and, when mutated, resulted in decreases in NHE1 activity. In addition, the A236C and V237C mutants were partially inhibited by reactivity with MTSET, suggesting that they are accessible from the cell exterior and pore lining or at least partially accessible to the pore. This extended region of TM VI appears to be critical to NHE1 function.
The crystal structure of the E. coli Na+/H+ antiporter NhaA revealed a novel fold consisting of two TMs, TMs IV and XI, that were assembled to form crossed extended chains (14). Modeling of NHE1 based on the structure of NhaA suggested that residues of TM VI (amino acids 227–249) might play a role equivalent to TM IV of NhaA. A comparison of these two segments is shown in Fig. 6. Although the two segments are not identical, they were similar in overall structure. It was proposed that Asp238 of NHE1 could play a critical role in stabilizing helix dipoles (13) similar to Asp133 of NhaA. When we mutated Asp238 to Cys, NHE1 was essentially inactive, supporting this suggestion. The location of Asp238, at the end of a helix, is also similar to that of Asp133 of NhaA (Fig. 6). One possibility is that Pro239 maintains the extended structure of this region by breaking the helix and allowing dipole formation, whereas Asp238 stabilizes the helix dipole that forms. Supporting this suggestion is the fact that TM VI of NHE1 is similar to that of TM IV of NhaA, more so than to other TMs of NhaA, including TM VI. A comparison of alignments between TM VI of NHE1 with TMs VI and IV of NhaA is shown in Fig. 6, A and B. Although the alignment of TM VI of NHE1 with TM VI of NhaA shows more sequence identity, alignment with TM IV of NhaA lines up the critical Asp238 residue and the extended regions from the structures of the two TMs. Further experiments are necessary to test if Pro239 and Asp238 are critical to formation of an extended helix and to dipole stabilization.
Comparison of the structure of NHE1 TMs with NhaA further supports the conclusion that TM VI of NHE1 is similar to TM IV of NhaA. TM IV of NHE1 (amino acids 155–177) has a general structure somewhat similar to that of TM IV of NhaA (3, 16), with an extended region followed by a helix; however, the extended region is not preceded by a helical region. Furthermore, unlike TM VI of NHE1 and TM IV of NhaA, TM IV of NHE1 does not contain a charged residue in the extended region of the NMR structure. Thus, TM IV of NHE1 does not compare as well to TM IV of NhaA as TM VI of NHE1 (Fig. 6C). TM VI of NhaA also does not compare well with TM VI of NHE1. TM VI of NhaA is an unbroken helix and is not similar to the structure of TM VI of NHE1 (13). It does not have an extended region similar to TM VI of NHE1, and these extended regions of TMs of NhaA are believed to be critical in protein function (see above).
We have earlier shown that TM XI plays a critical role in NHE1 function (19). Landau et al. (13) suggest that TM VI (amino acids 227–249, referred to as TM IV by them) plays a role equivalent to that of the TM IV-TM XI assembly in NhaA. These helices are suggested to form the core of the alternating access mechanism. Our results for both TMs are in agreement with their suggestion. We found that both of these TMs have a helix-extended-helix conformation, that is capable of forming dipoles similar to the TM IV-TM XI assembly in NhaA. Therefore, TM VI (amino acids 227–249) and TM XI may form the core of an alternating access mechanism, similar to TM IV and TM XI of NhaA. However, further experiments are necessary to test this hypothesis.
Fig. 4 illustrates the structure of TM VI of NHE1 with highlighting of some of the MTSET-reactive positions and Asp238. Some of the MTSET-reactive residues (Asn227, Ile233, and Val237) align along a similar face that is perhaps pore-lining. Asp238 does not, but it may be more involved in helix stabilization, as suggested above. Although the residues Leu243 and Glu247 line up on the same face of the C-terminal helix, they do not align on the same face as the N-terminal MTSET-reactive residues, although it must be remembered that they are separated from the more N-terminal amino acids by the extended flexible region. In addition, the TM probably has different positions in different parts of the reaction cycle, and it is unknown in which position the present segment is maintained.
The alignment of the sequences of TM VI of NHE1 and TM IV of NhaA might suggest that the residues found to be pore-lining in NHE1 may also be pore-lining in NhaA (Fig. 6B). Residues Leu243 and Glu247 in NHE1 would correspond to Leu138 and an Ala142 in NhaA based on this alignment. In agreement with the MTSET results in NHE1, these corresponding residues in NhaA are also pore-lining. This also supports the suggestion that these two helices play similar roles in each protein. The situation is less clear for the N-terminal region of the peptide. Residues Asn227 and Ile233 in NHE1 would correspond to Thr122 and Ile128 in NhaA. Both of these residues in the NhaA crystal structure are pointing away from the predicted pore region. However, conformational changes or flexibility in the protein (53, 54) may allow for access to these residues in NhaA, and this might also occur for NHE1.
The results from the paramagnetic relaxation experiments (Fig. 7) provide additional detail about the structure of the peptide and its interactions with the micelle. The periodic nature of the PRE values confirms the two helical regions in the NMR structure and suggests that they have one side facing the solvent and one side facing the micelle, with the smaller and more polar residues facing the solvent. There are two topologies of the peptide that might give these results: where the peptide is lying on the surface of the micelle or where two or more peptides interact to form a solvent-accessible pore within the micelle. Our experiments do not distinguish between these two possibilities. Hα PRE values for the Lys termini of the peptide are low. This could be a result of burial of the Lys residues in the micelle or the electrostatic repulsion of the Mn2+ by the positively charged side chains. The large increase in PRE values in the extended region of the peptide suggests that the region is binding Mn2+, probably through the attraction of Mn2+ by some combination of the negatively charged Asp238, exposed carbonyl groups, and a helix dipole (Fig. 5). This emphasizes the important role of this region in the function of NHE1. The results may also be reflective of the important role of this TM, which might be equivalent to the role of TM IV in NhaA, in cation binding and transport (13, 14). There is some correlation between the functional data and the PRE data. Some of the Mn2+-accessible residues (Gly231, Ala236, Asp238, Ala244, and Glu247) in the NMR studies are also accessible to MTSET when mutated to Cys or are critical in NHE1 function in the full-length protein. Other MTSET-accessible residues appear to be adjacent to the Mn2+-accessible regions. It is uncertain at this time whether the Mn2+ accessibility reflects a role Na+ or H+ coordination, but the possibility is intriguing. Further experimentation is necessary to determine if these NMR studies can be used to predict pore-lining residues in a TM.
Overall, our results have shown that TM VI (amino acids 227–249) is a pore-lining TM that is critical to NHE1 function. It has a characteristic helix-extended region-helix conformation that is conducive to formation of dipoles, with the extended middle region that is typically important in the function of transport proteins. The structure of TM VI of NHE1 has an overall similarity to that of TM IV of NhaA. Residue Asp238 has a location within the TM similar to that of the critical Asp133 of NhaA. We have recently developed an expression system for overproduction of the entire full-length NHE1 protein (38). Solving the structure of the full-length protein will provide more details of the TM and its position within the entire protein.
Supplementary Material
Acknowledgment
We thank the Canadian National High Field NMR Centre for assistance with and use of the 800-MHz spectrometer.
This work was supported by funding from the Canadian Institutes of Health Research (CIHR) (to L. F. and B. D. S.). The Canadian National High Field NMR Centre is funded by the Canadian Institutes of Health Research, the Natural Science and Engineering Research Council of Canada, and the University of Alberta.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4 and Figs. S1–S3.
The atomic coordinates and structure factors (code 2L0E) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TM
- transmembrane segment
- DPC
- dodecylphosphocholine
- MTSET
- (2-(trimethylammonium)ethyl)methanethiosulfonate
- MTSES
- (2-sulfonatoethyl)methanethiosulfonate
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- PRE
- paramagnetic relaxation enhancement
- cNHE1
- cysteineless NHE1
- sulfo-NHS-SS-biotin
- sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate.
REFERENCES
- 1.Karmazyn M., Sawyer M., Fliegel L. (2005) Curr. Drug Targets Cardiovasc. Haematol. Disord. 5, 323–335 [DOI] [PubMed] [Google Scholar]
- 2.Fliegel L. (2009) Expert Opin. Ther. Targets 13, 55–68 [DOI] [PubMed] [Google Scholar]
- 3.Slepkov E. R., Rainey J. K., Sykes B. D., Fliegel L. (2007) Biochem. J. 401, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grinstein S., Rotin D., Mason M. J. (1989) Biochim. Biophys. Acta 988, 73–97 [DOI] [PubMed] [Google Scholar]
- 5.Shrode L., Cabado A., Goss G., Grinstein S. (1996) in The Na+/H+ Exchanger (Fliegel L. ed) pp. 101–122, R.G. Landes Co., Austin, TX [Google Scholar]
- 6.Denker S. P., Barber D. L. (2002) J. Cell Biol. 159, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paradiso A., Cardone R. A., Bellizzi A., Bagorda A., Guerra L., Tommasino M., Casavola V., Reshkin S. J. (2004) Breast Cancer Res. 6, R616–R628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karmazyn M., Liu Q., Gan X. T., Brix B. J., Fliegel L. (2003) Hypertension 42, 1171–1176 [DOI] [PubMed] [Google Scholar]
- 9.Fliegel L. (2001) Basic Res. Cardiol. 96, 301–305 [DOI] [PubMed] [Google Scholar]
- 10.Mentzer R. M., Jr., Lasley R. D., Jessel A., Karmazyn M. (2003) Ann. Thorac. Surg. 75, S700–S708 [DOI] [PubMed] [Google Scholar]
- 11.Lang H. J. (2003) in The Na+/H+ Exchanger: From Molecule to Its Role in Disease (Karmazyn M., Avkiran M., Fliegel L. eds) pp. 239–253, Kluwer Academic Publishers, Boston [Google Scholar]
- 12.Wakabayashi S., Pang T., Su X., Shigekawa M. (2000) J. Biol. Chem. 275, 7942–7949 [DOI] [PubMed] [Google Scholar]
- 13.Landau M., Herz K., Padan E., Ben-Tal N. (2007) J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 14.Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. (2005) Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 15.Kemp G., Young H., Fliegel L. (2008) Channels 2, 329–336 [DOI] [PubMed] [Google Scholar]
- 16.Slepkov E. R., Rainey J. K., Li X., Liu Y., Cheng F. J., Lindhout D. A., Sykes B. D., Fliegel L. (2005) J. Biol. Chem. 280, 17863–17872 [DOI] [PubMed] [Google Scholar]
- 17.Ding J., Rainey J. K., Xu C., Sykes B. D., Fliegel L. (2006) J. Biol. Chem. 281, 29817–29829 [DOI] [PubMed] [Google Scholar]
- 18.Reddy T., Ding J., Li X., Sykes B. D., Rainey J. K., Fliegel L. (2008) J. Biol. Chem. 283, 22018–22030 [DOI] [PubMed] [Google Scholar]
- 19.Lee B. L., Li X., Liu Y., Sykes B. D., Fliegel L. (2009) J. Biol. Chem. 284, 11546–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B. L., Li X., Liu Y., Sykes B. D., Fliegel L. (2009) Biochim. Biophys. Acta 1788, 2481–2488 [DOI] [PubMed] [Google Scholar]
- 21.Zamoon J., Mascioni A., Thomas D. D., Veglia G. (2003) Biophys. J. 85, 2589–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zangger K., Respondek M., Göbl C., Hohlweg W., Rasmussen K., Grampp G., Madl T. (2009) J. Phys. Chem. B 113, 4400–4406 [DOI] [PubMed] [Google Scholar]
- 23.Cunningham F., Deber C. M. (2007) Methods 41, 370–380 [DOI] [PubMed] [Google Scholar]
- 24.Oblatt-Montal M., Reddy G. L., Iwamoto T., Tomich J. M., Montal M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt J. F., Earnest T. N., Bousché O., Kalghatgi K., Reilly K., Horváth C., Rothschild K. J., Engelman D. M. (1997) Biochemistry 36, 15156–15176 [DOI] [PubMed] [Google Scholar]
- 26.Katragadda M., Alderfer J. L., Yeagle P. L. (2001) Biophys. J. 81, 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naider F., Khare S., Arshava B., Severino B., Russo J., Becker J. M. (2005) Biopolymers 80, 199–213 [DOI] [PubMed] [Google Scholar]
- 28.Yeagle P. L., Choi G., Albert A. D. (2001) Biochemistry 40, 11932–11937 [DOI] [PubMed] [Google Scholar]
- 29.Damberg P., Jarvet J., Gräslund A. (2001) Methods Enzymol. 339, 271–285 [DOI] [PubMed] [Google Scholar]
- 30.Henry G. D., Sykes B. D. (1994) Methods Enzymol. 239, 515–535 [DOI] [PubMed] [Google Scholar]
- 31.Slepkov E. R., Chow S., Lemieux M. J., Fliegel L. (2004) Biochem. J. 379, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murtazina R., Booth B. J., Bullis B. L., Singh D. N., Fliegel L. (2001) Eur. J. Biochem. 268, 4674–4685 [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 34.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 35.Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 36.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 37.Rainey J. K., Fliegel L., Sykes B. D. (2006) Biochem. Cell Biol. 84, 918–929 [DOI] [PubMed] [Google Scholar]
- 38.Moncoq K., Kemp G., Li X., Fliegel L., Young H. S. (2008) J. Biol. Chem. 283, 4145–4154 [DOI] [PubMed] [Google Scholar]
- 39.Haworth R. S., Fröhlich O., Fliegel L. (1993) Biochem. J. 289, 637–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Ding J., Liu Y., Brix B. J., Fliegel L. (2004) Biochemistry 43, 16477–16486 [DOI] [PubMed] [Google Scholar]
- 41.Davis J. H., Clare D. M., Hodges R. S., Bloom M. (1983) Biochemistry 22, 5298–5305 [Google Scholar]
- 42.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. (1976) Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 44.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 45.Wishart D. S., Boyko R. F., Willard L., Richards F. M., Sykes B. D. (1994) Comput. Appl. Biosci. 10, 121–132 [DOI] [PubMed] [Google Scholar]
- 46.Arseniev A. S., Sobol A. G., Bystrov V. F. (1986) J. Magn. Reson. (1969) 70, 427–435 [Google Scholar]
- 47.Slepkov E., Ding J., Han J., Fliegel L. (2007) Biochim. Biophys. Acta 1768, 2882–2889 [DOI] [PubMed] [Google Scholar]
- 48.He M. M., Sun J., Kaback H. R. (1996) Biochemistry 35, 12909–12914 [DOI] [PubMed] [Google Scholar]
- 49.Sakmar T. P., Menon S. T., Marin E. P., Awad E. S. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 443–484 [DOI] [PubMed] [Google Scholar]
- 50.Kauko A., Illergård K., Elofsson A. (2008) J. Mol. Biol. 380, 170–180 [DOI] [PubMed] [Google Scholar]
- 51.Barlow D. J., Thornton J. M. (1988) J. Mol. Biol. 201, 601–619 [DOI] [PubMed] [Google Scholar]
- 52.Sansom M. S. (1992) Protein Eng. 5, 53–60 [DOI] [PubMed] [Google Scholar]
- 53.Appel M., Hizlan D., Vinothkumar K. R., Ziegler C., Kühlbrandt W. (2009) J. Mol. Biol. 388, 659–672 [DOI] [PubMed] [Google Scholar]
- 54.Arkin I. T., Xu H., Jensen M. Ø., Arbely E., Bennett E. R., Bowers K. J., Chow E., Dror R. O., Eastwood M. P., Flitman-Tene R., Gregersen B. A., Klepeis J. L., Kolossváry I., Shan Y., Shaw D. E. (2007) Science 317, 799–803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.