Abstract
Coat protein complex I (COPI) vesicles play a central role in the recycling of proteins in the early secretory pathway and transport of proteins within the Golgi stack. Vesicle formation is initiated by the exchange of GDP for GTP on ARF1 (ADP-ribosylation factor 1), which, in turn, recruits the coat protein coatomer to the membrane for selection of cargo and membrane deformation. ARFGAP1 (ARF1 GTPase-activating protein 1) regulates the dynamic cycling of ARF1 on the membrane that results in both cargo concentration and uncoating for the generation of a fusion-competent vesicle. Two human orthologues of the yeast ARFGAP Glo3p, termed ARFGAP2 and ARFGAP3, have been demonstrated to be present on COPI vesicles generated in vitro in the presence of guanosine 5′-3-O-(thio)triphosphate. Here, we investigate the function of these two proteins in living cells and compare it with that of ARFGAP1. We find that ARFGAP2 and ARFGAP3 follow the dynamic behavior of coatomer upon stimulation of vesicle budding in vivo more closely than does ARFGAP1. Electron microscopy of ARFGAP2 and ARFGAP3 knockdowns indicated Golgi unstacking and cisternal shortening similarly to conditions where vesicle uncoating was blocked. Furthermore, the knockdown of both ARFGAP2 and ARFGAP3 prevents proper assembly of the COPI coat lattice for which ARFGAP1 does not seem to play a major role. This suggests that ARFGAP2 and ARFGAP3 are key components of the COPI coat lattice and are necessary for proper vesicle formation.
Keywords: Cell Compartmentation, Cell Sorting, Golgi, Secretion, Vesicles, ARFGAP1, ARFGAP2, ARFGAP3, ARF1, COPI
Introduction
In the secretory pathway, proteins and lipids are transported between organelles by vesicles. Vesicles are generated by the recruitment of cytosolic protein coats to membranes, resulting in the concentration of protein cargo into the nascent bud and deformation of the donor membrane (1). In the early secretory pathway, coat protein complex II vesicles mediate export of newly synthesized proteins and lipids from the endoplasmic reticulum (ER).2 In contrast, COPI vesicles return proteins from the Golgi to the ER, such as those that have escaped the ER or are required to enter the ER for their functions (e.g. SNARE proteins, Erp44, and p24 proteins) (2). COPI vesicles also have a role in transport of proteins within the Golgi stack, although their precise role in intra-Golgi transport is debated (3). The formation of COPI vesicles is initiated by the small (21-kDa) GTP-binding protein ARF1 (4). When GDP is exchanged for GTP on ARF1, catalyzed by the ARF guanine-exchange factor (ARFGEF) GBF1, it associates tightly with Golgi membranes (5). ARF1 subsequently recruits the 800-kDa, seven-subunit, cytosolic coatomer complex to the Golgi membrane through direct interactions between the GTPase and coat subunits (6). In this way, ARF1 is able to promote the formation of COPI-coated vesicles from donor membranes even in the absence of other protein factors (7). Once the vesicle has budded from the membrane, it must be uncoated for fusion with its target membrane, as evidenced by the inability of coated vesicles to fuse (8). Uncoating of COPI vesicles is mediated by the hydrolysis of ARF1-bound GTP, rendering the coat unstable (9). Because the intrinsic GTPase activity of ARF1 is low, GTP-to-GDP conversion depends on the interaction with an ARF GTPase-activating protein (ARFGAP) (10). The prototypical member of this family of proteins, ARFGAP1, has been extensively investigated in the context of COPI vesicle formation and membrane traffic (11). ARFGAP1 is recruited by ARF1 and interacts with coatomer and is therefore a likely component of the COPI coat during vesicle formation (12–14). Premature stimulation of GTP hydrolysis by ARFGAP1 would prevent stable association of ARF1 and coatomer with the Golgi membrane and therefore counteract vesicle formation. Mechanisms for the temporal and spatial control of ARFGAP1 activity must therefore exist (15). Through one such mechanism, the ability of ARFGAP1 to induce GTP hydrolysis on ARF1 is strongly stimulated by increasing membrane curvature, a mechanism that would ensure that vesicles are rapidly uncoated after budding from the donor membrane (16). COPI vesicles generated in vitro are readily uncoated by the addition of ARFGAP1, demonstrating that this is a key function of ARFGAP1 (17). In addition, regulated GTP hydrolysis by ARFGAP1 is important for cargo concentration (18–20). This could occur through down-regulation of ARFGAP1 activity by cargo proteins, allowing for the formation of priming complexes that ensure cargo concentration through a kinetic “proofreading” mechanism (21, 22). Alternatively, cargo concentration could be promoted by the direct interaction between ARFGAP1 and cargo proteins through a “stochiometric binding” mechanism (13).
In Saccharomyces cerevisiae, the two ARFGAPs Gcs1p and Glo3p are thought to constitute an essential pair with partially redundant function for retrograde transport from the Golgi to the ER (23). Several observations suggest that Glo3p is more important than Gcs1p for COPI vesicle formation. First, deletion of Glo3p, in contrast to any other ARFGAP, causes an impairment in retrieval of dilysine-tagged protein from the Golgi to the ER (24). Second, deleting Glo3p has more severe phenotypes on the function of the secretory pathway than that of Gcs1p knockdown (23). Third, Glo3p but not Gcs1p, interacts with coatomer in vitro and in vivo (25, 26). Fourth, Glo3p, but not Gcs1p, is present on COPI vesicles generated in vitro and is required for their formation (26). Finally, Glo3p, but not Gcs1p, is able to suppress the temperature-sensitive growth of Sec26ts and Arf1pts mutants (27, 28). Strikingly, the ability of Glo3p to rescue temperature-sensitive mutants of coatomer and Arf1p is dependent on a well conserved motif, termed the Glo3 motif, in the C terminus of the protein (28). Through sequence analysis of the human genome, the Glo3 motif was identified in two ARFGAPs termed ARFGAP2 and ARFGAP3 (29). These human Glo3p orthologues are candidates for regulating ARF1 function on the Golgi membrane (30, 31) but have not been studied within the context of COPI function until recently. In support of such a role, ARFGAP2 has been found to interact in vivo with the γ-subunit of coatomer (32) and to co-localize with coatomer on Golgi and intermediate structures (29). The Glo3-type ARFGAPs accumulated on coated vesicles generated in vitro in the presence of a non-hydrolyzable analog of GTP, whereas ARFGAP1 is largely absent from these vesicles (29). The interaction of ARFGAP2 and ARFGAP3 with the Golgi membrane requires coatomer, which is not the case for ARFGAP1 (33). Here, we investigate the properties of ARFGAP2 and ARFGAP3 in vivo, as has been done for other core components of the COPI vesicle machinery (14, 34–37). Our results suggest that the two Glo3-type ARFGAPs play a more direct role for COPI vesicle formation than ARFGAP1, possibly as a structural part of the COPI coat. Importantly, ARFGAP2 and ARFGAP3 seem to be essential for the formation of the vesicle promoting coat lattice.
EXPERIMENTAL PROCEDURES
Reagents
BFA, propranolol, NaF, CaCl, MgCl, NaN3, NH4Cl, formaldehyde, puromycin, saponin, fish skin gelatin, and 1,4-diazabicyclo[2.2.2]octane were from Sigma-Aldrich. DMEM, glutamine, PEST, FBS, and G418 were from Invitrogen. Aluminum chloride was from ICN Biomedicals (Solon, OH).
Plasmids
Plasmids encoding wild-type and mutant (Q71L) ARF1 were described earlier (18). Plasmid encoding ARFGAP1-EGFP (ARFGAP1-GFP) was described earlier (34). Full-length ARFGAP2 (NM_032389) was generated by PCR using clone BC030148 as a template obtained through the IMAGE consortium (available on the World Wide Web). The PCR product was digested and inserted into pCMUIV using BamHI restriction sites (5′ and 3′). Plasmid encoding for ARFGAP2Δ147–521 was generated through PCR, digested, and inserted into pRSETA and pGEX-4T-3 using BamHI restriction sites (5′ and 3′). To generate ARFGAP2-EGFP, full-length ARFGAP2 was cut out of pCMUIV and inserted into pEGFPN3 (Clontech) using the BglII (5′) and BamHI (3′) restriction enzymes. In this orientation, the 5′ BamHI site is spoiled, whereas the 3′ site is kept intact. From the resulting plasmid a fragment was removed using XhoI (internally) and BamHI (3′) restriction enzymes. Simultaneously, full-length ARFGAP2 lacking the stop codon was generated by PCR. The resulting PCR product was cut using XhoI (internally) and BamHI (3′) to generate a 320-bp fragment for the 3′ part of ARFGAP2. This fragment replaced the part that was removed from the plasmid above to generate pEGFPN3-ARFGAP2. Full-length ARFGAP3 (NM_014570) was generated by PCR using pBAD-ARFGAP3 as a template (30) and inserted into pCMUIV using the BamHI restriction sites (5′ and 3′). Plasmid encoding for ARFGAP3Δ140–521 was generated by PCR, digested, and inserted into pRSETA and pGEX-4T-3 using BamHI restriction sites (5′ and 3′).
Protein Purification
Truncated GST-tagged ARFGAP2(147-521) and ARFGAP3(140–516) were purified from the BL21 strain of bacteria on glutathione beads according to the manufacturer's instructions (GE Healthcare) and used for generation of polyclonal antisera. Truncated His6-tagged ARFGAP2(147–521) and ARFGAP3(140–516) were purified under denaturing conditions on nickel beads according to the manufacturer's instructions (Qiagen, Germany). Protein was dialyzed for affinity purification of antibodies.
Antibodies
Truncated GST-tagged ARFGAP2(147–521) and ARFGAP3(140–516) were injected into rabbits to generate antisera (Sigma-Genosys). Polyclonal antisera were affinity-purified against His6-tagged truncated protein coupled to CnBr-activated Sepharose beads according to the manufacturer's instructions. Alexa Fluor 488-tagged and 594-tagged secondary antibodies were purchased from Invitrogen. Anti-α-tubulin was from Abcam (Cambridge, UK). Generation of polyclonal antibodies against ARFGAP1 has been described earlier (21). Monoclonal antibodies to native coatomer, CM1A10 (6), and β-COP, M3A5 (38), were kind gifts from Drs. Rothman and Kreis, respectively. Monoclonal antibody against β1,4-galactosyltransferase I was purchased from CellMab (Mölndal, Sweden).
Cell Culture, Treatments, and Transfections
HeLa cells were grown in DMEM supplemented with 10% FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 units/ml streptomycin. Generation of stable HeLa cell lines expressing ϵ-COP-GFP and ARF1-GFP was described earlier (34). BFA was stored in ethanol at −20 °C as a 1000× stock (5 mg/ml). Propranolol was prepared fresh for every experiment. For aluminum fluoride treatment of cells, 50 μm AlCl3 and 20 mm NaF were added separately to the culture media. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) or Fugene (Roche Applied Science) according to the manufacturer's instructions. Cells were analyzed 24 h after transfection.
siRNA
For RNA interference experiments, siRNA oligonucleotides were transfected with the RNAiMAX transfection reagent according to the manufacturer's instructions (Invitrogen). siRNA was purchased from MWG Eurofins Operon (Ebersberg, Germany). ARFGAP1 and ARFGAP3 were knocked down using previously published sequences (29). For ARFGAP2, efficient knockdown was obtained using the following target sequence: 5′-GGAGCAGGAAGTGTATCTCTG-3′. As a negative control, the nonspecific sequence (47% GC content) 5′-AGGUAGUGUAAUCGCCUUG-3′ was used. Efficiency and specificity of knockdown was evaluated by Western blot and immunofluorescence 48 h post-transfection. SDS-PAGE and Western blot were performed as before (18).
Immunocytochemistry
For all fluorescence imaging, an LSM 510 META confocal microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany) was employed. HeLa cells were seeded onto sterile coverslips and allowed to grow for 48 h prior to fixation. Following fixation with 4% formaldehyde in PBS, coverslips were incubated in 50 mm NH4Cl in PBS to quench remaining aldehyde groups. After washing three times with PBS, cells were permeabilized using 0.1% saponin in PBS. Nonspecific antibody binding sites were blocked using 0.2% fish skin gelatin in PBS. Cells were incubated with appropriate polyclonal or monoclonal primary antibody as indicated for the individual experiments. Primary antibodies were revealed using Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 chicken anti-mouse secondary antibodies. Cells were mounted on glass slides using homemade Mowiol mounting medium with anti-fade agent (1,4-diazabicyclo[2.2.2]octane) prior to microscopy. Alexa Fluor 488 was excited by the 488-nm argon ion laser line, and the fluorescence was collected using a BP505–530 emission filter, whereas Alexa Fluor 594 was excited by the 543-nm HeNe laser line, and the fluorescence was collected using an LP560 emission filter. All images were acquired using a Plan-Apochromat ×63/1.40 oil differential interference contrast objective in sequential scanning (multitrack) mode with the pinholes set to obtain an optical section of about 0.8 μm in both channels (∼1 Airy unit).
Live Cell Imaging
HeLa cells were grown on glass bottom dishes (MatTek Corp., Ashland, MA) and imaged in medium without phenol red. GFP was excited by the 488-nm argon ion laser line, and the fluorescence was collected using a LP505 emission filter using a C-apochromat ×40/1.20 W Corr objective or a numerical aperture 0.8 Neofluar ×25 variable immersion objective. For all quantitative imaging, the pinhole was fully opened, and the laser intensity was low enough to avoid bleaching during the imaging time period. Imaging time series monitoring the effect of drug treatments were acquired during 5 min (BFA treatment and BFA after AlF treatment), 10 min, or 30 min (AlF treatment). Images were acquired every 5–30 s, depending on the total length of the time series. The delay time was changed to 0 s for refocusing when required. Fluorescence recovery after photobleaching (FRAP) was performed. Initially, prebleach images were acquired. The Golgi area was then bleached by intense 488-nm excitation (100% transmission), followed by acquisition of images monitoring the recovery of Golgi fluorescence during ∼2–5 min. FRAP time series were performed with a 0.5–5-s image acquisition interval. Fluorescence loss in photobleaching (FLIP) was performed by taking time series of images during ∼10 min, monitoring the fluorescence loss in the Golgi apparatus due to bleaching of the cytosol pool of fluorescent molecules. Each image in the FLIP time series was followed by a bleach event, in which the whole cell apart from the Golgi area was bleached by intense 488-nm excitation (100% transmission).
Quantification and Evaluation of Data
All light microscopy quantification was performed in ImageJ (National Institutes of Health), where the mean intensities of the Golgi region and the whole cell were measured. The intensities, as functions of time, were imported to Sigma Plot (SPSS Inc.) or to Kaleidagraph (Synergy Software, Reading, PA). All curve fits employed the Marquardt-Levenberg algorithm. The FRAP analysis was performed by fitting the recovery curve with a single exponential function, I(t) = I0 + I∞ × (1 − e(−t/T), where I0 is the remaining fluorescence immediately after bleaching, I∞ is the finally recovered fluorescence, and T is the recovery time, which is related to the half-time for the recovery through t½ = ln 2 × T. The FLIP analysis was performed by fitting the decaying curve with a single exponential function, I(t) = I∞ + I0 × e(−t/T), where I∞ is the final remaining fluorescence at the end of the time series, I0 is the initial fluorescence, and T is the decay time, which is related to the half-time for the decay through t½ = ln 2 × T.
Electron Microscopy
For electron microscopy, cells were fixed using a double fixation protocol with osmium and tannic acid (39). Samples were dehydrated in graded ethanol series and embedded in Epon 812 (Serva). After 48 h at 60 °C, ultrathin sections (60 nm) were cut and mounted on grids. Samples were examined on a LEO 912 OMEGA energy filter transmission electron microscope (Zeiss) at 120 kV accelerating voltage. Digital images were obtained through a side-mounted MegaView III TEM CCD camera and quantified according to Ref. 40.
RESULTS
The Golgi Localization of ARFGAP2 and ARFGAP3 Is Dependent on ARFGTP
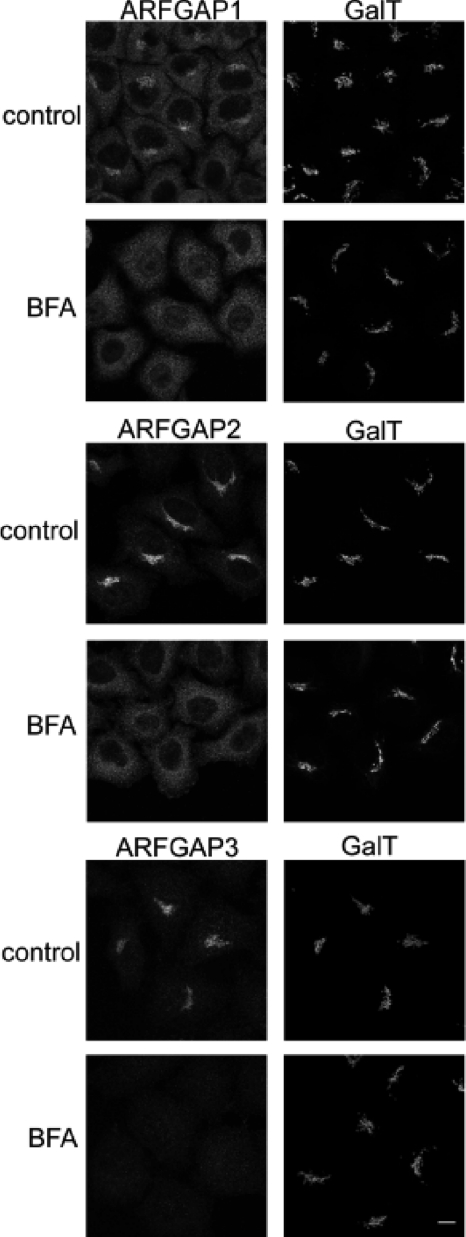
To examine the role of ARFGAP2 and ARFGAP3, we generated affinity-purified antibodies and fluorescent fusion protein constructs to monitor the behavior of the proteins upon manipulations of cells (see “Experimental Procedures”). As judged by immunofluorescence, we found that ARFGAP2 and ARFGAP3 co-localized with coatomer on the Golgi membrane, including some of the peripheral sites labeled with anti-coatomer antibodies (data not shown). This is in accordance with an earlier published report using independent antibodies to the different ARFGAPs (29). To probe the mechanism by which ARFGAP2 and ARFGAP3 associate with the Golgi membrane, we treated cells with the fungal metabolite BFA to inhibit the GDP for GTP exchange on ARF1 through ARFGEFs (41). This causes the dissociation of coatomer as well as ARFGAP1 from Golgi membranes (12, 42). This short treatment prevented any significant effect of BFA on Golgi structure (Fig. 1, right). This treatment therefore highlights the acute effect of BFA on peripheral membrane proteins that localize to the membrane by an ARFGEF-dependent mechanism, presumably involved in ARF1-dependent vesicle formation. Although the levels of β1,4-galactosyltransferase I are unperturbed, the Golgi pools of ARFGAP1, ARFGAP2, and ARFGAP3 are completely redistributed from the Golgi to the cytosol (Fig. 1, left). This suggests that all three ARFGAPs associate with the Golgi membrane as a consequence of GEF-mediated ARF activation. Furthermore, it highlights the highly dynamic nature of this Golgi localization because redistribution occurs within 1 min of BFA addition. To compare the effects of BFA on the GFP-tagged versions of the ARFGAPs, we performed the same experiment on HeLa cells expressing these as fusion proteins (data not shown). Here, we observed a BFA-resistant pool of ARFGAP1-GFP after 5 min of treatment with BFA, as demonstrated by a previous study (14). ARFGAP2-GFP and ARFGAP3-GFP, however, displayed similar sensitivity as endogenous protein toward BFA with no remaining pool on the Golgi after 5 min.
FIGURE 1.
The Golgi localization of the ARFGAPs is dependent on ARFGTP. HeLa wild-type cells were treated with BFA (5 μg/ml) for 1 min and stained with antibodies toward ARFGAP1–3 (left). To verify that this treatment did not affect Golgi structure, cells were co-stained with antibody against β1,4-galactosyltransferase I (GalT; right). Scale bar, 10 μm.
Stimulation of Vesicle Budding Causes Accumulation of Glo3-type ARFGAPs on the Golgi Membrane
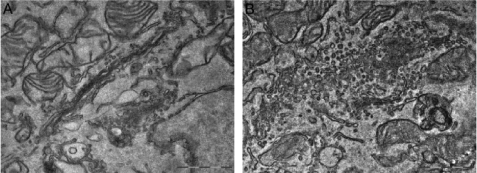
Having established that ARFGAP2 and ARFGAP3 dissociate from the Golgi upon inhibition of vesicle formation by BFA, we investigated the behavior of these proteins upon stimulation of vesicle formation. We employed aluminum fluoride (AlF) as a way of perturbing the GTP state of ARF1. For small G proteins, such as ARF1, the ionic complex of aluminum and fluoride mimics the terminal γ-phosphate of GTP, causing them to switch to a GTP-like conformation (43). This occurs only in the presence of the corresponding ARFGAP (44). The addition of this compound has been shown to induce the formation of a tripartite complex of ARF1-ARFGAP1-coatomer that drives vesicle formation both in vitro and in vivo (14, 16). In cells treated with AlF, coatomer is tightly associated with the Golgi membrane and cannot be dissociated by the addition of BFA (45–47). In vitro, AlF has been shown to cause the accumulation of coated and fusion-incompetent coated vesicles from purified membranes (48, 49). Based on these observations, we examined the effect of AlF on the ARFGAP2 and ARFGAP3 in vivo. To verify that AlF also stimulates vesicle budding in living cells, we treated cells with AlF and examined the morphology of the Golgi apparatus by electron microscopy (Fig. 2). Control cells have a typical Golgi stack consisting of 3–5 cisternae with few peri-Golgi vesicles (Fig. 2A). The addition of AlF for 10 min to the culture medium caused a massive accumulation of vesicles in the Golgi area, with significant consumption of cisternae (Fig. 2B). Incubation for a longer period of time (30 min) produced a similar phenotype (data not shown). Therefore, the effect of AlF is mediated not only by locking coatomer to the Golgi membrane but also by stimulating vesicle budding from the Golgi complex.
FIGURE 2.
The addition of AlF induces vesicle budding from the Golgi. Micrographs of HeLa WT control cells (A) have typical Golgi stacks. The addition of AlF for 10 min (B) causes massive accumulation of vesicles. Scale bar, 1 μm.
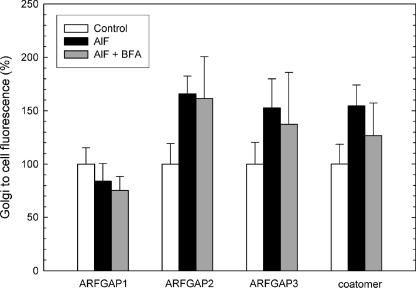
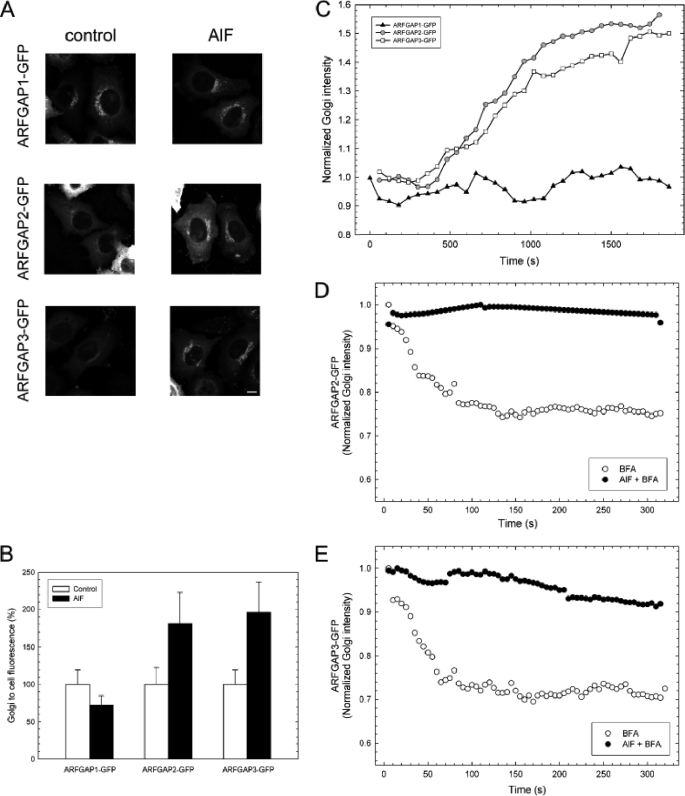
We took advantage of the ability of AlF to cause recruitment of coatomer and stimulate vesicle budding from the Golgi membranes in cells and monitored all three ARFGAPs proposed to be involved in COPI vesicle budding to compare their functions. HeLa cells were treated with AlF for 10 min or double-treated with AlF followed by BFA treatment (5 μg/ml) for 5 min. The levels of coatomer and ARFGAPs were revealed by immunofluorescence and quantified to indicate the level of protein associated with the Golgi. Strikingly, AlF addition causes Golgi accumulation of both coatomer and ARFGAP2 and ARFGAP3 but not of ARFGAP1 (Fig. 3). Coatomer, ARFGAP2, and ARFGAP3 increased their Golgi-to-cell ratios by ∼50%, whereas ARFGAP1 showed no increased Golgi association. Furthermore, recruited protein was incorporated into a BFA-resistant complex because subsequent addition of BFA did not remove the higher level of ARFGAP from the membrane (Fig. 3). To verify this, cells expressing ARFGAP1-GFP, ARFGAP2-GFP, and ARFGAP3-GFP were treated with AlF (Fig. 4). In these cells, recruitment of ARFGAP2-GFP and ARFGAP3-GFP was significant (about 75% higher than control levels), whereas ARFGAP1 demonstrated a slight decrease (about 30% lower than control levels, p < 0.05) (Fig. 4, A and B). To rule out the possibility that ARFGAP1 recruitment takes place with slower or delayed kinetics, we followed the accumulation of the ARFGAPs upon the addition of AlF in live cells for prolonged periods of time, up to 30 min (Fig. 4C). Whereas ARFGAP2-GFP and ARFGAP3-GFP continuously accumulated during this time period (consistent with continuous vesicle budding), the levels of ARFGAP1-GFP remained unchanged from initial levels. To establish that the GFP-tagged ARFGAP fusions are incorporated into a BFA-resistant complex, in a manner similar to the endogenous proteins, we treated cells expressing ARFGAP2-GFP and ARFGAP3-GFP with AlF and BFA (Fig. 4, D and E). Clearly, fusion proteins remain localized to the Golgi during BFA treatment if they are pretreated with AlF, indicating that they are protected against the effect of BFA, similar to the endogenous protein pool. This all suggests that ARFGAP2 and ARFGAP3 are recruited and incorporated into the growing COPI coat lattice upon stimulation of vesicle budding in vivo, whereas ARFGAP1 is excluded.
FIGURE 3.
AlF-induced association of ARFGAPs with the Golgi membrane. HeLa cells (n = 25) treated with AlF and BFA were stained with antibodies toward the ARFGAPs and coatomer. Quantification of the Golgi/cell ratio reveals Golgi accumulation of ARFGAP2, ARFGAP3, and coatomer upon the addition of AlF ± S.D. (error bars).
FIGURE 4.
AlF treatment promotes association of the Glo3-type ARFGAP-GFPs with the Golgi membrane. A, HeLa cells expressing GFP-tagged versions of ARFGAP1–3 (left) were treated with AlF (right) and imaged. The addition of AlF increases the association of ARFGAP2-GFP and ARFGAP3-GFP with the Golgi but does not increase the levels of ARFGAP1-GFP. Scale bar, 10 μm. B, Golgi-to-cell fluorescence was quantified on cells (n = 25) expressing the ARFGAPs ± S.D. (error bars). The increase of Golgi association is around 75% for ARFGAP2-GFP and ARFGAP3-GFP, whereas ARFGAP1-GFP decreases about 30% (p < 0.05). C, cells expressing GFP-tagged versions of the ARFGAPs were treated with AlF, and the Golgi intensity was continuously monitored. During a period of 30 min, recruitment of ARFGAP2-GFP (circles) and ARFGAP3-GFP (squares) occurs, whereas ARFGAP1-GFP (triangles) levels are stable. Pretreatment of cells expressing ARFGAP2-GFP (D) or ARFGAP3-GFP (E) with AlF (black circles) protects against BFA-induced redistribution of the Golgi pools.
Incorporation of ARFGAP2 and ARFGAP3 in a BFA-resistant Complex Is Driven by ARF1
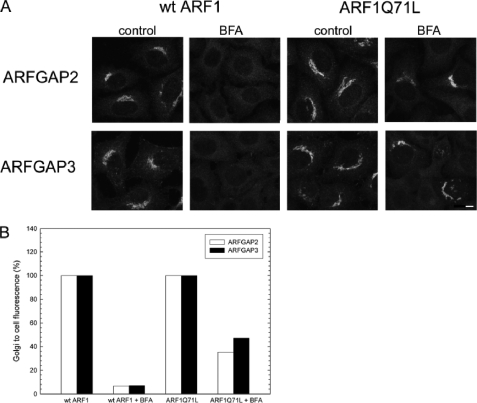
Because AlF is an activator of not only ARF1 but also trimeric G proteins, it has been suggested that some of its effects on coatomer function are mediated through this mechanism (45). Expression of the GTP-restricted mutant ARFQ71L is able to recruit coatomer to the Golgi even in the presence of BFA (50). In relation to the ARFGAPs, expression of ARFQ71L locks a significant portion (40%) of ARFGAP1 on the Golgi membrane (14). Because ARFGAP2 and ARFGAP3 require ARFGTP for proper Golgi localization (Fig. 1), the expression of ARF1Q71L should cause irreversible binding of these proteins on the Golgi. To test this, we expressed ARFWT and ARFQ71L in HeLa cells, treated the cells with BFA (5 μg/ml) for 1 min, and revealed the distribution of coatomer (data not shown), ARFGAP2, and ARFGAP3 by immunofluorescence (Fig. 5). As predicted, the expression of ARFQ71L caused BFA-resistant association of ARFGAP2 and ARFGAP3 to the Golgi membrane (Fig. 5A). Approximately 40% of cells displayed a BFA-resistant pool of ARFGAP2 or ARFGAP3 (Fig. 5B). This suggests that the AlF-induced accumulation of ARFGAP2 and ARFGAP3 is mediated by ARF1.
FIGURE 5.
Golgi association of ARFGAP2 and ARFGAP3 is promoted by ARF1-GTP. A, cells expressing ARF1WT or ARF1Q71L were treated with BFA (5 μg/ml) and stained with antibodies against ARFGAP2 (top) or ARFGAP3 (bottom). Scale bar, 10 μm. B, quantification of this effect. The proportion of cells (percentage of control) exhibiting typical Golgi staining of the ARFGAPs after expression of ARF1Q71L demonstrates the protective effect of this expression.
AlF Locks ARFGAP2 and ARFGAP3 on the Golgi Membrane
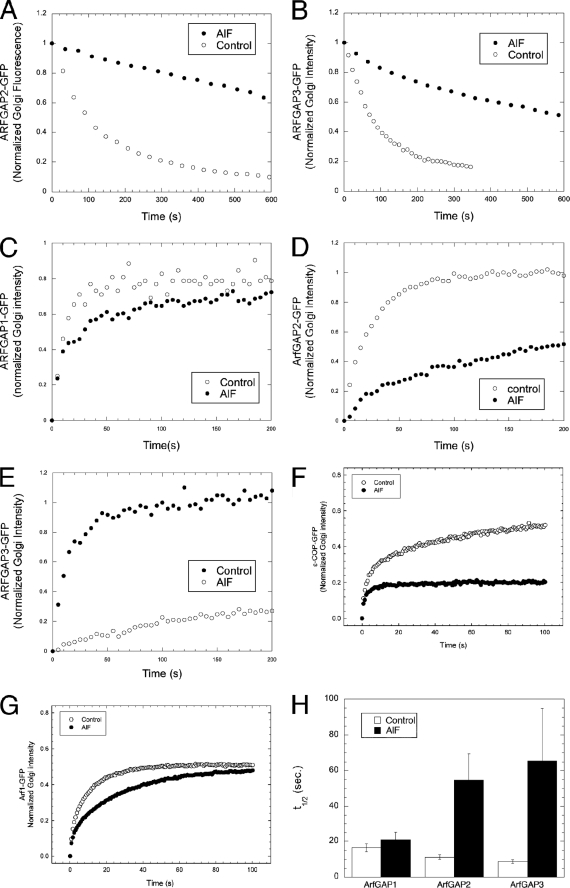
Next, we examined the dynamic nature of the binding and unbinding kinetics of ARFGAP2 and ARFGAP3 to the Golgi membrane, an approach that has yielded important insights into COPI coat assembly. For example, it has been shown that the addition of AlF to cells locks coatomer irreversibly on Golgi membranes, whereas ARF1 is still able to detach (albeit with altered kinetics) (34, 36). Therefore, the higher levels of ARFGAP2 and ARFGAP3 recruited to the Golgi upon the addition of AlF could be due to an increased number of binding sites and not due to irreversible association with the membrane. To discriminate between these possibilities and to quantify the extent of possible immobilization on the Golgi, we analyzed the membrane binding of the ARFGAPs by FRAP. If the hypothesis is correct that ARFGAP2 and ARFGAP3 are more closely linked with coatomer as coat components, then they should be irreversibly bound to the Golgi upon the addition of AlF, possibly to the same extent as coatomer (34, 36). First, we tested the nature of the association of ARFGAP2-GFP and ARFGAP3-GFP with the Golgi membrane to exclude the possibility that there is a stable pool binding to the Golgi that would interfere with the analysis. To do this, we used FLIP by bleaching the cytosol repetitively and analyzed the loss of Golgi-associated ARFGAP over time (Fig. 6, A and B). ARFGAP2 and ARFGAP3 are removed from the Golgi by repetitive bleaching and are eventually completely removed from the Golgi. The shapes of the recovery curves suggested a simple exponential decay. ARFGAP2 showed a t½ for removal from Golgi membranes of 64 ± 5.8 s (n = 12), and ARFGAP3 showed a t½ of 134 ± 30.3 s (n = 8), as compared with a t½ for ARFGAP1 of 68 ± 17.7 s (n = 13). This demonstrates that the entire Golgi-associated pool of ARFGAP2 and ARFGAP3 is dynamically exchanged with the cytosolic pool and that no stable association exists with the Golgi membrane. Next, we examined the effect of AlF on this dynamic association of ARFGAP2 and -3 with Golgi membranes using the same FLIP protocol. Loss of ARFGAP2 and -3 from Golgi membranes was greatly impeded (Fig. 6, A and B) with a substantial fraction remaining after 10 min (t½ for loss of ARFGAP2 of 444 ± 151 s (n = 13); t½ for loss of ARFGAP3 of 573 ± 138 s (n = 10)). This demonstrates substantial immobilization of ARFGAP2 and -3 on Golgi membranes by AlF, similar to the effect on coatomer. We used FRAP to further analyze the effect of AlF on the dynamics of ARFGAP association with Golgi membranes as well as ARF1 and coatomer. FRAP is more suitable than FLIP to accurately measure fast dynamics because FLIP requires repetitive bleach of the entire cellular cytoplasm. This can be slow due to laser energy being spread over the entire area of the cell, leading to bleaches of 30–45 s/bleach cycle on our system. Thus, bleaching is probably rate-limiting for processes on this time scale. With FRAP, in contrast, a single bleach over a smaller area suffices, and the recovery curve is obtained after bleaching is complete. For FRAP, we bleached the entire pool of Golgi-associated GFP-tagged protein and monitored and fitted the recovery (Fig. 6, C–G). The recovery curves of ARFGAP2-GFP (Fig. 6D) and ARFGAP3-GFP (Fig. 6E) were similar to those of ARFGAP1-GFP (Fig. 6C) and ARF1-GFP (Fig. 6G) with time constants ranging from 8 to 12 s. This demonstrates that these ARFGAPs associate with the Golgi membrane with approximately the same kinetics as previously shown for ARFGAP1 and ARF1 (T = 10 s) (34). Upon the addition of AlF, coatomer becomes locked on the Golgi to a significant extent, indicating irreversible binding (Fig. 6F), whereas ARF1 shows only a moderate increase in recovery time (Fig. 6G), in agreement with previous studies (34). For ARFGAP1, t½ for recovery in the FRAP experiment is not significantly changed relative to control (Fig. 6H). ARFGAP2 and -3 show strikingly slower recoveries, with t½ for the mobile fraction increasing 6-fold to around 60 s (Fig. 6H). These results demonstrate that ARFGAP2 and ARFGAP3 are immobilized by AlF to a much greater extent than ARF1 and approach the complete immobilization of coatomer. This suggests that during the process of vesicle formation, ARFGAP2 and ARFGAP3 become recruited to the COPI coat lattice in a manner that is similar to coatomer.
FIGURE 6.
Dynamic association of ARFGAP2 and ARFGAP3 with the Golgi membrane. Repetitive photobleaching of the cytoplasm causes the loss of the entire pool of fluorescence of ARFGAP2-GFP (A) and ARFGAP3-GFP (white circles) (B), demonstrating the dynamic association of these proteins with the Golgi membrane. AlF treatment (A and B) impairs loss of these proteins (black circles). C–G, recovery of Golgi fluorescence after a single bleach event in the absence (white circles) and presence of AlF (black circles) for ARFGAP1-GFP (C), ARFGAP2-GFP (D), ARFGAP3-GFP (E), ϵ-COP-GFP (F), and ARF1-GFP (G). H, average (n = 12 cells) t½ for ARFGAPs treated or not treated with AlF ± S.E. (error bars). The addition of AlF prolongs ARFGAP2-GFP and ARFGAP3-GFP association with Golgi membranes similarly to its effect on ϵ-COP-GFP.
ARFGAP2 and ARFGAP3 Are an Essential Pair for COPI Coat Assembly
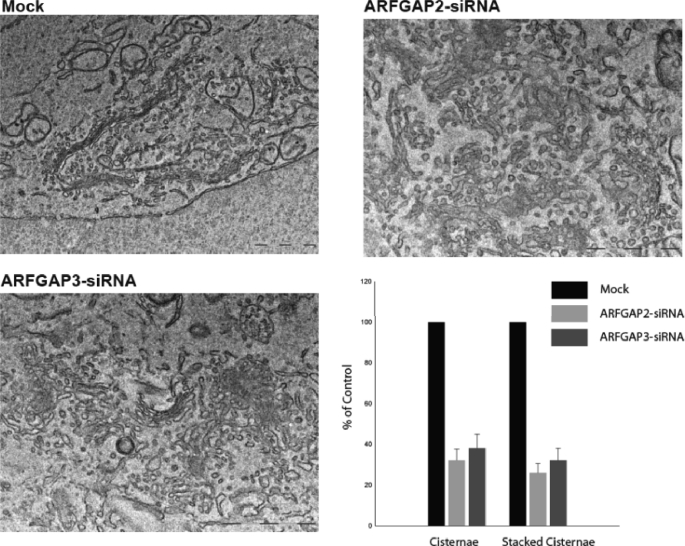
Given the role of the ARFGAPs to stimulate GTP hydrolysis by ARF, excess activity should destabilize the membrane association of ARF and its effector molecules In support of this, overexpression of ARFGAP1 causes dissociation of coatomer from the Golgi membrane and relocalization of Golgi enzymes to the ER (51). Therefore, we expressed ARFGAP2 and ARFGAP3 at high levels and examined the effect on Golgi structure by immunofluorescence. We did not see an obvious effect of such overexpression on Golgi structure or coatomer localization (data not shown). A more refined approach was therefore necessary for the investigation of the role of these ARFGAPs. The results presented in this study suggest that the Glo3-type ARFGAPs are actively recruited into the growing COPI coat lattice for vesicle budding, consistent with a structural role for these ARFGAPs. To test the importance of these ARFGAPs within this lattice, we analyzed the effects of reducing the expression of the individual ARFGAPs. Knockdown of individual ARFGAPs in mammalian cells does not cause lethality, indicating functional redundancy. Only when all three ARFGAPs are knocked down do cells exhibit growth arrest and die (29). To examine the phenotypes upon knockdown of each ARFGAP, we transfected cells with siRNA (as described under “Experimental Procedures”). Single or double knockdown of the ARFGAPs was found to be efficient, as determined by immunofluorescence (supplemental Fig. 1A) and Western blotting (supplemental Fig. 1B). Knockdown of one paralogue did not alter the level of the other ARFGAP, demonstrating the specificity of the antibodies. Knockdown of ARFGAP2 or ARFGAP3 caused a slight alteration of Golgi morphology at the level of light microscopy. The Golgi appeared slightly fragmented and compacted (supplemental Fig. 1A). At the ultrastructural level (Fig. 7), cells transfected with siRNA to either ARFGAP2 or ARFGAP3 revealed Golgi areas with considerably less stacked Golgi cisternae as well as a significant decrease in the number of observable Golgi cisternae, similar to our ultrastructural observations of AlF-treated cells (Fig. 2). Importantly however, the Golgi remained in its juxtanuclear localization in most cells.
FIGURE 7.
Electron microscopy of cells transfected with RNAiMock and RNAiARFGAP2 and RNAiARFGAP3. Thin plastic embedded sections (60 nm thick) of HeLa cells were examined at the ultrastructural level. Knockdown of ARFGAP2 and ARFGAP3 caused significant structural impairment of the Golgi stacks. Top left, cells transfected with RNAiMock have aligned stacks that are part of the Golgi ribbon. Top right and bottom left, cells transfected with RNAiARFGAP2 and RNAiARFGAP3 show Golgi regions composed of fewer cisternae and stacked structures. Bottom right, quantitation of number of cisternae and stacked cisternae as compared with control (Mock). Error bars, S.E.
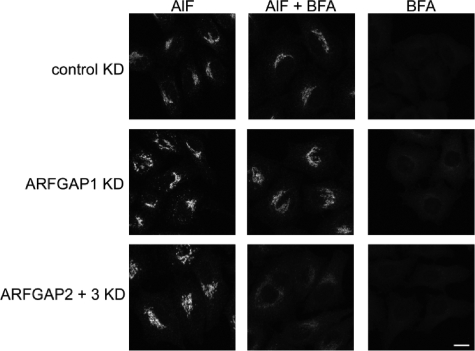
We examined the ability to generate the AlF-induced BFA-resistant COPI coat lattice in the absence of ARFGAP1–3. An absence of key structural factors of this complex (such as coatomer or relevant ARFGAPs) would prevent its efficient formation. Therefore, we examined the effect of ARFGAP knockdowns on the localization of coatomer to the Golgi upon double treatment with AlF and BFA. Knockdown using a control oligonucleotide or oligonucleotides against ARFGAP1 did not have any effect on the formation of this complex because coatomer remained localized to the Golgi upon the addition of BFA (Fig. 8). The single knockdown of ARFGAP2 or ARFGAP3 or combinations with ARFGAP1 did not have any effect either (data not shown). Interestingly however, knocking down both ARFGAP2 and ARFGAP3 restored BFA sensitivity of coatomer, even after pretreatment with AlF (Fig. 8). This suggests that these two ARFGAPs are essential components of the ARF1-ARFGAP-coatomer complex. In the absence of these two ARFGAPs, the complex that protects coatomer against the action of BFA is not efficiently formed, suggesting an essential contribution of these ARFGAPs to COPI vesicle formation, a function that cannot be supplied by ARFGAP1. It does suggest that one of the Glo3-type ARFGAPs is sufficient for this complex to be formed, consistent with a redundancy in function of ARFGAP2 and ARFGAP3.
FIGURE 8.
The ARFGAP2 and ARFGAP3 pair is essential for generation of the coat lattice. Cells treated with siRNA against the ARFGAPs or combinations thereof were treated with AlF or BFA or double-treated with AlF and BFA. Treated cells were stained for coatomer. Cells treated with control or siRNA against ARFGAP1 demonstrate BFA resistance after AlF treatment. After double knockdown (KD) of ARFGAP2 and ARFGAP3, coatomer becomes sensitive to BFA with redistribution of coatomer to the cytosol. Scale bar, 11 μm.
ARFGAP2 and ARFGAP3 Act at an Early Stage of Vesicle Formation
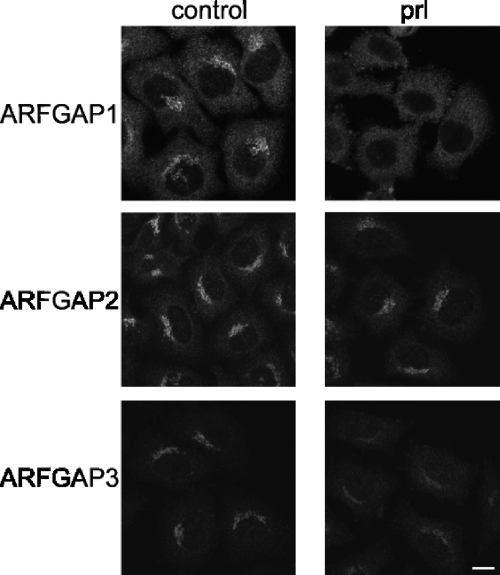
An important regulator of ARFGAP1 is the lipid diacylglycerol that stimulates binding and activity of the ARFGAP to the Golgi membrane (52). Lipid binding is mediated by the ARFGAP1 lipid-packing sensor domain present in ARFGAP1 (53). The role of lipid regulation of Glo3-type ARFGAPs remains an open question. ARFGAP3 activity shows some sensitivity to lipids, but the functional significance of this is not known (30). The importance of lipids for ARFGAP1 binding to the Golgi is highlighted by the rapid dissociation of ARFGAP1 upon the addition of propanolol, which prevents the formation of diacylglycerol (40). To examine the effect on ARFGAP2 and ARFGAP3, we added propranolol to cells (300 μm) for 3 min and monitored the levels of ARFGAPs on the Golgi membrane. Whereas ARFGAP1 dissociates quickly from the membrane, ARFGAP2 and ARFGAP3 both have propanolol-resistant pools (Fig. 9), suggesting that the lipid composition is not essential for their binding to Golgi membranes. This is consistent with recent findings showing that binding of ARFGAP2 and ARFGAP3 to liposomes does not depend on curvature (33). Instead, association of both ARFGAPs appears to depend more on the interaction with coatomer (33).
FIGURE 9.
The effect of propranolol treatment on Golgi association of the ARFGAPs. Cells treated with propranolol (prl; 300 μm for 3 min) were stained with antibodies for ARFGAP1 to -3. ARFGAP1 is highly sensitive to propranolol exposure. ARFGAP2 retains its Golgi localization after propranolol treatment, whereas ARFGAP3 has an intermediate sensitivity. Scale bar, 10 μm.
DISCUSSION
ARF1 regulates the membrane association of several coat proteins as well as lipid-modifying enzymes for the regulation of membrane traffic. To ensure that these diverse activities of ARF1 maintain spatial segregation, mechanisms must exist to ensure site-specific regulation of ARF1. One example of this is the compartment-specific localization of the ARFGEFs within the Golgi apparatus that restricts activation of ARF1 for certain processes to the cis-Golgi or the trans-Golgi network, respectively (5). In addition, the 24 genes containing an ARFGAP domain are candidates for providing GTPase-activating functions to the various roles performed by ARF proteins (54, 55).
One mechanism by which ARFGAPs could influence vesicle formation is as structural components of the protein coat of transport vesicles (15). With regard to COPI vesicle formation, ARFGAP1 has many characteristics consistent with a role as a coat protein, including interactions with cargo proteins (such as the KDEL-R and p24 proteins) and with the coatomer complex itself (reviewed in Ref. 11). ARFGAP2 and ARFGAP3 may contribute to vesicle formation through a coat protein mechanism, as suggested by studies in both yeast and mammalian cells. Here, we test this hypothesis by comparing the dynamic behavior of these three ARFGAPs in living cells. We find that ARFGAP2 and ARFGAP3 associate transiently and dynamically with Golgi membrane under the control of the GTPase cycle of ARF1, like other vesicle proteins. When ARF function is inhibited by the addition of BFA, the Golgi pool of ARFGAPs dissociates from the membrane, consistent with a role as a protein involved in coat lattice formation. By adding AlF to cells, we stimulate vesicle budding leading to the recruitment of coatomer and ARFGAP2 and ARFGAP3 to the Golgi. In contrast, ARFGAP1 does not show such an accumulation but rather a decrease in Golgi levels. Although the quality of the antibody does not allow for an accurate estimate of this effect, it strongly suggests that ARFGAP1 is excluded from the vesicles that are formed upon the addition of AlF. The decrease of Golgi-associated ARFGAP1-GFP upon the addition of AlF (about 30%) may indicate this (Fig. 5). This suggests a preferential incorporation of ARFGAP2 and ARFGAP3 over ARFGAP1 in vivo as well as in vitro. The presence of a high number of Glo3-type ARFGAPs in the coat lattice predicts an important role for assembly of the coat. The overexpression of the ARFGAPs did not remove coatomer from the Golgi membrane, as does overexpression of ARFGAP1. One possible explanation for this would be that coatomer is rate-limiting for the ability of Glo3-type ARFGAPs to induce GTP hydrolysis on ARF1 on the Golgi. This suggests coatomer-dependent recruitment of ARFGAP2 and ARFGAP3 to the Golgi membrane, as recently suggested (33). This study concludes that ARFGAP2 and ARFGAP3 are coatomer-dependent for their function with the membrane. To investigate the role within the coat lattice, we performed knockdown experiments of the three ARFGAPs. Strikingly, double knockdown of ARFGAP2 and ARFGAP3 prevented the AlF-dependent assembly of the coat lattice most efficiently. The fact that the single knockdown of either ARFGAP does not have an effect on the coat assembly suggests that there is an overlapping function between ARFGAP2 and ARFGAP3 but that they do perform a function not supplied by ARFGAP1. Redundant functions between the ARFGAPs involved in retrograde transport have been demonstrated in both yeast and humans (23, 29). Interestingly, our electron microscopic analysis showed significant disruption of Golgi structure upon knockdown of ARFGAP2 or ARFGAP3 alone, which could suggest some specialization of function. Electron microscopy can detect phenotypes that do not completely block a process or prevent cell survival and is potentially a more sensitive measure in this case than the abolition of AlF-induced resistance of COPI to AlF, where elimination of a single ARFGAP may leave a significant portion of COPI resistant to AlF and prevent BFA-induced loss of the total pool.
Lippincott-Schwartz and colleagues (14) reported that a significant portion (60%) of ARFGAP1-YFP associates with the Golgi in the presence of BFA (i.e. in an ARF1-independent way). The remaining pool of ARFGAP1-YFP (40% in their study) that does become locked on the Golgi after the addition of AlF could therefore represent the pool that associates with the Golgi by binding to ARF1 and is recruited into an ARF1 complex. In this paper, we present evidence that suggests that there is no ARF1-independent association of endogenous ARFGAP1 to -3 with the Golgi membrane. Instead, we propose an alternative explanation for the less drastic effect of AlF on ARFGAP1 association with the Golgi as compared with the Glo3-type ARFGAPs. ARFGAP1 function in the Golgi apparatus is not likely to be limited to coatomer function because ARFGAP1 is also implicated in AP-1 recruitment (56). This is also true in yeast, where Gcs1p appears to be involved in both retrograde transport and post-Golgi transport (57). Interestingly, AlF does not affect the association of the ARF1-dependent coat protein GGA with the Golgi membrane, suggesting a selective effect on the coatomer pathway (14). Because it is likely that only a portion of the total ARFGAP1 population is involved in COPI vesicle formation, this would be the pool that would respond to AlF. In contrast, the Glo3-type ARFGAPs are likely to be more specific for coatomer function. AlF would therefore be predicted to have a larger impact on ARFGAP2 and ARFGAP3 dynamics, which is what we observe in this study.
A current model favors “interdependent” function of the core proteins of the COPI vesicle machinery for generating vesicles (58). According to this model, the association of vesicle proteins, such as coatomer and ARFGAP1, with the Golgi membrane is only partially coupled to ARF1 hydrolysis. This is based on differences in residence time of coatomer and ARF1 on Golgi membranes in photobleaching experiments and the ARF1-independent association of ARFGAP1-YFP with the Golgi membrane (14, 36). Several observations bring this model into question. First, it was shown that differences in residence times between coatomer and ARF1 observed by photobleaching can be attributed to the diffusion-limited kinetics of coatomer due to its large size (34). Second, when comparing the effects of BFA on the steady-state distribution of endogenous ARFGAP1 to -3, we observe no stable pool existing on the Golgi, in agreement with the pioneering study on endogenous ARFGAP1 (12). This suggests that there is no ARF1-independent association of these ARFGAPs with the Golgi. Third, ARFGAP2 and ARFGAP3, predicted by studies both in yeast and mammalian cells to be an important part of the COPI coat, are affected by AlF similarly to coatomer. This suggests a high degree of dependence between these proteins. Although we do not exclude the existence of interdependent function of the components of the COPI vesicle machinery, we favor a model of functional hierarchy for vesicle formation where ARF1 regulates the association-dissociation cycles of both coatomer and the ARFGAPs.
Further studies of ARFGAP2 and ARFGAP3 should provide a more detailed understanding of how they function mechanistically to promote vesicle formation. Of particular interest will be to study the GAP activity of ARFGAP2 and ARFGAP3 and how they are regulated by interactions with other proteins. Strikingly, in an in vitro assay for GAP activity on Golgi membranes, GTP hydrolysis, supplied by yeast Glo3p, was increased 50-fold by the addition of coatomer (59). This suggests that coatomer-dependent localization of the Glo3-type ARFGAPs to the Golgi could be important for catalytic activity as well. Another pertinent question will be to establish the purpose of having two ARFGAPs of the Glo3 family with overlapping function present on the Golgi membrane. One such function could be to direct the formation of different subpopulations of vesicles, as is the case for the AGAP-type ARFGAPs in the endosomal pathway (60). Subpopulations could be the result of different affinities of the two ARFGAPs for various isoforms of coatomer subunits proposed to function within the Golgi apparatus (61). Knockdown of one ARFGAP would therefore only produce mild phenotypes in many assays because vesicle formation is intact in other levels of the stack. However, effects on Golgi morphology might be visible ultrastructurally, consistent with our observations (Fig. 7). In addition, the role of the non-catalytic domain, and especially the Glo3 motif, must be clarified because this is likely to be crucial for the specific targeting or regulation of ARFGAP2 and ARFGAP3, such as different sensitivity to lipids. These and other studies will be important for the future characterization of the individual contributions of different ARFGAPs to the process of COPI vesicle formation.
Supplementary Material
Acknowledgments
We thank the Centre of Cellular Imaging for use of the facility. We also thank Rainer Duden (Flensburg, Germany) for sharing both reagents and information prior to publication of his own work.
This work was supported in part by the Swedish Research Council and by the Canadian Institutes for Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ER
- endoplasmic reticulum
- ARF
- ADP-ribosylation factor
- ARFGAP
- ARF GTPase-activating protein
- ARFGEF
- ARF guanine-exchange factor
- COPI
- coat protein complex I
- FRAP
- fluorescence recovery after photobleaching
- FLIP
- fluorescence loss in photobleaching
- BFA
- brefeldin A
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Bonifacino J. S., Glick B. S. (2004) Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2.Elsner M., Hashimoto H., Nilsson T. (2003) Mol. Membr. Biol. 20, 221–229 [DOI] [PubMed] [Google Scholar]
- 3.Rabouille C., Klumperman J. (2005) Nat. Rev. Mol. Cell Biol. 6, 812–817 [DOI] [PubMed] [Google Scholar]
- 4.D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 5.Casanova J. E. (2007) Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 6.Palmer D. J., Helms J. B., Beckers C. J., Orci L., Rothman J. E. (1993) J. Biol. Chem. 268, 12083–12089 [PubMed] [Google Scholar]
- 7.Spang A., Matsuoka K., Hamamoto S., Schekman R., Orci L. (1998) Proc. Natl. Acad. Sci. 95, 11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra V., Serafini T., Orci L., Shepherd J. C., Rothman J. E. (1989) Cell 58, 329–336 [DOI] [PubMed] [Google Scholar]
- 9.Tanigawa G., Orci L., Amherdt M., Ravazzola M., Helms J. B., Rothman J. E. (1993) J. Cell Biol. 123, 1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn R. A., Gilman A. G. (1986) J. Biol. Chem. 261, 7906–7911 [PubMed] [Google Scholar]
- 11.Kartberg F., Elsner M., Fröderberg L., Asp L., Nilsson T. (2005) Biochim. Biophys. Acta 1744, 351–363 [DOI] [PubMed] [Google Scholar]
- 12.Cukierman E., Huber I., Rotman M., Cassel D. (1995) Science 270, 1999–2002 [DOI] [PubMed] [Google Scholar]
- 13.Lee S. Y., Yang J. S., Hong W., Premont R. T., Hsu V. W. (2005) J. Cell Biol. 168, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W., Duden R., Phair R. D., Lippincott-Schwartz J. (2005) J. Cell Biol. 168, 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Z., Randazzo P. A. (2006) J. Cell Sci. 119, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 16.Bigay J., Gounon P., Robineau S., Antonny B. (2003) Nature 426, 563–566 [DOI] [PubMed] [Google Scholar]
- 17.Reinhard C., Schweikert M., Wieland F. T., Nickel W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanoix J., Ouwendijk J., Lin C. C., Stark A., Love H. D., Ostermann J., Nilsson T. (1999) EMBO J. 18, 4935–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malsam J., Gommel D., Wieland F. T., Nickel W. (1999) FEBS Lett. 462, 267–272 [DOI] [PubMed] [Google Scholar]
- 20.Pepperkok R., Whitney J. A., Gomez M., Kreis T. E. (2000) J. Cell Sci. 113, 135–144 [DOI] [PubMed] [Google Scholar]
- 21.Lanoix J., Ouwendijk J., Stark A., Szafer E., Cassel D., Dejgaard K., Weiss M., Nilsson T. (2001) J. Cell Biol. 155, 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss M., Nilsson T. (2003) Traffic 4, 65–73 [DOI] [PubMed] [Google Scholar]
- 23.Poon P. P., Cassel D., Spang A., Rotman M., Pick E., Singer R. A., Johnston G. C. (1999) EMBO J. 18, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dogic D., de Chassey B., Pick E., Cassel D., Lefkir Y., Hennecke S., Cosson P., Letourneur F. (1999) Eur. J. Cell Biol. 78, 305–310 [DOI] [PubMed] [Google Scholar]
- 25.Eugster A., Frigerio G., Dale M., Duden R. (2000) EMBO J. 19, 3905–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis S. M., Poon P. P., Singer R. A., Johnston G. C., Spang A. (2004) Mol. Biol. Cell 15, 4064–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeRegis C. J., Rahl P. B., Hoffman G. R., Cerione R. A., Collins R. N. (2008) BMC Cell Biol. 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yahara N., Sato K., Nakano A. (2006) J. Cell Sci. 119, 2604–2612 [DOI] [PubMed] [Google Scholar]
- 29.Frigerio G., Grimsey N., Dale M., Majoul I., Duden R. (2007) Traffic 8, 1644–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Zhang C., Xing G., Chen Q., He F. (2001) FEBS Lett. 490, 79–83 [DOI] [PubMed] [Google Scholar]
- 31.Singh J., Itahana Y., Parrinello S., Murata K., Desprez P. Y. (2001) J. Biol. Chem. 276, 11852–11858 [DOI] [PubMed] [Google Scholar]
- 32.Watson P. J., Frigerio G., Collins B. M., Duden R., Owen D. J. (2004) Traffic 5, 79–88 [DOI] [PubMed] [Google Scholar]
- 33.Weimer C., Beck R., Eckert P., Reckmann I., Moelleken J., Brügger B., Wieland F. T. (2008) J. Cell Biol. 183, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elsner M., Hashimoto H., Simpson J. C., Cassel D., Nilsson T., Weiss M. (2003) EMBO Rep. 4, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. (2005) Mol. Biol. Cell 16, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Presley J. F., Ward T. H., Pfeifer A. C., Siggia E. D., Phair R. D., Lippincott-Schwartz J. (2002) Nature 417, 187–193 [DOI] [PubMed] [Google Scholar]
- 37.Zhao X., Claude A., Chun J., Shields D. J., Presley J. F., Melançon P. (2006) J. Cell Sci. 119, 3743–3753 [DOI] [PubMed] [Google Scholar]
- 38.Allan V. J., Kreis T. E. (1986) J. Cell Biol. 103, 2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simionescu N., Simionescu M. (1976) J. Cell Biol. 70, 608–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asp L., Kartberg F., Fernandez-Rodriguez J., Smedh M., Elsner M., Laporte F., Bárcena M., Jansen K. A., Valentijn J. A., Koster A. J., Bergeron J. J., Nilsson T. (2009) Mol. Biol. Cell 20, 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chardin P., McCormick F. (1999) Cell 97, 153–155 [DOI] [PubMed] [Google Scholar]
- 42.Donaldson J. G., Lippincott-Schwartz J., Bloom G. S., Kreis T. E., Klausner R. D. (1990) J. Cell Biol. 111, 2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bigay J., Deterre P., Pfister C., Chabre M. (1985) FEBS Lett. 191, 181–185 [DOI] [PubMed] [Google Scholar]
- 44.Mittal R., Ahmadian M. R., Goody R. S., Wittinghofer A. (1996) Science 273, 115–117 [DOI] [PubMed] [Google Scholar]
- 45.Donaldson J. G., Kahn R. A., Lippincott-Schwartz J., Klausner R. D. (1991) Science 254, 1197–1199 [DOI] [PubMed] [Google Scholar]
- 46.Donaldson J. G., Lippincott-Schwartz J., Klausner R. D. (1991) J. Cell Biol. 112, 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finazzi D., Cassel D., Donaldson J. G., Klausner R. D. (1994) J. Biol. Chem. 269, 13325–13330 [PubMed] [Google Scholar]
- 48.Melançon P., Glick B. S., Malhotra V., Weidman P. J., Serafini T., Gleason M. L., Orci L., Rothman J. E. (1987) Cell 51, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 49.Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. (1991) Cell 67, 239–253 [DOI] [PubMed] [Google Scholar]
- 50.Teal S. B., Hsu V. W., Peters P. J., Klausner R. D., Donaldson J. G. (1994) J. Biol. Chem. 269, 3135–3138 [PubMed] [Google Scholar]
- 51.Aoe T., Cukierman E., Lee A., Cassel D., Peters P. J., Hsu V. W. (1997) EMBO J. 16, 7305–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antonny B., Huber I., Paris S., Chabre M., Cassel D. (1997) J. Biol. Chem. 272, 30848–30851 [DOI] [PubMed] [Google Scholar]
- 53.Bigay J., Casella J. F., Drin G., Mesmin B., Antonny B. (2005) EMBO J. 24, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 55.Randazzo P. A., Hirsch D. S. (2004) Cell. Signal. 16, 401–413 [DOI] [PubMed] [Google Scholar]
- 56.Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2003) Mol. Biol. Cell 14, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M., Poon P. P., Schindler C., Murray L. E., Kama R., Gabriely G., Singer R. A., Spang A., Johnston G. C., Gerst J. E. (2006) Mol. Biol. Cell 17, 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lippincott-Schwartz J., Liu W. (2006) Trends Cell Biol. 16, e1–e4 [DOI] [PubMed] [Google Scholar]
- 59.Szafer E., Rotman M., Cassel D. (2001) J. Biol. Chem. 276, 47834–47839 [DOI] [PubMed] [Google Scholar]
- 60.Nie Z., Fei J., Premont R. T., Randazzo P. A. (2005) J. Cell Sci. 118, 3555–3566 [DOI] [PubMed] [Google Scholar]
- 61.Moelleken J., Malsam J., Betts M. J., Movafeghi A., Reckmann I., Meissner I., Hellwig A., Russell R. B., Söllner T., Brügger B., Wieland F. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4425–4430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.