Abstract
Cbl is an adaptor protein and an E3 ligase that plays both positive and negative roles in several signaling pathways that affect various cellular functions. Tyrosine 737 is unique to Cbl and is phosphorylated by Syk and Src family kinases. Phosphorylated Cbl Tyr737 creates a binding site for the p85 regulatory subunit of PI3K, which also plays an important role in the regulation of bone resorption by osteoclasts. To investigate the role of Cbl-PI3K interaction in bone homeostasis, we examined the knock-in mice (CblYF/YF) in which the PI3K binding site in Cbl is ablated due to the mutation in the regulatory tyrosine. We report that in CblYF/YF mice, despite increased numbers of osteoclasts, bone volume is increased due to defective osteoclast function. Additionally, in ex vivo cultures, mature CblYF/YF osteoclasts showed an increased ability to survive in the presence of RANKL due to delayed onset of apoptosis. RANKL-mediated signaling is perturbed in CblYF/YF osteoclasts, and most interestingly, AKT phosphorylation is up-regulated, suggesting that the lack of PI3K sequestration by Cbl results in increased survival and decreased bone resorption. Cumulatively, these in vivo and in vitro results show that, on one hand, binding of Cbl to PI3K negatively regulates osteoclast differentiation, survival, and signaling events (e.g. AKT phosphorylation), whereas on the other hand it positively influences osteoclast function.
Keywords: NFκB Transcription Factor, Phosphatidylinositol 3-Kinase, Protein Phosphorylation, Signal Transduction, Src, Bone Resorption, Cbl, RANK, Survival
Introduction
Bone is a dynamic tissue that maintains both physical integrity and calcium homeostasis and is continuously remodeled throughout the lifetime of mammals. Bone integrity is maintained by the coordinated regulation of two cell types, osteoblasts and osteoclasts. Osteoblasts are of mesenchymal origin and are responsible for mineralizing the bone matrix. Osteoclasts are of hematopoietic origin and are highly specialized multinucleated cells capable of resorbing bone during the normal and pathological conditions (1). Cbl and Cbl-b are mammalian members of a family of adaptor proteins with E3 ubiquitin ligase activity that down-regulate signaling from tyrosine kinases by targeting the ubiquitylating machinery to the activated kinases and associated proteins (2, 3). Cbl proteins, being multivalent adaptors, also promote assembling of signaling complexes downstream of several receptors, including the macrophage colony-stimulating factor receptor (c-Fms), receptor activator of NFκB (RANK),2 and integrins (2).
Tyrosine phosphorylation of Cbl upon receptor activation is one of the most common signaling phenomena shown to be crucial not only for its E3 ubiquitin ligase activity but also for its adaptor effects. All Cbl family members, including Cbl and Cbl-b, share a tyrosine kinase binding domain and RING domain that binds to ubiquitin-conjugating enzymes (E2s). The tyrosine kinase binding domain and the RING are highly conserved N-terminal halves of the Cbl family proteins. The C-terminal halves of Cbl and Cbl-b are much less conserved, with significant homology found primarily in short proline-rich motifs that bind to Src homology 3 domains. Based on sequence homologies and mutation of specific residues, specific roles for several of the SH2 binding sites have been determined. Phosphorylation of Tyr774 and 700 residues creates binding sites on Cbl for Crk/CrkL (4) and Vav (5). Phosphorylated tyrosine 731 (Tyr731 in humans; Tyr737 in mice), which binds to the p85 subunit of PI3K is unique to Cbl (6, 7), and no homologous site exists on other Cbl family members.
Cbl and Cbl-b are expressed in osteoclasts (8) and are associated with podosomes (9, 10), the highly dynamic F-actin-containing attachment structures (11–13), and also involved in microtubule organization (14). In osteoclasts, Cbl proteins are known to participate in integrin-, RANK-, and macrophage colony-stimulating factor (M-CSF)-mediated signaling pathways (2). Upon engagement of the vitronectin receptor, the predominant integrin expressed in osteoclasts, Cbl, is phosphorylated in a Src kinase-dependent manner (10). Cbl phosphorylation is significantly reduced in Src−/− osteoclast-like cells (OCLs), and reduction of Cbl expression with antisense oligonucleotides markedly inhibited in vitro bone resorption by OCLs (15). Overexpressing Cbl constructs with disabled binding sites for Src (16) and PI3K (17) decreased the in vitro pit formation capacity of osteoclasts. Cbl proteins also positively regulate osteoclast function by promoting survival by mediating ubiquitylation and degradation of the proapoptotic protein Bim (18).
PI3Ks are a class of enzymes that phosphorylate phosphatidylinositol and its derivatives. The importance of PI3K in osteoclast function has been established, although the details of its function(s) and the proteins with which it interacts are not well characterized. In osteoclasts, engagement of the vitronectin receptor induces an Src-dependent increase in PI3K activity and its association with Triton-insoluble gelsolin-containing complexes, presumably the podosomes (19, 20). Treatment with PI3K inhibitors disrupts the actin ring and inhibits attachment, spreading, and bone resorbing activity (20). In vivo deficiency of the p85 α subunit of PI3K results in increased bone volume due to decreased osteoclast function (21). The p85 subunit also regulates expression of multiple genes involved in osteoclast maturation (21). PI3K coordinately activates MEK/ERK and AKT/NFκB pathways to maintain osteoclast survival and also participates in cross-talk with Ras/Raf in promoting M-CSF-induced osteoclast survival (22). Downstream of RANK signaling, Cbl and Src form a complex with TRAF6 that couples RANK to the activation of PI3K and AKT (23, 24).
The single knockouts of Cbl and Cbl-b are viable (25–27), but embryonic lethality of Cbl/Cbl-b double knock out mice before embryonic day 10.5 (28) suggests that both Cbl and Cbl-b have important overlapping functions. It has been suggested that Cbl and Cbl-b play different roles in coupling RANK to downstream signaling events and in the down-regulation of RANK in dendritic cells and HEK 293 cells (23). Structural differences that could contribute to the unique functions of Cbl proteins include a tyrosine present only in Cbl (Cbl Tyr737) that when phosphorylated binds to the SH2 domains of the p85 subunit of PI3K (3). There are also sequence differences in the UBA domains at the C termini of these proteins, resulting in differing abilities to bind polyubiquitin chains and ubiquitylated proteins (29).
Because the cbl gene is globally knocked out, the Cbl−/− mice cannot be used to perform structure-function analyses to examine the role of Cbl-specific domains in regulating protein-protein interactions and thereby their influence on cellular functions. Therefore, to understand the role of tyrosine phosphorylation of Cbl proteins in bone remodeling, we undertook the characterization of the Cbl tyrosines that may be required for Cbl binding to critical signaling proteins that influence osteoclast function. In this context, to delineate the importance of Cbl-PI3K signaling in osteoclast function, we are employing CblYF/YF knock-in mice in which the Cbl-PI3K interaction is abrogated due to the substitution of the Tyr737 to Phe (30).
In this report, we show that in the skeletal system, abrogation of Cbl-PI3K interaction results in increased bone mass due to a cell-autonomous defect in osteoclast function. Furthermore, we found that in osteoclasts, the lack of Cbl-PI3K interaction promotes increased differentiation and survival, suggesting a novel role for Cbl protein in RANK-mediated signaling.
EXPERIMENTAL PROCEDURES
Materials
Minimum essential medium-α modification (α-MEM) and fetal bovine serum (FBS) were purchased from Sigma. Collagen gel was obtained from Nitta Gelatin Co. (Osaka, Japan). Bacterial collagenase and dispase were purchased from Calbiochem. Antibodies against phospho-Cbl Tyr737, phospho-p38, p38, phospho-AKT Thr308, AKT, phospho-JNK, JNK, phospho-ERK, phospho-IKKα/β, phospho-GSKα/β, GSK3β, phospho-AKT substrates, phospho-PLCγ2, PLCγ2, GAPDH, and Bim were purchased from Cell Signaling Technology (Danvers, MA). Anti-ERK1/2 and anti-p85 antibodies were obtained from Upstate Biotechnology, Inc. (Lake Placid, NY). Anti-ubiquitin, anti-Cbl, and IKKα antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-Cbl antibody was purchased from BD Biosciences. Rhodamine, DAPI, Dead End Fluorometric TUNEL, and Caspase Glo3/7 kits were obtained from Promega (Madison, WI). RANK ligand (RANKL) and MCSF were purchased from R&D Systems (Minneapolis, MN). 1,25-Dihydroxyvitamin D3 and prostaglandin E2 and the leukocyte acid phosphatase kit for tartrate-resistant acid phosphatase (TRAP) staining of osteoclasts were obtained from Sigma. Nuclear extract kit, TransAM NFATc1, and TransAM NFκB family transcription assay kits were purchased from Active Motif (Carlsbad, CA).
Mice
The generation of Cbl−/−, CblYF/YF mice has been described previously (27, 31). Mice were maintained on a mixed C57BL/6JX129SvJ background, and experiments were performed in compliance with the Institutional Animal Care and Use Committee at Temple University.
Histology and Histomorphometry
X-rays of long bones were taken (Eastman Kodak Co. MIN-R 2000) at 19 mV and exposed for 10 s using Faxitron. For histological and histomorphometric analysis, 6- or 12-week old littermates were sacrificed by CO2 inhalation. To measure dynamic bone formation parameters, mice were injected with calcein (30 mg/kg body weight) 10 and 3 days before sacrifice. Tibiae and femora were dissected and fixed in 3.7% formaldehyde in phosphate-buffered saline, preserved in 70% ethanol, and embedded in methlymethacrylate resin as described previously (32). Sections (5 μm) were deplasticized and stained with the von Kossa procedure as described elsewhere (33) or were left unstained for the measurements of the fluorochrome labels. Some sections were processed for TRAP staining as per the manufacturer's instructions (Sigma). For histomorphophometric analysis, to assess changes in bone structure and remodeling, tibial sections were measured in the proximal metaphysis beginning 340 μm below the chondro-osseous junction of the secondary spongiosa using image analysis software (BIOQUANT Image Analysis Corp., Nashville, TN) as described by Parfitt et al. (34). The number of TRAP+ cells was determined in secondary spongiosa, in contact with the trabecular bone.
Determination of Serum Collagen Telopeptide
Serum was prepared from blood collected by cardiac puncture. Concentrations of C-telopeptide, a degradation product of type-I collagen, in serum of 12-week-old mice were determined using the Rat Laps ELISA (Osteometer BioTech A/S, Herlev, Denmark).
Generation of Osteoclast-like Cells in Culture
For generation of OCLs, bone marrow was isolated from tibia and femur of 4–6-week-old WT and CblYF/YF mice. Following overnight incubation, the non-adherent cells were plated at 2.5 × 105/cm2 in α-MEM, 10% FBS, penicillin/streptomycin containing 20 ng/ml M-CSF. Subsequently, cells were treated with M-CSF (20 ng/ml) and RANKL (50 ng/ml) for an additional 5–6 days. For some experiments, OCLs were also generated by the co-culture method as described previously (35). Briefly, mouse primary osteoblastic cells were obtained from 1-day-old mouse calvaria by enzymatic digestion, and bone marrow cells were obtained from long bones of 4–6-week-old WT or CblYF/YF mice. Bone marrow cells (105 cells/cm2) were co-cultured with calvarial cells (2 × 104 cells/cm2) on tissue culture plates or collagen gel-coated plates in the presence of 10 nm 1,25-dihydroxyvitamin D3 and 1 μm prostaglandin E2 (Sigma).
Survival Assay
Following differentiation with MCSF and RANKL on day 5, one set of 96-well plates was fixed with 10% formaldehyde in PBS. Other sets were either kept in α-MEM or treated with RANKL (50 ng/ml) for 48 h. Cells were fixed and TRAP-stained using a commercial kit (Sigma). The total number of TRAP+ multinucleated cells was counted and expressed as percentage of the number of cells at the start of the experiment on day 5. In other experiments, cells were cultured on coverslips and were fixed with 10% formaldehyde in PBS or were treated with RANKL (50 ng/ml) for 6 or 12 h. A TUNEL assay was performed using the DeadEnd Fluorometric TUNEL kit (Promega) according to the manufacturer's protocol. TUNEL-positive cells were counted using a NIKON Eclipse E800 microscope (Melville, NY) at ×20 magnification.
Real-time PCR Analysis
The expression levels of osteoclast differentiation and fusion markers were analyzed by quantitative real-time PCR. RNA isolation was performed using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA was quantified using a spectrophotometer at 260/280 nm, 1 μg was used for each reaction performed using Super Script II reverse transcriptase (Invitrogen) following the manufacturer's protocol. Oligo(dT) and dNTPs were purchased from Promega. Quantitative real-time PCR was performed using 0.1 μl of cDNA and SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) on an Applied Biosystems7500 real-time PCR system. Gene expression levels were normalized to GAPDH and were calculated using the ΔΔCt method. Primers were designed using mRNA sequence obtained from Mouse Genomics Informatics (MGI) and the Primer3 program. The following primers were used: Atp6v0d2, CAATGAAGCGTCACCTCTGA (sense) and TCAGCTATTGAACGCTGGTG (antisense); CalcR, AGCCACAGCCTATCAGCACT (sense) and GACCCACAAGAGCCAGGTAA (antisense); CTSK, CAGCTTCCCCAAGATGTGAT (sense) and AAAAATGCCCTGTTGTGTCC (antisense); c-Fos, CCAGTCAAGAGCATCAGCAA (sense); antisense, AAGTAGTGCAGCCCGGAGTA; DCSTAMP, ACTAGAGGAGAAGTCCTGGGAGTC-3′ (sense) and CACCCACATGTAGAGATAGGTCAG (antisense); Fra-1, AGAGCTGCAGAAGCAGAAGG (sense) and CAAGTACGGGTCCTGGAGAA (antisense); Fra-2, CAAGTACGGGTCCTGGAGAA (sense) and GTTTCTCTCCCTCCGGATTC (antisense); GAPDH, TGTCTTCACCACCATGGAGAAG (sense) and GTGGATGCAGGGATGATGTTCTG (antisense); MMP9, TGAATCAGCTGGCTTTTGTG (sense) and GTGGATAGCTCGGTGGTGTT (antisense); NFATc1, GCCCACTGGATCAAAACACT (sense) and TAGGGCAGCCAGAAAAGCTA (antisense); OCSTAMP, TGGGCCTCCATATGACCTCGAGTAG (sense) and TCAAAGGCTTGTAAATTGGAGGAGT (antisense); PU.1, GGCAGCAAGAAAAAGATTCG (sense) and TTTCTTCACCTCGCCTGTCT (antisense); RANK, AAACCTTGGACCAACTGCAC (sense) and TCATTGACCCAATTCCACAA (antisense); TRAP, TCCTGGCTCAAAAAGCAGTT (sense) and ACATAGCCCACACCGTTCTC (antisense).
Flow Cytometry
Bone marrow (BM) was isolated from tibiae and femora of 4–6-week-old mice. Cells (107/sample) were fixed either immediately following BM isolation (D0) or after removal of adherent cells after 48 h in culture in the presence of 20 ng/ml M-CSF. Following lysis of RBCs using ammonium chloride lysis buffer, cells were collected by centrifugation and resuspended in 900 μl of PBS and fixed by the addition of 100 μl of 37% formaldehyde followed by incubation at 37 °C for 10 min and on ice for 1 min. Cells (106/staining sample) were resuspended in 3 ml of incubation buffer (0.5% BSA in PBS) and rinsed twice. Cells were blocked for 10 min at room temperature in incubation buffer and then incubated with 0.2 μg of phycoerythrin-conjugated anti-F4/80 antibody and FITC-conjugated anti-CD11b antibody or the appropriate isotype controls (BD Biosciences). Following incubation for 1 h at room temperature, cells were washed twice in incubation buffer and analyzed using flow cytometry. Following gating for background staining using isotype controls, the percentage of cells having the phenotype characteristic for osteoclast precursors (i.e. staining positive for both F4/80 and CD11b) was compared in the samples examined.
Immunofluorescence Microscopy
Cells were plated on sterile FBS-coated glass coverslips. OCLs were fixed in PBS containing 3.7% formaldehyde for 10 min and then permeabilized with ice-cold acetone for 5 min. Coverslips for actin labeling were incubated in a 1:40 dilution in PBS of rhodamine phalloidin stock solution (Invitrogen) for 20 min. Cells were examined using a confocal imaging system (Leica TCS FP5 X, Wetzlar, Germany).
In Vitro Pit Formation Assay
In vitro bone resorbing activity was assayed as described previously. Briefly, OCLs were generated in co-culture as described above in collagen gel. After 6–7 days of co-culture, collagen was removed by gentle digestion with 0.1% collagenase, and then cells were seeded onto sterile dentine slices (ImmunoDioagnostic Systems Ltd., Boldon, UK) in 96-well plates. Forty-eight hours later, dentine slices were immersed in 1 m ammonium hydroxide for 5 min, sonicated for 10 s, and then stained for 4 min with 1% toluidine blue in 1% sodium borate (Sigma) and briefly washed in water. Pit area was quantified with the measure tool in Adobe Photoshop CS3 Extended Edition and was normalized to the number of osteoclasts actually present in each sample, determined by counting OCLs present in a separate aliquot of OCL suspension.
Western Blot Analysis
Typically, 30–40 μg of total cell lysate protein was electrophoresed on 8 or 10% SDS-polyacrylamide gels. Proteins were then transferred to nitrocellulose membranes (Schleicher & Schuell). Transferred proteins were visualized by staining the membrane with 0.2% Ponceau S in 3% trichloroacetic acid (Sigma). To block nonspecific binding, the membranes were incubated for 2 h at room temperature in LICOR buffer (LICOR, Lincoln, NE). Antigens were visualized by immunoblotting with an appropriate primary antibody (1:1000 dilution) followed by an appropriate LICOR secondary antibody. To quantify changes, densitometry of the phosphorylated and total protein bands was performed using the LICOR analysis program. The values for the phosphorylated band were normalized to the density of the respective total protein band in the reprobed blot.
Measurement of Transcription Activity
OCLs were generated from bone marrow of WT and CblYF/YF mice using M-CSF and RANKL as described previously. Cells were either untreated or treated with RANKL (50 ng/ml) for 6 h. Nuclear extract (10 μg) was used to measure the transcriptional activity of NFκB or NFATc1 using TransAM transcription factor assay kits (Active Motif) following the manufacturer's instructions.
Measurement of Caspase Activity
OCLs were generated in triplicates in 96-well plates using M-CSF and RANKL as described above. On day 5 of differentiation, cells were either kept in α-MEM alone or treated with RANKL (50 ng/ml) for 3 h at 37 °C. Caspase activity was measured using the Caspase-Glo 3/7 kit (Promega, Madison, WI) as per the manufacturer's instructions.
Statistical Analysis
Each experiment was repeated at least three times. The results obtained from a typical experiment were expressed as the means ± S.D. Significant differences were determined using Student's t test. p < 0.05 was considered significant.
RESULTS
Loss of Cbl-PI3K Interaction in Mice Results in Increased Bone Mass
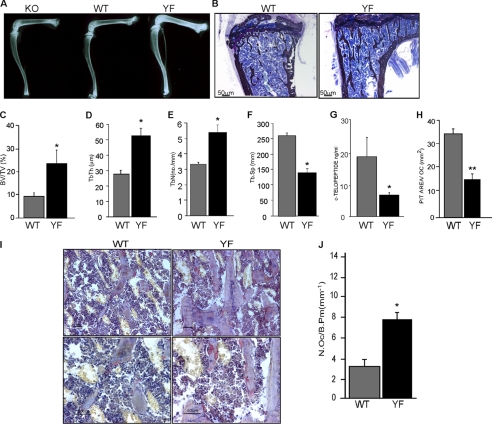
Gross radiological analysis of long bones of 12-week-old CblYF/YF mice showed increased bone density in the femur and tibia as compared with the age-matched control WT and Cbl−/− mice (Fig. 1A). Histological analysis of the tibiae from 6-week-old WT and CblYF/YF mice showed that cancellous bone increased in CblYF/YF relative to the age-matched control WT littermates (Fig. 1B). These results indicate that in contrast to adult Cbl−/− mice, which do not have any overt skeletal phenotype (8), abrogation of the interaction between Cbl and the p85 subunit of PI3K results in increased bone mass. This result was confirmed with histomorphometry of von Kossa-stained bone sections. Bone volume was significantly elevated in CblYF/YF mice compared with control WT mice (Fig. 1C). Also, whereas trabecular thickness and numbers were increased in CblYF/YF samples, trabecular separation was decreased (Fig. 1, D–F). No differences were observed in cortical thickness (data not shown).
FIGURE 1.
Abrogation of Cbl-PI3K interaction in mice results in increased bone volume due to decreased osteoclast activity. A, long bones from 12-week-old Cbl−/−, Cbl+/+, and CblYF/YF mice were subjected to x-ray to determine bone density. B, photomicrographs of the undecalcified section of tibiae from 6-week-old Cbl+/+ and CblYF/YF mice visualized at 4× magnification. Sections were stained with von Kossa and counterstained with toludine blue. C–F, histomorphometric analysis of cancellous region of the tibial metaphysis of 6-week-old WT and CblYF/YF mice. C, the bone volume in the metaphysis of the CblYF/YF mice was markedly increased. Histograms represent the bone volume expressed as the percentage of tissue volume (BV/TV). D, trabecular thickness (Tb.Th). E, trabecular number (Tb.N.). F, trabecular spacing (Tb.Sp). Data are presented as mean ± S.E. (error bars) (n = 5). *, p < 0.05 compared with the WT mice. G, measurement of serum collagen telopepetide demonstrated decreased OC activity in the CblYF/YF mice. H, pit formation activity of CblYF/YF OCLs was decreased. OCLs were generated by the co-culture method as described under “Experimental Procedures.” After removing adherent cells, the resorbed area on toluidine blue-stained dentine slices was quantified and normalized for the numbers of OCLs. Data are presented as mean ± S.E. (n = 9). **, p < 0.001 compared with the WT samples. I, histological analysis of 6-week-old WT and CblYF/YF bones. OCs were identified by a TRAP activity reaction product (red). Magnification was ×20 (upper panels) and ×40 (lower panels). J, histomorphometric analysis revealed increased numbers of OCs in CblYF/YF mice compared with WT mice.
Loss of Cbl-PI3K Interaction in Mice Results in Defective Osteoclast Function
We next examined the effect of this mutation on osteoclast function. Serum levels of the C-terminal collagen telopeptide (CTX), a marker for osteoclast in vivo activity, were 3-fold less in the CblYF/YF mice than that in the control WT mice (Fig. 1G), suggesting that the Cbl-PI3K interaction decreases the osteoclast function. To confirm this defect of CblYF/YF osteoclast function, we then examined bone resorption using an in vitro pit formation assay. CblYF/YF and WT OCLs were generated by co-culture with osteoblasts on collagen gel as described previously (17). After 5 days in culture, a portion of the crude OCL preparation was placed on dentine slices for an additional 48 h. The resorbed area was quantified and normalized to the number of OCLs. The CblYF/YF OCLs resorbed 2-fold less dentin surface area/cell than the WT OCLs (Fig. 1H), confirming the cell-autonomous nature of the observed defect in osteoclast function. Thus, increased bone mass in the adult CblYF/YF mice and decreased bone resorption in vivo and in vitro are, at least in part, due to impaired osteoclast function. Taken together, these results suggest that bone resorption under basal conditions is affected in CblYF/YF mice.
CblYF/YF Mice Have Increased Osteoclast Numbers and Enhanced Differentiation
Bone volume is maintained by the regulated action of bone-forming osteoblasts and bone-resorbing osteoclasts. CblYF/YF mice exhibited an increase in osteoblast numbers (Ob/Bpm WT 17.20 ± 0.31; CblYF/YF 20.59 ± 0.27; p < 0.05 versus WT); however, the lack of Cbl-PI3K interaction did not affect osteoblast differentiation and expression of differentiation markers, osterix and Runx2, (data not shown). To determine if the mutation affected numbers of osteoclasts, tibiae of 6-week-old CblYF/YF mice were decalcified and stained for an osteoclast marker enzyme TRAP (Fig. 1I). The bones of CblYF/YF mice exhibited a 2-fold increase in osteoclast numbers, suggesting that the lack of Cbl-PI3K interaction negatively regulates osteoclastogenesis (Fig. 1J).
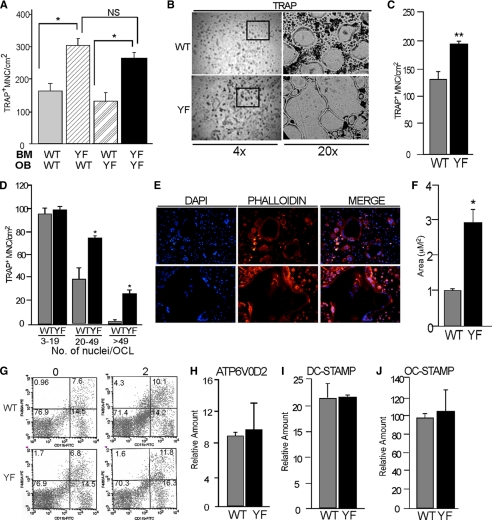
To further investigate if the lack of Cbl-PI3K interaction increased osteoclast numbers and whether this effect was intrinsic to the hematopoietic lineage or not, we next examined the development of OCLs using two standard culture methods: (a) co-culture of primary calvarial osteoblasts with BM monocyte precursors and (b) stromal cell-free BM monocyte precursors cultured with M-CSF and RANKL. In the co-culture system, irrespective of the genotype of osteoblasts, a 2-fold increase in osteoclast numbers was seen in CblYF/YF co-cultures when compared with WT cultures (Fig. 2A). Similar results were obtained when spleen was used as a source of the hematopoietic precursor cells (data not shown). To analyze how the absence of Cbl-PI3K interaction affects osteoclast formation, bone marrow from WT and CblYF/YF mice were induced to differentiate in the presence of M-CSF and RANKL. When the stromal cell-free BM precursors from CblYF/YF mice were cultured in the presence of M-CSF and RANKL, there was a 47% increase in the numbers of osteoclasts in the CblYF/YF cultures when compared with the control cultures (Fig. 2, B and C). Notably, the numbers of cells with more than 20–50 nuclei/cell were increased 2–2.5-fold in the CblYF/YF cultures (Fig. 2D), and these cells largely contributed to the observed increase in total numbers of OCLs. Also, as compared with the WT cells, the average size of CblYF/YF osteoclasts was 3-fold higher (Fig. 2, E and F). These observations in two different culture systems confirmed our finding that the number of osteoclasts in CblYF/YF mice was higher than in WT mice and suggested that the observed increase in the numbers of OCLs in CblYF/YF cultures is due to a cell-autonomous effect intrinsic to the bone marrow precursors and is not related to the osteoblasts.
FIGURE 2.
Absence of Cbl-PI3K interaction results in increased osteoclast numbers due to cell-autonomous enhanced differentiation. A, co-culture of calvarial osteoblasts with bone marrow cells was performed to determine the cell-autonomous differentiation of the CblYF/YF precursors. After 7 days of culture, cultures were TRAP-stained, and OCLs with three or more nuclei were counted. CblYF/YF cultures had increased numbers of OCLs irrespective of the source of osteoblasts. B–E, non-adherent bone marrow precursor cells were cultured on plastic or glass coverslips in the presence of M-CSF for 2 days and for an additional 3 days in the presence of MCSF and RANKL. B, photomicrographs at the indicated magnifications of TRAP-stained OCLs at day 5. TRAP staining showed increased numbers and larger cells in CblYF/YF cultures. The black box (within the left panels at ×4 magnification) indicates the region at ×20 magnification. C, the numbers of TRAP+ multinucleated osteoclasts (MNC) in the CblYF/YF cultures (black bars) were significantly greater than in WT cultures (gray bars) at day 5. D, cells with more than 20–50 nuclei predominantly contributed to the increased numbers in CblYF/YF cultures. E, cells that were cultured on glass coverslips were stained for F-actin using rhodamine phalloidin (red) and DAPI (blue). Cells with more than 20 nuclei predominantly contributed to the increased numbers in CblYF/YF cultures. F, cell size was measured by using the analysis tool in Adobe Photoshop PS3 Extended Version. The size of OCLs in CblYF/YF cultures was 3-fold larger in CblYF/YF cultures. G, detection of CD11b- and CD4/80-positive bone marrow precursor cells using flow cytometry. Bone marrow cells were either untreated (day 0) or cultured in the presence of M-CSF for 2 days (day 2). The fraction of double-positive cells (percentage) is indicated. H–J, the expression of osteoclast fusion markers (ATP6V0D2 (H), DC-STAMP (I), and OC-STAMP (J)) was determined by quantitative real-time PCR. *, p < 0.05 as compared with WT control. Error bars, S.D.
To exclude the possibility that increased numbers of OCLs were due to increased numbers of bone marrow precursors, we counted numbers of precursors that were CD11b+ and F4/80+ and found that they were comparable in both WT and CblYF/YF samples (Fig. 2G). Similarly, the rate of proliferation of precursors was also comparable between the WT and the CblYF/YF samples (data not shown). Osteoclasts are multinucleated cells, and the formation of large multinucleated cells is dependent on the fusion of the mononuclear precursors of the myeloid origin, a cellular event mediated by several cell surface fusion proteins (36–38). As demonstrated in Fig. 2, E and F, significant numbers of large multinucleated osteoclasts were present in CblYF/YF cultures. To determine the possibility that levels of DC-STAMP (36, 37), OC-STAMP (38), and ATP6V0D2 (39), each a marker of osteoclast fusion, were elevated in the CblYF/YF cells, real-time PCR was performed. The levels of the osteoclast fusion markers were found to be comparable between WT and CblYF/YF samples (Fig. 2, H–J).
CblYF/YF Osteoclast Precursors Are Hyperresponsive to RANKL
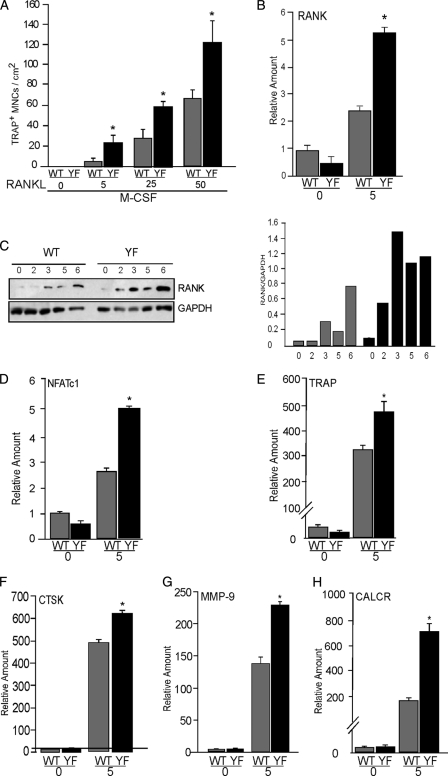
M-CSF and RANKL are both necessary and sufficient to induce in vitro osteoclastogenesis (1). The increase in numbers of osteoclasts in CblYF/YF mice in vivo and ex vivo suggests that BM precursors may differentially respond to these cytokines in the absence of the Cbl-PI3K interaction. To examine the differential response of BM monocytes, precursor cells were cultured in the presence of varying doses M-CSF and RANKL. In some experiments, the concentration of M-CSF was increased, whereas that of RANKL was kept constant. In other cases, RANKL levels but not M-CSF levels were varied. TRAP-positive multinucleated cells were counted after 5 days of culture. Numbers of osteoclasts in the WT and CblYF/YF cultures were independent of M-CSF concentration (data not shown) but increased following the increasing concentration of RANKL at a constant concentration of M-CSF (20 ng/ml). This increase was significantly more profound in CblYF/YF cultures than in WT cultures, suggesting hyper-responsiveness of the CblYF/YF precursors to RANKL (Fig. 3A). Furthermore, the mRNA and protein levels of RANK during the course of osteoclast differentiation were elevated in the CblYF/YF samples when compared with WT (Fig. 3, B and C). These data suggest that the lack of Cbl-PI3K interaction renders BM precursors hypersensitive to RANKL, due to higher expression of RANK during osteoclastogenesis in CblYF/YF cultures.
FIGURE 3.
Enhanced differentiation of CblYF/YF osteoclasts is due to hyperresponsiveness to RANKL. A, non-adherent bone marrow precursor cells were either untreated or cultured in the presence of M-CSF (20 μg/ml) and RANKL (μg/ml) at the indicated doses. After 5 days, cells were TRAP-stained, and OCLs with three or more nuclei were counted. B, bone marrow cells were either untreated (day 0) or cultured in the presence of MCSF (20 μg/ml) and RANKL (50 μg/ml) as described under “Experimental Procedures.” mRNA expression levels were measured by quantitative real-time PCR. C, total cell lysates were prepared at the indicated time points during the process of differentiation. The blot was probed using anti-RANK antibody (top). The blot was stripped and reprobed with GAPDH (bottom) to verify equal loading of protein. To correct for experimental variability, the amounts of total proteins in individual bands were quantified by using Odyssey Infrared Imaging Systems software 2.1 (LICOR Biosciences), and the ratio of RANK protein to GAPDH, a housekeeping protein, was calculated (gray bars, WT; black bars, CblYF/YF). D–F, bone marrow cells were differentiated in OCLs with M-CSF and RANKL. The expression of osteoclast differentiation markers on day 0 and 5 was determined by real-time PCR. D, NFATc1; E, TRAP; F, CTSK; G, MMP-9; H, CalcR. *, p < 0.05 as compared with WT control. Error bars, S.D.
To further examine the role of Cbl-PI3K interaction during osteoclastogenesis, real-time PCR analysis was used to determine the levels of known markers of osteoclast differentiation. Expression levels of early differentiation markers, PU.1 (40) Fra-1 (41), Fra-2 (42), and c-Fos (43), were comparable between WT and CblYF/YF samples (supplemental Fig. 1). However, expression levels of the differentiation markers that are induced upon treatment of RANKL, such as NFATc1 and TRAP, cathepsin K, MMP-9, and CalR (44), were significantly increased in CblYF/YF samples compared with WT (Fig. 3, D–H). Overall, these results indicate that the abrogation of Cbl-PI3K interaction results in increased numbers and enhanced differentiation of osteoclasts due to hyperresponsiveness of CblYF/YF precursors to RANKL. These results also demonstrate that the effect of mutating the PI3K binding site on Cbl protein is significantly different from that of Cbl−/−, in which there is no effect on osteoclast numbers or differentiation (data not shown) (8).
RANK-RANKL Signaling Is Up-regulated in the Absence of Cbl-PI3K Interaction
Because osteoclastogenesis is critically dependent on the activation of RANK by RANKL (45), and Cbl proteins are phosphorylated downstream of activated RANK (24) and have been reported to modulate RANK expression and signaling (23), the hyperresponsiveness to RANKL and the increased rate of OCL differentiation in the presence of RANKL suggest that RANKL-induced signaling might be altered in the CblYF/YF OCLs. Therefore, we next determined if enhanced RANKL responsiveness of CblYF/YF osteoclasts may be due to up-regulated signaling downstream of the RANK-RANKL interaction.
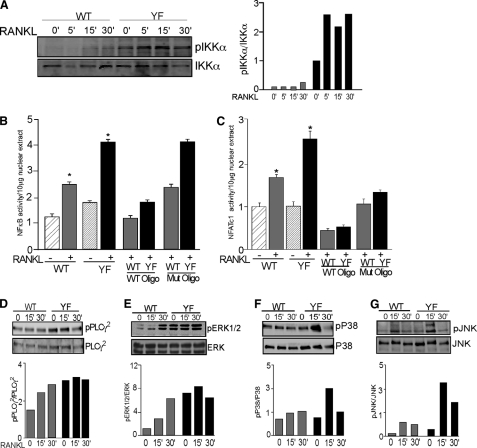
The transcription factor NFκB is the critical component of RANK-mediated signaling and function (46). NFκB is maintained in an inactive state in the cytosol in a complex with the inhibitory IκB proteins (47). Activation of RANK induces the phosphorylation of IκB-α on specific serine residues, which promotes the ubiquitylation and degradation of IκB-α and the consequent release and nuclear translocation of active NFκB. We observed that RANKL-induced phosphorylation of IKKα in CblYF/YF OCLs was higher and more sustained than in WT OCLs (Fig. 4A). In agreement with these data, RANKL-induced activity of NFκB was also elevated 2-fold in CblYF/YF OCLs compared with WT samples (Fig. 4B). The master transcription regulator NFATc1, which contains a promoter region for NFκB binding, is among several genes that are specifically up-regulated by NFκB (44). Treatment with RANKL up-regulated NFATc1 activity in the CblYF/YF OCLs to a higher extent than in WT samples (Fig. 4C); thus, expression of NFATc1 did not correlate with development of functional resorption by the osteoclast.
FIGURE 4.
RANK signaling is enhanced in CblYF/YF OCLs. A, OCLs were serum-starved for 1 h and then treated with RANKL (50 μg/ml) for the indicated times. Blots were probed with anti-phospho-IKKα antibodies (top). The blot was stripped and reprobed with total IKKα to demonstrate loading of proteins. The ratio of phosphorylated protein to total protein is represented in the graph (gray bars, WT; black bars, CblYF/YF). B, OCLs were treated with RANKL (50 μg/ml) for 6 h. A transcription assay was performed as described under “Experimental Procedures.” p65 transcription activity is expressed in relation to the untreated WT OCLs. Specificity of the assay was confirmed by competition of p65 activity with WT and mutant oligonucleotides. *, p < 0.05, as compared with WT control. C, OCLs were treated with RANKL as described above. ELISA-based transcription activity was determined by ELISA-based assay, as described under “Experimental Procedures.” NFATc1 transcription activity is expressed in relation to the untreated WT OCLs. Specificity of the assay was confirmed by competition with WT and mutant oligonucleotides. *, p < 0.05, as compared with WT untreated samples. D–G, OCLs were serum-starved for 30 min and treated with RANKL (50 μg/ml) for the indicated times. Cell lysates were processed for Western blot analysis. After blotting with the indicated phosphospecific antibodies (top panels), the blots were stripped and reprobed with antibodies to respective proteins to determine loading (bottom panels). To correct for experimental variability, the amounts of total proteins in individual bands were quantified by using Odyssey Infrared Imaging Systems software 2.1 (LICOR Biosciences), and the ratio of phosphorylated protein to total protein was calculated (gray bars, WT; black bars, CblYF/YF). A representative experiment of four repetitions is shown. Error bars, S.D.
PLCγ isoforms are known activators of the NFAT family of transcription factors downstream of ITAM-containing immune receptors (48). Because NFATc1 activity is up-regulated, we next examined whether activation of PLCγ2, the major isoform expressed in OCLs, was facilitated by CblYF/YF compared with WT Cbl. As indicated in Fig. 4D, in CblYF/YF OCLs, the basal level of PLC γ2 phosphorylation is up-regulated compared with WT OCLs to a level corresponding to that achieved in WT OCLs.
Stimulation of RANK leads to the activation of MAPKs. It has been demonstrated that members of all three MAPK families, JNK, p38, and ERK, are activated by RANK in osteoclasts or their precursors (46). We compared the RANKL-induced activities of these signaling effectors in WT and CblYF/YF OCLs. Western blot analysis showed that although the phosphorylation of all MAPKs was elevated in CblYF/YF OCLs, there were differences in the onset and duration of the phosphorylation of ERK, JNK, and p38. ERK phosphorylation was higher in CblYF/YF OCLs than in WT cells, both before and after stimulation with RANKL (Fig. 4E), whereas phosphorylation of p38 and JNK was significantly increased in CblYF/YF OCLs only after RANKL treatment (Fig. 4, F and G).
Loss of Cbl-PI3K Interaction Results in Increased AKT Activation
It has been established that activation of RANK induces direct or indirect association of TRAF-6, Src, Cbl, and PI3K, with concurrent increases in Src kinase activity and tyrosine phosphorylation of Cbl (24). We have shown that phosphorylation of Cbl Tyr737 is abrogated by treatment of cells with PP2, a Src kinase inhibitor (17), and others have shown that Cbl phosphorylation of Tyr737 is required for its binding to the p85 subunit of PI3K (6, 7, 49). We first established the phosphorylation of Cbl at Tyr737 in response to RANKL, by using anti-phospho-Tyr737 antibodies (Fig. 5A). In WT OCLs, Cbl was phosphorylated in response to RANKL in a time-dependent manner. Not surprisingly, phosphorylation of Tyr737 was not seen in the CblYF/YF OCLs due to the tyrosine to phenylalanine substitution, albeit the expression levels of mutant Cbl protein were comparable with those of WT protein. We also confirmed that the p85 subunit of PI3K did not co-immunoprecipitate with Cbl in CblYF/YF cells, whereas robust co-precipitation occurred in WT cells, as expected (data not shown). We also established that binding of Src, which binds to the proline-rich region of Cbl (16), was not altered in the mutant protein (data not shown).
FIGURE 5.
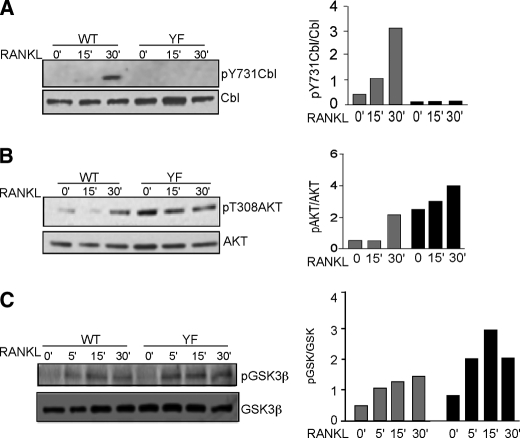
AKT phosphorylation and activity is increased in CblYF/YF OCLs. A, OCLs were serum-starved for 30 min and treated with RANKL (50 μg/ml) for the indicated times. The blot was probed with anti-phospho-Cbl Tyr737 (pY737Cbl) antibodies to detect the phosphorylation of Cbl Tyr737 (top). The blot was stripped and reprobed with anti-Cbl antibodies to detect total Cbl levels in WT and CblYF/YF OCLs. The amounts of total proteins in individual bands were quantified as described above. B, OCLs were treated with RANKL as described above. Blots were probed with anti-phospho-AKT Thr308 (pAkt) (top) and anti-AKT antibodies (bottom). Quantification of blots was performed as above. C, phosphorylation of GSK is enhanced in CblYF/YF OCLs. OCLs were treated with RANKL as described above. Western blot was performed, and the blots were probed with anti-phospho-GSK and anti-GSK antibodies. Quantification of blots was performed as above.
Of the several mechanisms that are activated downstream of RANK, only the activation of PI3K is known to involve Cbl proteins (24). Activation of PI3K results in phosphorylation of AKT downstream of RANK and in several other signaling pathways (50). It has been reported that in osteoclasts, the lack of either Cbl or Cbl-b alone does not affect RANKL-mediated AKT phosphorylation due to the existence of compensatory mechanisms, although RANK and CD40 ligand-mediated activation of AKT was defective in Cbl-b−/− dendritic cells and Cbl−/− B cells (23). Recently, it was demonstrated that in thymocytes, abrogation of Cbl-PI3K interaction in mice resulted in a significant decrease in AKT phosphorylation (51). Intriguingly, our results demonstrate that phosphorylation of AKT, both basal and RANKL-induced, was enhanced in CblYF/YF OCLs compared with WT cells, whereas the expression of AKT was similar in both cell types (Fig. 5B). These data suggest that in CblYF/YF osteoclasts, despite a lack of Cbl-PI3K interaction, PI3K/AKT signaling in response to RANKL was intact.
An increase in AKT activity should result in phosphorylation of AKT-specific substrates. In CblYF/YF OCLs in unstimulated OCLs, phosphorylation of IKKα, a substrate, is augmented as compared with the WT cells (Fig. 4A). In order to further examine the effect of Cbl-PI3K interaction on AKT activation, phosphorylation of AKT substrates in the WT and CblYF/YF cells under basal and RANKL-stimulated conditions was compared using Western blotting with a phosphospecific antibody recognizing AKT-specific phosphorylation sites. Phosphorylation of AKT substrates in CblYF/YF OCLs was higher than in WT OCLs under both basal and stimulated conditions (supplemental Fig. 2). Of several AKT substrates, phosphorylation of a protein with a molecular weight equivalent to that of glycogen synthase kinase (GSK3β) was increased. GSK3β is a well characterized substrate of AKT and exists in unphosphorylated form in resting cells. In CblYF/YF osteoclasts, similar to the enhanced activation of IKKα upon RANKL treatment, phosphorylation of GSK3β was also augmented in RANK-treated OCLs (Fig. 5C). These results suggest that in the absence of Cbl-PI3K interaction, AKT is capable of targeting its substrates and that RANKL treatment was sufficient to activate this pathway. Taken together, these data indicate that the absence of Cbl-PI3K interaction in osteoclasts increases AKT activity.
Increased PI3K Activity in CblYF/YF Osteoclasts Confers Protection from Cell Death
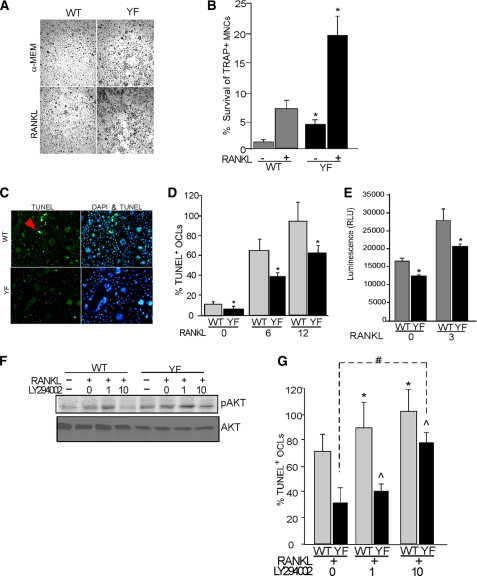
The PI3K-AKT axis of signaling is essential for survival of osteoclasts. Sustained phosphorylation of AKT in CblYF/YF OCLs may, in part, contribute to enhanced survival of osteoclasts in CblYF/YF mice and cell cultures and thus contribute to increased numbers of osteoclasts in mice and ex vivo cultures. Therefore, to examine the survival of OCLs, cells were cultured for 5 days and then either treated with RANKL for 48 h or left untreated. Cells were then fixed, and TRAP-positive multinucleated cells were counted. As compared with the WT samples, CblYF/YF OCLs demonstrated an enhanced ability to survive even in the absence of stimulation (Fig. 6, A and B). Additionally, RANKL increased the survival of CblYF/YF OCLs to a significantly higher extent than that of WT OCLs; ∼20% of the initial CblYF/YF population survived after RANKL treatment compared with ∼7% in WT cultures (Fig. 6B). In contrast, more TUNEL-positive OCLs were found in WT cultures than in CblYF/YF cultures (Fig. 6C). Because the enhanced survival of OCLs is likely to reflect their prolonged persistence in culture, which may be a result of a delay in the onset of apoptosis, we next examined the progression of apoptosis in these cultures. WT OCLs were found to be 64 and 95% TUNEL-positive following treatment with RANKL for 6 or 12 h, respectively. In contrast, CblYF/YF OCLs were only 37 and 61% TUNEL-positive under these conditions (Fig. 6D). In agreement with this result, a significant decrease in the caspase-3/7 activity was also found in CblYF/YF cultures under basal and RANKL-treated conditions (Fig. 6E). However, the levels of proapoptotic Bim were not altered under these experimental conditions (supplemental Fig. 3). These results indicate that progression toward apoptotic death was much slower in CblYF/YF cultures than in WT. Together, these findings further indicate that the lack of Cbl-PI3K interaction in CblYF/YF osteoclasts increased PI3K activity, which is known to enhance the survival of various cell types by protecting them from apoptosis.
FIGURE 6.
CblYF/YF OCLs demonstrate enhanced survival as a result of increased PI3K activity. OCLs were either untreated or were treated with RANKL (50 μg/ml) for 48 h. After treatment, cells were fixed and TRAP-stained. A, photomicrographs of the TRAP-stained OC culture with the indicated treatment. Magnification was ×10. B, histogram represents the data expressed as a percentage of the initial number of TRAP + MNCs surviving after 48 h. (gray bars, WT; black bars, CblYF/YF). *, p < 0.05 as compared with WT control. C, to visualize apoptotic OCLs on day 5 of differentiation, OCLs were fixed and stained with DAPI and TUNEL. The arrows indicate bright green TUNEL-positive cells showing apoptotic nuclei. D, to determine onset of apoptosis after 5 days of culture, cells were either left untreated or treated with RANKL (50 μg/ml) for indicated times. TUNEL-positive cells were counted, and data are expressed as percentage of TUNEL-positive OCLs (gray bars, WT; black bars, CblYF/YF). *, p < 0.05 as compared with WT control. E, to determine the caspase-3 activity, OCLs were either left untreated or treated with RANKL (50 μg/ml) for 3 h. Caspase activity was measured as described under “Experimental Procedures” (gray bars, WT; black bars, CblYF/YF). F and G, to determine the role of AKT in the prolonged survival of CblYF/YF OCLs, cells were treated with RANKL (50 μg/ml) or were treated with increasing concentrations of PI3K inhibitor LY294002 (1–10 μm) for 12 h. F, blots were probed with anti-phospho-AKT Thr308 (top) and anti-AKT antibodies (bottom). G, cells were fixed, and the TUNEL assay was performed as described above, and data are expressed as percentage of TUNEL-positive OCLs. *, p value between 0 and 1 or 10 mm in WT; #, p value between 0 and 10 mm in CblYF/YF OCLs; ^, p value showing significance between WT and CblYF/YF OCLs in the presence of RANKL. Data are representative of three independent experiments. RLU, relative luminescence units; Error bars, S.D.
To study if the lack of Cbl-PI3K interaction in CblYF/YF OCLs indeed promotes survival via the AKT pathway, we next examined the effect of decreasing AKT activity using LY294002, a specific PI3K inhibitor. We established that in both WT and CblYF/YF cultures treated with 10 μm LY294002, there was decreased AKT phosphorylation. This decrease was more pronounced in WT treated cultures compared with the CblYF/YF OCLs (Fig. 6F). We next examined the effect of different doses of LY294002 on the onset of apoptosis by a TUNEL assay. Treatment of WT OCLs with 1 μm LY294002 significantly increased the number of TUNEL-positive cells but had little or no effect in CblYF/YF cultures (Fig. 6G). A significant increase in numbers of TUNEL-positive cells in CblYF/YF cultures was achieved only after treatment of cells with 10 μm LY294002, although the number of TUNEL-positive WT OCLs was still higher than in CblYF/YF cultures under those conditions (Fig. 6G). These results also support an enhanced survival of CblYF/YF OCLs due to increased PI3K-mediated AKT activation.
Effect of Cbl Y737F on PI3K Is Not Mediated by the Disruption of Cbl-dependent Ubiquitylation of p85
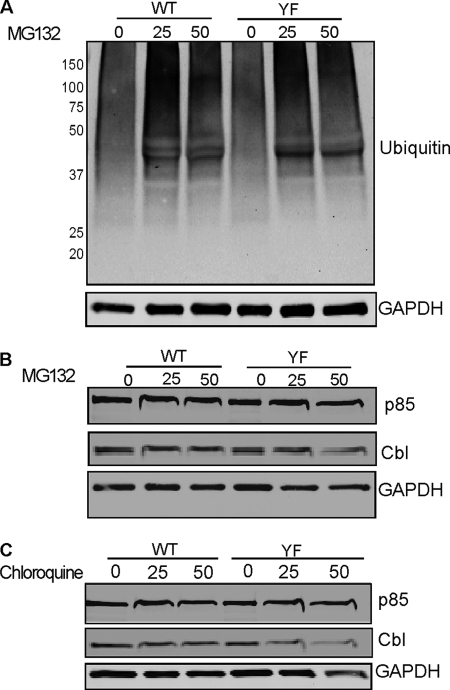
The Cbl family proteins function as negative regulators of many signaling proteins by acting as E3 ligases in the ubiquitylation system (3, 52). It is possible that Cbl regulates PI3K activity as an E3 ubiquitin ligase by mediating ubiquitylation of the p85 subunit. In this case, the lack of interaction between these proteins in CblYF/YF OCLs would result in up-regulation of p85 protein levels. Therefore, we next examined whether the increased PI3K activity was due to an increased level of PI3K in the CblYF/YF OCLs. WT and CblYF/YF OCL cultures were treated with RANKL, whereas proteasomal degradation was inhibited by increasing doses of MG132, a proteasomal inhibitor. Western blot analysis revealed that the total ubiquitylation of proteins was comparable between the WT and CblYF/YF cultures treated with different doses of MG132 (Fig. 7A). Also, the total levels of the p85 subunit of PI3K or Cbl were unaltered in the presence of increasing levels of MG132 in both YF and WT OCLs (Fig. 7B). In addition to ubiquitylation, the expression level of proteins is also modulated by lysomal degradation. Therefore, we also compared lysosomal degradation of proteins in WT and CblYF/YF OCLs. For these experiments, cells were treated with an increasing concentration of chloroquine. In both WT and CblYF/YF cultures, expression levels of Cbl and the p85 subunit of PI3K remained unchanged upon chloroquine treatment (Fig. 7C). Taken together, these data suggest that the effect of the PI3K-binding site mutation in Cbl on the overall PI3K activity was not caused by the disruption of Cbl-mediated PI3K ubiquitylation.
FIGURE 7.
PI3K levels are not altered in CblYF/YF OCLs. A, OCLs were treated with proteasome inhibitor MG132 at the indicated concentrations for 6 h. After treatment, total cell lysate was prepared and processed for Western blotting. Blot was probed with anti-ubiquitin antibody (top), and the membrane was stripped and reprobed with anti-GAPDH antibodies to determine equal protein loading. B, blots were also probed with anti-p85 and anti-Cbl antibodies as indicated. The blot was stripped and reprobed with anti-GAPDH antibodies to demonstrate protein loading. C, OCLs were also treated with chloroquine for 1 h at the indicated concentrations to prevent lysosome-mediated protein degradation. Blots were probed with anti-p85 or anti Cbl antibodies. The blot was stripped and reprobed with anti-GAPDH to verify equal protein loading. A representative experiment of three repetitions is shown.
DISCUSSION
Cbl phosphorylation on tyrosine 737 by Src family kinases and the subsequent binding of the p85 subunit to phosphorylated tyrosine 737 and activation of PI3K has been well documented (6, 49, 53). In this report, we demonstrate that the role of Cbl in osteoclast biology is predominantly dependent on its ability to bind and modulate PI3K activity. In keeping with this established role of Cbl, the skeletal analysis of mice in which Cbl-PI3K interaction is abolished showed increased bone volume due to a cell-autonomous defect in osteoclasts (Fig. 1). This result provides evidence that tyrosine phosphorylation of Cbl Tyr737 and subsequent recruitment of PI3K is required for normal bone resorption and thus plays a positive role in osteoclast function. Intriguingly, our results also demonstrate that recruitment of PI3K by Cbl has a negative role in regulating osteoclast survival because a lack of that interaction resulted in increased numbers of osteoclasts in mice (Fig. 1, I and J) and increased numbers and survival of osteoclasts in cell cultures (Figs. 2 and 6). In this case, we propose that the inability of Cbl to recruit and bind PI3K prevents its sequestration, thus increasing the pool of PI3K capable of interacting with other signaling proteins that regulate osteoclast differentiation and survival. Thus, in osteoclasts, Cbl affects PI3K-dependent events not so much by recruiting PI3K to the sites where PI3K exerts its effect on cellular activity but by modifying the spatio-temporal distribution of PI3K in such a way that PI3K is, to a significant degree, excluded from sites where it may positively affect cell function, such as bone resorption.
Interestingly, the CblYF/YF OCLs, which lack normal osteoclast functional resorption, differentiate in other ways with significant multinucleation, expression of mature osteoclast markers, spreading, and expression of NFATc1 (Figs. 2–4). The phenotype clearly demonstrates that expression of NFATc1 alone does not appear to be sufficient to generate a functional osteoclast.
Specifically, we show that abolishing Cbl-PI3K interaction perturbs RANKL-mediated signaling in mature osteoclasts. RANK signaling was previously shown to induce Src-mediated tyrosine phosphorylation of Cbl (24). Here we demonstrate that stimulation of WT OCLs with RANKL results in the phosphorylation of Tyr737 and correlates with AKT phosphorylation, suggesting that RANKL-induced phosphorylation of AKT could, in part, be mediated by the Cbl-PI3K interaction (Fig. 5, A and B). According to this notion, the loss of Cbl-PI3K interaction is expected to decrease AKT phosphorylation.
In contrast, we demonstrate that in osteoclasts, the lack of Cbl Tyr737 enhances AKT phosphorylation as well as other signaling events. Notably, AKT phosphorylation is significantly up-regulated in CblYF/YF OCLs under basal conditions (Fig. 5B). Indeed, AKT phosphorylation was previously linked to Cbl-PI3K interaction in thymocytes using a Cbl RING mutant (54). Recently, it was shown that the lack of Tyr737 in Cbl, which prevents Cbl-PI3K interaction, renders activation of AKT in response to T-cell receptor stimulation weak and transient, in contrast to a normally robust and sustained AKT activation (51). Therefore, it appears that in osteoclasts, the role of Cbl in the PI3K-mediated binding is distinct from that in thymocytes. Our data suggest that a major role of Cbl in osteoclasts is to modulate the availability of PI3K. In WT osteoclasts, binding of Cbl and PI3K would sequester PI3K. However, due to lack of interaction in CblYF/YF osteoclasts, it is possible that more PI3K becomes available for interactions with proteins other than Cbl, and this results in more robust activation compared with the WT OCLs. Interestingly, this increase in the amount of available PI3K in CblYF/YF is unrelated to changes in its Cbl-mediated down-regulation because inhibition of either ubiquitylation or lysosomal degradation exerts no effect on the level of PI3K in osteoclasts (Fig. 7, B and C). These findings further indicate that facilitation of the PI3K-mediated signaling, which occurs via activation of AKT, in CblYF/YF osteoclasts results from an increase in the level of non-Cbl-sequestered PI3K due to the loss of a regulatory mechanism, modulating availability of PI3K for other signaling proteins.
As mentioned above, this study also highlights a negative role for the Cbl-PI3K interaction in osteoclastogenesis and survival. Unexpectedly and in contrast to the findings made in Cbl−/− mice, which have normal osteoclast numbers (8), CblYF/YF mouse osteoclast numbers were elevated by 2-fold (Fig. 1, I and J), and the ex vivo differentiation of precursors into mature osteoclasts was enhanced (Fig. 2, A–E). These effects were observed along with hyperresponsiveness of CblYF/YF OCLs to RANKL, which was associated with increased RANK mRNA and protein levels (Fig. 3). M-CSF treatment of CblYF/YF precursors did not alter the proliferation of precursors, indicating that Cbl-PI3K interaction may not be involved in M-CSF-mediated effects on OC precursors (data not shown). In contrast, treatment of CblYF/YF OCLs with RANKL facilitated NFκB and NFATc1 activation (Fig. 4, A–C). Transcriptional activation of NFATc1 occurs subsequent to NFκB-mediated activation, and this leads to a positive feedback loop, resulting in increased transcription of osteoclast differentiation markers (44). Correspondingly, expression levels of TRAP, NFATc1, and CalcR, each a marker of osteoclast differentiation and maturation, were also increased in CblYF/YF OCLs (Fig. 3, D–H). RANKL-mediated signaling was also up-regulated, but a dramatic disparity in the effects of Y737F mutation on phosphorylation of individual signaling proteins was seen. There was a constitutive increase in phosphorylation of ERK proteins (Fig. 4E), whereas activation of p38 and JNK was induced to higher levels only upon stimulation with RANKL (Fig. 4, F and G). Our results suggest that Cbl-PI3K interaction is likely to modulate a subset of RANK-coupled signaling mechanisms to a greater degree. This differential effect of the Y737F mutation on various downstream elements of RANKL-induced signaling may lead to perturbed physiological outcomes because the optimal stimulation of osteoclasts is likely to require proper balance of activation between individual RANK-mediated signaling pathways. Detailed characterization of how Cbl-PI3K interaction regulates the coupling of RANK to NFκB and MAPKs and other RANK-activated signaling mechanisms will be the focus of future research.
Multinucleation and increased activity of NFATc1 is typically associated with differentiation of osteoclasts. In CblYF/YF OCLs, despite increased NFATc1 activity (Fig. 4C), the bone resorption capacity is diminished (Fig. 1, G and H). This phenotype clearly suggests that increased NFATc1 activity is not sufficient to generate osteoclast functional resorption despite enhancement of other markers of osteoclastogenesis. There are multiple mouse models where, despite the presence of multinucleated OCLs, the bone resorption capacity is compromised both in vivo and in vitro (17, 55–57). Furthermore, the decreased functionality of OCLs varies significantly in different osteopetrotic mouse models; thus, Src−/− mice have severe osteopetrosis and failure of tooth eruption (58), whereas gelsolin−/− (55), Pyk2−/− (56), β3−/− (57), and CblYF/YF mice have normal tooth eruption, and the osteopetrosis is relatively milder. However, gelsolin−/− Pyk2−/−, and β3−/− OCLs have abnormal spreading and/or cytoskeletal rearrangement, which is not apparent in the CblYF/YF OCLs, which are multinucleated and spread normally on plastic (Figs. 1 and 2) yet have defective bone resorption (Fig. 1). Although NFATc1 activation is augmented in CblYF/YF OCLs, they do not functionally resorb bone, thus demonstrating that NFATc1 expression is not sufficient for development of osteoclast function. Moreover, in CblYF/YF OCLs, although the NFATc1 levels are up-regulated, the expression levels of AP-1 transcription factors were comparable with those in WT OCLs (supplemental Fig. 1). Recently, it was reported that during osteoclast differentiation, Cbl and Cbl-b induced ubiquitylation and subsequent degradation of NFATc1 (59). However, as indicated in Fig. 7A, E3 ligase activity of the Cbl Tyr737 protein is not compromised, suggesting that other mechanism(s) may be responsible for increased activation of NFATc1 in CblYF/YF OCLs.
One likely mechanism for regulating NFATc1 activity is the PI3K-AKT pathway. This pathway is reported to regulate T cell differentiation and TCR responsiveness by modulating the cytoplasmic-nuclear localization of NFATc1 and NFκB proteins (60–62). Furthermore, in osteoclasts, it is documented that PI3K signaling coordinately regulates AKT/NFκB and MEK/ERK pathways to maintain osteoclast survival because inhibiting PI3K activity blocked these pathways (22). We propose that in CblYF/YF OCLs, defective spatial and temporal recruitment of PI3K by Cbl leads to enhanced AKT activity (Fig. 5), resulting in increased phosphorylation of AKT substrates, including IKK (Fig. 4A). In turn, increased IKK phosphorylation results in enhanced activation of NFκB and NFATc1 (Fig. 4). Thus, although the lack of Cbl-PI3K interaction augmented AKT-mediated NFκB and NFATc1 activation, failure of recruitment of the Cbl-PI3K complex to resorption sites inhibits bone resorption.
Our observation that the CblYF/YF mice have increased bone volume demonstrates that the ablation of Cbl-PI3K interaction has a distinct effect on bone. Histomorphometric analysis and decreased serum collagen telopeptide levels in the adult CblYF/YF mice indicated that the increased bone volume was, for the most part, a consequence of decreased bone resorption due to a cell-autonomous defect in osteoclast function (Fig. 1). The decreased functionality of CblYF/YF osteoclasts was evident by an in vitro pit formation assay, where we observed a 2-fold decrease in the pit-forming capacity of the CblYF/YF osteoclasts as compared with WT (Fig. 1H). The decreased bone resorption in the CblYF/YF mice is in contrast to the lack of overt skeletal phenotype in adult Cbl−/− mice and the osteopenic phenotype of the Cbl-b−/− mice (8, 63). Overexpression of Cbl in Cbl-b−/− OCLs had no effect on the elevated bone resorbing activity of Cbl-b−/− OCLs and appeared to involve E3 ligase activity because mutation in the RING domain abolished the ability of the recombinant Cbl-b to reduce the bone resorbing activity of Cbl-b−/− OCLs (63). In CblYF/YF mice, expression of Cbl-b is comparable with that in the wild type mice (data not shown). Thus, the loss of Cbl-PI3K interaction resulted in decreased osteoclast function notwithstanding the presence of normal amounts Cbl-b. These observations demonstrate that the interaction of Cbl with the p85 subunit of PI3K positively regulates osteoclast bone resorbing activity and support our previous in vitro observation that Src-mediated phosphorylation of Cbl Tyr737 is essential for bone resorption (17).
In many cells, including osteoclasts, PI3K is an important signaling molecule associated with the regulation of actin cytoskeletal components. p85 α−/− mice are osteopetrotic due to defective osteoclast function (21). Inhibition of PI3K activity interfered with the bone resorbing capability of osteoclasts (20, 64). In osteoclasts, PI3K is activated and translocated to the actin-rich cytoskeletal fraction upon adhesion to matrix proteins (20). In macrophages, Src family kinase-mediated phosphorylation of Cbl is required for Cbl-PI3K association and for the transfer of this complex to the actin-rich cytoskeleton (53). Cbl-PI3K interaction is also critical for spreading and motility of transformed fibroblasts (49). Overexpression of Cbl Y737F protein, which is unable to bind PI3K, reduces in vitro bone resorbing activity of osteoclasts by 80% (17). Lack of Cbl-PI3K interaction resulted in decreased bone resorption as demonstrated in this report. Whether or not this effect is due to the inability of the Cbl-PI3K complex to form and translocate remains unclear. As shown in Fig. 2E, CblYF/YF osteoclasts are able to form the peripheral actin ring, indicating that under basal conditions, lack of Cbl-PI3K interaction may not be necessary for the organization of podosomes and actin ring formation. Similarly, actin organization in the osteoclasts was not perturbed in the absence of either Cbl or Cbl-b (16, 63). It is possible that Cbl-b and/or other proteins that are capable of interacting with Cbl and actin cytoskeleton (e.g. Src) might be able to compensate for the lack of Cbl-PI3K interaction and preserve the actin organization in this situation. Recently, it has been reported that Cbl and Cbl-b act redundantly in the stabilization of microtubules and podosomes (14). In future studies, it will be important to characterize the actin organization in the CblYF/YF OCLs replated to different extracellular matrix proteins or treated with various other stimuli that induce cytoskeletal reorganization.
Taken together, our findings indicate that the loss of Cbl-PI3K interaction results in increased bone volume that is largely the consequence of decreased bone resorption. The CblYF/YF osteoclasts resorb bone less efficiently, although higher in number due to enhanced survival. Loss of Cbl-PI3K interaction leads to higher PI3K activity, facilitating this osteoclast signaling event. Overall, this study demonstrates that Cbl plays unique roles in both positive and negative regulation of osteoclast functions, which are not compensated by Cbl-b. Several signaling pathways depend on a step mediated by recruitment of proteins to Cbl. This commonality is restricted to the mode of interaction between Cbl and the recruited protein because the impact of such an event is dependent on the nature of the recruited molecule, the nature of the stimulus, and the cellular context. It has been shown previously that the phosphorylation-dependent binding of Cbl to a signaling protein may lead to its ubiquitylation and/or down-regulation (e.g. tyrosine kinases, Vav, and several other proteins) but may also facilitate signaling through formation of protein-protein complexes in the relevant locations at the appropriate time. This report provides evidence of further complexity of this system; interaction of Cbl with a signaling protein (PI3K, in this case) may have a negative regulatory effect on signaling by sequestering this protein. Importantly, the role of this sequestration is likely to be not just down-regulation of signaling but also providing the appropriate balance between activation of various pathways triggered by RANKL in order to achieve the optimal level of biological responses. Without such regulatory sequestration, various cellular functions may be affected differentially, thus leading to disjointed responses as seen in CblYF/YF mice, in which osteoclasts demonstrate multiple signs of hyperactive signals and are more resistant to apoptosis than their WT counterparts but are less active in bone resorption both in vitro and in vivo.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AR055601 (to A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- RANK
- receptor activator of NFκB
- M-CSF
- macrophage colony-stimulating factor
- α-MEM
- minimum essential medium-α modification
- OCL
- osteoclast-like cell
- OC
- osteoclast.
REFERENCES
- 1.Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 2.Horne W. C., Sanjay A., Bruzzaniti A., Baron R. (2005) Immunol. Rev. 208, 106–125 [DOI] [PubMed] [Google Scholar]
- 3.Swaminathan G., Tsygankov A. Y. (2006) J. Cell. Physiol. 209, 21–43 [DOI] [PubMed] [Google Scholar]
- 4.Andoniou C. E., Thien C. B., Langdon W. Y. (1996) Oncogene 12, 1981–1989 [PubMed] [Google Scholar]
- 5.Marengère L. E., Mirtsos C., Kozieradzki I., Veillette A., Mak T. W., Penninger J. M. (1997) J. Immunol. 159, 70–76 [PubMed] [Google Scholar]
- 6.Hunter S., Burton E. A., Wu S. C., Anderson S. M. (1999) J. Biol. Chem. 274, 2097–2106 [DOI] [PubMed] [Google Scholar]
- 7.Ueno H., Sasaki K., Honda H., Nakamoto T., Yamagata T., Miyagawa K., Mitani K., Yazaki Y., Hirai H. (1998) Blood 91, 46–53 [PubMed] [Google Scholar]
- 8.Chiusaroli R., Sanjay A., Henriksen K., Engsig M. T., Horne W. C., Gu H., Baron R. (2003) Dev. Biol. 261, 537–547 [DOI] [PubMed] [Google Scholar]
- 9.Bruzzaniti A., Neff L., Sanjay A., Horne W. C., De Camilli P., Baron R. (2005) Mol. Biol. Cell 16, 3301–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanjay A., Houghton A., Neff L., DiDomenico E., Bardelay C., Antoine E., Levy J., Gailit J., Bowtell D., Horne W. C., Baron R. (2001) J. Cell Biol. 152, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luxenburg C., Addadi L., Geiger B. (2006) Eur. J. Cell Biol. 85, 203–211 [DOI] [PubMed] [Google Scholar]
- 12.Luxenburg C., Parsons J. T., Addadi L., Geiger B. (2006) J. Cell Sci. 119, 4878–4888 [DOI] [PubMed] [Google Scholar]
- 13.Zambonin-Zallone A., Teti A., Grano M., Rubinacci A., Abbadini M., Gaboli M., Marchisio P. C. (1989) Exp. Cell Res. 182, 645–652 [DOI] [PubMed] [Google Scholar]
- 14.Purev E., Neff L., Horne W. C., Baron R. (2009) Mol. Biol. Cell 20, 4021–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka S., Amling M., Neff L., Peyman A., Uhlmann E., Levy J. B., Baron R. (1996) Nature 383, 528–531 [DOI] [PubMed] [Google Scholar]
- 16.Sanjay A., Miyazaki T., Itzstein C., Purev E., Horne W. C., Baron R. (2006) FEBS J. 273, 5442–5456 [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki T., Sanjay A., Neff L., Tanaka S., Horne W. C., Baron R. (2004) J. Biol. Chem. 279, 17660–17666 [DOI] [PubMed] [Google Scholar]
- 18.Akiyama T., Bouillet P., Miyazaki T., Kadono Y., Chikuda H., Chung U. I., Fukuda A., Hikita A., Seto H., Okada T., Inaba T., Sanjay A., Baron R., Kawaguchi H., Oda H., Nakamura K., Strasser A., Tanaka S. (2003) EMBO J. 22, 6653–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chellaiah M., Hruska K. (1996) Mol. Biol. Cell 7, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakkakorpi P. T., Wesolowski G., Zimolo Z., Rodan G. A., Rodan S. B. (1997) Exp. Cell Res. 237, 296–306 [DOI] [PubMed] [Google Scholar]
- 21.Munugalavadla V., Vemula S., Sims E. C., Krishnan S., Chen S., Yan J., Li H., Niziolek P. J., Takemoto C., Robling A. G., Yang F. C., Kapur R. (2008) Mol. Cell. Biol. 28, 7182–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gingery A., Bradley E., Shaw A., Oursler M. J. (2003) J. Cell. Biochem. 89, 165–179 [DOI] [PubMed] [Google Scholar]
- 23.Arron J. R., Vologodskaia M., Wong B. R., Naramura M., Kim N., Gu H., Choi Y. (2001) J. Biol. Chem. 276, 30011–30017 [DOI] [PubMed] [Google Scholar]
- 24.Wong B. R., Besser D., Kim N., Arron J. R., Vologodskaia M., Hanafusa H., Choi Y. (1999) Mol. Cell 4, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 25.Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y. Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., Le J., Ohashi P. S., Sarosi I., Nishina H., Lipkowitz S., Penninger J. M. (2000) Nature 403, 211–216 [DOI] [PubMed] [Google Scholar]
- 26.Chiang Y. J., Kole H. K., Brown K., Naramura M., Fukuhara S., Hu R. J., Jang I. K., Gutkind J. S., Shevach E., Gu H. (2000) Nature 403, 216–220 [DOI] [PubMed] [Google Scholar]
- 27.Naramura M., Kole H. K., Hu R. J., Gu H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15547–15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naramura M., Jang I. K., Kole H., Huang F., Haines D., Gu H. (2002) Nat. Immunol. 3, 1192–1199 [DOI] [PubMed] [Google Scholar]
- 29.Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravichandran S., Collins J., Nau M. M., Lipkowitz S. (2004) Oncogene 23, 7104–7115 [DOI] [PubMed] [Google Scholar]
- 30.Molero J. C., Jensen T. E., Withers P. C., Couzens M., Herzog H., Thien C. B., Langdon W. Y., Walder K., Murphy M. A., Bowtell D. D., James D. E., Cooney G. J. (2004) J. Clin. Invest. 114, 1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molero J. C., Turner N., Thien C. B., Langdon W. Y., James D. E., Cooney G. J. (2006) Diabetes 55, 3411–3417 [DOI] [PubMed] [Google Scholar]
- 32.Sims N. A., Clément-Lacroix P., Da Ponte F., Bouali Y., Binart N., Moriggl R., Goffin V., Coschigano K., Gaillard-Kelly M., Kopchick J., Baron R., Kelly P. A. (2000) J. Clin. Invest. 106, 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron R., Vignery A., Neff L., Silverglate A., Santa Maria A. (1983) in Bone Histomorphometry: Techniques and Interpretation (Recker R. R. ed) pp. 13–35, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 34.Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R. (1987) J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 35.Aoki K., Didomenico E., Sims N. A., Mukhopadhyay K., Neff L., Houghton A., Amling M., Levy J. B., Horne W. C., Baron R. (1999) Bone 25, 261–267 [DOI] [PubMed] [Google Scholar]
- 36.Kukita T., Wada N., Kukita A., Kakimoto T., Sandra F., Toh K., Nagata K., Iijima T., Horiuchi M., Matsusaki H., Hieshima K., Yoshie O., Nomiyama H. (2004) J. Exp. Med. 200, 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., Oike Y., Takeya M., Toyama Y., Suda T. (2005) J. Exp. Med. 202, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M., Birnbaum M. J., MacKay C. A., Mason-Savas A., Thompson B., Odgren P. R. (2008) J. Cell. Physiol. 215, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., Choi Y. (2006) Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 40.Tondravi M. M., McKercher S. R., Anderson K., Erdmann J. M., Quiroz M., Maki R., Teitelbaum S. L. (1997) Nature 386, 81–84 [DOI] [PubMed] [Google Scholar]
- 41.Matsuo K., Owens J. M., Tonko M., Elliott C., Chambers T. J., Wagner E. F. (2000) Nat. Genet. 24, 184–187 [DOI] [PubMed] [Google Scholar]
- 42.Bozec A., Bakiri L., Hoebertz A., Eferl R., Schilling A. F., Komnenovic V., Scheuch H., Priemel M., Stewart C. L., Amling M., Wagner E. F. (2008) Nature 454, 221–225 [DOI] [PubMed] [Google Scholar]
- 43.Grigoriadis A. E., Wang Z. Q., Cecchini M. G., Hofstetter W., Felix R., Fleisch H. A., Wagner E. F. (1994) Science 266, 443–448 [DOI] [PubMed] [Google Scholar]
- 44.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 45.Asagiri M., Takayanagi H. (2007) Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 46.Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 47.Baldwin A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683 [DOI] [PubMed] [Google Scholar]
- 48.Humphrey M. B., Lanier L. L., Nakamura M. C. (2005) Immunol. Rev. 208, 50–65 [DOI] [PubMed] [Google Scholar]
- 49.Feshchenko E. A., Shore S. K., Tsygankov A. Y. (1999) Oncogene 18, 3703–3715 [DOI] [PubMed] [Google Scholar]
- 50.Datta S. R., Brunet A., Greenberg M. E. (1999) Genes Dev. 13, 2905–2927 [DOI] [PubMed] [Google Scholar]
- 51.Thien C. B., Dagger S. A., Steer J. H., Koentgen F., Jansen E. S., Scott C. L., Langdon W. Y. (2010) J. Biol. Chem. 285, 10969–10981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanjay A., Horne W. C., Baron R. (2001) Science's STKE http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/110/pe40 [DOI] [PubMed]
- 53.Meng F., Lowell C. A. (1998) EMBO J. 17, 4391–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thien C. B., Blystad F. D., Zhan Y., Lew A. M., Voigt V., Andoniou C. E., Langdon W. Y. (2005) EMBO J. 24, 3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chellaiah M., Kizer N., Silva M., Alvarez U., Kwiatkowski D., Hruska K. A. (2000) J. Cell Biol. 148, 665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gil-Henn H., Destaing O., Sims N. A., Aoki K., Alles N., Neff L., Sanjay A., Bruzzaniti A., De Camilli P., Baron R., Schlessinger J. (2007) J. Cell Biol. 178, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McHugh K. P., Hodivala-Dilke K., Zheng M. H., Namba N., Lam J., Novack D., Feng X., Ross F. P., Hynes R. O., Teitelbaum S. L. (2000) J. Clin. Invest. 105, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soriano P., Montgomery C., Geske R., Bradley A. (1991) Cell 64, 693–702 [DOI] [PubMed] [Google Scholar]
- 59.Kim J. H., Kim K., Jin H. M., Song I., Youn B. U., Lee S. H., Choi Y., Kim N. (2010) J. Biol. Chem. 285, 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patra A. K., Drewes T., Engelmann S., Chuvpilo S., Kishi H., Hünig T., Serfling E., Bommhardt U. H. (2006) J. Immunol. 177, 4567–4576 [DOI] [PubMed] [Google Scholar]
- 61.Patra A. K., Na S. Y., Bommhardt U. (2004) J. Immunol. 172, 4812–4820 [DOI] [PubMed] [Google Scholar]
- 62.Xue L., Gyles S. L., Barrow A., Pettipher R. (2007) Biochem. Pharmacol. 73, 843–853 [DOI] [PubMed] [Google Scholar]
- 63.Nakajima A., Sanjay A., Chiusaroli R., Adapala N. S., Neff L., Itzsteink C., Horne W. C., Baron R. (2009) J. Bone Miner. Res. 24, 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura I., Takahashi N., Sasaki T., Tanaka S., Udagawa N., Murakami H., Kimura K., Kabuyama Y., Kurokawa T., Suda T., Fukui Y. (1995) FEBS Lett. 361, 79–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.