Abstract
Initial attachment of bacteriophage P22 to the Salmonella host cell is known to be mediated by interactions between lipopolysaccharide (LPS) and the phage tailspike proteins (TSP), but the events that subsequently lead to DNA injection into the bacterium are unknown. We used the binding of a fluorescent dye and DNA accessibility to DNase and restriction enzymes to analyze DNA ejection from phage particles in vitro. Ejection was specifically triggered by aggregates of purified Salmonella LPS but not by LPS with different O-antigen structure, by lipid A, phospholipids, or soluble O-antigen polysaccharide. This suggests that P22 does not use a secondary receptor at the bacterial outer membrane surface. Using phage particles reconstituted with purified mutant TSP in vitro, we found that the endorhamnosidase activity of TSP degrading the O-antigen polysaccharide was required prior to DNA ejection in vitro and DNA replication in vivo. If, however, LPS was pre-digested with soluble TSP, it was no longer able to trigger DNA ejection, even though it still contained five O-antigen oligosaccharide repeats. Together with known data on the structure of LPS and phage P22, our results suggest a molecular model. In this model, tailspikes position the phage particles on the outer membrane surface for DNA ejection. They force gp26, the central needle and plug protein of the phage tail machine, through the core oligosaccharide layer and into the hydrophobic portion of the outer membrane, leading to refolding of the gp26 lazo-domain, release of the plug, and ejection of DNA and pilot proteins.
Keywords: Bacteriophage, Biophysics, DNA Viruses, Lipopolysaccharide (LPS), Virus Assembly, DNA Injection, Endoglycosidase, O-antigen, Salmonella
Introduction
For nearly 6 decades, Salmonella enterica bacteriophage P22 has been used as model system in molecular biology (1). It belongs to the morphological class of Podoviridae, bacteriophages with short noncontractile tails and icosahedral heads filled with double-stranded DNA (2). The assembly and maturation pathway of phage P22 has been studied thoroughly (3). After assembly of empty procapsids, the 42-kbp genome is packaged. This process terminates via a pressure-dependent headful sensing mechanism. In the following, the head is sealed by gene products gp4, gp10, and gp26 to prevent DNA leakage. Finally, up to six trimers of gp9 bind to complete and stabilize the assembly (2). gp9 is the tailspike protein (TSP)5 required for host cell attachment. TSP has been studied as a model system for protein folding and protein carbohydrate interactions (4, 5). Three subunits with parallel right-handed β-helix fold make up the native trimer of 215 kDa (6, 7). A long surface-exposed groove on each subunit specifically recognizes the O-antigen portion of the Salmonella host LPS and harbors an endorhamnosidase activity, cleaving the α-(1→3) glycosidic linkages between rhamnose and galactose and producing dimers of 2 O-antigen repeat units (RU) as the main product (8, 9).
The phage genome and accompanying pilot proteins must cross the outer and inner membranes without affecting the vitality of the cell. This is a crucial event in phage infection. In the outer membrane of Gram-negative bacteria, proteins are embedded in an asymmetric bilayer with phospholipids on the periplasmic side and tightly packed lipid A molecules on the extracellular side (10). On the extracellular side, lipid A is decorated with a hydrophilic sugar core. This special architecture creates an effective LPS outer barrier repulsing large hydrophilic as well as small hydrophobic molecules (11). In Salmonella cells, about 50% of the LPS molecules may carry polysaccharide chains of varying length and composition, the O-antigen, which accounts for the high serological diversity of strains (11, 12). In many cases, phages use LPS as the first receptor initiating a series of molecular events that result in transmission of genetic material into the host cytosol. These infection mechanisms have been studied for different phage morphologies (13–16), but a general picture does not emerge. Apparently, phages developed individual strategies to surmount the two membranes and periplasmic space of Gram-negative hosts. Several phage receptors in the LPS-containing outer membrane have been studied in some detail. Escherichia coli phage T4, which undergoes tail sheath contraction during infection, uses its long tail fibers to recognize the host cell. This leads to the release of short tail fibers that fix the phage to the LPS core structure. In vitro, high LPS concentrations can promote T4 tail sheath contraction but not DNA ejection (15). Podovirus T7 tail fibers interact with LPS structures on different host strains and thus mediate early steps of infection (17). By contrast λ or T5, phages with long noncontractile tails, do not use LPS but membrane protein receptors. In vitro, the purified receptor proteins are sufficient to trigger DNA release (14, 18). In the case of phage P22, LPS recognition and cleavage are thought to be necessary for recognition of a putative secondary receptor (1). Interaction with this secondary receptor would then initiate a couple of events that enable the short tailed P22 phage to translocate material over two membranes and the periplasmic space. In vivo experiments with phage T7 showed that proteins gp15 and gp16 translocate DNA to the cytoplasm (13). However, neither the role of initial LPS binding and O-antigen hydrolysis for infection nor the dependence of subsequent infection steps on these initial events have been investigated in molecular detail in podoviruses.
In this study, we have developed an in vitro DNA ejection assay for phage P22. We find that incubation of P22 phage particles with purified Salmonella LPS is sufficient to trigger complete DNA ejection of phage P22. Thus, no secondary receptor is required. Using phage particles reconstituted with mutant TSP, we find that both TSP functions, LPS binding and hydrolysis, are required to trigger ejection. Based on our results, we propose a molecular model of the initial steps of the phage P22 infection mechanism.
EXPERIMENTAL PROCEDURES
Materials
Fluorescence dye YO-PRO-1 iodide (491/509) was obtained from Invitrogen; 3,5-dinitrosalicylic acid was obtained from Sigma. All other chemicals used during this study were of highest purity. Standard buffer in all experiments is 50 mm Tris/HCl, pH 7.6, 4 mm MgCl2. For fluorescence measurements, we used plastic cuvettes from Roth (Karlsruhe, Germany). LPS from E. coli IHE was received from Nina Lorenzen, Universität of Potsdam, Potsdam, Germany.
The clear plaque mutant H5 of P22 contains a wild-type gene 9 coding for the TSP and was used in all experiments with P22 phages unless indicated otherwise. It was kindly provided by Dr. Wolfgang Rabsch, Robert Koch Institut Wernigerode, Germany. The following strains of Salmonella enterica enterica serovar Typhimurium (S. typhimurium), were employed: DB7155 LT2 (19), SupE, amber suppressor strain; DB7136 LT2 (19); DB7136 c2ts30 containing the temperature-sensitive P22 prophage c2ts30 (13− am/9− am), a kind gift from Cameron Haase-Pettingell, Massachusetts Institute of Technology (20).
Preparation of P22 Taikspike Proteins, P22 Phage and P22 Heads
Purification of full-length and N-terminally shortened P22TSP has been described elsewhere (21). TSP mutant T307K was obtained using the QuikChange kit (Stratagene) for site-directed mutagenesis according to the manufacturer's instructions. TSP concentrations are given as molar concentrations of trimers.
P22 phages were purified from S. typhimurium DB7136 LT2 cell lysates. Cells debris was centrifuged at 5000 × g for 15 min, before phages were collected at 35,000 × g. Phages were resuspended in standard buffer and centrifuged in a CsCl gradient (1.3 to 1.7 g ml−1) for 2.5 h at 98,300 × g. Phage suspensions were harvested and dialyzed against standard buffer. Infectious particle concentrations as plaque-forming units were quantified by plating on S. typhimurium DB7136.
Phage heads for in vitro assembly assays were obtained from S. typhimurium DB7136 c2ts30. This strain contains a P22 mutant lysogen deficient in lysis (13− am) as well as in tailspike formation (9− am) and will accumulate TSP-less heads intracellularly upon induction (20). To reconstitute tailed phages, phage heads were incubated with excess TSP for 1 h at 37 °C (2), centrifuged in a CsCl gradient from 1.3 to 1.7 g·ml−1 for 2.5 h at 98,300 × g, and dialyzed against standard buffer. Infectious reconstituted phages were quantified by plating on amber suppressor strain S. typhimurium DB7155.
Determination of Phage Particle Concentration
Biochemical concentrations of particles were determined by 10% SDS-PAGE. Phages were heated to 100 °C for 5 min in 1.5% SDS and subjected to SDS-PAGE. After silver staining (22), the TSP bands were quantified densitometrically with the program Gelscan 5.1 (BioSciTec GmbH; Frankfurt, Germany), and phage concentration was calculated from a TSP standard curve under the assumption that every phage bound six TSP. Particle concentrations for every phage preparation were calculated from three independent experiments.
Lipopolysaccharide Samples from S. typhimurium
The preparation of LPS has been described previously (23, 24). After resuspension in standard buffer, purified LPS was present in small aggregates with an average Stokes radius of 90 nm, as determined by dynamic light scattering. It was free of nucleic acids and proteins, as shown by the absence of near-ultraviolet absorbance and by SDS-PAGE. Aliquots of 1 mg/ml LPS suspension were stored in standard buffer at −40 °C. Lipid A and O-antigen polysaccharide were obtained by the acid hydrolysis method of Freeman and Philpott (25), but using purified LPS as the starting material.
O-antigen digestion of 1 mg/ml purified LPS with 200 μg/ml TSP was performed in standard buffer at 37 °C overnight. TSP was removed by phenol/water extraction, and after excessive dialysis of the watery phase against distilled water, digested LPS was pelleted by ultracentrifugation (24). The product was lyophilized and stored at −20 °C in standard buffer. 15% SDS-PAGE of 750 ng of LPS and its TSP digestion products were performed after a published protocol, and gels were silver-stained (22).
Fluorescence DNA Ejection Assay
5 or 10 μg/ml LPS from S. typhimurium DB7155 and 1.1 μm Yo-Pro were equilibrated to 37 °C in standard buffer. Dye fluorescence was excited at 491 nm and detected at 509 nm. After addition of phages to the final concentration of 3.7 × 109 particles/ml (P22) or 7 × 109 particles/ml (reconstituted phages), ejection was followed for a total of 18,000 s. At the end of each ejection time course, DNase I (10 μg/ml) was added as a control for DNA accessibility. With the concentrations used, dye and LPS were not limiting signal changes.
Agarose Gel Electrophoresis of Phages
1.8 × 1011 particles/ml of phages P22 and P22tmutant (where P22tmutant indicates P22 particles reconstituted from phage heads with TSP variant) were incubated with 0.24 mg/ml LPS from S. typhimurium at 37 °C in standard buffer overnight. For a control, the same amount of phages was incubated at 40 °C for 2 h in 6 m guanidinium chloride, and chemically released DNA was purified with QIAprep spin miniprep kit (Qiagen GmbH, Hilden, Germany). Released DNA was either cleaved with 5–10 units of indicated restriction enzyme or digested with 100 μg/ml DNase I. Digestion products and assembled phages were analyzed on a 1% agarose gel and stained with ethidium bromide.
Saccharide Binding and Hydrolysis by TSP
Fluorescence titrations were performed and data fitted to an independent binding site model as described previously (8, 26). Hydrolytic activity of 0.09 μm TSP with 12 mg/ml O-antigen polysaccharide from S. typhimurium was measured at 37 °C in 50 mm phosphate buffer, pH 7. Initial rates of hydrolysis were determined by colorimetry with 3,5-dinitrosalicylic acid (27), where reducing ends are quantified using a glucose calibration curve. In this assay, the wild-type protein produced 207 ± 17 μm reducing ends per min, whereas the activities of the mutant TSP were reduced to 0.33 ± 0.02 μm/min for D392N, 0.19 ± 0.05 μm/min for D395N, and 27.5 ± 1.7 μm/min for T307K.
To determine the size of hydrolysis products, the enzymatic reaction at 10 °C was stopped with 0.125 m HCl, and the product oligosaccharides were analyzed on a Superdex Peptide HR 10/30 with refractive-index detection.
RESULTS
P22 DNA Is Released Specifically upon Contact with LPS from S. typhimurium
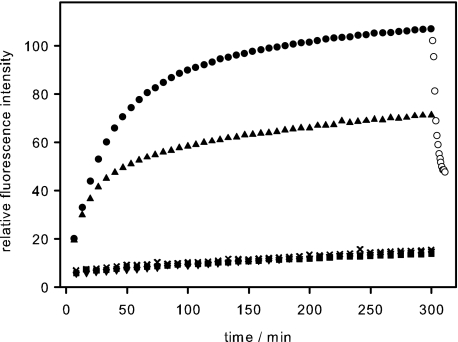
To study the elementary steps of the infection of Salmonella enterica by bacteriophage P22, we designed a fluorescence assay monitoring phage DNA ejection in vitro. We chose the fluorescent dye Yo-Pro that had been shown not to affect the stability of T5 phages but to bind DNA in solution rapidly (28). When purified phage P22 was incubated with purified LPS from S. typhimurium, we observed an increasing dye fluorescence signal that reached a plateau value after about 300 min at 37 °C (Fig. 1). We concluded that DNA became accessible to Yo-Pro staining upon ejection from the phage. The time course and the extent of the rise in fluorescence intensity did not change when the LPS concentration was increased 10-fold (data not shown). By contrast, the maximum signal scaled with the number of phage particles used in the assay. The gain in fluorescence was negligible, when either phages or LPS were incubated alone. DNA was specifically released of phage P22 only with its natural receptor S. typhimurium LPS. If phage P22 was incubated with LPS from E. coli or phospholipids extracted from S. typhimurium, no fluorescence increase was observed.
FIGURE 1.
In vitro DNA ejection from phage P22 particles. To follow DNA ejection at 37 °C, we added 3.7 × 109 phage P22 particles to 5 μg/ml S. typhimurium LPS and a fluorescent DNA-binding dye (●). Addition of DNase reversed the fluorescence increase, indicating DNA became released from the phage (○). When 11 nm free TSP were added 10 min after the phage particles, less DNA became ejected (▴). Neither LPS from E. coli (▾), digested S. typhimurium LPS (■), nor its lipid A mixed with O-antigen polysaccharide (×) was able to trigger DNA release from phage P22. Standard deviations from three independent experiments are not more than 4% of total fluorescence for every experiment.
To confirm that the fluorescence signal originated from DNA in solution, we added DNase I after the maximum signal was reached. As a result, the fluorescence increase was rapidly reversed (Fig. 1), consistent with the fact that the intercalating dye binds to double-stranded DNA with high affinity but does not stain nucleotide fragments produced by the enzyme. The rise in dye fluorescence indicating DNA ejection occurred with a half-time of about 30 min at 37 °C and without a resolvable lag phase. We refrained from using a mathematical model to describe the data, because multiple events, such as signal transduction in the ejection machinery and conformational changes during the ejection process, might contribute to the observed ejection kinetics.
When we added a large excess of purified TSP simultaneously with the phage P22 to the LPS in the assay, no fluorescence intensity increase was observed. TSP is an endorhamnosidase hydrolyzing the O-antigen part of LPS. Thus, in our assay, TSP either had destroyed all LPS receptors, before phage P22 was able to start DNA ejection, or excess TSP had rapidly blocked all binding sites for phage particles on the LPS aggregates. When TSP was added 10 min after phage P22, only about 60% of the maximum fluorescence signal was reached (Fig. 1). In this experiment, the signal increase observed after addition of TSP must be due to those phage particles already poised to eject their DNA at the time point of TSP addition, because free TSP completely blocked DNA ejection when added simultaneously with phage particles. The observations shed light on the origin of the time course of the fluorescence increase. If DNA ejection was triggered at the very beginning of the experiment in a large number of phages, but DNA release from a single particle was a slow process, the signal increase would persist even upon the sudden destruction or blockage of the LPS receptor by added TSP. The measured signal must therefore represent multiple fast events that add up to the final curve.
P22 Releases Its DNA Completely upon Contact with LPS
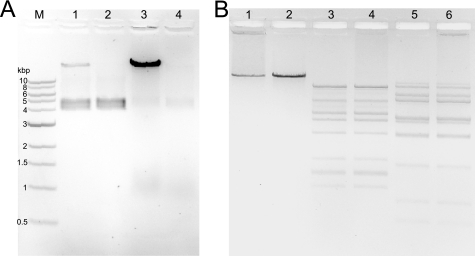
Quantifying released DNA from the fluorescence signal is not straightforward. For that reason, we analyzed phage preparations before and after incubation with LPS on ethidium bromide-stained agarose gels (Fig. 2A). The phage preparation itself has a net negative charge and therefore can migrate into the gel. DNA inside the capsids can be stained with ethidium bromide resulting in a fuzzy doublet band at a position between the 4- and 5-kbp markers. The doublet character of this band disappears at lower agarose concentrations (not shown) and appears to be an electrophoresis artifact rather than reflect a heterogeneity of the phage particles. When complete P22 DNA was liberated from phage capsids with guanidinium chloride, it only migrated very slowly in the gel, in agreement with its large size of 43 kbp. After incubation of phage P22 with LPS, the band corresponding to intact phages disappeared, and the amount of free phage DNA increased. DNA that was liberated by LPS could be fully digested with DNase I. Accordingly, phage particles incubated with LPS appeared intact but empty in negatively stained electron micrographs (data not shown). To further probe whether the LPS trigger provokes full or only partial ejection of phage DNA, we digested LPS-treated phage preparations with restriction endonucleases ClaI and AvaI (Fig. 2B). DNA was fully accessible to both enzymes and showed the same cleavage pattern when either released from phage particles upon incubation with LPS or purified after disruption of phage particles in 6 m guanidinium chloride. Fully assembled phages were not accessible to DNase I or restriction enzymes. From this, we conclude that every P22 phage that has been triggered to eject by contact with LPS releases its DNA completely.
FIGURE 2.
Agarose gel electrophoresis of phage P22 and its ejection products. A, EtBr stained 1% agarose gel electrophoresis with 1.75 × 109 complete P22 phages (lane 1) or complete phage particles with 10 μg/ml DNase (lane 2), 1.75 × 109 phage particles incubated with LPS over night (lane 3), and thereafter with 10 μg/ml DNase (lane 4). B, electrophoretic analysis of DNA released from intact phage particles upon incubation with LPS (lanes 1, 3, and 5) and of phage DNA purified from denatured particles (lanes 2, 4, and 6), either untreated (lanes 1 and 2) or digested with AvaI (lanes 3 and 4) or ClaI (lanes 5 and 6).
Endoglycosidase Activity of TSP Is Essential for Infection of Salmonella by Phage P22
Functional TSP are essential for phage P22, but it is not known at which steps of the infection cycle the binding and enzymatic functions of TSP are required. TSP-less heads can be accumulated in Salmonella cells infected with P22 carrying amber mutations in TSP gene 9 and in gene 13 required for cell lysis. When purified TSP-less heads are incubated with purified TSP, they bind irreversibly, and functional phages result (2). Because of the amber mutations in their genome, infectivity of the reconstituted phages has to be probed on a Salmonella strain producing an amber suppressor tRNA. Once the reconstituted phage has infected a cell, progeny with fully functional TSP is produced resulting in formation of clear plaques on the glutamine inserting amber suppressor strain S. typhimurium DB7155 (20). This provides us with a tailing assay to probe the role of TSP for infection.
To understand the role of LPS binding and hydrolysis by TSP in the infection process, we reconstituted phage head preparations with different mutant tailspikes (P22tmutant). Reconstituted particles were repurified by density gradient ultracentrifugation to remove excess TSP. The mutant TSP employed has reduced hydrolytic activity toward O-antigen polysaccharide as quantified from the amount of reducing ends released. In comparison with the wild-type protein, the activity toward O-antigen polysaccharide at 37 °C was reduced for TSP mutant D392N to 0.2% and for D395N to 0.1%, respectively (see “Experimental Procedures”). Interestingly, these two mutants also showed reduced infectivity in the tailing assay, where we observed 5.1 ± 0.7 × 107 plaques for P22tD392N and 2.4 ± 0.7 × 107 plaques for P22tD395N. As the same number of reconstituted particles containing wild-type TSP (P22tWT) formed 4.8 ± 1.0 × 1011 plaques, both mutants are about a thousand times less infective than P22tWT, in agreement with their reduced activity toward O-antigen.
Additionally, we probed a binding-deficient mutant in the tailing assay. Whereas oligosaccharide binding is unaffected by the mutations at Asp-392 or Asp-395 (5, 8), no binding of O-antigen octasaccharide to TSPT307K was detected by isothermal titration calorimetry at saccharide concentrations up to 0.5 mm. Despite the strongly reduced binding affinity to the short oligosaccharide, TSPT307K displayed high endorhamnosidase activity toward soluble polysaccharide substrate. The initial rate of reducing end formation measured for the mutant was 27.5 ± 1.7 μm/min, about 14% of the rate measured for the wild type under the same conditions. Although the polysaccharide binds to TSP much more tightly than short oligosaccharides (see below), the substrate concentration in the assay may not have been saturating for the mutant. Hence, the residual activity observed provides a lower estimate, and catalytic turnover appear largely unaffected by the T307K substitution. Accordingly, reconstituted P22tT307K particles formed the same number of plaques in the tailing assay (5.0 ± 1.7 × 1011) as particles reconstituted with wild-type TSP (4.8 ± 1.0 × 1011)). We conclude that significant catalytic activity toward O-antigen is required for P22 infection of Salmonella cells prior to phage DNA replication.
DNA Ejection Requires the Endorhamnosidase Activity of TSP
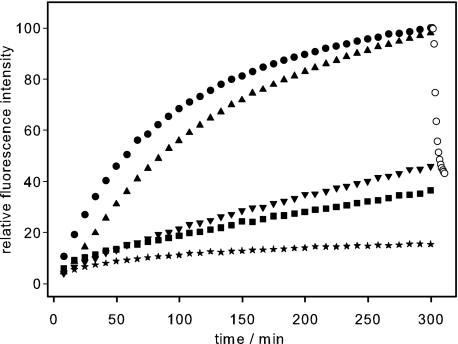
We wanted to elucidate whether the in vivo infection behavior directly and solely reflected interactions of TSP with LPS. For that reason, we measured in vitro DNA ejection kinetics of mutant reconstituted phages (Fig. 3). Because of the reduced plaque forming activity of particles containing mutant TSP, we calculated P22tWT, P22tT307K, P22tD392N, and P22tD395N particle concentration from a silver-stained 10% SDS-PAGE to ensure identical particle concentrations. When we compared plaque-forming units to biochemically determined concentrations of phage particles, we found that around 7% of the reconstituted P22tWT particles was able to form a plaque on S. enterica, in agreement with previous observations on the amount of TSP necessary to titrate P22 heads (29). The efficiency of plating determined in the same way for complete P22 particles isolated from cell lysates was 17%.
FIGURE 3.
TSP endorhamnosidase mutations delay DNA ejection from phage P22. The fluorescence ejection assay was used to follow DNA release from reconstituted phage particles carrying different TSP variants. To 10 μg/ml S. typhimurium LPS and the fluorescent dye, 7.2 × 109 reconstituted phage particles (P22tmutant) were added at 37 °C as follows: ●, P22tWT; ▴, P22tT307K; ▾, P22tD392N; ■, P22tD395N. ○, fluorescence decrease after addition of DNase I;  , P22tWT control without LPS. The standard deviation of fluorescence signals between repeated experiments was below 0.5%.
, P22tWT control without LPS. The standard deviation of fluorescence signals between repeated experiments was below 0.5%.
Therefore, we doubled the amount of reconstituted phages and LPS in the ejection assay to gain enough signal for our experimental set up. Upon incubation with LPS, phage particles reconstituted in vitro with wild-type TSP showed DNA ejection kinetics similar to those of particles assembled in vivo, and the fluorescence was equally sensitive to DNase digestion. For both P22tD392N and P22tD395N, the initial rate of the fluorescence increase was dramatically reduced (Fig. 3). If the final fluorescence signal observed after 300 min is set to 100% for P22tWT, the signal observed at this time for reconstituted phages P22tD392N and P22tD395N amounts to 44 and 34%, respectively. The smaller amount of DNA released with both endorhamnosidase-defective mutants is in agreement with their low infectivity. DNA ejection of phages carrying the low affinity TSP mutant T307K was somewhat decelerated compared with P22tWT, although their infectivity had been indistinguishable. For P22tT307K, ejection was delayed about 5 min, and the ejection half-time shifted to 87 min compared with 60 min for P22tWT. The lag observed with P22tT307K may be due to delayed initial adsorption or to the slightly reduced enzymatic activity of the mutant. From our observations on DNA ejection from phage particles carrying mutant TSP, we conclude that the hydrolytic activity of TSP is required for efficient DNA ejection in the infection cycle of phage P22.
Tailspike Proteins Are Necessary for Attachment and Direction of Phage toward the Membrane
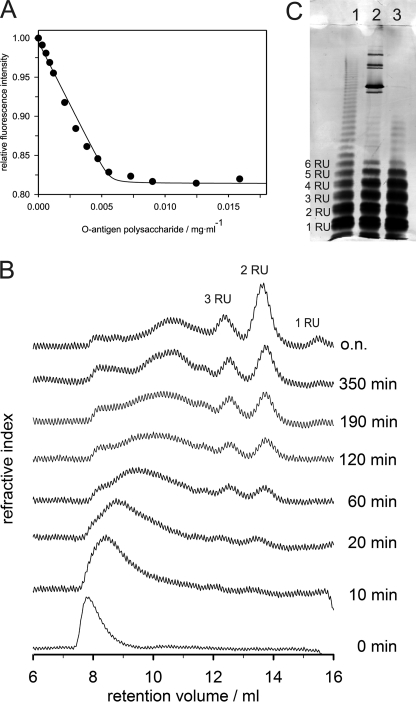
Affinity and activity of TSP had been extensively studied before with oligosaccharides (8) but less extensively with its natural substrate, which is O-antigen polysaccharide. P22TSP has three binding sites with micromolar affinity for O-antigen octasaccharide fragments. The binding affinity increases only slightly with increasing length of oligosaccharides (5). Unfortunately, the polydisperse character of O-antigen polysaccharide hampers the quantification of TSP affinity toward O-antigen. Nevertheless, we titrated a TSP mutant defective in polysaccharide cleavage (TSPD392N) with polysaccharide and measured protein fluorescence quenching. We obtained a kinked binding curve, characteristic for high affinity binding (Fig. 4A). Binding equilibrium was thus largely driven toward the TSP polysaccharide complex due to the multivalent nature of the polysaccharide ligand. For a phage with 6 TSP and 18 binding sites, this means strong fixation on the O-antigen receptor.
FIGURE 4.
Binding and hydrolysis activity of TSP. A, fluorescence binding titration of S. typhimurium O-antigen polysaccharide at 10 °C. Tryptophan fluorescence of 0.11 μm TSPD392N was excited at 295 nm and quenching upon binding followed at 350 nm. B, kinetics TSP O-antigen polysaccharide cleavage. Samples were analyzed after the indicated times on a Superdex Peptide HR 10/30. C, 15% SDS-PAGE of LPS cleavage products. 15% silver-stained SDS-PAGE of purified LPS fraction (lane 1), LPS incubated with phage P22 (lane 2), and LPS digested with TSP and purified (lane 3).
Hydrolysis of Salmonella O-antigen by P22 TSP produces oligosaccharide fragments comprising at least two RU (30). We incubated polysaccharide with TSP and analyzed the kinetics of product formation using gel filtration (Fig. 4B). Elution profiles of hydrolysis products showed large cleavage products after short incubation times and illustrated that TSP is an endoenzyme that is able to bind with high affinity on any site on its substrate. With incubation time, the exclusion peak broadens and is shifted toward smaller sizes before cleavage end products octa- and tetrasaccharides appear. These end products had also been observed earlier in preparations with the phage (30). They are a consequence of the special arrangement of catalytic residues that lie at the end of a high affinity binding pocket (Fig. 5A).
FIGURE 5.
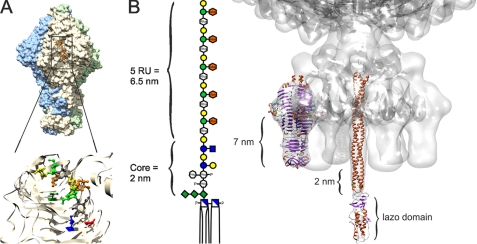
Putative DNA release mechanism of phage P22 triggered by LPS. A, tailspike binding and activity site. Binding cleft in the trimeric protein of P22 TSP (9) is shown. Residues carrying the mutations at Thr-307 (green) and in the active site Asp-392 (red) and Asp-395 (blue) are indicated. B, cell attachment apparatus of phage P22. TSP and gp26 plug crystal structures were modeled into the cryo-EM structure of phage P22 (7, 32, 40, 53). Dimensions of the LPS digestion product containing five O-antigen repeats (38) match the distances in the model and suggest insertion of the gp26 lazo-domain into the hydrophobic part of the outer membrane, presumably resulting in its refolding. Sugar icons are according to Varki et al. (54), an orange symbol was added for the abequose.
O-antigen Hydrolysis and DNA Ejection Are Not Separable Processes
We wanted to analyze whether phage P22 can destroy its LPS receptor completely. Therefore, we incubated it with purified LPS and analyzed chain distributions using SDS-PAGE (Fig. 4C). Undigested LPS showed a typical ladder pattern representing the distribution of different O-antigen chain lengths with different numbers of RUs. Upon incubation of LPS with phage P22 at 37 °C, LPS molecules with long O-antigen chains disappeared and short chains accumulated. Interestingly, only bands up to five RUs enriched upon incubation with phage and molecules carrying six repeat units appeared to be cleaved poorly. Moreover, we obtained the same cleavage pattern when we incubated LPS with purified TSP instead of whole phages. Prolonged incubation times and high TSP concentrations could not shift the observed pattern toward shorter chain lengths. Probably TSP access to the LPS core is sterically hindered. This is in agreement with our gel filtration experiment where we did not observe any short chained core saccharides.
We purified LPS after digesting long chains with TSP and used this short chain LPS in our in vitro ejection assay. Digested LPS is no longer able to trigger phage DNA ejection (Fig. 1). Moreover, we tested whether simultaneous enzymatic cleavage of soluble O-antigen polysaccharide in the presence of lipid A molecules was sufficient to promote DNA expulsion. No signal was obtained, independent of the order of addition of lipid A and polysaccharide (Fig. 1). This clearly showed that ejection of DNA does not occur solely upon hydrolysis of O-antigen polysaccharide. Apparently, O-antigen receptor and lipid A have to be assembled in one molecule and built up one LPS leaflet to promote DNA release. Cleavage of O-polysaccharide by TSP and DNA ejection appear to be intimately linked processes, and ejection was not triggered by a structure on LPS that merely became accessible upon digestion of O-antigen chains.
DISCUSSION
Although it has long been established that the interaction of tailspikes with LPS is responsible for initial adsorption of phage P22 and thus determines its host range (31), a putative secondary receptor that may trigger DNA ejection has remained elusive (32). Prompted by our observation that purified LPS inactivates phage P22 particles in vitro (26), we set up a DNA ejection assay using a fluorescent DNA binding dye. We found that DNA ejection from the phage particles was efficiently triggered when P22 virions were incubated with full-length LPS from S. typhimurium. The interactions resulting in DNA release are highly specific. No ejection was observed when P22 virions were exposed to LPS from a different enterobacterium, to Salmonella phospholipids, or to soluble S. typhimurium O-antigen polysaccharide. Under our in vitro conditions, LPS forms multilamellar and rigid structures the outer surfaces of which resemble the outer membrane of Gram-negative bacteria (10, 11). Neither upon incubation with EDTA-treated LPS that has lost its multilamellar character nor with LPS predigested with TSP or with lipid A aggregates do we observe any DNA release. Our results suggest that LPS is the only receptor for phage P22 at the Salmonella outer membrane.
Six TSP together form the cell attachment apparatus of phage P22. Its interaction with the outer membrane LPS is multivalent and thus essentially irreversible. Although TSP display endorhamnosidase activity and hydrolyze the LPS O-antigen chains, phages will not dissociate from LPS upon a single hydrolysis event. This confers processivity to O-antigen hydrolysis. Processive TSP have also been found in phages infecting encapsulated bacteria (33). Here, processivity appears to result from secondary polysaccharide-binding sites on the same protein, although there is no evidence for secondary LPS-binding sites on P22 TSP. Our in vitro tailing experiments shed light on the role of this hydrolytic activity in early steps of the infection cycle but does not allow us to access functions in later stages. This is because the mutant TSP used to complement the heads in vitro are not encoded in the phage genome, and hence, the phage progeny does not carry mutant TSP. We found that reconstituted phage particles are unable to initiate plaque formation, when they carry mutant TSP lacking endorhamnosidase activity. This proves that O-antigen hydrolysis function is required prior to phage DNA replication. Moreover, DNA ejection from reconstituted particles in vitro was dramatically slowed down when the particles carried mutant TSP and was correlated to the residual endorhamnosidase activity of the TSP mutants attached to the phage heads. Hence, O-antigen hydrolysis is prerequisite to DNA ejection. In addition to its role in the early phase of the infection cycle, the receptor destroying activity of TSP might also be important to prevent newly assembled P22 particles from sticking to cell wall debris upon lysis. This would be analogous to the role of influenza virus neuraminidase (34). We observed that free TSP can prevent LPS-triggered DNA ejection. Accordingly, the receptor inactivating activity of free TSP released upon cell lysis would avoid the loss of infectious phages due to interaction with membrane fragments. Indeed, TSP is encoded in a late structural gene and abundantly synthesized so that excess TSP not bound to phage particles accumulates in the cell (35) and is released upon cell lysis. Consequently, in our in vitro experiments, we observed that LPS pretreated with soluble TSP endorhamnosidase cannot trigger DNA ejection. Therefore, O-antigen hydrolysis appears important both in early and late phases of the infection cycle. Its function in the early phase by far surpasses the simple clearing of the polysaccharide layer to render the cell surface accessible for P22 particles. Rather, hydrolysis by TSP must be intimately connected to the whole triggering process.
From our experiments, we propose a molecular model of how infection of phage P22 might be started. As a first step, phage binding to LPS O-polysaccharide occurs via six trimeric TSP. Then, concomitantly with O-antigen cleavage by TSP, the phage particle descends toward the outer membrane surface. We observed that O-antigen cleavage reproducibly stops at a level of 5 RU attached to the lipid A portion and the LPS core saccharides, when LPS particles are treated with TSP or P22 phage. A similar number of remaining O-antigen repeats has been observed upon treatment of aldehyde-fixed Shigella cell with phage Sf6 TSP, a close functional and structural relative of P22 TSP (36, 37). To us, this suggests that the length of the remaining O-antigen is specific and functional and may be intimately linked to the signal transmitted to open the portal. According to molecular modeling studies combined with x-ray powder diffraction experiments, five O-antigen repeats on S. typhimurium LPS can be estimated to extend on average 7 nm from the lipid A-core portion (38). More extended conformations may protrude up to 8.5 nm from the outer membrane lipid A layer. The distance between the C-terminal tip of TSP and the O-antigen cleavage site also amounts to 7 nm (Fig. 5). Hence, phage TSP could cleave and slide down the O-antigen until encountering with their tips a barrier made of LPS core and lipid A regions. Indeed, these have been shown to be far more inflexible than the O-antigen portion (38). The C-terminal tips of the tailspikes, however, are not the outermost parts of P22 tail machines. It is plug protein gp26 that extends pronouncedly from the bottom of the tail structure (39, 40). In a position of the phage particle with TSP attached to the last O-antigen repeats susceptible to cleavage, gp26 would certainly penetrate the core structure that has a length of about 2 nm (38). We propose this gp26-membrane interaction to be the crucial event to trigger the ejection. Driven by the TSP interaction with the inner parts of the O-antigen polysaccharide, gp26 would have to insert into the lipid core region, which would then produce a conformational change in the rather flexible lazo-domain at the tip of gp26 (40–42). Together, TSP and gp26 may serve as a surface pressure sensor to initiate DNA release in analogy to the headful sensing during DNA packaging into the capsid. Here, a subtle conformational change in the portal protein induces release of DNA terminase and binding of gp4 that initiate the tail assembly, preventing DNA leakage from the phage head (39, 43, 44). Accordingly, a conformational change in the gp26 plug may lower the binding affinity toward gp10 or the more rigid part of the gp26 plug may be pulled out from the tail hub by the interaction forces exerted on the lazo-domain by the lipid parts of the outer membrane. Our model is compatible with the fact that at least three TSP are needed to form an infectious P22 particle, as the fixation at three polysaccharide chains would be minimally required to position the particle with the gp26 needle puncturing the outer membrane surface (43, 45). The steps following the trigger of ejection of pressurized DNA and ejection proteins remain ill-understood. Here, penetration of all three layers of the cell wall is the prerequisite for successful infection of Salmonella.
Once DNA release is triggered in our in vitro assay, the complete phage DNA is ejected without addition of external energy. Initially driven by the high pressure of DNA inside the phage head, release of linear double-stranded DNA from the particles is expected to slow down, so that the essentially complete release observed here on the time scale of the experiment may be due to solvent drag acting on the highly viscous DNA and Brownian motion of the particles. Pressure-induced ejection was observed for different phages like T5 (18) and λ (14), although complete DNA translocation during infection is likely driven by other mechanisms like diffusion, enzyme activity, or protein binding, and even osmotic gradients might contribute to the intrusion of DNA into the host cell (32, 46, 47). In the case of phage T7, ejection proteins have been proposed to build an extensible tail that supports the enzyme-driven transport of phage DNA across the two membranes into Gram-negative cells (48). DNA in phage P22 is circularly permuted and does not require RNA polymerase-binding sites for uptake, as the particles can carry alien-transducing DNA (49, 50). Together with DNA, P22 ejects four different proteins, the role of which remains largely unclear (51). From our in vitro ejection experiments, a picture emerges where during infection the outer LPS-containing membrane could be overcome initially by DNA ejection forces. Subsequently, ejection proteins help in a still unknown way to transport DNA across the peptidoglycan layer and the cytoplasmic membrane (51, 52).
There remains a discrepancy between the time scale of our in vitro ejection experiments and the duration of a lytic cycle of ∼1 h (1). Likely, it is connected to final conformational changes triggering DNA release. When comparing DNA release kinetics of reconstituted phage particles carrying wild-type or hydrolysis-defective mutants, it becomes obvious that O-antigen hydrolysis alone cannot be rate-limiting. DNA ejection is not blocked completely in particles carrying mutant TSP, although their endorhamnosidase activities are reduced to less than 0.15% of the wild-type activity. The difference between the rate-limiting step triggering DNA release in vitro upon contact with LPS aggregates and the obviously much faster reaction in vivo following adsorption onto bacterial cells might be related to the different geometries of the LPS surfaces. The aggregates formed from purified Salmonella LPS used here are rather small with an average radius of around 90 nm, as determined by dynamic light scattering. They are thus expected to exhibit significant surface curvature, whereas the surface of the much larger Salmonella cells is essentially flat. Although our model described above might predict an effect of the surface geometry on the triggering step, this hypothesis can only be tested in further experiments employing much larger LPS or lipid aggregates or flat bilayers carrying long O-antigen polysaccharides.
Acknowledgments
We thank Wolfgang Rabsch for helpful discussions; Klaus Gast for help with the characterization of LPS aggregates by light scattering; Cameron Haase-Pettingell for initial instructions on phage tailing assays, and Carolin Doering for excellent technical assistance.
This work was supported in part by Grants Se517/16 and Ba4046/1 from the Deutsche Forschungsgemeinschaft.
- TSP
- tailspike protein
- RU
- repeating unit.
REFERENCES
- 1.Prevelige P. E. (2006) in The Bacteriophages (Calendar R. ed) 2nd Ed., pp. 457–468, Oxford University Press, New York [Google Scholar]
- 2.Israel J. V., Anderson T. F., Levine M. (1967) Proc. Natl. Acad. Sci. U.S.A. 57, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teschke C. M., Parent K. N. (2010) Virology 401, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seckler R. (1998) J. Struct. Biol. 122, 216–222 [DOI] [PubMed] [Google Scholar]
- 5.Baxa U., Cooper A., Weintraub A., Pfeil W., Seckler R. (2001) Biochemistry 40, 5144–5150 [DOI] [PubMed] [Google Scholar]
- 6.Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. (1994) Science 265, 383–386 [DOI] [PubMed] [Google Scholar]
- 7.Steinbacher S., Miller S., Baxa U., Budisa N., Weintraub A., Seckler R., Huber R. (1997) J. Mol. Biol. 267, 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxa U., Steinbacher S., Miller S., Weintraub A., Huber R., Seckler R. (1996) Biophys. J. 71, 2040–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinbacher S., Baxa U., Miller S., Weintraub A., Seckler R., Huber R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10584–10588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder S., Kim D., McIntosh T. J. (1999) Biochemistry 38, 10758–10767 [DOI] [PubMed] [Google Scholar]
- 11.Nikaido H. (2003) Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C. Y., Kemp P., Molineux I. J. (2010) Virology 398, 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grayson P., Han L., Winther T., Phillips R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leiman P. G., Chipman P. R., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2004) Cell 118, 419–429 [DOI] [PubMed] [Google Scholar]
- 16.Roucourt B., Lavigne R. (2009) Environ. Microbiol. 11, 2789–2805 [DOI] [PubMed] [Google Scholar]
- 17.Molineux I. J. (2006) in The Bacteriophages (Calendar R., Abedon S. T. eds) 2nd Ed., pp. 277–301, Oxford University Press, Inc., New York [Google Scholar]
- 18.Mangenot S., Hochrein M., Rädler J., Letellier L. (2005) Curr. Biol. 15, 430–435 [DOI] [PubMed] [Google Scholar]
- 19.Winston F., Botstein D., Miller J. H. (1979) J. Bacteriol. 137, 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamei D. T., Liu C. L., Haase-Pettingell C., King J. A., Wang D. I., Blankschtein D. (2002) Biotechnol. Bioeng. 78, 190–202 [DOI] [PubMed] [Google Scholar]
- 21.Miller S., Schuler B., Seckler R. (1998) Protein Sci. 7, 2223–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heukeshoven J., Dernick R. (1988) Electrophoresis 9, 28–32 [DOI] [PubMed] [Google Scholar]
- 23.Darveau R. P., Hancock R. E. (1983) J. Bacteriol. 155, 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westphal O., Jann K. (1965) in Methods in Carbohydrate Chemistry (Whistler R. L. ed) pp. 83–91, Academic Press, New York [Google Scholar]
- 25.Freeman G. G., Philpot J. S. (1942) Biochem. J. 36, 340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andres D., Baxa U., Hanke C., Seckler R., Barbirz S. (2010) Biochem. Soc. Trans. 38, 1386–1389 [DOI] [PubMed] [Google Scholar]
- 27.Seckler R., Fuchs A., King J., Jaenicke R. (1989) J. Biol. Chem. 264, 11750–11753 [PubMed] [Google Scholar]
- 28.Eriksson M., Härdelin M., Larsson A., Bergenholtz J., Akerman B. (2007) J. Phys. Chem. B 111, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 29.Mitraki A., Danner M., King J., Seckler R. (1993) J. Biol. Chem. 268, 20071–20075 [PubMed] [Google Scholar]
- 30.Iwashita S., Kanegasaki S. (1976) Eur. J. Biochem. 65, 87–94 [DOI] [PubMed] [Google Scholar]
- 31.Lindberg A. A. (1977) in Surface Carbohydrates of the Procaryotic Cell (Sutherland I. ed) pp. 289–356, Academic Press, London [Google Scholar]
- 32.Chang J., Weigele P., King J., Chiu W., Jiang W. (2006) Structure 14, 1073–1082 [DOI] [PubMed] [Google Scholar]
- 33.Schwarzer D., Stummeyer K., Haselhorst T., Freiberger F., Rode B., Grove M., Scheper T., von Itzstein M., Mühlenhoff M., Gerardy-Schahn R. (2009) J. Biol. Chem. 284, 9465–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., Eichelberger M. C., Compans R. W., Air G. M. (1995) J. Virol. 69, 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berget P. B., Poteete A. R. (1980) J. Virol. 34, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chua J. E., Manning P. A., Morona R. (1999) Microbiology 145, 1649–1659 [DOI] [PubMed] [Google Scholar]
- 37.Müller J. J., Barbirz S., Heinle K., Freiberg A., Seckler R., Heinemann U. (2008) Structure 16, 766–775 [DOI] [PubMed] [Google Scholar]
- 38.Kastowsky M., Gutberlet T., Bradaczek H. (1992) J. Bacteriol. 174, 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lander G. C., Khayat R., Li R., Prevelige P. E., Potter C. S., Carragher B., Johnson J. E. (2009) Structure 17, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olia A. S., Casjens S., Cingolani G. (2007) Nat. Struct. Mol. Biol. 14, 1221–1226 [DOI] [PubMed] [Google Scholar]
- 41.Bhardwaj A., Olia A. S., Walker-Kopp N., Cingolani G. (2007) J. Mol. Biol. 371, 374–387 [DOI] [PubMed] [Google Scholar]
- 42.Olia A. S., Casjens S., Cingolani G. (2009) Protein Sci. 18, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lander G. C., Tang L., Casjens S. R., Gilcrease E. B., Prevelige P., Poliakov A., Potter C. S., Carragher B., Johnson J. E. (2006) Science 312, 1791–1795 [DOI] [PubMed] [Google Scholar]
- 44.Strauss H., King J. (1984) J. Mol. Biol. 172, 523–543 [DOI] [PubMed] [Google Scholar]
- 45.Israel V. (1978) J. Gen. Virol. 40, 669–673 [DOI] [PubMed] [Google Scholar]
- 46.Grayson P., Molineux I. J. (2007) Curr. Opin. Microbiol. 10, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inamdar M. M., Gelbart W. M., Phillips R. (2006) Biophys. J. 91, 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molineux I. J. (2001) Mol. Microbiol. 40, 1–8 [DOI] [PubMed] [Google Scholar]
- 49.Schmieger H. (1972) Mol. Gen. Genet. 119, 75–88 [DOI] [PubMed] [Google Scholar]
- 50.Casjens S., Hayden M. (1988) J. Mol. Biol. 199, 467–474 [DOI] [PubMed] [Google Scholar]
- 51.Israel V. (1977) J. Virol. 23, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Susskind M. M., Botstein D., Wright A. (1974) Virology 62, 350–366 [DOI] [PubMed] [Google Scholar]
- 53.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 54.Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Marth J. D., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Proteomics 9, 5398–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]