Abstract
TLR3 (Toll-like receptor 3) recognizes dsRNA, a potent indicator of viral infection. The extracellular domain of TLR3 dimerizes when it binds dsRNA, and the crystal structure of the dimeric complex reveals three sites of interaction on each extracellular domain, two that bind dsRNA and one that is responsible for dimer formation. The goal of this study was to determine which amino acid residues are essential for forming a stable receptor·ligand complex and whether dimerization of TLR3 is required for dsRNA binding. Using a novel ELISA to analyze dsRNA binding by mutant TLR3 constructs, we identified the essential interacting residues and determined that the simultaneous interaction of all three sites is required for ligand binding. In addition, we show that TLR3 is unable to bind dsRNA when dimerization is prevented by mutating residues in the dimerization site or by immobilizing TLR3 at low density. We conclude that dimerization of TLR3 is essential for ligand binding and that the three TLR3 contact sites individually interact weakly with their binding partners but together form a high affinity receptor·ligand complex.
Keywords: Immunochemistry, Innate Immunity, Ligand-binding Protein, Protein-Nucleic Acid Interaction, Toll-like Receptors (TLRs)
Introduction
Many viruses produce dsRNA at some stage in their replication cycle. Although short stretches of dsRNA are normally found in the microRNA, tRNA, and rRNA of most cells, only viruses synthesize long dsRNA molecules. Therefore, dsRNA longer than ∼30 bp can serve as a potent molecular signature of viral infection in higher organisms, including man. Consequently, dsRNA is recognized by a number of pattern recognition receptors of the innate immune system, including TLR3 (Toll-like receptor 3) (1–3). TLR3 is a type I transmembrane receptor with an N-terminal extracellular domain (ECD),2 a single transmembrane helix, and a C-terminal cytoplasmic signaling domain of the TIR (Toll/IL-1 receptor) family. The ECD of TLR3 is located in the interior of endosomes, where it encounters and binds dsRNA and transduces signals that initiate inflammatory and adaptive antiviral responses. dsRNA enters the endosomes either by direct uptake from the medium or by phagocytosis of virally infected cells (4), and as the dsRNA transits from early to late endosomes, the pH decreases progressively from pH ∼6.2 to 5.5 (5).
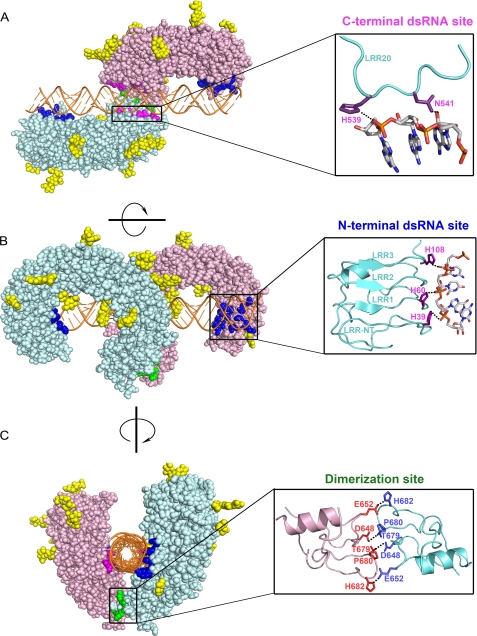
Recent crystallographic studies have shown how TLR3 recognizes dsRNA at the molecular level (for a review, see Ref. 6). The TLR3 ECD can be described as a coil comprising 23 tandem leucine-rich repeat (LRR) motifs bent into the shape of a horseshoe and capped at the N- and C-terminal ends by specialized structures known as the LRR-NT and LRR-CT domains, respectively (7, 8). The TLR3 ECD is decorated with 15 N-linked glycans and has one surface that is devoid of glycan and free to interact with dsRNA. Recombinant TLR3 ECD protein binds dsRNA under mildly acidic conditions, at pH values similar to those found in early and late endosomes (9). Although the TLR3 ECD is monomeric in solution, it binds as dimers to 45-bp segments of dsRNA, and several dimers can bind to long dsRNA strands (9). The x-ray structure of a single TLR3 ECD dimeric complex with 46-bp dsRNA (10) reveals that dsRNA interacts with two widely spaced sites on the glycan-free surface of each TLR3 ECD monomer (see Fig. 1, A and B), one near the N terminus and one close to the C terminus. In addition, the two TLR3 ECDs interact homotypically at their LRR-CT domains. This is the only contact between the two molecules of the dimer (see Fig. 1C) and is therefore responsible for dimer formation. In the intact TLR3 molecule, a short linker connects the LRR-CT domain to the transmembrane domain, and it is likely that dimerization of ECDs transduces a signal across the endosomal membrane by bringing the TIR domains together on the cytoplasmic side of the membrane. Importantly, no conformational change in the TLR3 ECD occurs upon ligand binding.
FIGURE 1.
The three sites of interaction on the TLR3 ECD. Three orthogonal views of the TLR3 ECD·dsRNA complex are shown. A, view looking down the 2-fold symmetry axis of the complex from the top. The C-terminal dsRNA-binding site is shown in detail. B, side view of the complex showing the “horseshoe” shape of the TLR3 ECD. The N-terminal dsRNA-binding site is shown in detail. C, view of the complex looking down the dsRNA helical axis. Note that the two TLR3 ECD molecules contact each other only at a single site near their C-terminal ends. The dimerization site is shown in detail. In A–C, the two TLR3 ECDs in the complex are colored pink and blue, and the dsRNA is colored orange and depicted in schematic form. N-Linked glycans are colored yellow. Contacting residues in the N- and C-terminal dsRNA-binding sites are colored dark blue and magenta, respectively, and the dimerization site is colored green. Coordinates are from Protein Data Bank code 3CIY, and interacting residues are identified in Ref. 10. The figures were generated using PyMOL.
The crystal structure identifies the amino acid residues that interact with either dsRNA or the other ECD in the TLR3 ECD·dsRNA complex but does not indicate which residues are essential for forming a stable complex. Previous studies (10–14) showed that a number of mutations in the N- and C-terminal dsRNA-binding site regions (summarized in supplemental Table S1) block dsRNA-dependent activation of TLR3, but it was not known if these mutations affect the binding of dsRNA to TLR3 or if an intact dimerization site is required for ligand binding or activation. In this study, we identified essential residues using a newly developed ELISA to measure dsRNA binding to mutant TLR3 proteins. We show that dimerization is required for ligand binding and that dsRNA recognition and signaling by TLR3 require the simultaneous interaction of the two dsRNA-binding sites and the dimerization site, each of which by itself interacts weakly with its binding partner.
EXPERIMENTAL PROCEDURES
Vector Construction and Site-directed Mutagenesis
Mutations of human TLR3 in pUNO (InvivoGen) were made using a QuikChange® site-directed mutagenesis kit (Stratagene) as described (10, 11). DNA encoding monomeric YFP (a gift from Dr. Susan Pierce, NIAID) with an N-terminal GGGGGG linker was inserted into BamHI and NheI sites at the 3′-end of each TLR3 construct. All constructs were verified by sequencing.
dsRNA Generation and Biotin Labeling
The synthesis and purification of dsRNA oligonucleotides were described previously (9). dsRNA was end-labeled with biotin as described (9) or was biotin-labeled using a LabelIT kit (Mirus Bio).
Transfection
HEK293 cells (8 × 106) were transfected with 20 μg of WT or mutant pUNO-TLR3-YFP plasmid DNA using Lipofectamine 2000 (Invitrogen) and harvested 48 h post-transfection. Pellets containing 107 cells were lysed with 1 ml of lysis buffer (1% Triton X-100, 10 mm Tris (pH 7.4), 150 mm NaCl, 5 mm EDTA, and protease inhibitors (Roche Applied Science)) on ice for 40 min. After centrifugation at 16,000 × g for 20 min at 4 °C, supernatants were stored at −80 °C. Control lysates were generated from untransfected cells.
dsRNA Binding Assay
This assay is shown schematically in supplemental Fig. S1. Corning/Costar 96-well microplates were coated with goat anti-mouse IgG2a (Fcγ-specific; Jackson ImmunoResearch Laboratories) at 4 μg/ml in PBS for 2 h at 37 °C. Plates were washed; blocked with 5% BSA in 10 mm Tris (pH 7.4), 150 mm NaCl, and 0.1% Tween 20 for 1.5 h at 37 °C; and coated with mouse anti-GFP mAb 3E6 (Invitrogen) at 0.5 μg/ml (except where stated otherwise) in PBS for 2 h at 37 °C. After washing, cell lysates (50 μl of a 1:10 dilution in lysis buffer of stock lysate, except where stated otherwise) were added to the wells and allowed to bind overnight at 4 °C. All subsequent steps were performed at room temperature. The wells were washed three times with lysis buffer and three times with PiBST (20 mm PIPES, 150 mm NaCl, and 0.1% Tween 20 at the indicated pH). Biotin-labeled 540-bp (unless stated otherwise) dsRNA (bio-dsRNA; 50 μl in PiBST at the indicated concentration and pH) was incubated with plate-bound TLR3-YFP at room temperature for 2 h, washed four times with PiBST, and labeled with horseradish peroxidase (HRP)-conjugated streptavidin (1:5000 in PiBST; Thermo Scientific). The relative amount of plate-bound TLR3-YFP was quantified for each lysate using rabbit anti-GFP polyclonal antibody (1 μg/ml, 50 μl/well; Invitrogen), followed by HRP-conjugated goat anti-rabbit IgG (1:5000, 50 μl/well; Jackson ImmunoResearch Laboratories). Bound bio-dsRNA and bound TLR3-YFP were detected using HRP substrate reagent (R&D Systems) and a FLUOstar OPTIMA plate reader (BMG Labtech). Duplicate wells were used for all samples. Data are presented as the mean ± S.D. of A450(dsRNA)/A450(TLR3-YFP). All TLR3 mutants were assayed in at least three independent experiments.
TLR3 Stimulation
TLR3 activation was followed using an NF-κB reporter system as described (11, 15). Briefly, HEK293 cells (2.5 × 104 in 100 μl/well) were plated in 96-well plates and cultured overnight. Cells were transfected using Lipofectamine 2000 with a mixture of pUNO-TLR3 (WT or mutant), pSV-β-galactosidase (12 ng), IgκB-firefly luciferase (6 ng), and empty vector (total DNA, 400 ng). At 16–18 h post-transfection, 540-bp dsRNA was added to a final concentration of 10 μg/ml. The β-galactosidase and luciferase activities were determined 6 h later. Each sample was measured in triplicate, and each mutant was assayed in a minimum of three separate experiments. Statistical comparisons between groups were made using analysis of variance and the Holm-Sidak method for multiple pairwise comparisons. p < 0.05 was considered to be significant. Data were analyzed using SigmaStat (SPSS, Inc.).
RESULTS
The C-terminal Dimerization Site Is Essential for TLR3 Signaling
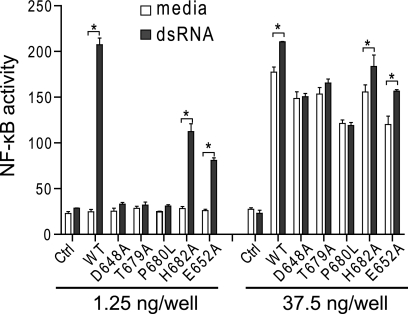
TLR3 dimerization is mediated by intermolecular contacts between LRR-CT domains. The site of dimerization is located on a 2-fold symmetry axis and contains two hydrogen bond pairs, Asp648/Thr679 and Glu652/His682, and a structural proline residue, Pro680 (Fig. 1C). Because it was not known if the dimerization interaction is necessary for signaling, we introduced a number of mutations designed to disrupt this interaction and tested their effects on the dsRNA-dependent activation of NF-κB. First, we found that all mutant proteins were expressed in transfected HEK293 cells (supplemental Fig. S2). We then tested their capacity to activate an NF-κB reporter plasmid in response to dsRNA in HEK293 cells. As shown in Fig. 2, three mutants, D648A, T679A, and P680L, were totally unresponsive to dsRNA, whereas H682A and E652A were partially active compared with WT TLR3. Cells transfected with high amounts of WT or mutant TLR3 were constitutively active, indicating that the mutants were still capable of signaling when forced to dimerize at high membrane densities. We conclude that the responsiveness of TLR3 to dsRNA requires an intact dimerization site. Whether this site also influences ligand binding is examined below.
FIGURE 2.
Mutations in the dimerization site render TLR3 nonresponsive to dsRNA. HEK293 cells were transfected with normal or high amounts of plasmid expressing WT or mutant TLR3 or empty vector (Ctrl) and then stimulated with 540-bp dsRNA and tested for NF-κB activation. Data are presented as the mean ± S.E. (n = 3). *, p < 0.001.
dsRNA Binding to TLR3
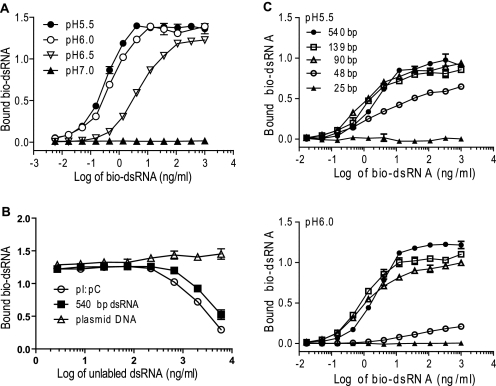
To address how amino acids in the TLR3 ECD contribute to the binding of dsRNA, we developed an ELISA that detects the binding of bio-dsRNA to immobilized TLR3 derived from transfected cell lysates (shown schematically in supplemental Fig. S1A). As shown in Fig. 3A, TLR3 bound bio-dsRNA at pH 5.5 and 6.0 and less at pH 6.5, and no binding was observed at pH 7.0. The binding of bio-dsRNA was saturable because it reached a plateau at higher dsRNA concentrations (Fig. 3A) and was inhibited by comparable concentrations of unlabeled 540-bp dsRNA (Fig. 3B). As expected, binding was also inhibited by poly(I)·poly(C), a high molecular weight dsRNA analog, but not by DNA, which does not bind to or activate TLR3 (1, 9). Moreover, a minimum length of ∼48 bp was required for binding, and the affinity of binding increased with dsRNA length (Fig. 3C), in agreement with previously reported surface plasmon resonance results (9). Because the ELISA detected a saturable binding site with the same specificity and pH dependence found previously for pure TLR3 ECD protein (9), we conclude that it provides a reliable semiquantitative method for measuring dsRNA binding to TLR3.
FIGURE 3.
Binding of dsRNA to TLR3 is saturable, specific, and depends upon pH and dsRNA length. A, serial dilutions of bio-dsRNA were added to immobilized WT TLR3-YFP at the indicated pH values. B, increasing concentrations of unlabeled 540-bp dsRNA, poly(I)·poly(C), or plasmid DNA were mixed with bio-dsRNA (0.5 μg/ml) and added to immobilized WT TLR3-YFP at pH 6.0. C, bio-dsRNA oligonucleotides of increasing lengths were tested for binding to immobilized WT TLR3-YFP at pH 5.5 and 6.0. Bound bio-dsRNA and TLR3-YFP were quantified as described under “Experimental Procedures.” At least three independent experiments were performed for each WT or mutant TLR3 protein. Data are presented as the mean ± S.D. from one representative experiment. Where error bars are not indicated, they are smaller than the symbol.
Binding Efficiency Increases with TLR3 Density
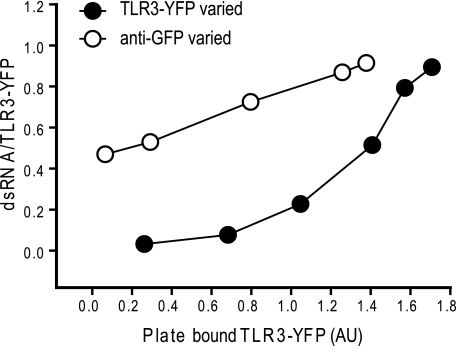
To determine whether multimerization of TLR3 is important for ligand binding, we measured the amount of dsRNA bound per TLR3 molecule at increasing levels of immobilized TLR3. We reasoned that if dsRNA binding required multiple TLR3 molecules, then no binding would occur until a critical density of TLR3 molecules was achieved. In the experiment shown in Fig. 4 (closed circles), increasing amounts of TLR3-YFP were added to immobilized anti-GFP mAb on the ELISA plate (supplemental Fig. S1B). A constant amount of bio-dsRNA was then added to each well, and binding was detected as described under “Experimental Procedures.” As seen, at low densities, TLR3 was unable to bind dsRNA; however, the dsRNA:TLR3 ratios increased sharply as the amount of immobilized TLR3 increased. This result strongly suggests that multiple TLR3 molecules are required to bind dsRNA. In a second experiment (Fig. 4, open circles), increasing amounts of anti-GFP mAb were immobilized and then treated with saturating concentrations of TLR3-YFP. In this case, because the anti-GFP mAb is divalent, we would expect TLR3-YFP molecules to bind to the plate in dimeric clusters (supplemental Fig. S1C). Interestingly, when bound to the plates in this way, even low densities of immobilized TLR3 were able to bind dsRNA, suggesting that two closely spaced TLR3 molecules are sufficient for ligand binding.
FIGURE 4.
dsRNA binding depends upon the density of immobilized TLR3-YFP. ●, ELISA plates were coated with anti-mouse Fc, followed by anti-GFP mAb (which cross-reacts with YFP) and increasing concentrations of cell lysate containing WT TLR3-YFP; ○, anti-mouse Fc-coated plates were incubated with increasing amounts of anti-GFP mAb, followed by a saturating concentration of WT TLR3-YFP. In both cases, the amount of bound bio-dsRNA/TLR3-YFP was determined as indicated under “Experimental Procedures.” See also supplemental Fig. S1 AU, absorbance units.
An Intact Dimerization Site Is Required for dsRNA Binding
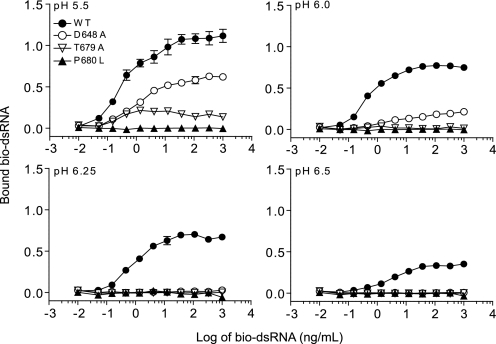
Because the data of Fig. 4 suggest that TLR3 oligomerization is essential for dsRNA binding, we asked whether the LRR-CT dimerization site is also essential for ligand binding. For these experiments, we tested mutants used in the experiment of Fig. 2 for dsRNA binding. As shown in Fig. 5, P680L failed to bind dsRNA at all pH values tested, whereas T679A and D648A bound weakly at pH 5.5 but did not bind dsRNA at higher pH. Pro680 appears to stabilize a prominent loop in the dimerization site that includes Thr679 (Fig. 1C) but does not itself interact with other amino acids. Interestingly, this Pro residue is conserved in all human TLR paralogs, but in contrast to TLR3, it is not required for activation of other TLRs.3 We conclude that the TLR3 C-terminal dimerization site is essential for dsRNA binding even though it does not interact directly with dsRNA (Fig. 1C).
FIGURE 5.
An intact dimerization site is required for dsRNA binding. Shown is the binding of bio-dsRNA to WT TLR3 and mutants D648A, T679A, and P680L in the dimerization site of TLR3 at pH 5.5, 6.0, 6.25, and 6.5. These mutants were inactive in signaling experiments (Fig. 2). A diagram of this site is shown in Fig. 1C.
Both dsRNA-binding Sites Are Required for Ligand Binding
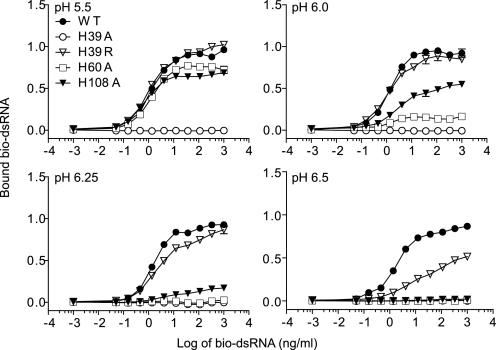
Previous studies (10–14) showed that mutations in the two dsRNA-binding site regions of TLR3 abrogate signaling, but it is not known how these mutations affect ligand binding. In the N-terminal dsRNA-binding site, three conserved histidine residues, His39, His60, and His108, form salt bridges with consecutive dsRNA phosphate groups (Fig. 1B). In signaling studies, the H39A and H60A mutants were inactive, whereas H108A still responded to dsRNA stimulation (supplemental Table S1). In agreement with the signaling results, H39A lost the capacity to bind dsRNA at all pH values tested (Fig. 6), indicating that the imidazole group of His39 is essential for forming a stable TLR3·dsRNA complex. However, the H60A mutant retained binding capacity similar to WT levels at pH 5.5 but showed diminished binding relative to WT TLR3 only at pH 6.0 and above. Because H60A bound dsRNA at pH 5.5, the inability of H60A-transfected reporter cells to respond to dsRNA suggests that the pH of the intracellular compartment in which TLR3 encounters dsRNA is higher than 5.5, consistent with binding occurring in early, but not late endosomes. Interestingly, Pirher et al. (12) found that H39R, a mutation that replaces a positively charged imidazole side chain with a positively charged guanidinium group, retained almost full signaling capacity. This was also reflected in binding capacity because the H39R mutant bound dsRNA equivalently to WT TLR3 at pH below 6.5 (Fig. 6).
FIGURE 6.
Relative importance of histidine residues in the N-terminal dsRNA-binding site. Shown is the binding of dsRNA to WT TLR3 and N-terminal dsRNA-binding site mutants H39A, H39R, H60A, and H108A at pH 5.5–6.5. A diagram of this site is shown in Fig. 1B.
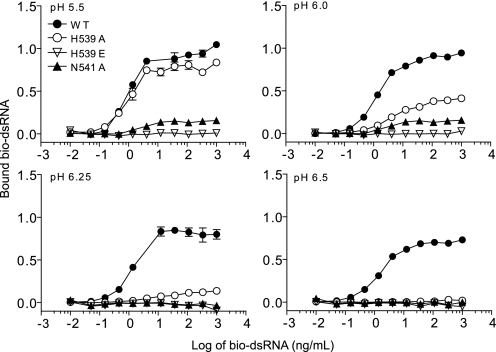
In the C-terminal dsRNA-binding site, two residues that are important for signaling, His539 and Asn541, contact a phosphate group and a 2′-hydroxyl, respectively, in dsRNA (Fig. 1A). H539E and N541A were inactive in signaling assays (11, 14), and as shown in Fig. 7, these mutants were unable to bind dsRNA at all pH values tested. A third mutant, H539A, retained partial signaling activity, which was also reflected in its binding capacity. At pH 5.5, the H539A mutant bound dsRNA equivalently to WT TLR3, but at higher pH, binding was markedly less compared with WT TLR3. We conclude that the amide group of Asn541 is essential for dsRNA binding and that the imidazole group of His539 contributes to binding but is not essential for signaling. The substitution of a negatively charged carboxyl group for a positively charged imidazole in H539E most probably leads to electrostatic repulsion of a phosphate group on the dsRNA and total loss of binding. The observation that Asn541 is essential for dsRNA binding and interacts with a 2′-hydroxyl provides one explanation for why TLR3 binds dsRNA but not dsDNA.
FIGURE 7.
The C-terminal dsRNA-binding site is required for ligand binding. Shown is the binding of dsRNA to C-terminal binding site mutants H539A, H539E, and N541A. A diagram of this site is shown in Fig. 1A.
DISCUSSION
Our previous studies showed that the TLR3 ECD binds as dimers to dsRNA and that dimerization is mediated by intermolecular interactions between the LRR-CT domains (9, 10). However, it was not known whether dimerization is essential for either ligand binding or signaling. Here, we have shown that both ligand binding and signaling require receptor dimerization. First, we found that if dimerization was prevented by immobilizing TLR3 at low density, TLR3 was unable to bind dsRNA (Fig. 4). However, TLR3 molecules that were immobilized in dimeric clusters at the same low overall density readily bound dsRNA. Second, mutation of several contacting residues in the dimer interface resulted in a loss of dsRNA binding and signaling by TLR3 (Figs. 2 and 5). These residues are far removed from dsRNA in the crystal structure and could not have interacted directly with the ligand, implying that the interaction responsible for dimerization is essential for ligand binding and signaling. From these results, we conclude that TLR3 dimerization is not only a result of ligand binding but also plays an essential role in forming the receptor·ligand complex.
Mutational analyses also showed that stable dsRNA binding requires functional C-terminal dsRNA-binding sites. These findings were consistent with previous results (Refs. 10–14; summarized in supplemental Table S1) in which several N and C-terminal dsRNA-binding site mutations resulted in loss of signaling capacity of TLR3 in reporter cell experiments. Whereas some of these mutations prevented dsRNA binding at all pH values tested (H39A and N541A), others (H60A and T648A) retained partial binding capacity, particularly at lower pH. We also identified other mutations (H108A and H539A) that showed decreased ligand binding but were nevertheless active in signaling experiments. Thus, results from signaling experiments do not strictly correlate with binding in all situations. The loss of binding and signaling capacities induced by a single mutation at one site in the TLR3 ECD implies that together the other two sites interact too weakly to form a stable complex. Because mutations at each of the three interacting sites render TLR3 inactive, we conclude that no two sites together provide sufficient energy for stable complex formation, but instead, a stable TLR3·dsRNA complex requires the cooperation of three weakly interacting, widely spaced sites. Previous studies (9) indicated that the binding of dsRNA to TLR3 is positively cooperative. This could best be explained if a TLR3 molecule first binds weakly to dsRNA by its N- and C-terminal dsRNA-binding sites and then forms a stable complex by interacting with a second molecule at its dimerization site perhaps by sliding or jumping along a dsRNA strand.
The requirement for the simultaneous interaction of the three TLR3 binding sites for stable complex formation provides several advantages for dsRNA recognition. First, the weak interaction of the two dsRNA-binding sites with dsRNA prevents the formation of stable complexes in which single TLR3 molecules are located far apart from each other on a long dsRNA strand and are therefore unable to trigger downstream signaling pathways. Stable binding occurs only when two molecules come together. In addition, because the homotypic dimerization interaction by itself is weak, TLR3 does not dimerize and signal in the absence of dsRNA, except at abnormally high membrane densities. Finally, short dsRNA segments, such as those present in self-tRNA, microRNA, and ribosomes, cannot activate TLR3 because dsRNA can form a stable complex with TLR3 only by interacting with both dsRNA-binding sites on both molecules of the dimer, which requires a minimum length of ∼45 bp.
The low affinity contributed by each site is likely due to the fact that the interactions involved in complex formation consist for the most part of hydrogen bonds or salt bridges (10), which are intrinsically weak in an aqueous environment. These types of interactions are quite distinct from those stabilizing the other known TLR·ligand complexes, LPS with TLR4·MD-2, and lipopeptides with TLR1·TLR2 or TLR6·TLR2 (16–18). In those complexes, fatty acids from the ligand cross-link TLRs by forming strong interactions with hydrophobic amino acid side chains in the TLR ECD or in MD-2, often in hydrophobic pockets or cavities. It is remarkable that although the contact residues in TLR3 are nonstructural, hydrophilic, and exposed to the medium, they are nevertheless strictly conserved in all known TLR3 orthologs (10). This suggests that the cooperation of three widely spaced, weak binding sites represents an ancient strategy used by most vertebrates for the detection of viral infection.
Supplementary Material
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health (NCI and NIDDK) and by a National Institutes of Health/Food and Drug Administration intramural biodefense award from NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Table S1, and additional references.
Y. Wang, unpublished data.
- ECD
- extracellular domain
- LRR
- leucine-rich repeat
- bio-dsRNA
- biotin-labeled 540-bp dsRNA
- TLR
- Toll-like receptor.
REFERENCES
- 1.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 2.Kumar H., Kawai T., Akira S. (2009) Biochem. J. 420, 1–16 [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O., Akira S. (2009) Immunol. Rev. 227, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz O., Diebold S. S., Chen M., Näslund T. I., Nolte M. A., Alexopoulou L., Azuma Y. T., Flavell R. A., Liljeström P., Reis e Sousa C. (2005) Nature 433, 887–892 [DOI] [PubMed] [Google Scholar]
- 5.Gruenberg J., Maxfield F. R. (1995) Curr. Opin. Cell Biol. 7, 552–563 [DOI] [PubMed] [Google Scholar]
- 6.Botos I., Liu L., Wang Y., Segal D. M., Davies D. R. (2009) Biochim. Biophys. Acta. 1789, 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell J. K., Botos I., Hall P. R., Askins J., Shiloach J., Segal D. M., Davies D. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10976–10980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe J., Kelker M. S., Wilson I. A. (2005) Science 309, 581–585 [DOI] [PubMed] [Google Scholar]
- 9.Leonard J. N., Ghirlando R., Askins J., Bell J. K., Margulies D. H., Davies D. R., Segal D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Botos I., Wang Y., Leonard J. N., Shiloach J., Segal D. M., Davies D. R. (2008) Science 320, 379–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell J. K., Askins J., Hall P. R., Davies D. R., Segal D. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8792–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirher N., Ivicak K., Pohar J., Bencina M., Jerala R. (2008) Nat. Struct. Mol. Biol. 15, 761–763 [DOI] [PubMed] [Google Scholar]
- 13.Fukuda K., Watanabe T., Tokisue T., Tsujita T., Nishikawa S., Hasegawa T., Seya T., Matsumoto M. (2008) J. Biol. Chem. 283, 22787–22794 [DOI] [PubMed] [Google Scholar]
- 14.Ranjith-Kumar C. T., Miller W., Xiong J., Russell W. K., Lamb R., Santos J., Duffy K. E., Cleveland L., Park M., Bhardwaj K., Wu Z., Russell D. H., Sarisky R. T., Mbow M. L., Kao C. C. (2007) J. Biol. Chem. 282, 7668–7678 [DOI] [PubMed] [Google Scholar]
- 15.Mullen G. E., Kennedy M. N., Visintin A., Mazzoni A., Leifer C. A., Davies D. R., Segal D. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 17.Kang J. Y., Nan X., Jin M. S., Youn S. J., Ryu Y. H., Mah S., Han S. H., Lee H., Paik S. G., Lee J. O. (2009) Immunity 31, 871–884 [DOI] [PubMed] [Google Scholar]
- 18.Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.