Abstract
Solid tumor development is frequently accompanied by energy-deficient conditions such as glucose deprivation and hypoxia. Follistatin (FST), a secretory protein originally identified from ovarian follicular fluid, has been suggested to be involved in tumor development. However, whether it plays a role in cancer cell survival under energy-deprived conditions remains elusive. In this study, we demonstrated that glucose deprivation markedly enhanced the expression and nucleolar localization of FST in HeLa cells. The nucleolar localization of FST relied on its nuclear localization signal (NLS) comprising the residues 64–87. Localization of FST to the nucleolus attenuated rRNA synthesis, a key process for cellular energy homeostasis and cell survival. Overexpression of FST delayed glucose deprivation-induced apoptosis, whereas down-regulation of FST exerted the opposite effect. These functions depended on the presence of an intact NLS because the NLS-deleted mutant of FST lost the rRNA inhibition effect and the cell protective effect. Altogether, we identified a novel nucleolar function of FST, which is of importance in the modulation of cancer cell survival in response to glucose deprivation.
Keywords: Cell Death, Gene Expression, Nucleolus, Ribosomal RNA (rRNA), RNA Polymerase I, Follistatin, Glucose Deprivation
Introduction
Solid tumors over a certain size are continuously exposed to glucose deficiency and hypoxia microenvironments because of the inadequate vascular supply. The cancer cells comprising these tumors thus have to limit energy expenditure to survive under such energy-deprived conditions (1, 2). An effective way is to reduce ribosome biogenesis, the most energy-consuming process in eukaryotic cells. As a matter of fact, the rate of cellular rRNA transcription is tightly regulated in response to metabolic changes (3–5).
Transcription of rRNA is catalyzed by polymerase I. Energy deprivation activates AMP-activated protein kinase, which then inactivates the polymerase I-associated transcription factor TIF-IA and thereby impairs rRNA synthesis (6). Energy depletion also triggers heterochromatin formation and rRNA gene silencing by activating the nucleolar remodeling complex (7) or the energy-dependent nucleolar silencing complex (eNoSC) (8). Inhibition of rRNA transcription by eNoSC suppresses energy expenditure and protects cells from energy deprivation-induced apoptosis (8). Meanwhile, increased production of H+ under hypoxia promotes the interactions between VHL and rRNA gene (rDNA) to reduce rRNA synthesis (9). Overall, cells repress rRNA transcription through multiple pathways to preserve energy and survive under energy-deprived conditions.
FST3 was originally identified from ovarian follicular fluid, with a function to suppress follicle-stimulating hormone secretion (10, 11). Later, FST was found to be expressed in a variety of tissues and organisms, participating in various processes such as cell growth, development, differentiation, and secretion (12). FST-deficient mice exhibit numerous phenotypes, including musculoskeletal and cutaneous abnormalities, and die within hours of birth because of respiratory failure (13). Recently, several reports have shown that FST regulates a variety of processes during tumor progression including angiogenesis (14), metastasis (15), and cell apoptosis (16). Although it is believed that FST exerts most of its functions through extracellularly inactivating TGF-β-like molecules (17–19), we have recently identified the nuclear localization ability of this protein (20), suggesting a possible nuclear mechanism.
In this study, we report that FST was up-regulated and translocated to the nucleoli in HeLa cells in response to glucose deprivation. Nucleolar accumulation of FST caused cells to reduce rRNA transcription, which in turn protected them from glucose deprivation-induced apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
HeLa human cervical carcinoma cells were obtained from ATCC and maintained in DMEM containing 25 mm glucose (Invitrogen) supplemented with 10% fetal bovine serum (Thermo Fisher) and penicillin-streptomycin mixed solution (Invitrogen). The cells were maintained at 37 °C in an atmosphere containing 5% CO2 and 100% humidity. To achieve glucose deprivation, the cells were washed with PBS and transferred to glucose-free DMEM (Invitrogen).
Protein Extraction and Fractionation
The whole cell lysate was extracted with radioimmune precipitation assay buffer (100 mm Tris at pH 8.0, 1% Triton X-100, 100 mm NaCl, 0.5 mm EDTA) with freshly added complete protease inhibitor mixture (Roche Applied Science) and cleared by centrifugation (13,200 × g, 30 min, 4 °C). To detect the subcellular distribution of FST, cytoplasmic and nuclear fractions were prepared according to the previously reported method (21) with modifications. Briefly, the cells were washed with PBS and resuspended in hypotonic buffer (10 mm HEPES, pH 8.0, 10 mm KCl, 3 mm MgCl2, 0.5 mm DTT, and protease inhibitors). The suspension was incubated on ice for 10 min before adding Triton X-100 to a final concentration of 0.3% to allow a sufficient breakage of the cytoplasmic membranes. After centrifugation at 500 × g for 5 min at 4 °C, the resultant supernatant was used as the cytoplasmic fraction. The nuclear pellet was washed twice with hypotonic buffer and reconstituted in radioimmune precipitation assay buffer. The resultant supernatant was used as the nuclear lysate. The proteins were qualified using a modified Bradford protocol (Bio-Rad) and applied to immunoblotting analysis as described previously (22).
Immunoblotting Analysis
Equal amounts of proteins were subjected to SDS-PAGE electrophoresis and transferred to nitrocellulose membrane (Whatman, Clifton, NJ). The membrane was blocked with 3% bone serum album in TBS-T buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.05% Tween 20), probed with antibodies targeting to FST (Santa Cruz Biotechnology, Santa Cruz, CA), α-tubulin (Cell Signaling Technology, Beverly, MA), Lamin B (Santa Cruz Biotechnology), or PARP (Cell Signaling Technology), incubated with horseradish-conjugated secondary antibodies, detected with the SuperSignal West Pico chemiluminescence substrate (Thermo Fisher Scientific), and finally exposed to an x-ray film.
Immunofluorescence Detection
Cells grown on glass coverslips were rinsed with PBS and fixed in 4% formaldehyde in PBS for 15 min. After rinsing twice with PBS, the cells were permeabilized in 0.2% Triton X-100 in PBS and blocked with goat serum for 1 h at room temperature. The cells were then incubated with monoclonal anti-FST monoclonal antibodies (R & D Systems, Minneapolis, MN) together with anti-nucleolin antibodies (Santa Cruz Biotechnology) for 1 h, stained with the appropriate FITC- or TRITC-conjugated secondary antibody in PBS for 1 h and mounted on microscope slides. Confocal microscopic images were obtained with a Zeiss confocal imaging system (Carl Zeiss MicroImaging, Thornwood, NY). The relative fluorescence intensities were quantified with Image J software.
Plasmid Construction
The construction of pECFP-FST, in which a FST gene lacking its signal sequence has been fused to the C terminus of the enhanced CFP gene, was described previously (20).
To construct CFP-tagged FST deletion mutants, the corresponding DNA domains (supplemental Table S1) were amplified by PCR from plasmid pFS288mycHis (kindly provided by Henry T. Keutmann, Massachusetts General Hospital, Boston, MA) using the primers listed in supplemental Table S2. PCR products were cloned into the pECFP-C1 vector to produce fusion proteins with the CFP in the N terminus.
To generate FST eukaryotic expression plasmid pcDNA-FST, full FST gene with its signal sequence was amplified by PCR from plasmid pFS288mycHis using primer set sigFST-F/sigFST-R (supplemental Table S2) and cloned into the EcoRI and XhoI sites of vector pcDNA3.1(+). To generate the plasmid pcDNA-FST/ΔNLS for eukaryotic expression of NLS-deleted FST, cDNAs encoding the N-terminal (residues −29 to 63) and C-terminal (residues 88–288) of the FST gene were amplified (supplemental Table S1) and cloned to the EcoRI and XhoI sites of vector pcDNA3.1(+). All of the primer sequences are listed in supplemental Table S2.
Chromatin Immunoprecipitation and Quantitative PCR (qPCR) Measurement
ChIP assays were performed using the ChIP assay kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, HeLa cells were cross-linked with 1% formaldehyde for 10 min at 37 °C. Cross-linking was stopped with 0.125 m glycine. The cells were collected and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.1, 1% SDS, 10 mm EDTA, and protease inhibitors). After sonication to yield DNA fragments of 300–1000 base pairs, the lysates were cleared by centrifugation, diluted 10-fold with ChIP dilution buffer (16.7 mm Tris-HCl, pH 8.1, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm NaCl, and protease inhibitors), and precleared with salmon sperm DNA/protein A-agarose at 4 °C for 1 h. For each immunoprecipitation assay, the lysates were incubated with 5 μg of anti-FST (Abcam, Cambridge, UK), anti-K4-dimethylated histone H3 (H3K4me2) (Thermo Fisher Scientific), anti-acetylated histone H3 (acetyl-H3) (Thermo Fisher Scientific), or control IgGs (Santa Cruz Biotechnology) overnight at 4 °C with rotation. The immunocomplexes were then collected with protein G-agarose slurry, eluted, and de-cross-linked at 65 °C. After RNase digestion and proteinase digestion, immunoprecipitated DNA was extracted. The purified DNA was amplified by real time PCR with the ABI7900 (Applied Biosystems, Foster City, CA) and SYBR GREEN PCR Master Mix (Applied Biosystems).
The primer sequences for rDNA were as reported (23): 5′-CCCGGGGGAGGTATATCTTT-3′ and 5′-CCAACCTCTCCGACGACA-3′ for H42.9; 5′-GGCGGTTTGAGTGAGACGAGA-3′ and 5′-ACGTGCGCTCACCGAGAGCAG-3′ for H1; 5′-AGTCGGGTTGCTTGGGAATGC-3′ and 5′-CCCTTACGGTACTTGTTGACT-3′ for H8; and 5′-GTTGACGTACAGGGTGGACTG-3′ and 5′-GGAAGTTGTCTTCACGCCTGA-3′ for H18. The primer sequences for the GAPDH gene were as follows: 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′.
RNA Interference
To knock down the endogenous FST, HeLa cells were transiently transfected with 10 nm of the chemically synthesized siRNAs targeting FST or with the negative control siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The cells were harvested or treated for further experiments 48 h after transfection. siRNA sequences used in the present study are designed as follows: FST siRNA1 (FSTi-1): forward, GAUCUAUUGGAUUAGCCUATT, and reverse, UAGGCUAAUCCAAUAGAUCTG; FST siRNA2 (FSTi-2): forward, GGUCCUGUACAAGACCGAATT, and reverse, UUCGGUCUUGUACAGGACCTG; and negative control siRNA: forward, UUCUCCGAACGUGUCACGUTT, and reverse, ACGUGACACGUUCGGAGAATT. siRNAs were synthesized by GenePhama (Shanghai, China).
RNA Purification and RT-qPCR Measurement
Total RNA was isolated with TRIzol reagent (Invitrogen) following the manufacturer's protocol. 0.5 μg of total RNA was reverse transcribed using random hexamers and a high capacity cDNA reverse transcription kit (Applied Biosystems). Real time quantitative PCR analysis was performed in 10-μl reactions using the ABI7900 (Applied Biosystems) and SYBR GREEN PCR Master Mix (Applied Biosystems). Primers 5′-CCTGCTGTTCTCTCGCGCGTCCGAG-3′ and 5′-GCGTCTCGTCTCGTCTCACT-3′ were used for PCR amplification of 5′ external transcribed spacer of the human pre-rRNA as described (23). Primers 5′-TTGCGTTACACCCTTTCTTG-3′ and 5′-CACCTTCACCGTTCCAGTTT-3′ were used for β-actin gene amplification. Primers 5′-GGGAACTGCTGGCTCC-3′ and 5′-TTTACAGGGGATGCAG-3′ were used for FST gene amplification. Expression of pre-rRNA and FST gene was normalized relative to β-actin endogenous control using the 2−ΔΔCT method (24).
Northern Hybridization
RNA was separated on the agarose/formaldehyde gel and transferred onto a nylon membrane. Hybridization was carried out using a DIG Northern starter kit (Roche Applied Science). The digoxigenin-labeled probes were synthesized by TaKaRa Biotechnology Company (Dalian, China). The probe sequence for 47 S rRNA is 5′-GGTCGCCAGAGGACAGCGTGTCAG-3′. The probe sequence for β-actin mRNA is 5′-AGGGATAGCACAGCCTGGATAGCAAC-3′.
Stable Transfection
The plasmid pcDNA-FST, pcDNA-FST/ΔNLS, or the null vector pcDNA3.1 was transfected into HeLa cells with Lipofectamine 2000 following the manufacturer's protocol (Invitrogen). The cells were then selected with 500 μg/ml of G-418. Cell clones were subsequently screened by immunoblotting analysis with anti-FST polyclonal antibody. FST- or FST/ΔNLS-expressing clones were then obtained for further experiments.
Assays for Apoptosis Detection
Both floating (dead) and attached cells were harvested and applied to different assays for apoptosis detection. The cells were incubated with 5 μl of annexin V-FITC (Biovision, Mountain View, CA) and 10 μl of PI for 10 min and analyzed by flow cytometry (Beckman Coulter, Brea, CA). Annexin V-FITC staining was detected in the FL1 channel, whereas PI staining was monitored in the FL3 channel. Alternatively, total proteins were extracted, and PARP cleavage was visualized by immunoblotting.
RESULTS
Glucose Deprivation Enhances FST Expression and Nucleolar Localization in HeLa Cells
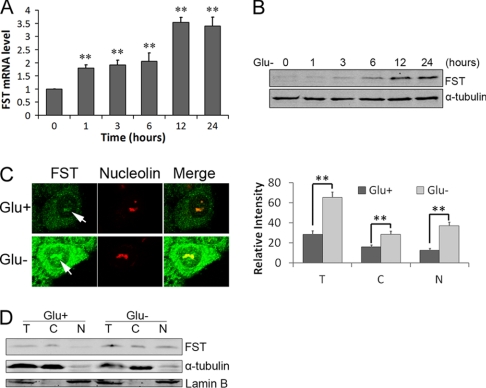
Because survival of cancer cells under energy deficiency conditions is an important aspect for tumor progression, it is of interest to study whether FST is involved in this process. Therefore, we first measured FST expression in HeLa cells when glucose in the medium was withdrawn. The data showed that the mRNA level of FST started to increase 1 h after glucose deprivation and reached the peak at 12 h (Fig. 1A). The protein level started to significantly increase 6 h after glucose removal (Fig. 1B).
FIGURE 1.
Glucose deprivation enhances FST expression and nucleolar localization. HeLa cells were incubated in glucose-free medium and harvested at the indicated time. A, the FST mRNA level was measured with real time qPCR. B, FST protein level was detected by immunoblotting. The RT-qPCR data are shown as arbitrary values after normalizing to β-actin mRNA level, and the data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus normal glucose controls. C, HeLa cells were incubated with (Glu+) or without (Glu−) glucose for 24 h and then stained with FST monoclonal antibodies. Nucleolin was used as the nucleolar marker. The arrows indicate the nucleolar FST (left panel). The relative fluorescence intensities in the entire cell (columns T), cytoplasm (columns C), and nucleus (columns N) were measured. The data shown are the means ± S.D. of ∼50 cells (right panel). **, p < 0.01. D, FST protein expressions in total (lanes T), cytoplasmic (lanes C), and nuclear (lanes N) fractions were analyzed by immunoblotting. Lamin B and α-tubulin were used as nuclear and cytoplasm protein marker, respectively.
Because overexpressed FST-CFP was reported to localize in the nuclei of HeLa cells (20), we therefore analyze the subcellular localization of the endogenous FST in response to glucose deprivation. HeLa cells cultured with or without glucose were stained with FST monoclonal antibodies using the indirect immunofluorescence method and observed under a confocal microscope. Surprisingly, the endogenous FST showed clear nucleolar localization in addition to its commonly reported cytoplasm distribution (21, 25). Glucose deprivation enhanced the FST expression, and a great fraction was in the nucleus (44% in normal conditions versus 57% under glucose deprivation; Fig. 1C). The nucleolar localization was confirmed by simultaneous staining of the nucleolar protein nucleolin (Fig. 1C). To exclude artifacts that might have occurred in the staining, nuclear and cytoplasmic proteins were fractioned and applied to immunoblotting analysis. The data showed that FST was detected in both the cytoplasm and the nucleus, whereas the nuclear FST increased when glucose was deprived (Fig. 1D). Taken together, our data revealed that FST is a nucleolar protein and that glucose deprivation promotes its expression and nucleolar localization.
Residues 64–87 Are Essential for the Nucleolar Localization of FST
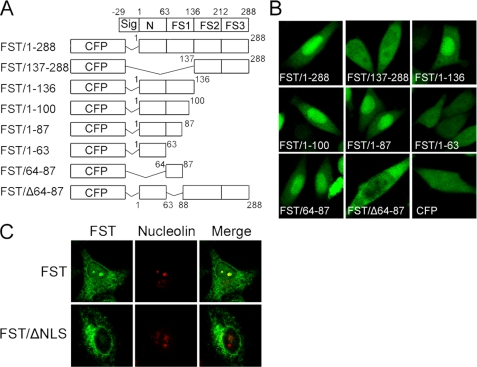
Nuclear import of a protein generally relies on the presence of a NLS (26). However, bioinformatics searching of FST sequence by the programs PROSITE (27) and PredictNLS (28) failed to detect any traditional NLSs in it. To map a potential NLS, a series of FST truncated fragments were fused to the C-terminal of CFP, and subcellular localizations of the fusion proteins were observed with a confocal microscope. Our results showed that FST mutants containing residues 64–87 accumulated in the nucleus, whereas FST mutants lacking these residues did not (Fig. 2, A and B). The data demonstrated that residues 64–87 are both essential and sufficient for FST nuclear localization. Sequence analysis revealed that it contained several basic amino acids (75KKCRMNKKNKPR86), which is in accordance with the common characteristic of a NLS (29).
FIGURE 2.
Residues 64–87 are essential for the nucleolar localization of FST. A, schematic illustration of the truncated FST fragments tagged to CFP. The numbers refer to the first and last residues of each construct in full-length FST sequence. B, HeLa cells were transfected with FST truncated plasmids as indicated, and the subcellular localization of FST mutants were examined by a confocal microscope. C, HeLa cells were transfected with plasmids expressing FST or FST/ΔNLS, and the subcellular localizations of FST and FST/ΔNLS were examined by immunofluorescence method with FST monoclonal antibody. Nucleolin was used as the nucleolar marker.
To further explore whether those residues could control the nucleolar localization of FST as well, we expressed full-length FST and NLS-deleted FST (FST/ΔNLS) in HeLa cells and detected their localizations using the immunofluorescence method. The data showed that the full-length FST could accumulate in the nucleolus but the NLS-deleted FST could not (Fig. 2C), demonstrating that residues 64–87 in FST indeed control its nucleolar localization. It should be noted that full-length FST was mainly localized in the nucleolus (Fig. 2C), whereas the signal peptide-deleted FST-CFP was diffused in the nucleus (Fig. 2B, FST/1–288), suggesting that the signal peptide might play a role in controlling FST nucleolar translocation, or the CFP might influence the localization.
FST Binds to rDNA Locus and Inhibits rRNA Synthesis
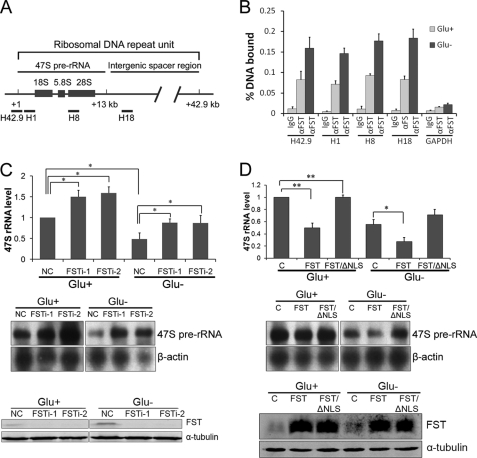
Ribosomal RNA genes (rDNA) reside in the nucleoli, and nucleolar localization of FST suggests that FST might bind to rDNA. To test this possibility, we performed ChIP assays in HeLa cells with anti-FST antibody. DNA enrichment was measured by real time PCR using sets of primer pairs spanning the rRNA gene promoter (H42.9), the 47 S rRNA coding region (H1), the 28 S rRNA coding region (H8), or the intergenic spacer region (H18), respectively (Fig. 3A). The data revealed that endogenous FST was bound throughout the rDNA loci, and glucose deprivation significantly increased the binding of FST to rDNA (Fig. 3B).
FIGURE 3.
FST binds to rDNA and inhibits 47 S pre-rRNA synthesis. A, schematic illustration of a ribosomal DNA repeat unit and locations of the primers used in ChIP-qPCR experiment. B, HeLa cells were cultured with (Glu+) or without (Glu−) glucose for 24 h and applied to ChIP experiments. The data shown are the means ± S.D. of three independent experiments. C, HeLa cells transfected with siRNAs targeting FST were cultured with (Glu+) or without (Glu−) glucose for 24 h. Total RNA was then isolated, and 47 S pre-rRNA level was analyzed by RT-qPCR (top panel) and Northern blot (middle panel). The RT-qPCR data are the means ± S.D. of three independent experiments. The asterisk indicates p < 0.05. Reduced FST expression by siRNAs was detected by immunoblotting analysis (bottom panel). D, HeLa cells were transiently transfected with a null vector pcDNA3.1 (column C) or constructs encoding FST or FST/ΔNLS, as indicated and then cultured with (Glu+) glucose or without (Glu−) glucose for 24 h. The 47 S pre-rRNA level was determined by RT-qPCR (top panel) and Northern blot (middle panel). The RT-qPCR data are the means ± S.D. of three independent experiments. **, p < 0.01; *, p < 0.05. The expression of FST by siRNAs was detected by immunoblotting analysis (bottom panel).
Association of FST with rDNA clusters prompted us to test whether FST could regulate rRNA transcription. For this purpose, the levels of 47 S pre-rRNA were measured by both RT quantitative PCR (RT-qPCR) and Northern blot when FST expression was down-regulated or overexpressed. The silencing of FST with siRNAs (FSTi-1 and FSTi-2) resulted in an increased expression of pre-rRNA compared with the control siRNA under normal glucose conditions (Fig. 3C), suggesting that FST is a repressor of rRNA transcription. Glucose deprivation decreased pre-rRNA expression, and knockdown of FST under glucose-deprived conditions also increased the pre-rRNA level (Fig. 3C). However, the pre-rRNA level was lower than the one in FST down-regulated but normal glucose-cultured cells, suggesting that other mediators might participate in glucose deprivation-repressed rRNA synthesis as well (Fig. 3C). Consistently, cells overexpressing FST displayed a marked decrease in pre-rRNA synthesis compared with null vector-transfected cells under both normal and glucose-deprived conditions (Fig. 3D). To investigate whether FST-mediated rRNA repression relies on its nucleolar localization, pre-rRNA levels in cells expressing NLS-deleted FST (FST/ΔNLS) were also measured. The data revealed that the loss of NLS prevented FST from repressing rRNA synthesis (Fig. 3D). Taken together, our data demonstrated that nucleolar FST directly binds to rDNA and inhibits pre-rRNA synthesis.
FST Affects Histone Modifications at rDNA Locus
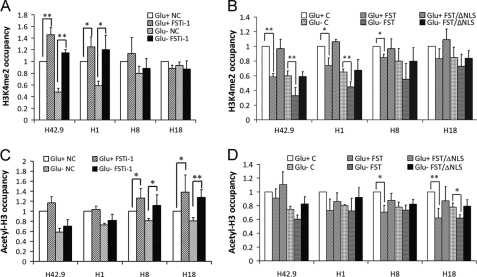
Epigenetic modifications of histones play key roles in regulating rRNA transcription (30, 31). To explore whether the nucleolar FST attenuates rRNA synthesis through affecting histone modifications, we employed ChIP assays to analyze the H3K4me2 and acetyl-H3 status when FST was down-regulated. The data showed that knockdown of FST with FSTi-1 significantly increased the H3K4me2 level at the rDNA region, especially near the transcription start site (H42.9; Fig. 4A), and the acetyl-H3 level at the intergenic spacer region (H18; Fig. 4B). The same histone modification pattern was observed under glucose deprivation (Fig. 4, A and B).
FIGURE 4.
FST influences histone modifications at rDNA region. HeLa cells transfected with siRNA targeting FST (FSTi-1) or control siRNA (NC) were cultured with (Glu+) or without (Glu−) glucose for 24 h. ChIP analysis was performed with antibodies against H3K4me2 (A) or acetyl-H3 (B) and analyzed by qPCR. Cell lines stably expressing FST, FST/ΔNLS, or control vector were cultured with (Glu+) or without (Glu−) glucose for 24 h. ChIP analysis was then performed with antibodies against H3K4me2 (C) or acetyl-H3 (D) and analyzed by qPCR. The occupancies of H3K4me2 and acetyl-H3 at rDNA regions were normalized to GAPDH. The values are the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01.
To further confirm the above results, we stably expressed FST in HeLa cells. Real time PCR confirmed that nucleolar accumulation of FST resulted in a reduction of pre-rRNA synthesis (supplemental Fig. S1). ChIP analysis revealed that overexpressed FST decreased the H3K4me2 and acetyl-H3 levels at the rDNA region under both normal and glucose-deprived conditions (Fig. 4, C and D). The NLS-deleted mutant served as non-nucleolar control and had no effect on these modifications. Because both H3K4me2 and acetyl-H3 are recognized as active gene markers (32, 33), our results suggested that nucleolar FST might change the ratio of euchromatin to heterochromatin and then decrease the rRNA transcription.
FST Protects Cells from Glucose Deprivation-induced Apoptosis
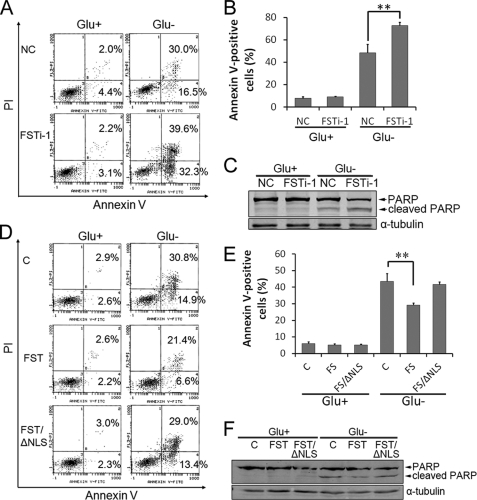
Apoptosis is a common cellular response to metabolic stress in mammalian cells. Cellular energy reservation has been reported to protect cells from glucose deprivation-induced apoptosis (34, 35), and down-regulating rRNA transcription is one of the key mechanisms (8). Because FST repressed rRNA transcription and its expression was up-regulated in response to glucose deprivation, we suspected that FST has an anti-apoptotic role. To test this hypothesis, the expression of FST was down-regulated with the siRNA FSTi-1, and apoptosis was determined by an annexin V-FITC/PI staining method. The fractions of annexin V-positive cells were low in both groups (6.2 ± 0.4 and 5.9 ± 0.6%, respectively) under normal glucose conditions. Glucose deprivation markedly increased cell apoptosis in control siRNA-transfected HeLa cells (48.6 ± 7.5%), and elimination of FST enhanced the apoptotic tendency to 72.9 ± 2.9% (p < 0.01 compared with control) (Fig. 5, A and B), indicating that FST has a protective role on apoptosis. The cleavage of PARP is recognized as an apoptosis marker. Our data revealed that PARP was cleaved after glucose deprivation and that down-regulation of FST increased PARP cleavage (Fig. 5C).
FIGURE 5.
FST protects cells from glucose deprivation-induced apoptosis. A, HeLa cells were transfected with siRNAs targeting FST gene or control siRNAs and then grew with glucose (Glu+) or without glucose (Glu−) for 48 h. Apoptotic cells were monitored by annexin V-FITC/PI staining and flow cytometry analysis. The right lower quadrant of each plot shows early apoptotic cells, whereas the right upper quadrant shows late apoptotic cells. Each experiment was performed in triplicate, and similar results were obtained each time. B, quantification of apoptotic cells by flow cytometry analysis. The values are the means ± S.D. **, p < 0.01 versus control siRNA-transfected control. C, PARP cleavage was detected by immunoblotting. D, HeLa cells stably expressing FST, FST/ΔNLS, or control vector were cultured with glucose (Glu+) or without glucose (Glu−) for 48 h. Apoptotic cells were monitored by annexin V-FITC/PI staining and flow cytometry analysis. E, quantification of apoptotic cells by flow cytometry analysis. The values are the means ± S.D. **, p < 0.01 versus null vector-transfected control. F, PARP cleavage was detected by immunoblotting.
To further confirm the protective effect of FST on glucose deprivation-induced apoptosis and to determine the role of nucleolar FST in this process, the apoptotic rates of HeLa cells stably transfected with FST, FST/ΔNLS, or null vector were determined. The apoptotic tendency induced by glucose deprivation was significantly reduced in FST-expressing cells compared with control (29.2 ± 1.2% versus 43.5 ± 4.7%, p < 0.01), but not in FST/ΔNLS-expressing cells (41.6 ± 1.7%) (Fig. 5, D and E), indicating that the protective effect of FST relied on its nucleolar localization. PARP cleavage detection also revealed a consistent result (Fig. 5F).
Meanwhile, we performed a TUNEL assay to determine cell apoptosis and found that overexpression of FST inhibited glucose deprivation-induced apoptosis (20.5% versus 42.5%), whereas knockdown of FST promoted glucose deprivation-induced apoptosis (61.1% versus 41.1%), further confirming the protective effect of FST (data not shown).
DISCUSSION
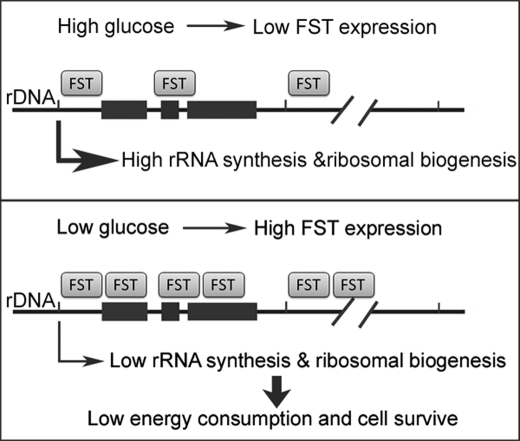
FST was traditionally recognized as a secretory protein and has been reported to exert its biological functions through extracellularly inactivating activin (36), myostatin (18), and bone morphogenic proteins (19). However, nuclear localization of FST has been observed in epithelial cells of breast tissue (37) and HeLa cells (20), suggesting a possible nuclear function of FST. In this study, we provide experimental evidence to support a nucleolar function of FST-inhibiting rRNA synthesis and promoting cancer cell survival. The major findings of our study include: 1) glucose deprivation significantly increases endogenous expression and nucleolar accumulation of FST; 2) nucleolar translocation of FST is dependent on its NLS comprised of residues 64–87; 3) the nucleolar FST negatively regulates rRNA synthesis; and 4) the nucleolar FST inhibits glucose deprivation-induced apoptosis. These findings demonstrate that repression of rRNA synthesis by the nucleolar FST plays a vital role in inhibiting glucose deprivation-induced cell apoptosis, as illustrated in Fig. 6.
FIGURE 6.
A schematic model for FST-repressed rRNA synthesis in preserving cell survival under glucose deprivation condition. Glucose deprivation promotes FST expression and nucleolar accumulation. Nucleolar FST binds to rDNA and represses rRNA synthesis to lower energy consumption in cancer cells, thus promoting cancer cell survival under glucose-deprived conditions.
The regulation of rRNA transcription is complicated and not restricted to the promoter region. One human rDNA transcription unit contains a pre-rRNA coding sequence separated by a long intergenic spacer. Regulatory elements, including gene promoters, spacer promoters, repetitive enhancer elements, and transcription terminators, are located in the long intergenic spacer (3). Many rRNA transcription regulators have been reported to be associated with the entire rDNA region. The upstream binding factor binds throughout the long intergenic spacer, and the transcribed region of rDNA repeats with low sequence specificity of DNA binding (38) and plays an important structural and functional role in the formation of active nucleolar organizer regions (39). c-Myc was reported to bind to the E-box sequence within the promoter region, the coding region as well as the long intergenic spacer region of rDNA. The binding of c-Myc with rDNA influences polymerase I machinery formation and histone acetylation at rDNA (23). Histone-modifying proteins such as the eNoSC (8) and the plant homeo domain finger protein 8 (40) were also reported to be associated with the entire rDNA. Here we found that FST could also bind throughout the rDNA loci and function as an rRNA transcription repressor. Further studies are needed to determine how FST discriminates rDNA from the remainder of the genome and whether there are conserved DNA elements within FST-binding regions.
Ribosomal RNA genes are present in hundreds of copies in the genome, and its transcription level could be controlled by regulating the transcription rate per gene or by varying the number of active genes epigenetically (30, 31). A key player in the epigenetic state establishment at the rDNA region is TTF-I. TTF-I binds to the promoter-proximal terminator T0 and recruits the nucleolar remodeling complex to the rDNA promoter. The nucleolar remodeling complex is composed of two subunits: the ATPase SNF2h and TTF-I-interacting protein 5. TTF-I-interacting protein 5 recruits histone-modifying enzymes to induce histone methylations at H3K9, and H3K27, histone demethylations at H3K4, as well as histone deacetylations. The ATPase SNF2h may senses the signals of heterochromatic histone modifications and induces rDNA silencing (30). We found that the euchromatin marker H3K4me2 levels were reduced at the rDNA promoter region in FST down-regulated cells (Fig. 4B), suggesting that FST represses rRNA levels partially through reducing the number of active rRNA genes. Further studies on the relationships between FST and the nucleolar remodeling complex and other histone-modifying enzymes will improve our knowledge on the mechanisms of FST-regulated rRNA transcription.
The synthesis of rRNA is tightly tuned to match environmental nutrition conditions. Because of insufficient nutrition supply, cells thus have to down-regulate rRNA synthesis to reduce energy utilization and make a survival. Our data demonstrated that up-regulation of FST expression is one of the mechanisms for cancer cells to restrict rRNA synthesis under glucose-deprived conditions (Fig. 3C) and then protect cells from apoptosis (Fig. 5). However, the data also showed that FST down-regulated cells express lower levels of pre-rRNA under glucose-deprived conditions than under normal conditions (Fig. 3C), suggesting that other mediators were synchronously involved in glucose deprivation-repressed rRNA synthesis. Recently, eNoSC (8) has been reported to induce rDNA silencing on glucose deprivation. Therefore, FST might work in concert with eNoSC to achieve rRNA gene silencing and thus low energy consumption.
Because the increased resistance of cancer cells to glucose deficiency contributes positively to tumor progression (2), it is reasonable to speculate that FST could promote tumor development. As a matter of fact, FST has been implied to facilitate tumorigenesis. Up-regulated expression of FST was observed in experimentally induced rodent liver tumors (41), human hepatocellular carcinomas (41), and mouse gastric tumors (42). It was also reported that the deficiency of the tumor suppressor gene parkin caused the up-regulation of endogenous FST and rendered hepatocytes resistant to apoptosis in a FST-dependent manner, which resulted in hepatic tumor development (16). However, it should be noted that an inhibitory role of FST in tumor development has also been reported (14, 15). Hence, FST might play different roles in different types of tumors and in different stages during tumor progression.
In summary, FST has an anti-apoptotic effect in glucose deprivation condition through the inhibition of rRNA and ribosome biogenesis. Nucleolar translocation of FST is required to exert the effect. This activity of FST represents a novel molecular mechanism of tumor cell survival under conditions of glucose deprivation and may be a possible target for tumor therapy.
Supplementary Material
Acknowledgments
We thank Wei Liu (Department of Biochemistry, Zhejiang University School of Medicine) and Yuehai Ke (Department of Pathology, Zhejiang University School of Medicine) for helpful discussions and Min Zhao (Department of Dermatology, University of California at Davis) and Larenda Mielke (English Language Programs, Washington University) for critical reading of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 30171035, 30770470, and 30900235 and Zhejiang Provincial Natural Science Foundation of China Grant Y2090095.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
- FST
- follistatin
- NLS
- nuclear localization signal
- CFP
- cyan fluorescent protein
- H3K4me2
- K4-dimethylated histone H3
- acetyl-H3
- acetylated histone H3
- eNoSC
- energy-dependent nucleolar silencing complex
- PARP
- poly(ADP-ribose) polymerase
- TRITC
- tetramethylrhodamine isothiocyanate
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative PCR.
REFERENCES
- 1.Dewhirst M. W., Cao Y., Moeller B. (2008) Nat. Rev. Cancer 8, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grummt I. (2003) Genes Dev. 17, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 4.Russell J., Zomerdijk J. C. (2005) Trends Biochem. Sci. 30, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. (2007) Cell Mol. Life Sci. 64, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoppe S., Bierhoff H., Cado I., Weber A., Tiebe M., Grummt I., Voit R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17781–17786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Schmitz K. M., Mayer C., Yuan X., Akhtar A., Grummt I. (2009) Nat. Cell Biol. 11, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 8.Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., Fukamizu A., Kimura K., Shimizu T., Yanagisawa J. (2008) Cell 133, 627–639 [DOI] [PubMed] [Google Scholar]
- 9.Mekhail K., Rivero-Lopez L., Khacho M., Lee S. (2006) Cell Cycle 5, 2401–2413 [DOI] [PubMed] [Google Scholar]
- 10.Robertson D. M., Klein R., de Vos F. L., McLachlan R. I., Wettenhall R. E., Hearn M. T., Burger H. G., de Kretser D. M. (1987) Biochem. Biophys. Res. Commun. 149, 744–749 [DOI] [PubMed] [Google Scholar]
- 11.Ueno N., Ling N., Ying S. Y., Esch F., Shimasaki S., Guillemin R. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8282–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmati-Brivanlou A., Kelly O. G., Melton D. A. (1994) Cell 77, 283–295 [DOI] [PubMed] [Google Scholar]
- 13.Matzuk M. M., Lu N., Vogel H., Sellheyer K., Roop D. R., Bradley A. (1995) Nature 374, 360–363 [DOI] [PubMed] [Google Scholar]
- 14.Krneta J., Kroll J., Alves F., Prahst C., Sananbenesi F., Dullin C., Kimmina S., Phillips D. J., Augustin H. G. (2006) Cancer Res. 66, 5686–5695 [DOI] [PubMed] [Google Scholar]
- 15.Ogino H., Yano S., Kakiuchi S., Muguruma H., Ikuta K., Hanibuchi M., Uehara H., Tsuchida K., Sugino H., Sone S. (2008) Clin. Cancer Res. 14, 660–667 [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara M., Marusawa H., Wang H. Q., Iwai A., Ikeuchi K., Imai Y., Kataoka A., Nukina N., Takahashi R., Chiba T. (2008) Oncogene 27, 6002–6011 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T., Takio K., Eto Y., Shibai H., Titani K., Sugino H. (1990) Science 247, 836–838 [DOI] [PubMed] [Google Scholar]
- 18.Amthor H., Nicholas G., McKinnell I., Kemp C. F., Sharma M., Kambadur R., Patel K. (2004) Dev. Biol. 270, 19–30 [DOI] [PubMed] [Google Scholar]
- 19.Glister C., Kemp C. F., Knight P. G. (2004) Reproduction 127, 239–254 [DOI] [PubMed] [Google Scholar]
- 20.Gao X., Hu H., Zhu J., Xu Z. (2007) FEBS Lett. 581, 5505–5510 [DOI] [PubMed] [Google Scholar]
- 21.Saito S., Sidis Y., Mukherjee A., Xia Y., Schneyer A. (2005) Endocrinology 146, 5052–5062 [DOI] [PubMed] [Google Scholar]
- 22.Hu H., Gao X., Sun Y., Zhou J., Yang M., Xu Z. (2005) Biochem. Biophys. Res. Commun. 329, 661–667 [DOI] [PubMed] [Google Scholar]
- 23.Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005) Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 24.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 25.Tortoriello D. V., Sidis Y., Holtzman D. A., Holmes W. E., Schneyer A. L. (2001) Endocrinology 142, 3426–3434 [DOI] [PubMed] [Google Scholar]
- 26.Jans D. A., Hassan G. (1998) Bioessays 20, 400–411 [DOI] [PubMed] [Google Scholar]
- 27.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C. J., Hofmann K., Bairoch A. (2002) Nucleic Acids Res. 30, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cokol M., Nair R., Rost B. (2000) EMBO Rep. 1, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. (2003) J. Biol. Chem. 278, 27981–27987 [DOI] [PubMed] [Google Scholar]
- 30.McStay B., Grummt I. (2008) Annu. Rev. Cell Dev. Biol. 24, 131–157 [DOI] [PubMed] [Google Scholar]
- 31.Grummt I., Ladurner A. G. (2008) Cell 133, 577–580 [DOI] [PubMed] [Google Scholar]
- 32.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 33.Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 34.Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 35.Shaw R. J., Kosmatka M., Bardeesy N., Hurley R. L., Witters L. A., DePinho R. A., Cantley L. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips D. J., de Kretser D. M. (1998) Front. Neuroendocrinol. 19, 287–322 [DOI] [PubMed] [Google Scholar]
- 37.Bloise E., Couto H. L., Massai L., Ciarmela P., Mencarelli M., Borges L. E., Muscettola M., Grasso G., Amaral V. F., Cassali G. D., Petraglia F., Reis F. M. (2009) BMC Cancer 9, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan A. C., Sullivan G. J., McStay B. (2002) Mol. Cell. Biol. 22, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mais C., Wright J. E., Prieto J. L., Raggett S. L., McStay B. (2005) Genes Dev. 19, 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. (2010) Nat. Struct. Mol. Biol. 17, 445–450 [DOI] [PubMed] [Google Scholar]
- 41.Grusch M., Drucker C., Peter-Vörösmarty B., Erlach N., Lackner A., Losert A., Macheiner D., Schneider W. J., Hermann M., Groome N. P., Parzefall W., Berger W., Grasl-Kraupp B., Schulte-Hermann R. (2006) J. Hepatol. 45, 673–680 [DOI] [PubMed] [Google Scholar]
- 42.Kang W., Saqui-Salces M., Zavros Y., Merchant J. L. (2008) Biochem. Biophys. Res. Commun. 376, 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.