Abstract
Toll-like receptors (TLRs) act as primary sensors of the immune system by recognizing specific microbial motifs and inducing proinflammatory genes that facilitate innate and adaptive immunity. TLRs regulate gene expression by activating transcription factors, such as NF-κB and interferon-regulatory factors. Dysregulation of these pathways can lead to inflammatory diseases, and thus they are subject to stringent control by negative regulators of innate immune signaling. Cactin (Cactus interactor) was initially discovered as a novel interactor of Drosophila Cactus, a regulator of Drosophila Toll signaling. We now describe the first functional characterization of the human ortholog of Cactin (hCactin) and show that it acts as a negative regulator of TLRs. Overexpression of hCactin suppresses TLR-induced activation of NF-κB and interferon-regulatory factor transcription factors and induction of TLR-responsive genes, whereas knockdown of endogenous hCactin augments TLR induction of these responses. hCactin also interacts with IκB-like protein and targets other proteins that are encoded by genes in the MHC Class III region of chromosome 6. We demonstrate that hCactin localizes to the nucleus, and this nuclear localization is critical for manifesting its inhibitory effects on TLR signaling. This study thus defines hCactin as a novel negative regulator of TLR signaling and reveals its capacity to target MHC Class III genes at the molecular and functional level.
Keywords: Innate Immunity, MHC, NF-kappaB, Signal Transduction, Toll-like Receptors (TLR)
Introduction
Toll-like receptors (TLRs)2 are type I transmembrane proteins that possess an N-terminal leucine-rich repeat domain that recognizes pathogen-associated molecular patterns, a single transmembrane domain, and a C-terminal intracellular signaling domain (1–3). The TLR C terminus is homologous to the intracellular domain of the type I IL-1 receptor (IL-1R) and is termed the Toll/IL-1R (TIR) domain (4). To date, 13 murine and 10 human TLRs have been identified (5). The engagement of TLRs by pathogen-associated molecular patterns promotes dimerization of the receptors, followed by recruitment of TIR domain-containing adaptors and the initiation of downstream signal transduction cascades (6–8). MyD88 (myeloid differentiation primary response protein 88) is the universal TIR adapter protein recruited by all TLRs except TLR3, and thus the signaling pathways activated by TLRs are classified into MyD88-dependent and MyD88-independent pathways (9–12). The binding of MyD88 to TLRs recruits members of the IL-1R-associated kinase family (13–15). The IL-1R-associated kinase-MyD88 association triggers hyperphosphorylation of IL-1R-associated kinase, leading to its dissociation from MyD88 and interaction with the downstream adaptor TRAF-6 (tumor necrosis factor receptor-associated factor 6) (16). This results in activation of TAK-1 (transforming growth factor-β-activating kinase 1), leading to stimulation of IκB kinases (IKKs). IKKα and IKKβ phosphorylate members of the IκB family (17), resulting in their polyubiquitination and proteasome-dependent degradation (18). Members of the canonical cytoplasmic IκB family include IκBα, IκBβ, and IκBϵ, and they function to retain Rel subunits of NF-κB in their inactive form in the cytosol. Following degradation of the IκB proteins, NF-κB is released, leading to exposure of a nuclear localization signal and translocation of NF-κB to the nucleus (19). Once in the nucleus, NF-κB binds to a consensus decameric κB-responsive DNA element and induces the up-regulation of a plethora of genes encoding proinflammatory proteins and co-stimulatory molecules (20). In addition to this MyD88-dependent pathway, TLR4 can also employ another TIR adaptor, termed TIR domain-containing adaptor inducing IFN-β (TRIF), to trigger activation of NF-κB in a MyD88-independent pathway (21). TLR3 is unique among members of the TLRs in that it exclusively employs TRIF as its receptor proximal adaptor protein. In addition to activation of NF-κB, TRIF can also form a complex with TBK1 (TRAF family member-associated NF-κB activator (TANK)-binding kinase) and IKKi, which induce specific phosphorylation and activation of the transcription factors IRF3 (interferon-regulatory factor 3) and IRF7 to induce type I interferons (IFNs) (22).
A striking similarity of insect and vertebrate immunity lies in the conserved role of Toll and TLRs in driving activation of the Rel/NF-κB transcription factor family in response to a perceived pathogenic challenge. Thus, activation of NF-κB and the subsequent induction of immune response genes is a core defensive mechanism of both fly and mammalian innate immune responses. Moreover, NF-κB fills dual roles in early development and adult immunity in both insects and mammals. In addition, consideration of the pathways leading to the activation of NF-κB factor reveals homology at the level of the intracellular signaling components utilized. Given such conservation, the identification of novel proteins in the NF-κB pathway in the fly has proved valuable in adding to our understanding of the molecular components of the NF-κB signaling system in mammalian cells. In 2000, Cactus, the Drosophila ortholog of IκBα, was employed as bait in a yeast two-hybrid screen, leading to identification of a novel Cactus-interacting protein termed Cactin (23). Drosophila Cactin was shown to be maternally inherited, and its overexpression promoted a ventralized phenotype in the early Drosophila embryo. Given that the NF-κB family member Dorsal is subject to inhibition by Cactus and that Dorsal is an important trigger of ventralization, these studies suggested that Cactin positively regulates Dorsal function in early Drosophila development. More recent studies have confirmed early developmental roles for Cactin in zebrafish and Caenorhabditis elegans (24, 25). However, there are no existing data on its function in higher organisms. In addition, an immune function for Cactin has not yet been investigated. The parallels between the fly and human TLR pathways at the molecular level prompted us to perform the first functional characterization of the human ortholog of Cactin (hCactin).
Given that IκBα, the mammalian ortholog of Cactus, is central in control of NF-κB activation and regulation of inflammatory signaling, we were especially interested in exploring the potential of human Cactin to act as a novel regulator of innate immune signaling. We thus cloned hCactin and demonstrated it to be a negative regulator of TLR signaling. Intriguingly, hCactin fails to interact with the cytoplasmic IκB proteins but instead localizes to the nucleus of the cell, where it interacts with IκB-like (IκBL) protein and some other proteins that are encoded by genes in the MHC Class III region of chromosome 6. The strong association of the genes in this genomic region with inflammatory diseases coupled to the inhibitory effects of hCactin on innate immune signaling promotes hCactin as a highly novel and crucially important regulator of immunity-mediated disease.
EXPERIMENTAL PROCEDURES
Biological Reagents and Cell Culture
The anti-IRF3 antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal anti-Myc and anti-phospho-IRF3 antibodies were from Cell Signaling Technology Inc. (Danvers, MA). Anti-β-actin antibody was supplied by Sigma-Aldrich. The plasmids pCMV-dR 8.91 and VSV-G and constructs encoding IRF3/7 Gal4 reporter and IFN-β-luciferase reporter were gifts from Dr. Kate Fitzgerald (University of Massachusetts Medical School, Worcester, MA). The pFR-luciferase Gal4 reporter construct was a gift from Prof. Andrew Bowie (Trinity College, Dublin, Ireland). The NF-κB-luciferase reporter construct was a gift from Prof. Luke O'Neill (Trinity College). GFP-tagged IκB-L was a present from Dr. Ross McManus (Trinity College). The human embryonic kidney 293 (HEK293) cells that stably express TLR3 and TLR4 receptors were from InvivoGen (San Diego, CA), and U373 cells were a gift from Dr. Sinead Miggin (National University of Ireland, Maynooth, Ireland). THP-1 suspension cells were a gift from Prof. Catherine Godson (Conway Institute, University College Dublin).
HEK and U373 cells were maintained in Dulbecco's modified Eagle's medium (DMEM), which was supplemented with 10% (v/v) fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. G418 (500 μg/ml) was used to select for the stably transfected TLR cell lines. THP-1 suspension cells were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 2 mm l-glutamine, 10% (v/v) FBS, and penicillin/streptomycin (100 units/ml and 100 μg/ml, respectively). Cells were maintained in a 37 °C humidified atmosphere with 5% CO2.
Cloning and Expression Vectors
hCactin (nucleotides 40–2330 of GenBankTM entry NP_067054) was initially TOPO-cloned (TOPO TA cloning kit, Invitrogen) from 1321N1 astrocytoma, and this was used as a template for PCR amplification, with the PCR product being ligated into pcDNA3.1/Zeo (Invitrogen), pEGFP-N1, and pIRES2-DsRed (BD Biosciences) mammalian expression vectors. The primers 5′-GATCGAATTCCACTGGCCCAGCCGATGGGTC-3′ and 5′-TACTCTTCACTTGGCCTCCTTCTTGGCCAGCC-3′ were used in the PCR for subsequent ligation with pcDNA3.1 cloning (with the c-Myc epitope tag at the C terminus); primers 5′-GATCCTCGAGGCCACCATGGGTCGGGACACACGCTCGC-3′ and 5′-GATCGAATTCCCCGCCGATAGCGGTAGCGCTT-3′ were used for cloning into pEGFP-N1. PCR products and vectors were double-digested with 30 units each of EcoRI and XhoI at 37 °C overnight and then mixed in ligation reactions. The integrity of all generated clones was confirmed by sequencing. The truncation mutant of hCactin was generated by PCR using the GC-RICH PCR system (Roche Applied Science) and TOPO cloned DNA template with the forward primer 5′-ATCCTCGAGGATGAACCAGCTGCAGGTCAT-3′ and reverse primer 5′-ATCGGATCCGCCGCCGATAGCGGTAGCGCTT-3′. These primers contain restriction sites for XhoI and BamHI, respectively. The PCR products were cloned into the pcDNA3.1/Zeo and pEGFP-N1 mammalian expression vectors as described above.
Lentiviral Production and Transduction
HEK293 T cells were seeded (2 × 105 cells/ml; 3 ml) in 6-well plates and grown for 24 h to ∼80% confluence. The cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions with the packaging plasmid pCMV-dR 8.91 (900 ng), envelop plasmid VSV-G (100 ng), sh-pLKO.1 vector (1 μg), and hCactin shRNA or control shRNA (Sigma-Aldrich): hCactin shRNA1, 5′-CAGGGATACAAGTTCAACATC-3′; hCactin shRNA2, 5′-AGGTCAGATTCAGAGGAAGAG-3′. The control shRNA (CT) is a non-targeting shRNA vector that encodes for shRNA that does not match to any known human or mouse gene. The conditioned medium was changed 24 h post-transfection and replaced with fresh growth medium containing 30% (v/v) FBS. The cells were then incubated for a further 24 h. The lentivirus-containing medium was harvested and stored at −20 °C. Fresh growth medium containing 30% (v/v) FBS was again added to cells and incubated for another 24 h. The virus-containing medium was again harvested and stored for use.
THP-1 and U373 cells were seeded (2 × 105 cells/ml; 3 ml) in 6-well plates and grown for 24 h. The growth medium was then removed and replaced with fresh medium containing Polybrene (8 μg/ml) and 600 μl of lentivirus-containing medium from above. The plates were incubated at 37 °C for 24 h, after which the medium was removed and replaced with fresh growth medium containing puromycin (5 μg/ml) to select for cells transduced with shRNA. Prior to experiments, cells were selected for another 2–3 weeks in the presence of puromycin.
Transfection and Luciferase Reporter Systems
HEK293 cells, stably expressing TLR3 or TLR4, were seeded (1.5 × 105 cells/ml; 200 μl) in 96-well plates and grown for 24 h. Cells were then transfected, using Lipofectamine 2000 (Invitrogen), with NF-κB firefly luciferase reporter construct (80 ng) or IFN-β promoter-regulated firefly luciferase (80 ng), constitutively expressed TK Renilla luciferase reporter construct (phRL-TK, 20 ng; Promega), and varying amounts of expression constructs. In some experiments, transfections also included siRNAs (25 nm) (Invitrogen) targeting hCactin (sense sequence, 5′-UCUUGAAGUGCUCUGCCUCCUUCUC-3′) and a corresponding scrambled siRNA (sense sequence, 5′-GAGAAGGAGGCAGAGCACUUCAAGA-3′). In all transfections, total DNA was kept constant (310 ng/well) using the appropriate empty vector. The activation of IRF3, IRF7, and c-Jun was determined by performing similar transfections using Gal4-firefly luciferase reporter plasmid pFR-Luc (60 ng) with the trans-activator plasmids pFA-IRF3, pFA-IRF7, or pFA-Jun (activation domain of IRF3, IRF7, or Jun is fused with the yeast Gal4 DNA binding domain) (30 ng), phRL-TK plasmid (20 ng), and varying amounts of expression constructs. Cell extracts were generated 24 h post-transfection using the reporter lysis buffer (Promega), and extracts were assayed for firefly luciferase and Renilla luciferase activity using the luciferase assay system (Promega) and coelenterazine (0.1 μg/ml Insight Biotechnology Ltd.), respectively. Luminescence was monitored with a Glomax microplate luminometer (Promega).
Cytokine Immunoassays
Conditioned medium from cells was assayed for levels of IL-8, TNF, and RANTES by sandwich ELISA (DuoSet kits (R&D Systems)) and for IFNβ using a Singleplex kit (Meso Scale Discovery). All kits were used according to the manufacturers' instructions.
Co-immunoprecipitation Analysis
Cells were seeded (2 × 105 cells/ml; 3 ml) in 6-well plates and transfected, using Lipofectamine 2000, with defined amounts of various expression constructs and/or siRNAs as described above. Cells were washed with prechilled PBS (1 ml) and then lysed with prechilled lysis buffer (0.5 ml) (50 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, 0.5% (v/v) IGEPAL, 50 mm NaF, 1 mm Na3VO4, 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mixture (leupeptin (25 μg/ml), aprotinin (25 μg/ml), benzamidine (1 mm), trypsin inhibitor (10 μg/ml)) for 30 min on a rocker at 4 °C. Lysates were centrifuged at 12,000 × g for 10 min at 4 °C. Supernatants were removed to new tubes, and protein concentrations were determined by the method of Bradford (26). Aliquots (50 μl) were retained for Western immunoblotting, whereas the remaining supernatant was incubated with mixing for 30 min at 4 °C with rabbit IgG (1 μg) and Protein A/G-agarose beads (10 μl). Samples were centrifuged at 1000 × g for 5 min at 4 °C to pellet non-specifically bound proteins. Supernatants were removed to fresh prechilled tubes. Samples were incubated overnight with anti-Myc antibody (2 μg) at 4 °C with rocking. This was followed by the addition of protein A/G-agarose beads (20 μl/sample). Incubations were placed at 4 °C with rocking for 4 h. Immunoprecipitates were collected by centrifugation at 1000 × g for 5 min at 4 °C, and the beads were then washed four times with lysis buffer (500 μl) lacking the protease inhibitor mixture. The beads were then resuspended in 2× SDS-PAGE sample buffer (40 μl) and incubated for 30 min at room temperature. Samples were centrifuged at 16,000 × g for 2 min, subsequently boiled at 100 °C for 5 min, and subjected to Western immunoblotting as described previously (27). Nitrocellulose blots were blocked for 1 h in 20 mm Tris-HCl, pH 7.5, containing 0.05% (v/v) Tween 20 and 0.5 m NaCl (TBS) and 5% (w/v) skimmed milk powder. Blots were incubated overnight at 4 °C with primary antibodies (at recommended concentrations) in blocking buffer followed by a 1-h incubation with IRDye secondary antibodies (Licor Biosciences) (1:5000 dilution in blocking buffer). The immunoreactive bands were detected, and images were captured using the Odyssey infrared imaging system from Licor Bioscience.
Electrophoretic Mobility Shift Assay (EMSA)
THP-1 and U373 cells, stably transduced with control shRNA or Cactin-specific shRNAs, were grown in 6-well plates for 24 h. Cells were then stimulated with LPS or poly(I-C) for various times. Nuclear extracts were generated as described previously (28). Nuclear protein (10 μg) was incubated with LiCor IRDye 700-labeled oligonucleotide containing the NF-κB binding site 5-AGTTGAGGGGACTTTCCCAGGC-3′ (κB site underlined) according to the manufacturer's instructions. For supershift analysis, polyclonal antibody (1 μg) against the NF-κB subunit p65 or C-Rel or nonimmune IgG was added to the extracts and chilled for 30 min in ice prior to incubation with labeled oligonucleotide. All incubations were subjected to electrophoresis on a 4% native polyacrylamide gel for 2 h at 110 V and subsequently analyzed and images captured using the Odyssey infrared imaging system.
Quantitative Real-time PCR
Cells were seeded (2 × 105 cells/ml; 3 ml) into 6-well plates and grown for 24 h. After transfection, cells were treated with poly(I-C) (25 μg/ml) for various time periods. Cells were washed in PBS, and RNA was extracted using Tri-Reagent (Sigma). After DNase I digestion, cDNA was generated from normalized RNA using avian myeloblastosis virus reverse transcriptase (Promega). Samples were assayed by quantitative real-time PCR for levels of IFN-β or hCactin cDNA using Brilliant SYBR® Green QPCR Master Mix (Stratagene). PCR was conducted with the CFB-322001G Opticon thermal cycler (Bio-Rad). Reactions were performed using prevalidated primers (Eurofins MWG Operon): forward hCactin, 5′-TACACCAACACCGACAACCC-3′; reverse hCactin, 5′-TTCAGCTCCTTCTCCTCCAG-3′; forward IFN-β, 5′-GACCAACAAGTGTCTCCTCCAAA-3′; reverse IFN-β, 5′-CTCCTCAGGGATGTCAAAGTTCA-3′; forward HRPT, 5′-GGTGAAAAGGACCCCACGAA-3′; reverse HPRT, 5′-GGCGATGTCAATAGGACTCCAGAT-3′.
Screening of Tissue Distribution of hCactin
RNA was extracted from various murine tissues using TRIzol according to the manufacturer's instructions (Sigma). cDNA was generated as above and used as template for PCR using primers for hCactin and GAPDH: hCactin primers as above; forward GAPDH, 5′-CCATGCCATCACTGCCACCCAGAA-3′; reverse GAPDH, 5′-GTCCACACCCTGTTGCTGTAGCCG-3′. PCR fragments were analyzed by agarose gel electrophoresis.
Confocal Microscopy
HEK293 TLR4 cells were seeded (2 × 105 cells/ml) in 4-well chamber slides (Lab-Tek, Nunc A/S, Roskilde, Denmark) and grown for 24 h to ∼80% confluence. Cells were transfected using Lipofectamine 2000 with plasmids encoding truncated hCactin-pEGFP or hCactin-pRFP and IκBL-pEGFP (400 ng). Processing of samples was performed on ice. Medium was removed, and the cells were gently washed three times in chilled 1× PBS (500 μl). Cells were then fixed by the addition of 2% (v/v) paraformaldehyde (500 μl) for 20 min. Cells were washed three times with PBS. An aliquot (500 μl) of 4′,6-diamidino-2-phenylindole (DAPI) (1.5 μg/ml) in water was added to each well for 1 min. Cells were mounted with Vectashield hard set mounting medium (Molecular Probes, Inc.). Confocal images were captured using the ×40 or ×63 objective lens on the Olympus FluoView FV1000 confocal laser-scanning microscope or UV Zeiss 510 Meta System laser-scanning microscope equipped with the appropriate filter sets. Acquired images were analyzed using the Olympus FV-10 ASW imaging software or LSM 5 browser imaging software, respectively.
Statistical Analysis
Data are expressed relative to untreated cells transfected with empty vector and are the means ± S.E. of triplicate determinations from three independent experiments. For comparison between two groups, paired Student's t test (one tail) was used. A p value of <0.05 was considered significant.
RESULTS
hCactin Is Widely Expressed and Acts as a Negative Regulator of the NF-κB Pathway
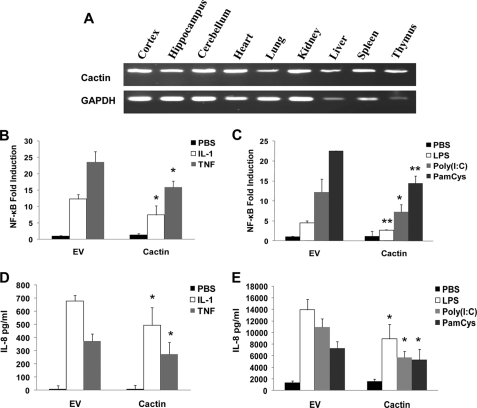
hCactin was initially assessed for its tissue distribution using RT-PCR to measure mRNA levels in various murine tissues. hCactin is ubiquitously expressed with detectable expression in samples from various parts of the brain and a wide variety of peripheral organs (Fig. 1A). Given that Cactin was discovered in Drosophila as a Cactus-interacting protein and Cactus is a homologue of the IκB family that regulates NF-κB (23), hCactin was initially probed for regulatory effects on NF-κB signaling. In order to perform this analysis, HEK293 cells were transfected with an expression plasmid encoding Myc-tagged hCactin and assessed for its ability to induce the expression of a cotransfected NF-κB-regulated luciferase gene. Overexpression of hCactin failed to activate NF-κB (Fig. 1B). hCactin was next examined for regulatory effects on inflammatory signaling pathways known to activate NF-κB. These studies demonstrated that overexpression of hCactin inhibits the activation of NF-κB in response to endogenous proinflammatory stimuli, such as IL-1β and TNF-α (Fig. 1B), and exogenous ligands for TLR2, TLR3, and TLR4, namely PamCys, poly(I-C), and LPS, respectively (Fig. 1C). The functional consequences of such hCactin-mediated down-regulation of NF-κB was confirmed by showing that overexpression of hCactin reduced the ability of these proinflammatory stimuli to induce expression of the NF-κB-responsive gene IL-8 (Fig. 1, D and E). The inhibitory effects of hCactin were not due to nonspecific cytotoxicity because the overexpression of hCactin had no effect on cell viability (data not shown).
FIGURE 1.
hCactin is ubiquitously expressed and inhibits IL-1β, TNF-α, PamCys, poly(I-C), and LPS-induced activation of NF-κB. A, tissue distribution of Cactin. Total RNA was extracted from various tissues and subjected to RT-PCR using Cactin- and GAPDH-specific primers. B–E, HEK293 cells stably transfected with TLR2, TLR3, and TLR4 were cotransfected with plasmids encoding NF-κB-regulated firefly luciferase (80 ng) and constitutively expressed TK Renilla luciferase (40 ng) in the absence or presence of Cactin (90 ng). Empty vector (EV) pcDNA3.1 was used to normalize the amount of total DNA transfected. Transfected cells were left overnight and then treated with or without various ligands: IL-1β (10 ng/ml) and TNF-α (50 ng/ml) (B and D) or PamCys (100 ng/ml), Poly(I-C) (25 μg/ml), and LPS (100 ng/ml) (C and E) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity (B and C). Data are represented relative to untreated cells that had been transfected in the absence of hCactin. Conditioned media from cells were also assayed for IL-8 levels by sandwich ELISA with data being expressed in absolute concentrations of IL-8 (D and E). Data are presented as the mean ± S.E. (error bars) of triplicate determinations from three independent experiments and analyzed by paired Student's t test. *, p < 0.05; **, p < 0.01; ligand-stimulated cells transfected with empty vector versus ligand stimulated cells transfected with hCactin.
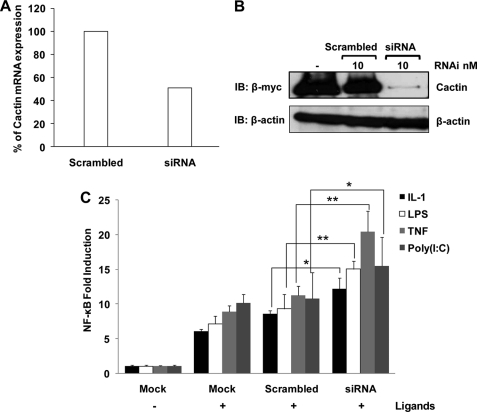
In order to further confirm the inhibitory effects of hCactin on NF-κB, the overexpression studies were complemented by use of hCactin-specific siRNAs to suppress endogenous expression of hCactin. The efficacy and specificity of the hCactin siRNA was initially proved by showing that transfection of HEK293 cells, stably expressing TLR4, with hCactin siRNA suppressed hCactin expression at the mRNA (Fig. 2A) and protein (Fig. 2B) levels, whereas a sequence-scrambled version of this siRNA failed to affect expression. These validated siRNAs were then examined for their regulatory effects on the various proinflammatory stimuli and their capacity to activate NF-κB as assessed by induction of the NF-κB-regulated luciferase gene. HEK293-TLR4 cells (or HEK293-TLR3 cells in the case of poly(I-C)) were cotransfected with the NF-κB luciferase reporter gene with or without hCactin-specific siRNA or scrambled siRNA. Cells were then treated with a variety of stimuli, including IL-1β, TNF-α, LPS, or poly(I-C). The transfection of cells with scrambled siRNA failed to affect the activation of NF-κB by these various ligands. However, hCactin-specific siRNA augmented the activation of NF-κB in response to each of the proinflammatory stimuli (Fig. 2C), thus complementing the overexpression studies and further confirming the inhibitory regulatory effects of hCactin on the NF-κB signaling pathway.
FIGURE 2.
Knockdown of hCactin by siRNA augments IL-1β, TNF-α, LPS, and poly(I-C)-induced activation of NF-κB. A, HEK293 cells, stably expressing TLR4, were transfected with hCactin-specific siRNA or a sequence-scrambled version of this siRNA (25 nm). Total RNA was extracted 48 h post-transfection. The expression level of hCactin was normalized relative to expression of the housekeeping gene HPRT. Data represent the mean from three independent experiments. B, HEK293 TLR4 cells were transfected with Myc-tagged Cactin (2 μg) with or without Cactin siRNA or scrambled siRNA (10 nm). Cell lysates were generated 48 h post-transfection and analyzed by Western immunoblotting (IB) using anti-Myc and anti-β-actin antibodies. Results are representative of two independent experiments. C, HEK293-TLR4 or -TLR3 (in the case of poly(I-C)) cells were cotransfected with plasmids encoding NF-κB-regulated firefly luciferase (80 ng) and constitutively expressed TK Renilla luciferase (40 ng) in the absence (Mock) or presence of Cactin-specific siRNA or scrambled siRNA (25 nm). 48 h post-transfection cells were treated with or without various ligands: IL-1β (10 ng/ml), TNF-α (50 ng/ml), LPS (100 ng/ml), or poly(I-C) (25 μg/ml) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are presented relative to unstimulated mock transfected cells and represent the mean ± S.E. (error bars) of triplicate determinations from three independent experiments and analyzed by paired Student's t test *, p < 0.05; **, p < 0.01; ligand-stimulated cells transfected with scrambled siRNA versus ligand-stimulated cells transfected with hCactin-specific siRNA.
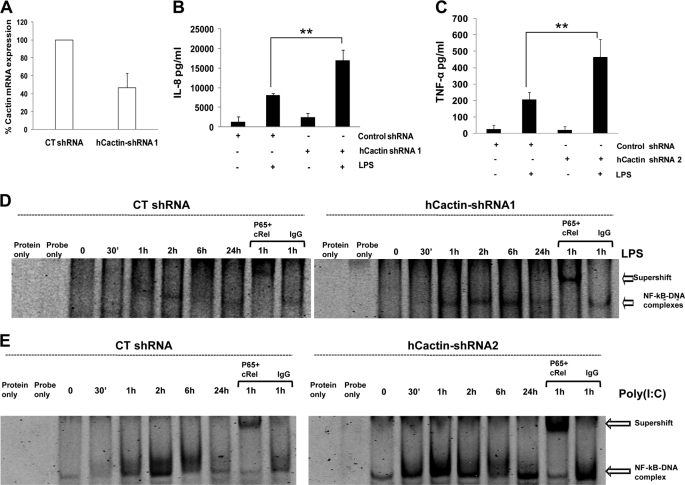
Given that the above studies were performed in HEK293 cells that were engineered to express some of the TLRs, it was next important to demonstrate a functional role for hCactin in a more physiologically relevant system that was naturally responsive to these various proinflammatory stimuli. We were also keen to assess the cell type specificity of hCactin function, and we thus examined its role in two different cell types that naturally express TLRs. THP-1 is a human, acute monocytic, leukemia cell line that naturally expresses TLR4 and responds to LPS (29), whereas the U373 human astrocytoma cell responds to the TLR3 ligand poly(I-C) (30). Because these cells are relatively refractory to transfection, we transduced them with lentiviral particles containing a plasmid encoding hCactin-specific shRNA. Given that this construct also encodes a puromycin resistance gene, the approach also offered the considerable advantage of selecting populations of cells exhibiting stable integration of the construct and thus stable knockdown of hCactin expression. Quantitative RT-PCR was used to confirm hCactin knockdown in cells transduced with hCactin shRNA versus cells that had been transduced with control shRNA that did not target hCactin (Fig. 3A). Both populations of THP-1 cells that had been transduced with control or hCactin shRNA were stimulated with LPS and assessed for induction IL-8. Although hCactin shRNA had no effect on basal expression of IL-8, it strongly augmented the efficacy of LPS in inducing IL-8 (Fig. 3B). The potentiation of LPS signaling by hCactin knockdown is not restricted to monocytic cells and is not specific for expression of IL-8 because U373 cells that were transduced with hCactin shRNA similarly demonstrated enhanced LPS-induced expression of TNF (Fig. 3C). Such augmentation of IL-8 and TNF expression is probably due to potentiation of LPS-induced activation of NF-κB because THP-1 cells transduced with hCactin shRNA displayed earlier, stronger, and more prolonged activation of NF-κB in response to LPS, as judged by an electrophoretic mobility shift assay (Fig. 3D), relative to cells transduced by control shRNA. The NF-κB-DNA complexes contain the strongly transactivating Rel subunits p65 and c-Rel, as evidenced by supershift analysis using antibodies that specifically recognize these subunits. Again the augmentation of NF-κB activation in response to LPS was not limited to THP-1 cells and was not specific for TLR4 because U373 cells that were transduced with hCactin shRNA similarly demonstrated greatly enhanced and more prolonged activation of NF-κB in response to the TLR3 ligand poly(I-C) relative to cells transduced with control shRNA (Fig. 3E).
FIGURE 3.
Cactin-specific shRNA augments TLR activation of NF-κB and induction of IL-8 in THP-1 and U373 cells. A, total RNA was extracted from THP-1 cells, grown under puromycin selection, after transduction with hCactin shRNA1 or control shRNA. Samples were assayed by quantitative real-time PCR for levels of hCactin mRNA and normalized relative to the housekeeping gene HPRT. Data represent the mean from three independent experiments. Puromycin-selected THP-1 (B) and U373 (C) cells, previously transduced with control or hCactin shRNA, were seeded into 96-well plates and stimulated with LPS (100 ng/ml) for 24 h. Supernatants were analyzed for IL-8 (B) and TNF production (C) using sandwich ELISA. Data are presented as the mean ± S.E. (error bars) of triplicate determinations from three independent experiments and analyzed by paired Student's t test. **, p < 0.01; LPS-treated cells transduced with control shRNA versus LPS-treated cells transduced with Cactin-specific shRNA. THP-1 (D) and U373 (E) cells stably transduced with Control or Cactin-specific shRNA were grown in 6-well plates for 24 h. (Quantitative real-time PCR confirmed ∼50% suppression of hCactin expression in U373 cells by Cactin-specific shRNA2.) Cells were then stimulated with LPS (100 ng/ml) (D) or poly(I-C) (25 μg/ml) (E) for the indicated time periods. Nuclear extracts (10 μg of protein) were generated and assayed for binding to an oligonucleotide containing a consensus NF-κB-binding motif by EMSA. Nuclear extracts from cells stimulated for 1 h with LPS or poly(I-C) were preincubated with anti-p65 and c-Rel or nonimmune (IgG) antibody before assaying NF-κB-DNA binding activity. These results are each representative of two independent experiments.
hCactin Inhibits the IRF Pathways and Induction of IRF-responsive Genes
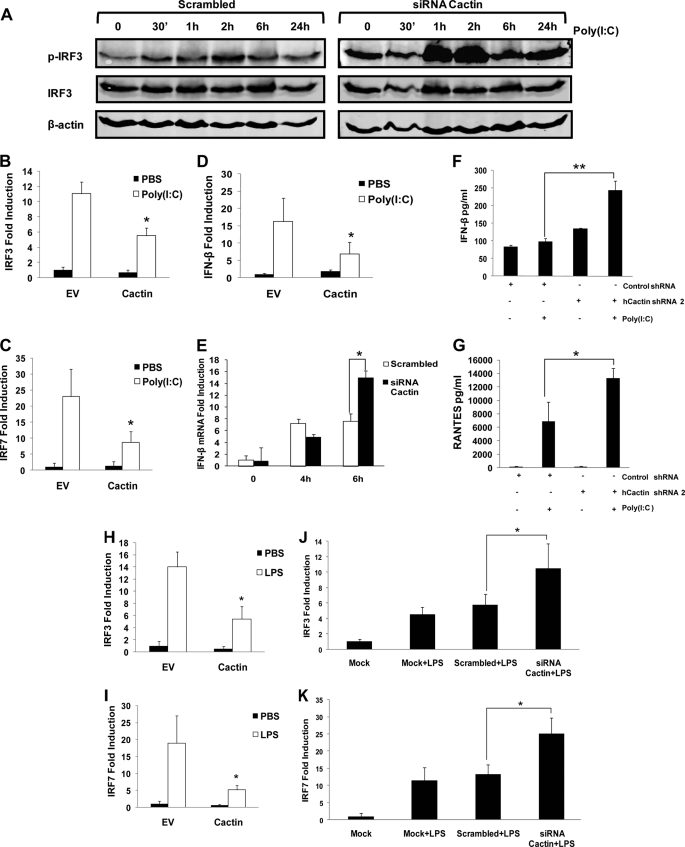
The inhibitory effects of hCactin on IL-1, LPS, and poly(I-C)-induced activation of NF-κB and induction of NF-κB-responsive genes implicate hCactin as a negative regulator of both MyD88-dependent and MyD88-independent activation of NF-κB. However, the MyD88-independent pathway also triggers activation of the IRF transcription factors and induction of type I interferons. It was thus of interest to probe the potential of hCactin to regulate activation of the IRF pathway. Because activation of the IRF pathway initially requires phosphorylation of IRFs, followed by their dimerization and nuclear translocation, hCactin was first assessed for its regulatory effects on phosphorylation of IRFs. In HEK293 cells expressing TLR3, poly(I-C) induced time-dependent phosphorylation of IRF as judged by immunoblotting using phospho-IRF3-specific antibodies (Fig. 4A). The knockdown of hCactin by specific siRNA caused stronger and more prolonged phosphorylation of IRF3 in response to poly(I-C) without affecting the overall levels of IRF3. Interestingly, the levels of basal phosphorylated IRF3 were also slightly higher in the hCactin knockdown cells. These data implicate an important inhibitory role for hCactin in TLR-induced phosphorylation of IRF3. This is further confirmed by studies showing that overexpression of hCactin inhibits Poly(I-C)-induced expression of an IRF3 reporter gene (Fig. 4B). The inhibitory effect of hCactin on TLR3 activation of IRFs also extends to regulation of IRF7 because activation of the latter is strongly inhibited by hCactin (Fig. 4C). The regulatory effects of hCactin on TLR3 activation of IRFs have important functional consequences for downstream gene expression as judged by the inhibitory effects of hCactin on TLR3 activation of the IFNβ promoter (Fig. 4D). Furthermore, knockdown of hCactin by siRNA in HEK-TLR3 cells augmented the induction of IFNβ mRNA in response to poly(I-C) (Fig. 4E). The negative regulatory effects of hCactin on the IRF/IFNβ pathways are also relevant to cells that naturally express TLR3. Thus, U373 cells, transduced with hCactin shRNA, show greatly enhanced induction of IFNβ protein in response to poly(I-C) relative to cells transduced with control shRNA (Fig. 4F). Furthermore, hCactin knockdown cells also demonstrate augmented induction of another IRF-responsive gene, RANTES, in response to poly(I-C) (Fig. 4G), indicating that the negative effects of hCactin on IRFs have functional consequences for downstream expression of multiple IRF-responsive genes. In addition, the regulation of the IRF pathways by hCactin is not restricted to the TLR3 system because overexpression of hCactin strongly inhibits LPS-induced activation of IRF3 (Fig. 4H) and IRF7 (Fig. 4I), and conversely, knockdown of endogenous expression of hCactin potentiates the activation of IRF3 (Fig. 4J) and IRF7 (Fig. 4K) in response to LPS.
FIGURE 4.
hCactin negatively regulates TLR-mediated activation of IRF3 and IRF7 and induction of IRF-responsive genes. A, HEK293-TLR3 cells were cotransfected with Cactin-specific siRNA or a sequence-scrambled siRNA (25 nm) and IRF3 (2 μg) and grown for 48 h. Cells were stimulated with poly(I-C) (25 μg/ml) for the indicated time periods, and cell lysates were generated and subjected to Western blotting using anti-phospho-IRF3, anti-IRF3, and anti-β-actin antibodies. Immunoreactivity was visualized using the Odyssey infrared imaging system. Results are indicative of three independent experiments. B–D, HEK293-TLR3 cells were cotransfected with pFA-IRF3 (30 ng) and pFR-regulated firefly luciferase (60 ng) (B), pFA-IRF7 (25 ng) and pFR-regulated firefly luciferase (60 ng) (C), and IFN-β promoter-regulated firefly luciferase (80 ng) and constitutively expressed TK Renilla luciferase (20 ng) (D) in the absence or presence of Cactin (90 ng). Empty vector pcDNA3.1 (EV) was used to normalize the amount of total DNA transfected. Transfected cells were left overnight and then treated with or without poly(I-C) (25 μg/ml) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are represented relative to untreated cells that had been transfected in the absence of hCactin. E, HEK293 cells, stably expressing TLR3, were transfected with hCactin-specific siRNA or a sequence-scrambled version of this siRNA (25 nm) and grown for 48 h. Cells were stimulated with poly(I-C) (25 μg/ml) for 0–6 h. Total cDNA was generated and assayed by quantitative real-time PCR for levels of IFN-β mRNA. The expression level of hCactin was normalized relative to expression of the housekeeping gene HPRT. Data are presented relative to untreated cells transfected with scrambled siRNA. F and G, U373 cells, stably transduced with control or Cactin-specific shRNA were stimulated with poly(I-C) (25 μg/ml) for 24 h. Supernatants were analyzed for levels of IFN-β (F) and RANTES protein (G) by Mesoscale Singleplex and sandwich ELISA, respectively. H and I, HEK293-TLR3 cells were transfected with IRF3 (H) and IRF7 (I) reporter constructs as described for B and C. Cells were then treated with LPS (100 ng/ml) for 6 h and processed as above. J and K, HEK293-TLR4 cells were cotransfected with pFA-IRF3 (30 ng) (J) or pFA-IRF7 (25 ng) (K), pFR-regulated firefly luciferase (60 ng), and constitutively expressed TK Renilla luciferase (40 ng) in the absence (Mock) or presence of Cactin-specific siRNA or scrambled siRNA (25 nm). 48 h post-transfection, cells were treated with or without LPS (100 ng/ml) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are presented relative to unstimulated mock-transfected cells. All data are presented as the mean ± S.E. (error bars) of triplicate determinations from three independent experiments and analyzed by paired Student's t test. *, p < 0.05; **, p < 0.01. B–D, H, and I, ligand-stimulated cells transfected with empty vector versus ligand-stimulated cells transfected with hCactin.
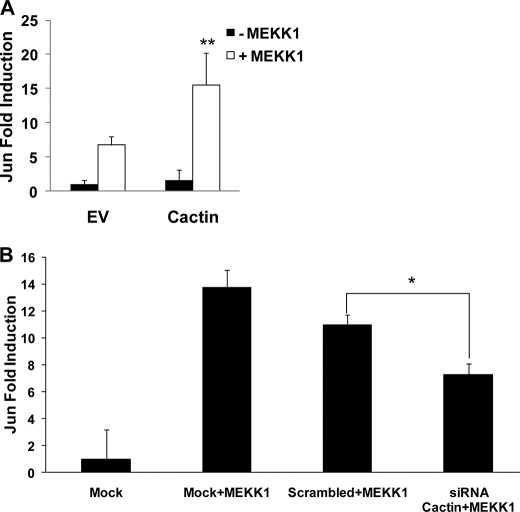
Given the inhibitory effects of hCactin on multiple transcription factors, including NF-κB, IRF3, and IRF7, it was important to exclude the possibility that these effects are nonspecific in nature and represent suppression of both global transcription factor activation and general gene expression. This scenario does not apply because we have also observed that overexpression of hCactin enhances the activation of the transcription factor c-Jun by its upstream regulator MEKK1 (Fig. 5A), whereas siRNA-mediated knockdown of endogenous expression of hCactin inhibits MEKK1-driven activation of c-Jun (Fig. 5B). This indicates specificity of action for hCactin in that it exerts differential effects on different transcription factor pathways.
FIGURE 5.
hCactin positively regulates activation of c-Jun. A, HEK293-TLR4 cells were cotransfected with pFA-Jun (30 ng), pFR-regulated firefly luciferase (60 ng), and constitutively expressed TK Renilla luciferase (20 ng) with or without a construct encoding MEKK1 (30 ng) in the absence or presence of hCactin (90 ng). Empty vector pcDNA3.1 (EV) was used to normalize the amount of total DNA transfected. B, cells were similarly transfected with the reporter constructs and MEKK1 in the absence or presence of Cactin-specific siRNA or scrambled siRNA (25 nm). Transfected cells were left for 48 h, and cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are represented relative to cells that had been transfected in the absence of MEKK1 and hCactin. Data are presented as the mean ± S.E. (error bars) of triplicate determinations from three independent experiments and analyzed by paired Student's t test. *, p < 0.05; **, p < 0.01. A, cells transfected with MEKK1 and EV versus cells transfected with MEKK1 and hCactin.
hCactin Fails to Interact with Canonical Cytoplasmic IκB Proteins but Interacts with Nuclear IκBL
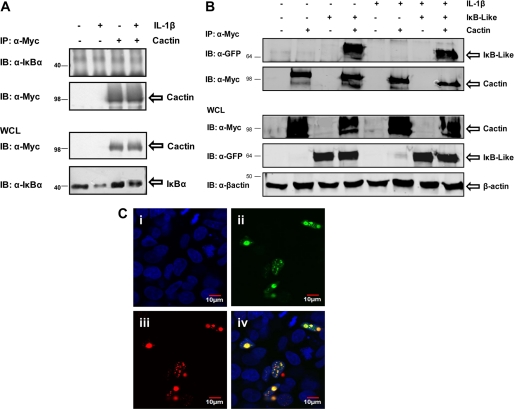
We next attempted to address the mechanistic basis to the above described inhibitory effects of hCactin on TLR signaling. Given that Cactin was initially described in Drosophila as a protein-binding partner for the IκBα ortholog Cactus, we investigated the capacity of hCactin to interact with IκBα. Myc-tagged hCactin was thus overexpressed in HEK293 cells and subsequently immunoprecipitated, and immunoprecipitates were probed for the presence of IκBα by Western immunoblotting (Fig. 6A). This approach failed to show association of hCactin with IκBα. Cells were also stimulated with the potent inflammatory stimulus IL-1 to examine if hCactin could interact with IκBα under proinflammatory conditions. Again there was no detectable association of hCactin with IκBα (Fig. 6A). Using the same approach, we also explored the potential of hCactin to associate with the other cytoplasmic IκB proteins, namely IκBβ and IκBϵ. However, there was no detectable interaction with either of these proteins (data not shown).
FIGURE 6.
hCactin fails to interact with IκBα but interacts with nuclear IκBL. A, HEK293 cells were transfected with Myc-tagged hCactin (1 μg) or pcDNA3.1 (1 μg) (−) and grown for 24 h. Cells were treated with or without IL-1 (10 ng/ml) for 15 min. Cell lysates (WCL) were immunoprecipitated (IP) using anti-Myc antibodies. Immunoprecipitates and cell lysates were subsequently subjected to Western immunoblotting (IB) using anti-Myc and anti-IκBα antibodies. B, HEK293 cells were cotransfected in the absence and presence of Myc-tagged hCactin (1 μg) and GFP-tagged IκBL (1 μg) and grown for 24 h. Cells were treated with or without IL-1 (10 ng/ml) for 15 min. Cell lysates were immunoprecipitated using anti-Myc antibodies. Immunoprecipitates and cell lysates were subsequently subjected to Western immunoblotting using anti-Myc and anti-GFP and anti-β-actin antibodies. C, HEK293 cells were grown in chamber slides and transfected with and without hCactin-RFP (400 ng) (ii and iv) and GFP-tagged IκBL (400 ng) (iii and iv) and grown for 24 h. Cells were harvested 24 h post-transfection and mounted in anti-fade medium with DAPI. Slides were visualized using confocal microscopy. Confocal images were captured using a ×63 objective lens (oil immersion) on the UV Zeiss 510 Meta System laser-scanning microscope equipped with the appropriate filter sets. Data analysis was performed using the LSM 5 browser imaging software. All data are representative of three independent experiments.
Bioinformatic analysis of hCactin using PROSITE, a protein family and domain data base, predicts two bipartite nuclear localization sequences in the N-terminal region of hCactin. We thus next investigated the ability of hCactin to interact with IκBL, a nuclear member of the IκB family (31). Myc-tagged hCactin and GFP-tagged IκBL were co-expressed in HEK293 cells, and the two proteins were shown to interact by co-immunoprecipitation analysis (Fig. 6B). This interaction was apparent in the absence of any proinflammatory stimulus and was not regulated by treatment of cells with the inflammatory cytokine IL-1. In order to further confirm the association of hCactin and IκBL, hCactin was subcloned into a red fluorescent protein (RFP) plasmid construct, pIRES2-DsRed, and transfected into HEK293 cells with or without IκBL-GFP. DAPI was used in the mounting medium to enable visualization of nuclei. hCactin showed a strong nuclear localization with a speckled expression pattern in some cells (Fig. 6C). Interestingly, IκBL showed a very similar nuclear expression pattern. This strongly suggests nuclear co-localization of hCactin-RFP and IκBL-GFP as evidenced by the overlapping red and green formation of yellow/orange immunofluorescence (Fig. 6C).
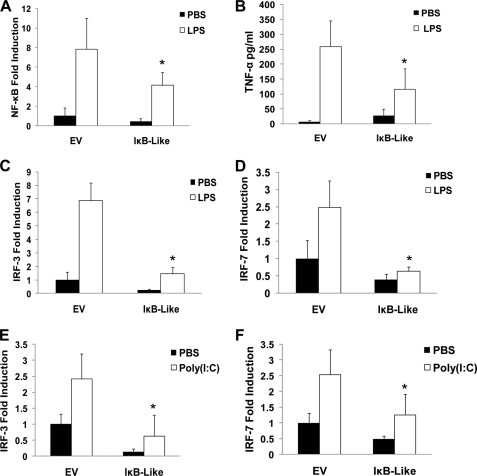
IκBL Inhibits TLR-induced Activation of NF-κB and IRFs
Given the co-localization and interaction of hCactin and IκBL, the latter was next probed for its ability to regulate the TLR signaling pathways that are sensitive to inhibition by hCactin. Like hCactin, IκBL was shown to negatively regulate LPS-induced activation of NF-κB (Fig. 7A) and expression of TNF (Fig. 7B). IκBL also negatively affected the IRF pathways as evidenced by its inhibitory effects on LPS-induced activation of IRF3 (Fig. 7C) and IRF7 (Fig. 7D) and poly(I-C)-induced activation of IRF3 (Fig. 7E) and IRF7 (Fig. 7F). The magnitudes of the inhibitory effects of IκBL were comparable with those shown by hCactin, and co-expression of both IκBL and hCactin failed to effect any additional inhibition (data not shown), suggesting that the two proteins may manifest their regulatory effects by the same mechanism.
FIGURE 7.
IκBL inhibits TLR-induced activation of NF-κB and IRFs. A and B, HEK293 cells stably transfected with TLR4 were cotransfected with plasmids encoding NF-κB-regulated firefly luciferase (80 ng) and constitutively expressed TK Renilla luciferase (40 ng) in the absence or presence of an expression construct encoding IκBL (90 ng). B, supernatants were analyzed for TNF production using sandwich ELISA. HEK293-TLR4 (C and D) or HEK293-TLR3 cells (E and F) were cotransfected with pFA-IRF3 (30 ng) (C and E), pFA-IRF7 (25 ng) (D and F), and pFR-regulated firefly luciferase (60 ng) and TK Renilla luciferase (20 ng) in the absence or presence of IκBL (90 ng). Empty vector pcDNA3.1 (EV) was used to normalize the amount of total DNA transfected. Transfected cells were left overnight and then treated with or without LPS (100 ng/ml) (A–D) or poly(I-C) (25 μg/ml) (E and F) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are presented relative to untreated cells that had been transfected in the absence of IκBL and represent the mean ± S.E. (error bars) of triplicate determinations from three independent experiments analyzed by paired Student's t test. *, p < 0.05; **, p < 0.01; ligand-stimulated cells transfected with empty vector versus ligand stimulated cells transfected with IκBL.
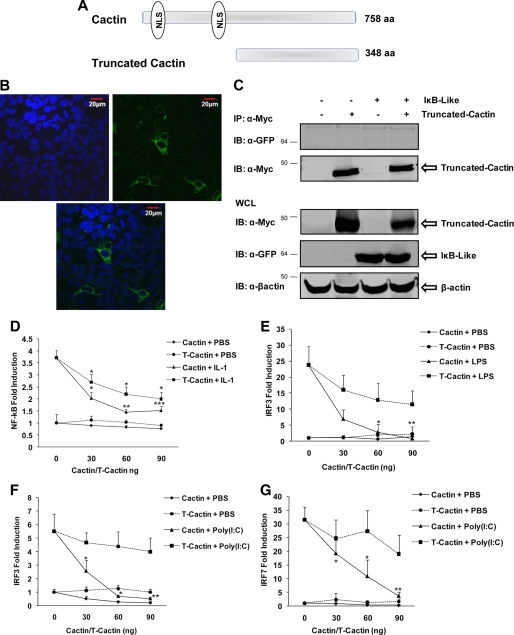
The Nuclear Localization of hCactin Is Critical for Manifesting Its Inhibitory Effects on TLR Signaling
The microscopy studies described above clearly demonstrate a nuclear subcellular localization for hCactin. In order to assess the importance of nuclear localization for the inhibitory effects of hCactin, a truncated form of hCactin was cloned that lacks both nuclear localization sequences (depicted in Fig. 8A), and its regulatory effects on TLR signaling were compared with those of full-length hCactin. It was first necessary to confirm that the truncated form of hCactin was excluded from the nucleus. Confocal microscopic analysis of HEK293 cells transfected with a GFP-tagged form of truncated hCactin revealed an exclusive cytoplasmic localization for this mutant (Fig. 8B). This mutant also offered an opportunity to evaluate the importance of IκBL binding for manifesting the inhibitory effects of hCactin because co-immunoprecipitation analysis demonstrates that truncated hCactin fails to interact with IκBL (Fig. 8C). Although the truncated form of hCactin showed some modest inhibitory effects on activation of NF-κB (Fig. 8D) and on LPS-induced (Fig. 8E) and poly(I-C)-induced (Fig. 8, F and G) activation of IRFs, the magnitudes of these effects are greatly reduced relative to the inhibitory effects shown by full-length hCactin. These findings provide strong evidence that the regulatory effects of hCactin in innate immune signaling are heavily dependent on its nuclear localization where it associates with a number of proteins, including IκBL. This study thus provides the first functional characterization of hCactin and defines it as a novel negative regulator of innate immune signaling.
FIGURE 8.
The nuclear localization of hCactin is important for manifesting its inhibitory effects on TLR signaling. A, schematic representation of full-length and truncated hCactin with location of nuclear localization sequences (NLS) depicted. B, HEK293 cells were grown in chamber slides, transfected with GFP-truncated hCactin (800 ng), and grown for 24 h. Cells were harvested 24 h post-transfection and mounted in anti-fade medium with DAPI. Slides were visualized using confocal microscopy. Confocal images were captured using a ×40 objective lens (oil immersion) on the UV Zeiss 510 Meta System laser-scanning microscope equipped with the appropriate filter sets. Data analysis was performed using the LSM 5 browser imaging software. i, DAPI staining channel; ii, GFP channel; iii, overlay. C, HEK293-TLR4 cells were cotransfected with or without expression constructs encoding Myc-tagged truncated hCactin (1 μg) in the presence or absence of GFP-tagged IκBL (1 μg) and grown for 24 h. Cell lysates were immunoprecipitated (IP) with anti-Myc antibodies. Immunoprecipitates and cell lysates were subsequently subjected to Western immunoblotting (IB) using anti-Myc, anti-GFP, and anti-β-actin antibodies. Immunoreactivity was visualized using the Odyssey infrared imaging system. D, HEK293 cells stably transfected with TLR4 were cotransfected with plasmids encoding NF-κB-regulated firefly luciferase (80 ng) and constitutively expressed TK Renilla luciferase (40 ng) in the absence or presence of an expression construct encoding full-length Cactin or truncated Cactin (T-Cactin) (0–90 ng). HEK293-TLR4 (E) or HEK293-TLR3 cells (F and G) were cotransfected with pFA-IRF3 (30 ng) (E and F), pFA-IRF7 (25 ng) (G), and pFR-regulated firefly luciferase (60 ng) and TK Renilla luciferase (20 ng) in the absence or presence of full-length Cactin or truncated Cactin (0–90 ng). Empty vector pcDNA3.1 was used to normalize the amount of total DNA transfected. Transfected cells were left overnight and then treated with or without IL-1 (10 ng/ml) (D), LPS (100 ng/ml) (E), or poly(I-C) (25 μg/ml) (F and G) for 6 h. Cell lysates were assayed for firefly luciferase activity and normalized for transfection efficiency using Renilla luciferase activity. Data are presented relative to untreated cells that had been transfected in the absence of either Cactin construct and represent the mean ± S.E. (error bars) of triplicate determinations from three independent experiments analyzed by paired Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ligand-stimulated cells transfected with empty vector versus ligand stimulated cells transfected with hCactin/truncated Cactin.
DISCUSSION
The activation of NF-κB and the subsequent induction of immune response genes is a core defense mechanism of both fly and mammalian innate immune responses. In addition, consideration of the pathways leading to the activation of this ubiquitous transcription factor reveals homology at the level of the intracellular signaling components utilized, albeit not always of precisely interchangeable function. Such a consideration of the similarities or parallels between insect and mammalian innate immunity highlights the significance of novel protein identification in model organisms. In 2000, Lin et al. (23) identified Cactin, a conserved protein that interacts with Drosophila Cactus. Because the mammalian homolog of cactus, IκBα, is central in control of NF-κB activation, the identification of a highly conserved component with the potential to interact with and possibly modify the function of this inhibitory protein was exciting and inspired the presently described studies.
The Rel/NF-κB pathway is essential in the early determination of Drosophila embryonic dorsoventral polarity, and Cactus acts as a negative regulator of this pathway. Overexpression of Cactin in a Cactus heterozygous background results in the enhancement of the Cactus haploinsufficient phenotype, strongly increasing both the embryonic lethality and the ventralized phenotype observed. This suggests that Cactin in Drosophila may function in the dorsal-ventral pathway to positively regulate the nuclear targeting of Dorsal. Given this regulation of the NF-κB in Drosophila, it seemed pertinent to initiate functional characterization of hCactin with an analysis of its effect on the NF-κB pathway. Intriguingly, the present studies demonstrate that hCactin acts as a negative regulator of NF-κB. The evidence for this inhibitory role is especially compelling based on the findings from a number of complementary approaches, including overexpression studies and the suppression of endogenous expression of hCactin by two independent modalities, namely siRNA- and shRNA-mediated suppression. Indeed, the prolonged nature of NF-κB in response to LPS and poly(I-C) in cells where hCactin expression is suppressed suggests that hCactin may be an endogenous braking system for NF-κB activation by innate immune stimuli. hCactin appears to exert its inhibitory influence on a wide array of inflammatory stimuli that employ the NF-κB pathway and in a cell type-independent manner. Such inhibitory effects of hCactin contrast with the proposed stimulatory effects of Cactin on Dorsal in Drosophila. Although homology relationships are frequently exploited as an aid in assignation of novel gene function, it is clear that homology need not always correspond to conserved function (32). Thus, Drosophila expresses genes encoding members of the TRAF (TNF receptor-associated factor) family and homologues of IKKβ and IKKγ/NEMO, and yet genetic studies have failed so far to demonstrate involvement of any of these genes downstream of Toll, unlike in mammalian Toll signaling. Instead, genetic data point to the involvement of the Drosophila equivalent of the mammalian signalosome comprising IKK-β and IKK-γ/NEMO homologues (33–35) in the Imd pathway. It has therefore been observed that although homologues of the human Toll signaling pathway exist in Drosophila, they need not necessarily occupy an identical functional niche. Indeed, in the present study, we show that the role of hCactin extends beyond regulation of the NF-κB pathway in that it also acts as a strong inhibitor of IRF signaling and the induction of IRF-responsive genes, such as IFNβ.
In an effort to gain insight into the mechanistic basis of the inhibitory effects of hCactin on NF-κB, we initially assessed its ability to interact with the cytoplasmic IκB proteins based on their homology to the Cactin-interacting protein Cactus. There was no detectable interaction of hCactin with any of the cytoplasmic IκBs. However, hCactin associates with IκBL, a nucleus-localized member of the IκB family. This interaction with IκBL is likely to be of functional relevance because our findings to date suggest that IκBL mimics the regulatory effects of hCactin on the NF-κB and IRF pathways. The interaction of hCactin with IκBL is especially interesting given that the IκBL gene is encoded by the MHC Class III region of the human genome (31), and an earlier study had reported hCactin as a binding partner for other proteins that are encoded by this region of the genome (36). The latter study had used a high throughput yeast two-hybrid screen to document interaction partners for intracellular proteins encoded by genes in the MHC Class III region of the human genome, and hCactin was identified as an interaction partner for two of these proteins, namely RD RNA-binding protein (RDBP) (also called negative elongation factor subunit E (NELF_E)) and leukocyte-specific transcript 1 protein (LST1) (36). Intriguingly, the LST1 gene is located at a locus that is only one gene removed from the TNF that in turn is only one gene removed from the IKBL gene (31). The findings thus propose a model where hCactin is capable of directly interacting with three proteins (LST1, RDBP, and IκBL) that are encoded by genes clustered in the MHC Class III region, whereas it is also capable of functionally targeting TNF that is also encoded by a gene in this cluster region. Thus, hCactin may emerge to be a key regulator of this genomic region and an important modulator of disease states given that the MHC Class III region contains immunity-related proteins that affect susceptibility to numerous immunopathological disorders, including rheumatoid arthritis. More specifically and relevant to the present study, an earlier study aimed to identify non-HLA genes in the chromosome 6p21.3 region that influence the development of rheumatoid arthritis (37). This effort revealed an HLA-independent susceptibility locus at the telomeric end of the Class III region, which was eventually identified as a single nucleotide polymorphism in the promoter/enhancer region of the IκBL gene that conferred transcriptional repression on the promoter. Furthermore a single nucleotide polymorphism giving rise to structural alteration at amino acid 224 in a predicted protein kinase C phosphorylation site of IκBL defines a propensity for more extensive and severe disease states in ulcerative colitis (38). The interaction of hCactin with IκBL in conjunction with their similar functional properties in the present study clearly implicates hCactin as a potential regulator of immunity-mediated diseases previously associated with genes that are encoded by the MHC Class III region of chromosome 6.
In addition to acting as regulators of the NF-κB pathway, it is also interesting to note that hCactin and IκBL demonstrate strong inhibitory effects on IRFs. Although the mechanism underlying their inhibitory effects on NF-κB is somewhat intuitive, given that IκBL belongs to a family of proteins that interact with and inhibit NF-κB, the molecular basis of their modulation of IRFs is less clear. The nuclear localization of hCactin appears to be very important for its function, at least with respect to regulation of NF-κB and IRFs, because a truncation mutant of hCactin that localizes to the cytoplasm fails to replicate the inhibitory effects of nucleus-resident hCactin. The design of this truncation mutant was dictated by the need to remove the two nuclear localization motifs in the N-terminal region but also by the mutant corresponding to that region of Drosophila Cactin that strongly interacts with Cactus. Interestingly, the truncation mutant fails to interact with IκBα, demonstrating that the interaction of Drosophila Cactin with cytoplasmic Cactus is not conserved in humans.
It would appear that hCactin exemplifies the capacity of evolution to diversify and functionally expand the core biological processes embodied in a highly conserved protein. By extension, this may translate into multiple roles for hCactin in a number of diseases. Indeed, although the potential role of hCactin in inflammatory-based disorders has been discussed above, it is worth emphasizing that hCactin was first observed as a renal tumor antigen, NY-REN-24 (39), and is expressed in multiple myeloma (40). This may implicate a role for hCactin in regulating cell transformation, and although such a function remains speculative, it is interesting to note that hCactin negatively regulates the prosurvival transcription factor NF-κB. However, the regulatory effects of hCactin clearly extend beyond the NF-κB pathway. More specifically, hCactin is conserved across a broad phylogenetic range and is also found in primitive organisms, such as C. elegans, in which there is no NF-κB/Rel homologue (41), and its TLR gene, TOL-1, fails to play a direct role in defense against fungal or bacterial pathogens (41, 42). Although C. elegans has apparently lost Rel-based inducible defenses and is not dependent on a Toll pathway for resistance to pathogenic challenge, it is clear that a homolog of Cactin, displaying striking conservation, has been retained, suggesting some additional targets for Cactin and a core biological function for this protein. The process of evolution and speciation may have permitted the protein to adopt new functions when encountering the increasingly complex biological systems and pathways of higher organisms. Although Cactin was originally identified in the Toll dorsoventral regulatory pathway in Drosophila, as a protein that regulates Dorsal translocation, it is now apparent that this function may have evolved in higher organisms, and the present study strongly promotes hCactin as an important regulator of innate immune signaling. More importantly, it also targets a number of proteins, directly or functionally, that are encoded by the MHC Class III region of the genome and have been strongly associated with an array of diseases. It is thus vitally important to gain an appreciation of the physiological and pathological roles of hCactin. The present study offers the first important step in this process.
Acknowledgments
We thank Drs. Mark Mellett and Eric Downer for technical assistance.
This work was funded by Science Foundation Ireland Grant SFI/RFP/BIC1501, the Health Research Board of Ireland, and the European Union Marie Curie Action (MNIEST).
- TLR
- Toll-Like receptor
- TIR
- Toll/IL-1R homology
- IRF
- interferon-regulatory factor
- IL-1R
- IL-1 type 1 receptor
- RANTES
- regulated upon activation, normal T cell expressed and secreted
- PamCys
- palmitoyl-cysteine
- HPRT
- hypoxanthine phosphoribosyltransferase
- HEK
- human embryonic kidney
- hCactin
- human Cactin
- EGFP
- enhanced GFP
- RFP
- red fluorescent protein
- NF-κB
- nuclear factor κB
- IκB
- inhibitor of κB
- TRIF
- TIR domain-containing adaptor inducing IFN-β
- TK
- thymidine kinase.
REFERENCES
- 1.Medzhitov R., Preston-Hurlburt P., Janeway C. A., Jr. (1997) Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 4.O'Neill L. A., Greene C. (1998) J. Leukoc. Biol. 63, 650–657 [PubMed] [Google Scholar]
- 5.Roach J. C., Glusman G., Rowen L., Kaur A., Purcell M. K., Smith K. D., Hood L. E., Aderem A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 7.Horng T., Barton G. M., Medzhitov R. (2001) Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 9.Kaisho T., Takeuchi O., Kawai T., Hoshino K., Akira S. (2001) J. Immunol. 166, 5688–5694 [DOI] [PubMed] [Google Scholar]
- 10.Kawai T., Takeuchi O., Fujita T., Inoue J., Mühlradt P. F., Sato S., Hoshino K., Akira S. (2001) J. Immunol. 167, 5887–5894 [DOI] [PubMed] [Google Scholar]
- 11.Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 13.Li S., Strelow A., Fontana E. J., Wesche H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D. V. (1996) Nature 383, 443–446 [DOI] [PubMed] [Google Scholar]
- 15.Suzuki N., Suzuki S., Duncan G. S., Millar D. G., Wada T., Mirtsos C., Takada H., Wakeham A., Itie A., Li S., Penninger J. M., Wesche H., Ohashi P. S., Mak T. W., Yeh W. C. (2002) Nature 416, 750–756 [DOI] [PubMed] [Google Scholar]
- 16.Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. (2000) Nat. Cell Biol. 2, 346–351 [DOI] [PubMed] [Google Scholar]
- 17.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 18.Traenckner E. B., Pahl H. L., Henkel T., Schmidt K. N., Wilk S., Baeuerle P. A. (1995) EMBO J. 14, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beg A. A., Ruben S. M., Scheinman R. I., Haskill S., Rosen C. A., Baldwin A. S., Jr. (1992) Genes Dev. 6, 1899–1913 [DOI] [PubMed] [Google Scholar]
- 20.Moynagh P. N. (2005) J. Cell Sci. 118, 4589–4592 [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 22.Moynagh P. N. (2005) Trends Immunol. 26, 469–476 [DOI] [PubMed] [Google Scholar]
- 23.Lin P., Huang L. H., Steward R. (2000) Mech. Dev. 94, 57–65 [DOI] [PubMed] [Google Scholar]
- 24.Atzei P., Yang F., Collery R., Kennedy B. N., Moynagh P. N. (2010) Gene Expr. Patterns 10, 199–206 [DOI] [PubMed] [Google Scholar]
- 25.Tannoury H., Rodriguez V., Kovacevic I., Ibourk M., Lee M., Cram E. J. (2010) Dev. Biol. 341, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Bourke E., Kennedy E. J., Moynagh P. N. (2000) J. Biol. Chem. 275, 39996–40002 [DOI] [PubMed] [Google Scholar]
- 28.Curran N. M., Griffin B. D., O'Toole D., Brady K. J., Fitzgerald S. N., Moynagh P. N. (2005) J. Biol. Chem. 280, 35797–35806 [DOI] [PubMed] [Google Scholar]
- 29.Janeway C. A., Jr., Medzhitov R. (2002) Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 30.Kurt-Jones E. A., Sandor F., Ortiz Y., Bowen G. N., Counter S. L., Wang T. C., Finberg R. W. (2004) J. Endotoxin. Res. 10, 419–424 [DOI] [PubMed] [Google Scholar]
- 31.Xie T., Rowen L., Aguado B., Ahearn M. E., Madan A., Qin S., Campbell R. D., Hood L. (2003) Genome Res. 13, 2621–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen R. A. (2001) Genome Biol. 2, INTERACTIONS1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y., Wu L. P., Anderson K. V. (2001) Genes Dev. 15, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutschmann S., Jung A. C., Zhou R., Silverman N., Hoffmann J. A., Ferrandon D. (2000) Nat. Immunol. 1, 342–347 [DOI] [PubMed] [Google Scholar]
- 35.Silverman N., Zhou R., Stöven S., Pandey N., Hultmark D., Maniatis T. (2000) Genes Dev. 14, 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehner B., Semple J. I., Brown S. E., Counsell D., Campbell R. D., Sanderson C. M. (2004) Genomics 83, 153–167 [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K., Makino S., Yoshikawa Y., Takaki A., Nagatsuka Y., Ota M., Tamiya G., Kimura A., Bahram S., Inoko H. (2003) Am. J. Hum. Genet. 72, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Concha E. G., Fernandez-Arquero M., Lopez-Nava G., Martin E., Allcock R. J., Conejero L., Paredes J. G., Diaz-Rubio M. (2000) Gastroenterology 119, 1491–1495 [DOI] [PubMed] [Google Scholar]
- 39.Scanlan M. J., Gordan J. D., Williamson B., Stockert E., Bander N. H., Jongeneel V., Gure A. O., Jäger D., Jäger E., Knuth A., Chen Y. T., Old L. J. (1999) Int. J. Cancer 83, 456–464 [DOI] [PubMed] [Google Scholar]
- 40.Davies F. E., Dring A. M., Li C., Rawstron A. C., Shammas M. A., O'Connor S. M., Fenton J. A., Hideshima T., Chauhan D., Tai I. T., Robinson E., Auclair D., Rees K., Gonzalez D., Ashcroft A. J., Dasgupta R., Mitsiades C., Mitsiades N., Chen L. B., Wong W. H., Munshi N. C., Morgan G. J., Anderson K. C. (2003) Blood 102, 4504–4511 [DOI] [PubMed] [Google Scholar]
- 41.Pujol N., Link E. M., Liu L. X., Kurz C. L., Alloing G., Tan M. W., Ray K. P., Solari R., Johnson C. D., Ewbank J. J. (2001) Curr. Biol. 11, 809–821 [DOI] [PubMed] [Google Scholar]
- 42.Aballay A., Drenkard E., Hilbun L. R., Ausubel F. M. (2003) Curr. Biol. 13, 47–52 [DOI] [PubMed] [Google Scholar]