Abstract
The PLMItRNA database for mitochondrial tRNA molecules and genes in Viridiplantae (green plants) [Volpetti,V., Gallerani,R., DeBenedetto,C., Liuni,S., Licciulli,F. and Ceci,L.R. (2000) Nucleic Acids Res., 28, 159–162] has been enlarged to include algae. The database now contains 436 genes and 16 tRNA entries relative to 25 higher plants, eight green algae, four red algae (Rhodophytae) and two Stramenopiles. The PLMItRNA database is accessible via the WWW at http://bio-www.ba.cnr.it:8000/PLMItRNA.
DATABASE DESCRIPTION
Algae are photosynthetic eukaryotic organisms that can be found in very different habitats (sea and freshwater, ice and snow and desert soils) showing an enormous variety of cell morphologies and life cycles. Nowadays algae constitute an artificial group of organisms with polyphyletic origins within the crown-eukaryotes radiation. Phylogenetic relationships among algae are still a matter of controversy and several attempts are in progress to also elucidate relationships by using molecular data from chloroplasts, mitochondria and nuclei (1–6). We therefore thought it useful to enlarge the PLMItRNA database established for green plants (higher plants and green algae) mitochondrial tRNA molecules and genes (7) also to include algae mitochondria.
A list of algae mitochondrial tRNA genes has been obtained essentially as already described for green plants in the previous version of PLMItRNA (7). A set of master sequences, containing a single gene sequence for each tRNA species has been produced for both Rodophytae and Stramenopiles thanks to the SRS (Sequence Retrieval System) service at the European Bioinformatics Institute (EBI), http://srs.ebi.ac.uk/. No other algae lineages were found for which mitochondrial tRNA genes have been described. Master sequences were then used to re-analyse the EMBL database with the FastA program for retrieval of similar sequences (http://www2.ebi.ac .uk/fasta3/). The database was updated in July 2000 using each set of master sequences in Viridiplantae, Rodophytae and Stramenopiles. Sugar beet sequences have been downloaded from the EMBLNEW database (accession no. AP000396 and AP000397).
The organisation of PLMItRNA is similar to that described for the previous version (7). Multialignments for Viridiplantae, Rhodophytae and Stramenopiles genes are reported separately.
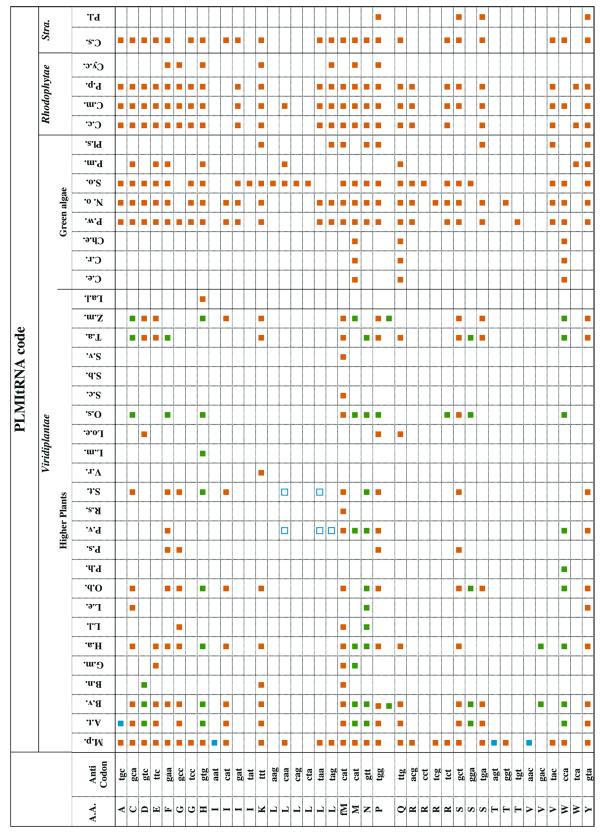
Table 1 shows all complete genes present in PLMItRNA, highlighted according to their genetic origin. For more details on the different origin of genes coding for plant mitochondrial tRNAs see http://bio-www.ba.cnr.it:8000/PLMItRNA. A hypertext link from any coloured square in Table 1 to the related PLMItRNA entry is also available at the database web site. Other links in the site are:
Table 1. Genes coding for mitochondrial tRNAs in Viridiplantae and algae .
Full names for plants and algae are reported in Table S1 (Supplementary Material).
Stra., Stramenopiles; orange square, mitochondrial gene; blue square, nuclear gene; green square, chloroplast gene; blue open square, imported cytoplasmic tRNA found.
• http://bio-WWW.ba.cnr.it:8000/srs6/ for retrieval of entries sharing common features;
• http://bigarea.area.ba.cnr .it:8000/Bi oWWW/fasta.htm where sequences in PLMItRNA are available for database searching.
Users of the database should cite the present publication as a reference. Comments, corrections and new entries are welcome.
SUPPLEMENTARY MATERIAL
Supplementary tables on the database are available at NAR Online.
References
- 1.Bhattacharya D. and Medlin,L. (1998) Algal phylogeny and the origin of land plants. Plant Physiol., 116, 9–15. [Google Scholar]
- 2.Burger G., Saint-Louis,D., Gray,M.W. and Lang,B.F. (1999) Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea: cyanobacterial introns and shared ancestry of red and green algae. Plant Cell, 11, 1675–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreira D., Le Guyader,H. and Philippe,H. (2000) The origin of red algae and the evolution of chloroplast. Nature, 405, 69–72. [DOI] [PubMed] [Google Scholar]
- 4.Nedelcu A.M., Lee,R.W., Lemieux,C., Gray,M.W. and Burger,G. (2000) The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res., 10, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesnick J.M., Goff,M., Graham,J., Ocampo,C., Lang,B.F., Seif,E. and Burger,G. (2000) The mitochondrial genome of the stramenopile alga Chrysodidymus synuroideus. Complete sequence, gene content and genome organization. Nucleic Acids Res., 28, 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stiller J.W. and Hall,B.D. (1997) The origin of red algae: implications for plastid evolution. Proc. Natl Acad. Sci. USA, 94, 4520–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volpetti V., Gallerani,R., De Benedetto,C., Liuni,S., Licciulli,F. and Ceci,L.R. (2000) PLMItRNA, a database for tRNAs and tRNA genes in plant mitochondria: enlargement and updating. Nucleic Acids Res., 28, 159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.