Abstract
Msc1, a member of the Jarid1 family of putative histone demethylases, is required for chromosome stability in fission yeast. Msc1 associates with the Swr1 complex that facilitates deposition of histone H2A.Z into chromatin. To assess the function of Msc1 in the Swr1 complex, domains of Msc1 necessary for interaction with Swr1 were identified. The C-terminal plant homeodomain (PHD) 2 and PHD3 of Msc1 are sufficient to confer association with Swr1 and allow Msc1 to function in the context of kinetochore mutants. On the other hand, a mutant with a single amino acid substitution in PHD2 within the full-length Msc1 protein retains the ability to bind to Swr1 but eliminates the function of Msc1 in combination with kinetochore mutants. Thus, Swr1 association is critical but not sufficient for Msc1 function. An activity of Msc1 that depends on the cysteine residue within PHD2 of Msc1 is likewise critical for function. On the basis of our observation that the PHDs of Msc1 act as E3 ubiquitin ligases and that mutations of cysteine residues within those domains abolish ligase activity, we speculate that the ability of Msc1 to facilitate ubiquitin transfer is critical for the function it mediates through its association with Swr1.
Keywords: Centromeres, Chromatin, Histone Modification, Ubiquitin Ligase, Yeast, PHD Finger, S. pombe, Jarid Protein, JmjC, Msc1
Introduction
The fission yeast msc1 gene was first isolated as a multicopy suppressor of loss of Chk1 function (1), a phenotype that requires the histone H2A variant H2A.Z to be intact (2). The absence of Msc1 function results in chromosome loss, suggesting that Msc1 plays a critical role in maintaining genomic stability (2). Furthermore, the viability of conditional mutants with defective components of the kinetochore is compromised in the absence of Msc1 function. In addition, phenotypes of cells deleted for msc1 are similar to those of cells deleted for pht1, the gene encoding histone H2A.Z in Schizosaccharomyces pombe (2). Studies from multiple groups demonstrate that the Msc1 protein is a component of the Swr1 chromatin-remodeling complex (3–6), confirming genetic and proteomic predictions (7, 8). Swr1 complexes from a variety of organisms facilitate the incorporation of the histone variant H2A.Z into chromatin in exchange for histone H2A (9, 10).
Msc1 has multiple conserved domains that suggest it plays a role in chromatin structure and/or function, including three plant homeodomains (PHDs)3 and a Jumonji C (JmjC) domain. Structural studies of isolated PHDs suggest that the two Zn2+-binding domains could fold in a manner similar to that of RING domain-containing E3 ubiquitin ligases (12). Indeed, PHDs, including those of Msc1 (13), have been demonstrated to possess E3 ubiquitin protein ligase activity (14–18). Additionally, PHDs of several proteins demonstrate the ability to bind in vitro to methylated residues in histone tails (19–22) and to bind phosphoinositide (23).
Msc1 also possesses a JmjC domain, a functional unit that can catalyze the demethylation of histone residues (24). Indeed, when overexpressed, the RBP2 and PLU-1 proteins bring about the demethylation of di- and trimethylated Lys4 of histone H3 (25–28). Deletion of the msc1 gene can result in elevation of methylated Lys4 of histone H3 (13), an effect most readily observed in cells that are deficient for the E2 ubiquitin ligase Rhp6. Cells lacking rhp6 are extremely slow growing and have essentially no detectable H3 methyl-Lys4 as determined by Western blot analysis. When msc1 is simultaneously deleted in an rhp6 deletion strain, cells are restored to normal growth rates and have readily detectable H3 methyl-Lys4. Thus, the cellular behavior of Msc1 is consistent with a possible role as a histone demethylase.
As a protein characterized by PHDs, a JmjC domain, and association with Swr1, Msc1 may play an important regulatory role in the modification of histone tails at transcribed genes or at other locations in the genome. Both H3K4 methylation and histone H2A.Z are found at the 5′-end of transcribed and non-transcribed genes (3, 29, 30). In addition, some reports suggest that H2A.Z may localize to centromeric heterochromatin in S. pombe (31), as is true for mammalian chromosomes (32), although other studies using genome-wide analysis in S. pombe do not support this localization (4, 5). We have previously demonstrated that Msc1 facilitates the incorporation of the histone H3 variant CENP-A at centromeres (although this result is controversial; see Refs. 5 and 6); we have also shown genetically that multicopy Msc1 compensates for a conditionally defective CENP-A mutant (2). In sum, these results suggest that Msc1 may influence stability of the specialized nucleosomes found at the centromere of eukaryotic chromosomes.
Given the association of Msc1 with the Swr1 complex and the critical role of Swr1 in mediating chromosome stability and histone modifications, it is of interest to determine how Msc1 interacts with Swr1 and the functional consequences of that interaction. Here, we demonstrate that the C-terminal PHDs of Msc1 are necessary for association with Swr1 and that a truncated Msc1 protein consisting of the C-terminal PHDs alone is sufficient to associate with Swr1. Significantly, point mutations of cysteine residues within the PHDs of Msc1 retain the ability of Msc1 to associate with Swr1. However, a cysteine-to-alanine mutation in PHD2 eliminates the function of Msc1 as assayed by the ability to maintain viability of kinetochore mutants. Such mutations have been shown previously to compromise the E3 ubiquitin ligase activity of the Msc1 PHDs (13). Thus, we hypothesize that, in addition to associating with Swr1, the activity of Msc1 as a ubiquitin ligase is critical for its function in mediating chromosome stability and segregation.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Cells were grown at 30 °C unless indicated otherwise. The strains used are listed in supplemental Table 1. For spotting assays, cells were grown to mid-log phase, and 10-fold serial dilutions were made. Aliquots of 5 μl of each dilution were spotted on plates. Incorporation of epitope tags at the C termini of Msc1, Swr1, Ino80, and Mst1 was carried out using primers homologous to the 3′-end of the genes to amplify C-terminal tagging cassettes from the Bähler plasmid collection (33). The functionality of tagged Msc1, Swr1, and Pht1 was confirmed by crossing the tagged strains with a mis6-302 strain. Growth at 30 °C was used as evidence that the tagged alleles were functional because mis6-302 in combination with deletion alleles of any of these is inviable at 30 °C (Ref. 2 and this study). The genes encoding Ino80 and Mst1 are essential. Thus, the viability of the tagged strains was used as evidence of protein function.
Protein Purification and Mass Spectrometry Analysis
Affinity purification of the Msc1-TAP complex was performed as described (34). 5, 15, and 25 μl of purified protein complexes resuspended in lithium dodecyl sulfate-PAGE buffer were resolved on a 4–12% NuPAGE gel using Mops buffer (Novex). Silver staining was performed using a PlusOne silver staining kit (GE Healthcare). Mass spectrometry was done in the laboratory of Dr. Peter Lobel (Center for Advanced Biotechnology and Medicine and UMDNJ-Robert Wood Johnson Medical School).
Chromatin Immunoprecipitation
Assays were performed as described (2), except that a mouse monoclonal antibody to the HA tag (F7, Santa Cruz Biotechnology, Inc.) on H2A.Z was used.
Co-immunoprecipitations
Immunoprecipitation was performed as described previously (13) with modifications. 60 μl of antibody F7 was used to bind HA-tagged protein, or 60 μl of antibody M2 (Sigma) was used to bind FLAG-tagged protein. For detection of proteins by Western blot analysis of nitrocellulose filters following separation of immune complexes by SDS-PAGE, antibody F7 or M2 was used at a dilution of 1:500, followed by detection with HRP-coupled secondary antibodies.
Immunofluorescence
Immunofluorescence was performed using mid-log phase growing cells as described previously (1).
Reverse Transcription-PCR
50-ml cultures were grown to mid-log phase, and a RiboPure-Yeast kit (Ambion) was used to extract RNA according to the manufacturer's protocol. SuperScript II reverse transcriptase (Invitrogen) was used to perform RT-PCR. 1 μl of RNase H (New England BioLabs) was used to generate cDNA at 37 °C for 20 min. A Sprint Advantage single shot strip (Clontech) was used for regular PCR. The primers used were as follows: Swr1, 5′-GCT TCA TCG TGT ACT CAG AC-3′ and 5′-GGC AGA TCT TCG CAA GGC GA-3′; Msc1: 5′-GCA TCT AGA AAG TCC TTC GG-3′ and 5′-CTA TAG CCA AAC ATT CCT CG-3′; and Fbp1, 5′-AAT GAC AAT TCC CCA CTA GCC-3′ and 5′-ACT TCA GCT AGG ATT CAC CT-3′.
RESULTS
Msc1 Associates with Swr1, and Its Stability Is Dependent on the Presence of Swr1
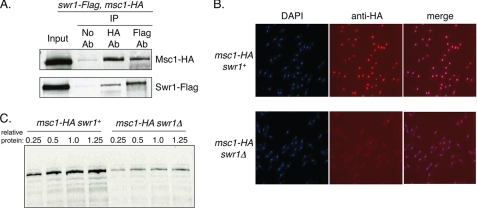
The Msc1 protein of fission yeast was originally isolated as a dosage suppressor capable of conferring cell survival on cells defective for the checkpoint kinase chk1 (1). Loss of function of Msc1 results in elevated rates of chromosome loss and genetic interactions with components of the fission yeast kinetochore and centromere (2). To further investigate the role of Msc1 in cells, we introduced a TAP tag into the genomic copy of msc1, purified TAP-tagged Msc1, and identified associated proteins. We detected components of the fission yeast Swr1 complex (supplemental Fig. 1), consistent with recent reports from other investigators that Msc1 is a stoichiometric component of this complex (3–6). Some components of the Swr1 complex are also components of other chromatin-modifying complexes in yeast, including the NuA4 histone acetyltransferase complex and the Ino80 complex (9). Therefore, we tested whether Msc1 could be reciprocally co-immunoprecipitated with the catalytic subunit of the Swr1 complex, Swr1, the Mst1 catalytic subunit of NuA4, or Ino80. Msc1, Swr1, Ino80, and Mst1 were tagged at their chromosomal loci with HA (Msc1) or FLAG (Swr1, Ino80, and Mst1). Strains bearing tagged alleles of Msc1 were crossed with strains of each of the FLAG-tagged genes. As shown in Fig. 1B, immunoprecipitation of HA-tagged Msc1 coprecipitated Swr1-FLAG, whereas immunoprecipitation of Swr1-FLAG coprecipitated Msc1-HA. In contrast, no co-immunoprecipitation of either Ino80 or Mst1 could be detected with Msc1 (supplemental Fig. 1). We conclude that Msc1 specifically associates with the Swr1 complex.
FIGURE 1.
Msc1 associates with Swr1, and its stability is compromised in cells lacking Swr1. A, Msc1 co-immunoprecipitated with Swr1. NW2632 cells with HA-tagged Msc1 and FLAG-tagged Swr1, each at their respective chromosomal locus, were grown to mid-log phase, lysed, and immunoprecipitated (IP) with antibody (Ab) to either the HA or the FLAG epitope. Captured immune complexes were separated by SDS-PAGE and subjected to Western blot analysis with antibody to HA or FLAG. B, cells with endogenous HA-tagged Msc1 and either wild-type (NW1562) or deleted (NW2633) Swr1 were grown to mid-log phase, fixed, and incubated with antibody to the HA epitope. Cells were stained with secondary antibody coupled to Cy3 and with DAPI to visualize nuclei. C, lysates from mid-log phase cells with (NW1562) or without (NW2633) Swr1 were loaded as serial dilutions, separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibody to detect the HA epitope on Msc1.
Msc1 localizes to the nucleus and is associated with chromatin (1). To determine whether localization is dependent on Swr1, we examined the nuclear localization of Msc1-HA in cells lacking swr1, which are viable. As shown in Fig. 1C, localization of Msc1 to the nucleus was retained, although the signal of Msc1-HA as detected by immunofluorescence with antibody to the HA tag was much weaker in the swr1Δ cells. We postulated that this reduced signal could be due to instability of the Msc1 protein in cells lacking Swr1. Indeed, by Western blot analysis of serially diluted protein extracts, we determined that the Msc1-HA protein was less abundant at steady state in swr1Δ cells than in swr1+ cells (supplemental Fig. 2). As Swr1 has been implicated in transcriptional regulation, we performed RT-PCR to determine whether the message level of Msc1 is altered when Swr1 is absent, but we found that the mRNA level of the msc1 gene was constant in wild-type cells and cells deleted for swr1 (supplemental Fig. 2). We conclude that it is the protein level of Msc1 that is compromised when Swr1 is absent. Thus, phenotypes associated with loss of swr1 function must also assume diminished msc1 function as well. In contrast to the dependence of the Msc1 protein level on Swr1, Swr1 levels do not vary in cells lacking Msc1 (data not shown).
Loss of Swr1 Function, Like Loss of Msc1 Function, Compromises Viability of Kinetochore Mutants and Affects Silencing at Centromeres
We have shown previously that loss of Msc1 function compromises the viability of mutants with conditional alleles of genes encoding kinetochore proteins (2). Because Msc1 is present in the Swr1 complex, we sought to determine whether loss of Swr1 function has similar phenotypes. Consistent with their physical association, the absence of swr1 compromised the viability of mis12 and mis6 mutants, genes that encode conserved components of the outer and inner kinetochores of fission yeast, respectively (supplemental Fig. 3). No additive effects were apparent upon simultaneous deletion of msc1 and swr1, consistent with a shared functional role at kinetochores.
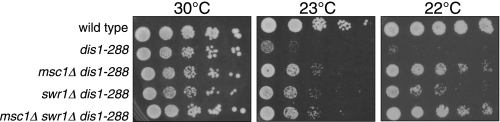
We had shown previously that loss of msc1 function has little effect on viability of cnp1-1, which bears a temperature-sensitive mutation in the gene encoding the CENP-A histone H3 variant found in eukaryotic centromeres, although multicopy expression of the msc1 gene allows cnp1-1 cells to grow at restrictive temperature (2). Likewise, deletion of swr1 did not further compromise the phenotype of the cnp1-1 mutant (supplemental Fig. 3). The dis1 gene of fission yeast encodes a microtubule-associated protein that localizes to the kinetochore and mediates the attachment of chromosomes to the mitotic spindle (35–37). Mutation of dis1, homologs of which regulate microtubule dynamics in Xenopus extracts (XMAP215), mammalian cells (TOG), and budding yeast (STU2) (38–40), confers a cold-sensitive phenotype, as chromosomes fail to disjoin (41). Interestingly, deletion of msc1, swr1 or both resulted in suppression of the cold-sensitive lethality associated with dis1-288 (Fig. 2). These genetic interactions with the kinetochore mutants and dis1 suggest that Swr1, like Msc1, may play a role in mediating centromere structure or function.
FIGURE 2.
Deletion of Swr1 or Msc1 suppresses cold sensitivity of dis1-288. The absence of Msc1 or Swr1 improved growth at restrictive temperature for the cold-sensitive mutant dis1-288. The indicated strains (SP6, NW1624, NW2638, NW2639, and NW2640 from top to bottom) were grown at 30 °C to mid-log phase, and serial dilutions were prepared for spotting on YEA plates and incubation at the indicated temperatures.
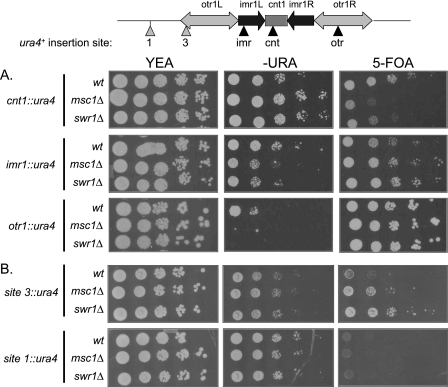
Fission yeast centromeres are characterized by a central core region that is flanked by regions of heterochromatin that confer silencing on genes inserted within them. Evaluating the expression state of reporter genes in these regions is an indirect assay of alterations in centromere structure (42). Consistent with the similar phenotypes of swr1Δ and msc1Δ in genetic interactions with kinetochore and centromere mutants, deletion of either gene confers similar phenotypes in silencing assays using the ura4+ reporter gene inserted at various locations within the centromere of chromosome I. As shown in Fig. 3A, deletion of msc1 or swr1 resulted in increased expression of the ura4+ reporter gene inserted in the central core region (cnt1), consistent with observations reported by others (5). Extending this analysis beyond the central core, we found that deletion of msc1 or swr1 resulted in decreased expression of the ura4+ reporter gene inserted either at imr1 or otr1, indicating increased repression in these regions. When the ura4+ gene was inserted at the boundary of the heterochromatic otr1 region, increased repression was also apparent (Fig. 3B, site 3). However, no effect resulting from loss of msc1 or swr1 was seen on expression of ura4+ when the reporter was inserted outside of the otr1 region (Fig. 3B, site 1). Thus, although increased repression due to the absence of msc1 or swr1 is apparent in the already heterochromatic regions of imr1 and otr1, increased repression does not spread to euchromatic regions.
FIGURE 3.
Swr1, like Msc1, affects chromatin structure at the centromere. Strains with ura4+ genes inserted at the locations indicated in the diagram were grown in YEA at 30 °C, and serial dilutions were spotted on YEA plates, minimal media plates lacking uracil, and YEA plates with 5-fluoroorotic acid (5-FOA), which is toxic to cells expressing the ura4+ gene. A, strains with reporters integrated in the centromere of chromosome I. Upper panels, the ura4+ gene is integrated in the central core (cnt1) of the centromere (NW1764, NW1586, and NW2643). Middle panels, the ura4+ gene is integrated at the imr1 region (NW1767, NW1584, and NW2644). Lower panels, the ura4+ gene is integrated in the heterochromatic otr1 region (NW1768, NW1585, and NW2645). B, reporter gene expression at the boundary and outside of the centromere. Upper panels, ura4+ expression at the boundary of otr1 (site 3) was tested in strains NW2648, NW2652, and NW2650. Lower panels, ura4+ expression outside of otr1 (site 1) was tested in strains NW2647, NW2651, and NW2649.
Histone H2A.Z Localizes to the Heterochromatic Outer Repeats of the Fission Yeast Centromere
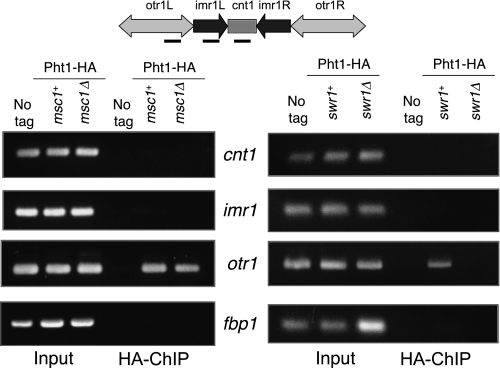
Using imaging methods, histone H2A.Z has been reported to be found discontinuously interspersed with nucleosomes containing CENP-A in mammalian centromeres (32). Centromeric localization of H2A.Z is consistent with a role for it in mediating chromosome stability (43, 44). Recent studies of H2A.Z localization along fission yeast chromosomes on a genome-wide scale describe the absence of H2A.Z from fission yeast centromeres (4, 5), in contrast to other reports of H2A.Z localization to the centromere in fission yeast based on studies using location-specific chromatin immunoprecipitation (6, 31). To assess whether H2A.Z localizes to the centromere using HA-tagged H2A.Z, which we know to be functional (see “Experimental Procedures”), we performed chromatin immunoprecipitations with antibody to the HA tag using wild-type cells and cells lacking Msc1 or Swr1. As shown in Fig. 4, we found that HA-tagged H2A.Z localized to the heterochromatic outer repeat (otr1) region of centromere I, although not to the central core (cnt1) or inner repeat (imr1) regions. Association of H2A.Z with the otr1 region was compromised in cells lacking swr1 (Fig. 4, right panel), although it remained associated in cells lacking msc1 (left panel).
FIGURE 4.
Histone H2A.Z localization to the otr1 region of centromere I is dependent on Swr1 but not on Msc1. Cells with an integrated copy of HA-tagged pht1 (NW1776) and HA-tagged pht1 with deletion of either msc1 (NW1959) or swr1 (NW2048) were grown to mid-log phase and processed as described under “Experimental Procedures” for chromatin immunoprecipitation. Input and precipitated samples were probed with primers for the cnt1, imr1, and otr1 regions of the centromere and for the fbp1 gene.
C-terminal PHDs of Msc1 Are Necessary and Sufficient for Association with Swr1
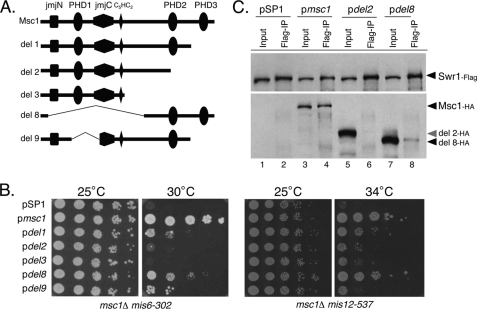
To gain insight into the domains necessary for interaction between Msc1 and Swr1, we made use of truncated alleles of Msc1 previously characterized for the ability to coprecipitate histone deacetylase activity (1). As shown in Fig. 5A, these truncations had deletions of one or more of the conserved domains of Msc1, including the PHDs and the JmjN and JmjC domains. The truncated alleles were expressed from the endogenous msc1 promoter on a pSP1 plasmid (45), which has the S. pombe ars1 sequence to allow replication. Such plasmids are thought to be present at 5–10 copies/cell, consistent with analysis of protein expression levels (supplemental Fig. 4). Plasmids were transformed into strains with deletions of msc1 and temperature-sensitive alleles of either mis6 or mis12. Transformation of an empty vector and a plasmid expressing wild-type msc1 served as negative and positive controls, respectively. As shown in Fig. 5B, the wild-type msc1 gene allowed the msc1::kanR mis6-302 strain to grow at 30 °C, whereas the empty vector allowed no growth at this temperature. A plasmid lacking the C-terminal PHD3 domain (del1) allowed some growth, but a plasmid lacking both the C-terminal PHD2 and PHD3 domains allowed no growth. Likewise, a plasmid expressing only the N-terminal half of Msc1 (del3) was nonfunctional in this assay. In addition, deletion of the PHD1 and PHD3 domains (del9) was nonfunctional. In contrast, a plasmid that lacks the JmjN and JmjC domains as well as the N-terminal PHD1 domain but that contains the C-terminal PHD2 and PHD3 domains was largely functional in this assay. Similar behaviors were observed using the same plasmids in the msc1::kanR mis12-537 strain (Fig. 5B, right panels). Thus, it would appear that the C-terminal PHD2 and PHD3 domains are necessary and surprisingly sufficient to confer viability on cells lacking Msc1 that have compromised kinetochore function by virtue of mutations in mis6 and mis12.
FIGURE 5.
C-terminal PHDs of Msc1 are important for function and for association with Swr1. A, shown is schematic representation of the deletion mutants of Msc1 carried by the pSP1 plasmid. All constructs also had a 3×HA tag at the C terminus. B, the indicated plasmids were introduced into the msc1Δ mis6-302 (NW1703; left panels) and msc1Δ mis12-537 (NW1617; right panels) strains and grown at 25 °C to mid-log phase. Serial dilutions were spotted on YEA plates and incubated at the indicated temperatures for 3 days. C, a strain with an integrated FLAG-tagged allele of swr1 and deletion of the msc1 gene (NW2646) was transformed with the indicated plasmids. Cells were grown to mid-log phase, lysed, and subjected to immunoprecipitation with antibody to FLAG-tagged Swr1. Precipitates were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with antibodies to detect Swr1-FLAG (upper panel) and Msc1-HA (lower panel).
We then tested which domains of Msc1 are necessary for association with Swr1. The truncation mutants described in Fig. 5A were introduced into a strain with a deletion of msc1 and a FLAG-tagged allele of Swr1. The plasmid-expressed mutants each contained an HA tag. Cells were grown to mid-log phase, lysed, and subjected to immunoprecipitation with anti-FLAG antibody. As shown in Fig. 5C, wild-type Msc1-HA co-immunoprecipitated with Swr1-FLAG. In contrast, the truncation mutant del2, which lacked the C-terminal PHDs, did not coprecipitate with Swr1-FLAG (Fig. 5C, lane 6), although the del2 protein was efficiently expressed (lane 5). However, the del8 protein, which had the C-terminal PHDs but lacked the bulk of the N-terminal half of the protein, did associate with Swr1-FLAG (Fig. 5C, lane 8). These interactions (and lack thereof) were confirmed by immunoprecipitation of Msc1 with an anti-HA antibody and probing of the precipitates with anti-FLAG antibody to detect Swr1 (data not shown). We conclude that the C-terminal PHDs of Msc1 are important for association with Swr1 and that the C terminus of the protein is sufficient to confer this association. We speculate that the ability of Msc1 to confer viability on mutants with compromised kinetochore function requires association with Swr1.
Point Mutations in the C-terminal PHDs of Msc1 Retain Association with Swr1 but Are Nonfunctional in the Kinetochore Function Assay
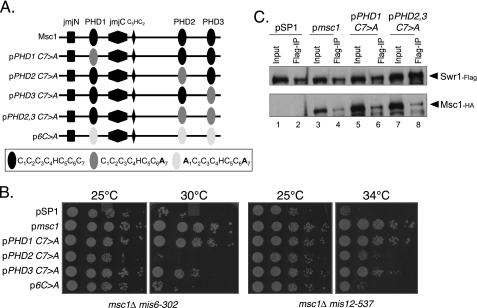
Although PHDs have been described in recent years as methyl histone-binding domains, a subset of PHDs was previously described as capable of acting as E3 ubiquitin protein ligases (14–18). Indeed, isolated PHD fingers of Msc1 have the capacity to facilitate ubiquitin transfer in an in vitro assay (13). PHDs have a characteristic pattern of four spaced cysteine residues, followed by one histidine and three additional cysteine residues that in crystal structures coordinate a pair of zinc ions (12). The function of the Msc1 PHDs as E3 ligases is compromised by mutation of either the first or seventh cysteine residue (13). Furthermore, the E3 ligase activity of the full-length Msc1 protein is compromised, although not abolished, by single point mutations in any single PHD (13). We tested whether such mutants, described schematically in Fig. 6A, could sustain viability when kinetochore function is compromised in the msc1::kanR double mutants with either mis6-302 or mis12-537. As shown in Fig. 6B, although a point mutation in the first PHD had essentially no effect on viability, mutation of the second PHD severely compromised viability, particularly in the msc1::kanR mis6-302 strain. Similarly, mutation of the first and seventh cysteine residues in each of the three PHDs compromised viability. A single point mutation of the third PHD had an intermediate phenotype. We conclude that the second PHD, which resides in the C terminus, is of particular functional importance for survival when kinetochore function is compromised by mutation of mis6 or mis12.
FIGURE 6.
Mutation of PHD2 retains Swr1 binding but abolishes Msc1 function. A, shown is a schematic of mutations in the PHDs of Msc1. B, the indicated plasmids were introduced into NW1703 and NW1617, and cells were treated as described for Fig. 5B. C, the indicated plasmids were introduced into NW2646, and cells were treated as described for Fig. 5C.
We then tested whether point mutations within the PHDs affect the ability of Msc1 to associate with Swr1. As shown in Fig. 6C, single point mutations in PHD1 or in each of the C-terminal PHDs, PHD2 and PHD3, did not affect the ability of Msc1 to associate with Swr1. Thus, although mutation of PHD2 does not eliminate binding to Swr1, it does compromise the ability of Msc1 to confer survival in strains with compromised kinetochore function. On the basis of these results and those shown in Fig. 5, we speculate that the C-terminal PHDs are important for association of Msc1 with Swr1 and for mediating survival of strains with compromised kinetochore function. Furthermore, PHD2 must be intact for Msc1 to function properly. Thus, the association of Msc1 with Swr1 is critical but not sufficient for kinetochore function. An activity that is disrupted by mutation of Cys7 in PHD2 is likewise important for function.
DISCUSSION
The fission yeast Msc1 protein was first identified in an effort to find new proteins that function to confer survival in response to DNA damage when the DNA damage checkpoint is compromised by mutation of the chk1 gene (1). Subsequent studies revealed that Msc1 is important for maintaining genomic stability in cooperation with proteins that are important for centromere function, including Mis6 and Mis12 (2). Cells deficient for Msc1 exhibit alterations in histone modifications, specifically an elevated level of histone H3 acetylation (1) and H3 methylation (13). Msc1 has a domain structure similar to that of mammalian RBP2 and PLU-1, all of which are members of the Jarid1 family of proteins (46). The mammalian proteins of the Jarid1 family have demonstrable histone demethylase activity targeted to H3K4 (25–28). In accord with these observations, cells lacking Msc1 exhibit an elevated level of H3K4 methylation if rhp6 is also absent from the cell, which, on its own, results in dramatically reduced levels of methylation both in fission yeast (13) and in budding yeast (47, 48). Thus, loss of Msc1 function can restore histone methylation, consistent with the possibility that it functions as a demethylase. However, upon overexpression of Msc1, we did not detect a decrease in H3K4 methylation (data not shown). Furthermore, the JmjC domain of Msc1 does not possess the catalytic residues thought to be critical for demethylase activity. Thus, it is equally plausible that the absence of Msc1 restores methylation in an rhp6 deletion strain by derepressing the activity of a histone methyltransferase, for example. Further studies will be needed to establish the mechanism by which Msc1 impacts the methylation status of histone H3.
Msc1 is clearly a component of the Swr1 complex in fission yeast, as multiple groups have detected copurification of Msc1 with Swr1, and genetically, mutation of msc1 or swr1 behaves similarly (Fig. 2 and supplemental Fig. 3) (7). Nonetheless, no ortholog of Msc1 is found in budding yeast despite the conservation of the Swr1 complex in this organism. Perhaps the function of the Swr1 complex evolved differently in the two yeasts such that fission yeast incorporated a requirement for the Msc1 protein. Whether or not the Msc1 domain structure homologs RBP2 and PLU-1 are components of the mammalian Swr1 complex remains to be determined. The Swr1 complex has been demonstrated to incorporate the histone H2A variant H2A.Z into specific regions of chromosomes, namely the 5′-end of transcribed and non-transcribed genes (9). On the basis of studies in budding yeast, several investigators have postulated that H2A.Z may function as a switch to allow or repress transcription (30, 49, 50). Two recent genome-wide studies in fission yeast are consistent with the localization of histone H2A.Z to the promoter regions of transcribed genes (3, 5). Both of these reports suggest that localization of H2A.Z to the promoter region of genes is dependent on msc1 and swr1 (3, 5). Another report suggests similar localization of H2A.Z genome-wide and indicates that Swr1 is required for localization, whereas Msc1 plays a lesser role (4).
Several results support a role for Msc1 and Swr1 in centromere structure and/or function. Genetically, the absence of msc1 (2) or swr1 (supplemental Fig. 3) compromises survival of mis6 and mis12 mutants. Whether or not Msc1 or Swr1 affects the localization of CENP-A to centromeric regions remains controversial. In our hands, CENP-A localization to the central core was reduced in cells lacking msc1, consistent with two observations: multicopy expression of msc1 very effectively restores viability to a strain with a temperature-sensitive mutant of cnp1-1, and the absence of msc1 compromises survival of cells with a temperature-sensitive allele of the CENP-A loader, Mis6 (2). In contrast, two recent reports (5, 6) do not indicate that Msc1 is required for localization of CENP-A to the central core. Different antibodies were used for the chromatin immunoprecipitation experiments, and there are some differences in the auxotrophic markers in the strains. Nonetheless, there is no clear explanation for the discrepancy in these results.
Several groups recently reported the localization of histone H2A.Z in fission yeast, with rather variable results. In our hands, we readily detected histone H2A.Z at the heterochromatic outer repeats of the centromere (Fig. 4), consistent, at least in part, with two reports (6, 31) but in contradiction to two others (4, 5). Why such disparate results have been generated is unclear. We have tested the tagged versions of the pht1 gene used by these different groups, and all are capable of function as determined by the ability to support growth at 30 °C of a mis6-302 strain (although a pht1Δ strain is viable, a pht1Δ mis6-302 strain is inviable at 30 °C). Localization of H2A.Z to the region of otr1 detected in our experiments is dependent on Swr1 but independent of Msc1. In this regard, cells lacking msc1 or swr1 behave differently, yet genetically, the absence of either one similarly affects centromere silencing and compromises viability of kinetochore mutants. Thus, unlike Swr1 itself, the mechanism by which Msc1 influences these behaviors may not rely on histone H2A.Z localization per se. Perhaps there are important dynamic modifications to H2A.Z or other histones that are affected by the absence of Msc1.
We previously showed that the PHDs of Msc1 are capable of acting as E3 ubiquitin protein ligases that can facilitate the transfer of ubiquitin to an in vitro substrate (13). Mutations of cysteine residues within these domains compromise that ability. In the present study, we found that the C-terminal PHDs are important for association of Msc1 with Swr1, although cysteine mutations within those domains do not compromise the ability of Msc1 to associate with Swr1. Nonetheless, mutation of PHD2, in particular, eliminates the ability of Msc1 to promote survival of cells temperature-sensitive for mis6 or mis12. Thus, we postulate that association of Msc1 with Swr1 is important but not sufficient for centromere function; the ability of Msc1 to act as an E3 ligase may be required for the Msc1-Swr1 interaction to be productive in supporting centromere function. The target of the E3 ligase activity of Msc1 necessary to confer centromere function and chromosome stability is as yet unknown. Because PHDs of some proteins have been shown to bind to methylated lysine residues within the N-terminal tails of histone H3 or H4 (11, 19–22), it is possible that such an activity of PHD2 may be important for its function. Although we were unable to detect histone binding to a fusion protein with isolated PHD2 when incubated with calf thymus histones, we cannot rule out that it interacts with methylated lysine residues in the context of the whole protein. To our knowledge, there are no reports of cysteine residue mutations within PHDs that affect their binding to methyllysine residues, although it is not inconceivable that such mutations could affect binding. Further studies will need to address this and other possibilities to establish the precise mechanism by which Msc1 influences the activity of Swr1 and affects chromosome stability.
Supplementary Material
Acknowledgments
We are grateful to Jason Tanny and David Allis (The Rockefeller University), Robin Allshire (University of Edinburgh), and Mitsuhiro Yanagida (Kyoto University) for providing strains. We thank Marc Gartenberg, X. F. Steven Zheng, and Thomas Kusch for helpful discussions during the course of this work.
This work was supported, in whole or in part, by United States Public Health Service Grant GM53194 from NIGMS, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- PHD
- plant homeodomain
- JmjC
- Jumonji C
- YEA
- yeast extract adenine.
REFERENCES
- 1.Ahmed S., Palermo C., Wan S., Walworth N. C. (2004) Mol. Cell. Biol. 24, 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S., Dul B., Qiu X., Walworth N. C. (2007) Genetics 177, 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zofall M., Fischer T., Zhang K., Zhou M., Cui B., Veenstra T. D., Grewal S. I. (2009) Nature 461, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan L., Durand-Dubief M., Roguev A., Sakalar C., Wilhelm B., Strålfors A., Shevchenko A., Aasland R., Shevchenko A., Ekwall K., Francis Stewart A. (2009) PLoS Genet. 5, e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou H., Wang Y., Kallgren S. P., Thompson J., Yates J. R., 3rd, Jia S. (2010) J. Biol. Chem. 285, 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H. S., Vanoosthuyse V., Fillingham J., Roguev A., Watt S., Kislinger T., Treyer A., Carpenter L. R., Bennett C. S., Emili A., Greenblatt J. F., Hardwick K. G., Krogan N. J., Bähler J., Keogh M. C. (2009) Nat. Struct. Mol. Biol. 16, 1286–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roguev A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., Collins S. R., Qu H., Shales M., Park H. O., Hayles J., Hoe K. L., Kim D. U., Ideker T., Grewal S. I., Weissman J. S., Krogan N. J. (2008) Science 322, 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevchenko A., Roguev A., Schaft D., Buchanan L., Habermann B., Sakalar C., Thomas H., Krogan N. J., Shevchenko A., Stewart A. F. (2008) Genome Biol. 9, R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. (2004) PLoS Biol. 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altaf M., Auger A., Monnet-Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R. T., Gaudreau L., Côté J. (2010) J. Biol. Chem. 285, 15966–15977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin D. G., Baetz K., Shi X., Walter K. L., MacDonald V. E., Wlodarski M. J., Gozani O., Hieter P., Howe L. (2006) Mol. Cell. Biol. 26, 7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pascual J., Martinez-Yamout M., Dyson H. J., Wright P. E. (2000) J. Mol. Biol. 304, 723–729 [DOI] [PubMed] [Google Scholar]
- 13.Dul B. E., Walworth N. C. (2007) J. Biol. Chem. 282, 18397–18406 [DOI] [PubMed] [Google Scholar]
- 14.Lu Z., Xu S., Joazeiro C., Cobb M. H., Hunter T. (2002) Mol. Cell 9, 945–956 [DOI] [PubMed] [Google Scholar]
- 15.Uchida D., Hatakeyama S., Matsushima A., Han H., Ishido S., Hotta H., Kudoh J., Shimizu N., Doucas V., Nakayama K. I., Kuroda N., Matsumoto M. (2004) J. Exp. Med. 199, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., Yanagi S. (2006) EMBO J. 25, 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto E., Ishido S., Sato Y., Ohgimoto S., Ohgimoto K., Nagano-Fujii M., Hotta H. (2003) J. Biol. Chem. 278, 14657–14668 [DOI] [PubMed] [Google Scholar]
- 18.Coscoy L., Sanchez D. J., Ganem D. (2001) J. Cell Biol. 155, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peña P. V., Davrazou F., Shi X., Walter K. L., Verkhusha V. V., Gozani O., Zhao R., Kutateladze T. G. (2006) Nature 442, 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., Patel D. J. (2006) Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y., Landry J., Kauer M., Tackett A. J., Chait B. T., Badenhorst P., Wu C., Allis C. D. (2006) Nature 442, 86–90 [DOI] [PubMed] [Google Scholar]
- 23.Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., Baird C. L., Zhu H., Field S. J., Lessnick S. L., Villasenor J., Mehrotra B., Chen J., Rao V. R., Brugge J. S., Ferguson C. G., Payrastre B., Myszka D. G., Cantley L. C., Wagner G., Divecha N., Prestwich G. D., Yuan J. (2003) Cell 114, 99–111 [DOI] [PubMed] [Google Scholar]
- 24.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 25.Christensen J., Agger K., Cloos P. A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K. H., Salcini A. E., Helin K. (2007) Cell 128, 1063–1076 [DOI] [PubMed] [Google Scholar]
- 26.Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D. G., Zhang Y., Kaelin W. G., Jr. (2007) Cell 128, 889–900 [DOI] [PubMed] [Google Scholar]
- 27.Yamane K., Tateishi K., Klose R. J., Fang J., Fabrizio L. A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. (2007) Mol. Cell 25, 801–812 [DOI] [PubMed] [Google Scholar]
- 28.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H. H., Whetstine J. R., Bonni A., Roberts T. M., Shi Y. (2007) Cell 128, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 29.Schneider R., Bannister A. J., Myers F. A., Thorne A. W., Crane-Robinson C., Kouzarides T. (2004) Nat. Cell Biol. 6, 73–77 [DOI] [PubMed] [Google Scholar]
- 30.Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. (2005) Cell 123, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence R. J., Volpe T. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves I. K., Rangasamy D., Ridgway P., Tremethick D. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 34.Gould K. L., Ren L., Feoktistova A. S., Jennings J. L., Link A. J. (2004) Methods 33, 239–244 [DOI] [PubMed] [Google Scholar]
- 35.Nabeshima K., Kurooka H., Takeuchi M., Kinoshita K., Nakaseko Y., Yanagida M. (1995) Genes Dev. 9, 1572–1585 [DOI] [PubMed] [Google Scholar]
- 36.Nakaseko Y., Nabeshima K., Kinoshita K., Yanagida M. (1996) Genes Cells 1, 633–644 [DOI] [PubMed] [Google Scholar]
- 37.Nakaseko Y., Goshima G., Morishita J., Yanagida M. (2001) Curr. Biol. 11, 537–549 [DOI] [PubMed] [Google Scholar]
- 38.Slep K. C. (2010) Curr. Opin. Cell Biol. 22, 88–95 [DOI] [PubMed] [Google Scholar]
- 39.Spittle C., Charrasse S., Larroque C., Cassimeris L. (2000) J. Biol. Chem. 275, 20748–20753 [DOI] [PubMed] [Google Scholar]
- 40.Wang P. J., Huffaker T. C. (1997) J. Cell Biol. 139, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohkura H., Adachi Y., Kinoshita N., Niwa O., Toda T., Yanagida M. (1988) EMBO J. 7, 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G. (1994) Cell 76, 157–169 [DOI] [PubMed] [Google Scholar]
- 43.Rangasamy D., Greaves I., Tremethick D. J. (2004) Nat. Struct. Mol. Biol. 11, 650–655 [DOI] [PubMed] [Google Scholar]
- 44.Carr A. M., Dorrington S. M., Hindley J., Phear G. A., Aves S. J., Nurse P. (1994) Mol. Gen. Genet. 245, 628–635 [DOI] [PubMed] [Google Scholar]
- 45.Cottarel G., Beach D., Deuschle U. (1993) Curr. Genet. 23, 547–548 [DOI] [PubMed] [Google Scholar]
- 46.Klose R. J., Kallin E. M., Zhang Y. (2006) Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 47.Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 48.Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 49.Li B., Pattenden S. G., Lee D., Gutiérrez J., Chen J., Seidel C., Gerton J., Workman J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Roberts D. N., Cairns B. R. (2005) Cell 123, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.