Abstract
Of the 54 human keratins, five members have, at present, only been characterized at the gene level. In this study we have investigated the expression patterns of keratin K80, whose gene is located at the centromeric end of the type II keratin gene domain. K80 possesses a number of highly unusual properties. Structurally, it is distinctly closer to type II hair keratins than to type II epithelial keratins. Nonetheless, it is found in virtually all types of epithelia (stratified keratinizing/non-keratinizing, hard-keratinizing, as well as non-stratified tissues, and cell cultures thereof). This conspicuously broad expression range implies an unprecedented in vivo promiscuity of K80, which involves more than 20 different type I partners for intermediate filament (IF) formation. Throughout, K80 expression is related to advanced tissue or cell differentiation. However, instead of being part of the cytoplasmic IF network, K80 containing IFs are located at the cell margins close to the desmosomal plaques, where they are tightly interlaced with the cytoplasmic IF bundles abutting there. In contrast, in cells entering terminal differentiation, K80 adopts the “conventional” cytoplasmic distribution. In evolutionary terms, K80 is one of the oldest keratins, demonstrable down to fish. In addition, KRT80 mRNA is subject to alternative splicing. Besides K80, we describe a smaller but fully functional splice variant K80.1, which arose only during mammalian evolution. Remarkably, unlike the widely expressed K80, the expression of K80.1 is restricted to soft and hard keratinizing epithelial structures of the hair follicle and the filiform tongue papilla.
Keywords: Cytoskeleton, Differentiation, Gene Expression, Intermediate filaments, Skin, Hair
Introduction
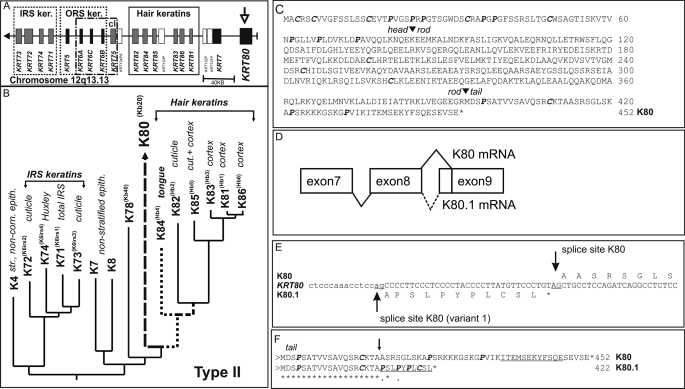
At present, of the 54 human keratins, only five members, i.e. the type I keratins K23, K24, and the type II keratins K78, K79, and K80 (1–3), have been characterized essentially at the gene/cDNA level, whereas detailed studies on the expression of the respective mRNAs and their proteins in human epithelia are still missing. To close this gap, we set out to investigate the expression characteristics of the type II keratin K80 (previously designated Kb20 (4)). Earlier bioinformatic analyses have shown that the KRT80 gene is located at the centromeric end of the human type II keratin gene domain on chromosome 12q13.13 (Fig. 1A). The isolated K80 cDNA encodes a 452-amino acid protein with a calculated molecular mass of 50.5 kDa and an isoelectric point of 5.47 (2).
FIGURE 1.
Analysis of the human K80 gene. A, localization of at the centromeric end of the type II keratin gene domain on chromosome 12q13.13. B, partial phylogenetic tree analysis of the α-helical rod domains of human type II keratins. C, amino acid sequence of K80. Arrowheads mark the beginning and end of the α-helical rod domain. Cysteine and proline residues in the head and tail domains are given in bold. D, schematic presentation of alternative splicing of exons 8 and 9. E, nucleotide sequence of the 5′-end of exon 9. The acceptor splice sites are underlined. An asterisk denotes an in-frame stop codon of the alternative variant. F, amino acid sequence alignment of K80 and K80.1 C-terminal ends. Underlined, peptide sequences used for specific antibody production; arrow, splice site-generated unique C-terminal peptide sequence of K80.1. Accession numbers of sequences used are given in bold under supplemental Table S1.
Remarkably, α-helical rod domain-based evolutionary analyses of the 26 type II keratins revealed a conspicuously long branch length for K80, thus indicating a relatively low homology of its α-helix to those of the remaining type II epithelial keratins, whereas its sequence affinity to type II hair keratins was relatively high (Fig. 1B, and Ref. 2). This was also reflected in the non-α-helical K80 end domains that contained a relatively high number of proline and cysteine residues along with the complete absence of GGG or GGX repeats (2), typically found in the head and tail domains of most type II epithelial keratins (Fig. 1C, bold italics).
At present, very few data exist regarding the expression profile of K80 in human epithelia. Northern blot analyses indicated an extremely weak expression of the K80 mRNA in tongue epithelium, whereas in scalp extracts, signals were not detectable, despite the fact that the complete cDNA was originally isolated from a scalp cDNA library (2). Moreover, RT-PCR analyses suggested the presence of K80 transcripts in corneal epithelium, however, the authors failed to demonstrate the respective protein (5). In contrast, we were recently able to localize the K80 protein in the upper region of the medulla of beard hairs (6). Although very preliminary, these fragmentary data insinuated rather unusual expression properties of the keratin.
The high diversity of keratins appears to have evolved by gene duplication and sequence diversification of the duplication products (4). Interestingly, another mechanism that generates multiple related gene products, namely alternative mRNA splicing, has not been reported for keratins so far. However, in the case of K80, the sequence databases UniProtKB/Swiss-Prot and NCBI contained cDNAs corresponding to alternatively spliced mRNAs that may encode functional K80 protein variants differing in their carboxyl termini. (For accession numbers see, supplemental Table S1.) The large protein variant corresponded to that described by Hesse et al. (4) and Rogers et al. (2). The smaller variant encompassed only 422 amino acids and had a calculated molecular mass of 47.2 kDa and an isoelectric point of 5.08. The striking peculiarities of the available K80 expression data as well as the prospect of functional alternative splicing of the KRT80 mRNA prompted us to perform an evolutionary analysis of the KRT80 gene along with a comprehensive expression study in epithelia with a special emphasis on the roles of the two alternative gene products.
EXPERIMENTAL PROCEDURES
Tissues and Cultured Cells
Human tissues were obtained either during surgery for medical reasons or from cadavers during pathological investigations (National Tissue Collection, University of Heidelberg, Germany) under institutional approval and included adherence to the Declaration of Helsinki. Immediately after excision, fresh tissues were snap-frozen in liquid nitrogen pre-cooled isopentane and stored at −70 °C (for details, see Ref. 7). Analyzed skin from various body sites (i.e. scalp, forearm, footsole), glandular tissues (e.g. eccrine sweat, mammary, and submandibular salivary glands), and additional epithelial tissues (dorsal tongue, gingiva, palate, esophagus, vagina, cervix, stomach, colon, duodenum, and bovine liver) as well as plucked beard hairs, obtained from volunteers in the laboratory, were used. Also investigated were primary cultures of normal human epidermal keratinocytes and HaCaT2 (immortalized human keratinocytes) cells grown confluently and overconfluently with focal onset of stratification under high calcium conditions, as well as polar epithelial cells of cell lines PLC (liver carcinoma, ATCC CRL-8024), MCF-7 (mammary carcinoma; ATTC, HTB-22), and CaCo2 (colon carcinoma; ATTC, HTB-37).
Primary Antibodies/Antisera
Antisera against the human keratin K80 and its splice variant K80.1 were raised in the laboratory by injection of synthetic peptides derived from the respective carboxyl termini of the variants into two to three series of two guinea pigs each: K80 (IKITEMSEKYFSQE-C; lab designation K27.2; indirect immunofluorescence (IIF) 1:500, Western blot 1:5000), and K80.1 (TAPSLPYPLCSL; lab designation K80b; IIF 1:1000; Western blot 1:5000). The peptides are underlined in Fig. 1F. Endogenous cysteine residues and those added at the carboxyl-terminal end (italics) were used for coupling to keyhole limpet protein (Peptide Specialty Laboratories, Heidelberg, Germany). The specificity of the two antisera was verified by (i) dot blot detection of the two respective antigen peptides used for immunization and those of unrelated proteins; (ii) testing the appropriate antiserum concentration by dilution until the disappearance of the respective staining in IIF and Western blotting; (iii) Western blotting analysis of various tissues to check molecular weight and occurrence of additional bands; (iv) comparison of tissue localization of the gene products by two independent methods: in situ hybridization (mRNA) and IIF (protein); and (v) absorption of the antisera with their antigens used for immunization in IIF and Western blotting. Mouse monoclonal antibodies against keratin K10 (DEK10; DAKO, IIF 1:100), K6 (KA12, 1:10, Progen), K2 (Ks2.342.7.1, IIF, undiluted, Progen), K31 (clone LH-Tric1, 1:50) and K32 (clone LH-Tric17, 1:20, both generous gifts of I. Leigh, London), desmoplakin I/II (dp antibody mixture, IIF 1:50, Progen), and antiserum from guinea pig (dp 495, IIF 1:400, generous gift of J. Koeser, German Cancer Research Center) were used in this study.
Secondary Antibodies
IgG or IgG+IgM used for IIF were: goat anti-guinea pig, anti-mouse, coupled to Cy3 (1:500, red fluorescence) or Alexa 488 (1:500, green fluorescence). These antibodies (Molecular Probes) were used at a dilution of 1:200. Sections were incubated for 20 min each. For chemiluminescence detection (ECL), horseradish peroxidase-coupled rabbit anti-guinea pig IgG (H+L) (Dianova, 1:10,000) were used.
IIF Microscopy
After briefly rinsing in PBS, cryostat sections were fixed in acetone (−20 °C; 5 min), blocked, and permeabilized with 5% normal goat serum in Tris-buffered saline with Triton X-100 (TBST; TBS + 0.001% Triton X-100). Primary antibodies were applied for 1 h, and after rinsing in PBS, incubation with secondary antibodies was done for 30 min. After washing in PBS, sections were dried and mounted. Visualization and documentation were performed with a photomicroscope (Axiophot II; Carl Zeiss, Oberkochen, Germany). For confocal laser scanning microscopy, a Zeiss LSM 510Meta microscope (Carl Zeiss) operating with an argon ion laser (488 nm) and a HeNe laser (543 nm) was used (for details see Ref. 7).
Electron Microscopy (EM)
For EM, scalp and plucked hair were briefly rinsed with PBS and fixed in 2.5% glutaraldehyde in sodium cacodylate buffer. After three rinses in sodium cacodylate buffer, they were postfixed in 2% OsO4 on ice, followed by washes with distilled water. The specimens were then stained overnight in 0.5% uranyl acetate, dehydrated, and embedded in Epon. Micrographs were taken with an electron microscope EM910 (LEO, Oberkochen, Germany) (for details see, Ref. 7).
Immunoelectron Microscopy (IEM)
For IEM, ∼5-μm thick cryosections of snap-frozen tissues mounted on coverslips were fixed with 2% formaldehyde in PBS, freshly prepared from paraformaldehyde. Reactivities of residual free aldehyde groups were quenched by three washes with PBS containing 50 mm NH4Cl. Specimens were permeabilized with 0.1% Triton X-100 in PBS for 1–2 min, incubated with the specific primary antibodies for 2 h, washed with PBS, and incubated overnight with secondary antibodies (in most cases, goat anti-guinea pig IgG + IgM coupled to 1.4-nm gold particles; Nanogold, Biotrend, Cologne, Germany). After washes with PBS, the samples were fixed with glutaraldehyde, rinsed with cacodylate buffer, followed by two rinsings with Hepes buffer (pH 5.8) containing 200 mm sucrose. The primary gold particles were silver-enhanced, using the HQ-Silver Enhancement Kit (Biotrend, enhancement times 3–6 min). Specimens were washed with distilled water, and postfixed with 0.2% OsO4. After several washes with water, the samples were dehydrated and flat-embedded in Epon as described. Ultrathin sections were prepared with a Reichert-Jung ultramicrotome (Ultracut; Leica, Bensheim, Germany) (for details see Ref. 7).
In Situ Hybridization (ISH)
A 245-bp PCR product (annealing temperature 58 °C; sense primer: gtatgggtgagtcggaatta; reverse primer: cttcccaggaattccagataca) of the 3′ non-coding region of the KRT80 gene (for details, see Ref. 1) was cloned into the pCR4.1 vector (Invitrogen) and used to prepare a 35S-labeled riboprobe. ISH on cryostat sections of human scalp and plucked beard hairs was carried out as described previously in detail (8). Following overnight hybridization at 42 °C, sections were repeatedly washed with SSC/formamide at 50 °C, digested with RNase A at 37 °C, followed by washing with SSC/formamide/DTT at 50 °C. Sections were then dehydrated and dried. To estimate the appropriate time of exposure, sections were covered with an x-ray film (X-Omat, Kodak) and exposed overnight. After dipping in photoemulsion (NTB-2; Kodak) and drying, sections were exposed for 2 to 3 days, stained with hematoxylin, and embedded. For the recording of the ISH signals by reflection microscopy, the confocal laser scanning microscope ZEISS LSM 510 Meta was used, which allows simultaneous visualization of ISH in epi-illumination for the detection of reflection signals and transmitted light in bright field for hematoxylin staining and the combination of the two signal channels by an overlay (for details see Refs. 7 and 8).
Extraction of Keratins, Gel Electrophoresis, and Western Blots
Cytoskeletal extracts were prepared from about 4 to 5 cryosections (10–15 μm thick) as previously reported (7). For keratin analysis, the extracts were resolved by SDS-PAGE (10% polyacrylamide). For Western blots, gels were transferred to PVDF membranes (Immobilon-P, Millipore, Eschborn, Germany) by wet blotting. After staining (0.1% Coomassie Blue R-250), destaining, and blocking with 5% nonfat milk powder in Tris-buffered saline, membranes were incubated with the respective primary antibodies (see “Antibodies”; for all details see Refs. 7 and 8).
RESULTS
KRT80 mRNA Splice Variants and Designation of the Encoded Keratins
Screening of the GenBankTM for cDNAs with sequences corresponding to the human KRT80 gene revealed the existence of 2 types of K80 transcripts that differed by the utilization of alternative splice acceptor sites at the 5′-end of exon 9, i.e. the last exon of the KRT80 gene (Fig. 1D). The mRNA variant encoding the larger protein was generated by splicing at a consensus splice acceptor site 138 nucleotides downstream of the end of exon 8, whereas the other variant was generated by splicing at another consensus splice acceptor site only 101 nucleotides downstream of this site (Fig. 1E). Translation of the second variant resulted in a protein with a truncated carboxyl terminus that lacked sequence similarity to that of the larger K80 variant (Fig. 1F).
An appropriate designation of keratin splice variants needed the consideration of the new nomenclature system for mammalian keratins. In this system, it was agreed to name keratin gene isoforms and their encoded proteins alphabetically, i.e. K6a, K6b, K6c, K33a, and K33b, respectively (3). To distinguish genuine keratin isoforms from alternative keratin splice variants, we advocate to keep the now valid database designation of a given keratin unchanged (i.e. KX) and to designate variants generated by alternative splicing by the addition of consecutive numbers (i.e. KX.1, KX.2, etc.). Accordingly, the protein encoded by conventionally spliced mRNA of the human KRT80 gene remains K80, and the protein encoded by the alternatively spliced mRNA variant becomes K80.1.
Demonstration of KRT80 Transcripts
Radioactive ISH was first carried out on scalp sections by means of a cDNA probe of the 3′ non-coding region of the KRT80 gene. The probe detected the mRNAs of variant K80 and K80.1, respectively. As shown in Fig. 2A, KRT80 transcripts were seen in the granular layer of the interfollicular epidermis. The signal intensity seemed to increase toward the infundibulum of the hair follicles and remained strong within the differentiating cells of the infundibular region (Fig. 2B, open triangle). In hair follicles, we noticed an extraordinary complex expression profile that comprised the companion layer (cl) and all three compartments of the inner root sheath (IRS), i.e. the Henle and Huxley layers, as well as the IRS cuticle. The mRNA synthesis in the cl and IRS layers started almost concomitantly at the level of the lowermost cortex region and terminated first in the abruptly differentiating Henle layer (Fig. 2, C and D, lower red arrows), followed by the cl (Fig. 2, C and D, upper red arrows). Conversely, transcripts in the Huxley layer and the IRS cuticle could be seen up to their terminal differentiation below the isthmus region (Fig. 2D, double red arrow). Moreover, the upper portion of the cuticle of the hair also seemed to be weakly labeled by the cDNA probe (Fig. 2, C and D). Throughout, the outer root sheath of the hair follicle remained unlabeled.
FIGURE 2.
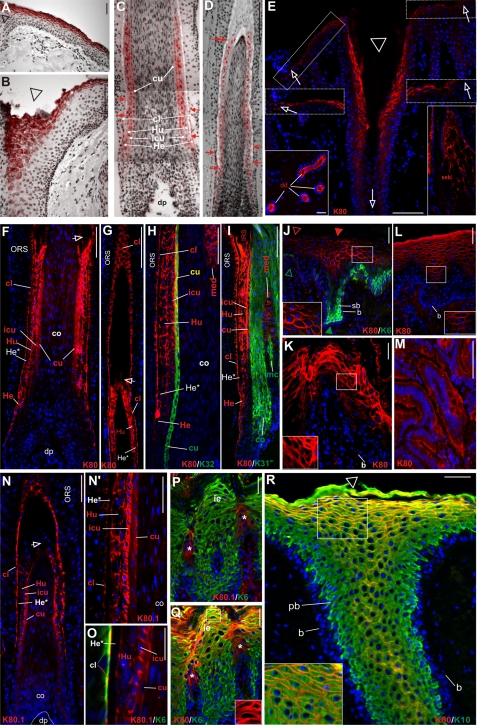
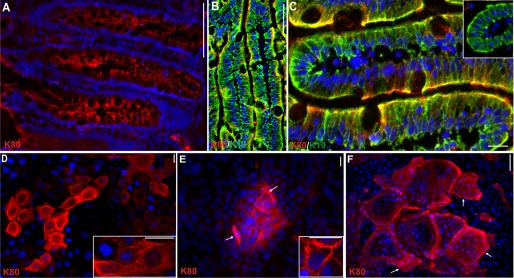
Expression pattern of K80 and K80.1. A–D, radioactive in situ hybridization. Note the cRNA probe used detects both splice variants of the mRNA (ISH signal in red). A, interfollicular epidermis. B, transition of the interfollicular epidermis with increasing labeling toward the infundibulum (triangle). C and D, follicle of plucked beard (C) and scalp (D) hair. Note the complex labeling pattern in the companion layer (cl), the three IRS compartments, Henle (He), and Huxley layer (Hu), and IRS-cuticle (icu), as well as the hair cuticle (cu). Red arrows, termination of RNA synthesis in the respective compartments. E-M, IIF staining of the larger K80 variant. K80 synthesis is strongest in upper infundibular cells (E, triangle) and decreases toward and along the interfollicular epidermis (E, upper inserts, open arrows), better visible after digital contrast enhancement (middle inserts, open arrows). K80 is also present in luminal cells of the eccrine sweat gland ducts (lower left insert, dd) and in sebaceous glands (lower right insert, seb). F, in the hair follicle, K80 is expressed in the companion layer (cl), the three IRS layers (He, Hu, and icu), and also in the hair cuticle (cu). G, note K80 expression in the cl persists considerably longer than in the IRS layers. H, double staining of K80 (red) and the hair cuticle keratin K32 (green) confirms coexpression in the upper hair cuticle (merged yellow). Note, K80 staining in the upper hair medulla (med). I, double staining of K80 (red) and the cortex keratin K31 (green; K31m, mouse monoclonal antibody against K31) confirms the absence of K80 in the hair cortex, but presence in the upper medulla (med). mc, medullary cortex cells (see Langbein and Schweizer (16)). J, double staining of K80 (red) and K6 (green) of foot sole epidermis. K80 staining occurs along the contours of upper suprabasal cells (see also inset) and is strongest (closed triangle) above the areas of K6 staining in the lower suprabasal (sb) cells. b, unstained basal cells. Prominent K80 staining of the cell margins of upper suprabasal layers of the epithelia of the tongue (K) and vagina (L) (and insets). M, cells of non-stratified, simple colon epithelium exhibit an apical cytoplasmic staining of K80. N–Q, IIF staining of the smaller K80 variant, K80.1. N and higher magnification, N′, the expression of K80.1 is restricted to the hair follicle and occurs in the middle to upper region of the cl, the IRS (Hu, icu) and strongly, in the hair cuticle (cu). In this region, the Henle layer is terminally differentiated (He*) and not labeled (N′). O, double staining of K80.1 (red) and K6 (green) shows that in cl cells, K80.1 is strictly concentrated at the side facing the Henle cells (merged yellow). P and Q, the hard-keratinizing upper cells of the filiform papillae of the tongue epithelium express both K80.1 (P, red, *) and K80 (Q, red, *). In contrast to K80, which is expressed together with K6 (green) also at the cell margins in the upper layers of the interpapillary tongue epithelium (Q and inset; serial section of P), K80.1 is absent from the interpapillary tongue epithelium (P). R, much better than in E, double staining of K80 (red) and K10 (green) of scalp emphasizes the cell margin staining of K80 in the infundibulum (merged yellow, see also inset), whereas except for basal (b) and parabasal cells (pb) K10 is cytoplasmic throughout. Blue label, DAPI staining. Bars represent 25 (A, B, E, J–M, and P–R) and 100 μm (C, D, F–I, N, and O).
K80 Expression in Scalp
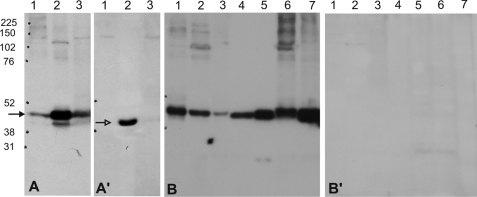
To investigate expression at the protein level and to be able to discriminate between the two splice variants, we raised specific antibodies by using synthetic peptides derived from the individual carboxyl-terminal sequences of the two keratins (Fig. 1F, underlined). Western blots of keratin extracts of forearm skin, plucked beard hairs, and scalp, resolved by one-dimensional SDS-PAGE showed that, in all tissues the K80 antibody detected a protein that migrated close to the calculated molecular mass of the large K80 variant at ∼50 kDa (Fig. 3A, lanes 1–3). In accordance with the ISH data, the protein was strongest in extracts of plucked beard hairs (Fig. 3A, lane 2), weaker in scalp (Fig. 3A, lane 3), and distinctly weaker in body skin (Fig. 3A, lane 1). In contrast, the antiserum against the second splice variant K80.1 reacted only with extracts of plucked beard hairs in which it revealed a single protein band at ∼47 kDa, i.e. a value that corresponded to the calculated molecular mass of K80.1 (Fig. 3A′, lane 2).
FIGURE 3.
Western blot analysis of K80 and K80.1 in epithelial tissues and cell lines. A, K80 is found most strongly at ∼50 kDa (arrow) in protein extracts of plucked beard hairs (lane 2) and to a lower extend in scalp (lane 3) and arm skin (lane 1) (A′) K80.1 is only detectable at ∼47 kDa (open arrow) in plucked hair extracts (lane 2). In A, lane 2 contains a weak band below that of K80, which is exclusively seen in extracts of plucked hairs and runs at a height comparable with that of the K80.1 variant, also present in plucked hair extracts (A′, lane 2). (Most probably the staining of this putative K80.1 band is due to a contamination with residual K80.) B, K80 is also demonstrable in extracts of the epithelia of the foot sole (lane 1), tongue (lane 2), vagina (lane 3), duodenum (lane 4), as well as in the liver carcinoma cell line PLC (lane 5), keratinocytes of the HaCaT line (lane 6) and in the mamma carcinoma cell line MCF-7 (line 7). B′, throughout, keratin extracts of the same tissues and cell lines were negative for K80.1. Dots on left sides, rainbow size markers in kDa (Amersham Biosciences).
IIF studies using the K80-specific antibody on scalp sections not only confirmed, but also extended, the ISH data. Thus K80 was strongly expressed in the upper layers of the entire infundibular epithelium down to the isthmus (Fig. 2E, open triangle and open arrows). Its expression gradually diminished toward and within the interfollicular epidermis (Fig. 2E, open arrows) in which it was also restricted to the uppermost living cell layers (Fig. 2E). This weak epidermal label outside the infundibulum (Fig. 2E, upper inserts) was much better seen after digital contrast enhancement (Fig. 2E, middle inserts). In addition, a strong label was seen specifically in luminal cells of the intradermal duct of eccrine sweat glands (Fig. 2E, lower left insert, dd) as well as in sebaceous glands (Fig. 2E, lower right insert, seb). Other glandular tissues such as mammary glands and submandibular salivary glands were essentially negative in their secretory part and only labeled in their ducts (not shown).
In the hair follicle, K80 expression mirrored the pattern revealed by ISH with some modifications. Its expression in the abruptly differentiating Henle layer was mainly visible in the cytoplasm of the last undifferentiated cell, whereas it was marginal in its immediate precursors (Fig. 2, F and H, He) and lacked in the terminally differentiated Henle cells (Fig. 2F, He*). Likewise, the extended K80 pattern in Huxley cells (Fig. 2, F and G, Hu) and the IRS cuticle was identical to that revealed by ISH (compare Fig. 2, C and D, and F and G, icu). In contrast, K80 could be seen in virtually the entire length of the companion layer (Fig. 2, F and G, cl), which, remarkably, exceeded that in the Huxley layer (Fig. 2G, open arrow) and reached the isthmus region.
More importantly, the evidence for the presence of transcripts in the hard keratinizing hair cuticle could unambiguously be confirmed by double label IIF using the K80 guinea pig antiserum in combination with the mouse antibody against the hair cuticle-specific type I hair keratin K32 (8). As shown in Fig. 2H (K80 in red; K32 in green), K80 was clearly expressed in the uppermost portion of the hair cuticle (merged yellow). Moreover, confirming previous results (8), Fig. 2H also showed that in medullated beard hairs, K80 was expressed in the upper medulla (med). This was further corroborated by double label studies using the K80 antibody (in red) and a mouse antibody against the type I hair cortex keratin K31 (K31m, in green), which also helped to confirm the strong expression of K80 in the upper hair cuticle (Fig. 2I).
K80 Expression in Other Epithelial Tissues
In the course of expression studies in stratified cornified and non-cornified epithelia as well as “simple” non-stratified epithelia and epithelial cell lines, we detected K80 in the granular layers of the prominent foot sole epidermis (Fig. 2J, in red). Surprisingly, however, in these cells the K80 antiserum did not stain a cytoplasmic IF network but specifically decorated their margins (Fig. 2J, see also inset). In addition, the K80 expression in foot sole epidermis was heterogeneous (Fig. 2J, open and closed red triangles) and widely correlated with the degree of the likewise heterogeneous, but cytoplasmic keratin K6 expression in the lower suprabasal layers (Fig. 2J, in green, open and closed green triangles). The uncommon accumulation of K80 along the cell borders was also seen in the suprabasal layers of non-cornified stratified epithelia such as the papillary and interpapillary areas of the dorsal tongue epithelium (Fig. 2K), the vaginal epithelium (Fig. 2L) as well as the epithelia of the esophagus, gingiva, palate, and cervix (results not shown). In contrast to the terminally keratinizing foot sole epidermis with its uppermost layers of densely packed, fully cornified and unstained cells (Fig. 2J), the corresponding essentially non-cornified cells of the vaginal mucosa appeared as a homogenously stained band (Fig. 2L, see also below).
Surprisingly, besides stratified epithelia and glands/ducts, keratin K80 was also detected in cells of non-stratified, simple epithelia such as the colon (Fig. 2M, see below) and duodenum (not shown, but see below). Remarkably, in these polarized epithelial cells, K80 was most strongly detected in the cytoplasm of the apical part, leaving the ventral pole essentially unstained (see also below).
To further confirm the described scenario of K80 expression, we performed Western blots using keratin extracts of the various types of epithelia. As shown in Fig. 3B, the 50-kDa band could be shown in foot sole epidermis (lane 1), tongue (lane 2), vagina (lane 3), and duodenum (lane 4).
K80.1 Expression in Scalp
In accordance with Western blot analyses that revealed the K80.1 variant only in extracts of plucked beard hairs (Fig. 3A′, lane 2), IIF studies on scalp sections with the K80.1 antibody did not lead to a positive label in the interfollicular epidermis and the infundibulum of hair follicles (results not shown). Clearly visible, however, was a labeling of the Huxley layer and notably, the IRS cuticle of hair follicles (Fig. 2, N and N′). The labeled area was restricted to a short region of these tissue compartments situated at the level of the upper cortex, thus explaining the absence of K80.1 label in the differentiated Henle layer (Fig. 2, N and N′, He*). Higher magnification showed that K80.1 was also expressed in the cuticle of the hair (Fig. 2N′, cu). Likewise, this variant was also present in the entire upper companion layer up to the isthmus (Fig. 2N). Double label IIF studies (K80.1, in red; K6, in green) showed that in cl cells both K80.1 and, even more pronounced, K6 strongly accumulated at the side facing the here terminally differentiated Henle layer (Fig. 2O, merged yellow, cl, He*).
K80.1 Expression in Other Epithelial Tissues
With one exception, all further epithelia exhibiting positive expression of the K80 variant were negative for K80.1 in both IIF studies (results not shown) and Western blot analyses (Fig. 3B′, lanes 1–4). The exception was the dorsal tongue epithelium, in which K80.1 was clearly expressed in upper cells of the hard keratinizing, posterior compartment of the filiform papillae, as shown by a double label study with the K80.1 antibody (in red) and the K6 antibody (in green) (Fig. 2P, asterisks). It is conceivable that due to the extremely small K80.1 expressing area within the tongue epithelium, this variant was strongly underrepresented in the respective keratin extracts and escaped detection by the K80.1-specific antibody in Western blots (Fig. 3B′, lane 2). The same reasoning held true for the lack of label in keratin extracts of scalp (Fig. 3A′, lane 3). A serial section, double stained with the K6 antibody (in green) and the antibody against the large variant K80 (in red), confirmed colocalization of K80 and K80.1 in cells of the upper posterior compartment of the filiform papillae (compare Fig. 2, P and Q, asterisks). Furthermore, and better visible than in Fig. 2K, the K6/K80 double label study also demonstrated localization of K80 at the cell margins of the upper interpapillary tongue epithelium (Fig. 2Q). This could be better shown in the inset of this figure by selecting for the red K80 label alone (Fig. 2Q, inset).
As such a localization was also difficult to see in the upper cells of the infundibular funnel of Fig. 2E, we used a slightly oblique section through the infundibulum of a hair follicle for a double label IIF study with antibodies against K80 to clearly show cell margin staining, and the suprabasal keratin K10 for conventional cytoplasmic staining (Fig. 2R, inset).
Special Cellular Localization of K80
In the preceding sections we have observed that K80 was remarkable by its expression in a hitherto unprecedented wide spectrum of epithelia, which comprised keratinizing and non-keratinizing stratified (mucous) epithelia as well as non-stratified simple epithelia and, in addition, the highly complex hair follicle.
More importantly and best seen in differentiating cells of the stratified epithelia, K80 accumulated along the cell margins instead of exhibiting the conventional cytoplasmic IF network. To better define this unusual IF localization, we performed double label IIF using the K80 antibody (in red) and an antibody against desmoplakin (in green) as a ubiquitous marker of desmosomal cell-cell junctions of epithelial cells (9–11). Analysis of scalp sections by confocal laser scan microscopy revealed a typically dotted, green desmoplakin staining at all cell boundaries, except for infundibular cells and the adjacent keratinocytes of the upper epidermal strata just before their terminal differentiation (Fig. 4A, triangle). In the latter, the lateral staining appeared essentially red, with, however, intervening more or less yellow and, less frequently, green patches (Fig. 4A, inset). Neither higher magnifications of areas of the deep infundibulum (Fig. 4B) and the uppermost interfollicular epidermis (supplemental Fig. S1A) nor of the Huxley layer of the IRS of hair follicles (supplemental Fig. S1, B and C) showed a K80/desmoplakin co-localization, rather they suggested a close side by side grouping of the two proteins (Fig. 4B and supplemental Fig. S1C).
FIGURE 4.
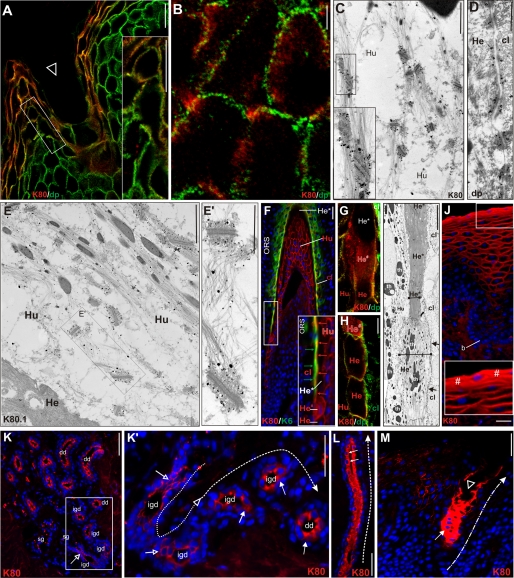
Cellular localization of K80 and K80.1. A and B, confocal laser scan microscopy of K80 (red) and desmoplakin (dp, green) double labeling. A, uppermost cells of the infundibulum exhibit K80 staining at the cell borders. Higher magnification (A, inset) and particularly, the larger cells in the deeper infundibulum (B) clearly reveal an internal K80 location (red) close to but not at desmosomes (green), as seen by the paucity of merged yellow areas. Immunogold labeling of K80 confirms that it is specifically concentrated close to desmosomes. K80 is labeled at the broadening ends of cytoplasmic keratin bundles streaming toward the desmosomes of upper cells of the Huxley layer (C, and inset, Hu). As a control, immunogold staining of desmoplakin clearly labels the desmosomal plaques between cells of the companion layer and Henle layer (D, cl/He) leaving filaments unstained. Immunogold labeling of K80.1 shows also that this keratin is preferentially located in close vicinity to desmosomes, but also occurs, albeit weaker, in the cytoplasm of Huxley layer cells (E, Hu/Hu, higher magnification E′). F, IIF double staining of K80 (red) and K6 (green) on a strongly oblique hair follicle section reveals cell margin staining in Huxley cells (Hu, red) and the concentration of K80 in the inner portion of cl cells facing differentiated Henle cells (He*, merged yellow and red arrows in inset). Note, the localization of K80 (red) at cell margins of lower Henle cells (He), but a strong cytoplasmic staining of the last living cell (He#) undergoing abrupt terminal differentiation (He*) and excluding further staining of the Henle layer (He*) well detectable by double label IIF with desmoplakin (dp, in green) and confocal microscopy (G and H). This process is accompanied by dramatic morphological changes from the large and broad pre-terminal Henle cells (I, He#) to elongated and very slender terminally differentiated cells (I, He*, horizontal bars). Note that the loose keratin filaments bundles are transformed into an amorphous mass in terminally differentiated He* cells. Arrows, compact keratin belt in cl cells adjacent to Henle cells. J, IIF staining of K80 in the non-cornified stratified vaginal epithelial also shows a shift during epithelial differentiation from a marginal staining in the upper suprabasal strata to a clear cytoplasmic staining in the superficial differentiated layer (J, #). K–M, a comparable shift is observed in the eccrine sweat gland. K and K′, idg, open arrows, staining of the margins of luminal cells of the intraglandular duct; and K and K′, dd, closed arrows, subsequent cytoplasmic staining of the cells in the dermal duct. In the latter, the pronounced cytoplasmic label occurs toward the lumen (H, arrows) and is maintained along the entire dermal duct (L, dashed arrow, dd). K80 can be followed up (M, dashed-dotted arrow) to the acrosyringium (M, open triangle). Bars represent 20 (A), 10 (B, G, and H), 25 (J–M), 50 (F), and 1 μm (C, E, and I).
This assumption could be essentially substantiated by IEM. This showed that the K80 staining at the cell margins in the various epithelia clearly corresponded to areas where rather thin cytoplasmic IF bundles formed by other keratins broadened and became clearly denser and more compact before abutting onto the desmosomal complexes (Fig. 4C and supplemental Fig. S1, D–G). This scenario occurred in vaginal epithelium (supplemental Fig. S1D) and was best seen in Huxley cells (Fig. 4E and inset and supplemental Fig. S1E, Hu) or the middle and upper cells of the IRS cuticle (supplemental Fig. S1F, icu) as well as the companion layer (supplemental Fig. S1G, cl). The latter was particularly interesting as in the area shown, the adjacent Henle cells were fully keratinized so that the K80 label could only be demonstrated in the opposite cl cells (supplemental Fig. S1G, He*). It is known that the latter exhibit both an accumulation of keratin IFs and a high desmosomal density at the side facing Henle cells (12–13). Supplemental Fig. S1G demonstrates that this was also true for K80. The labeling showed up within a fuzzy vertical belt of IFs, which was so firmly anchored to the closely spaced desmosomes that part of it remained attached to the Henle cells when occasionally, in plucked hair follicles, the complete ORS and the major part of the cl cells were ripped off (see upper portion in supplemental Fig. S1G, arrow). The particularly strong asymmetric K80 accumulation in cl cells could also be observed at lower magnifications by means of double immunolabeling using antibodies against K80 (in red) and K6 (in green) (Fig. 4F, insert). This confirmed that both K6 and K80 were restricted to the portion of cl cells that was opposed to Henle cells, with K80 being located visibly closer to the differentiated Henle cells than K6 (Fig. 4F, insert, red and green arrows).
Not unexpected, also the K80.1 variant clearly accumulated along the areas where cytoplasmic keratin IF bundles in cells of the various IRS layers terminated at desmosomes (Fig. 4, E and E′, and supplemental Fig. S1, H and H′). However, unlike its larger counterpart K80 (see Fig. 4C and supplemental Fig. S1, D, H, and I), this variant also displayed a weak labeling within the loose cytoplasmic IF network. The two different localizations of K80.1 were particularly apparent in higher magnifications of Huxley cells (Fig. 4E′) or the junction of cells of the IRS cuticle and the Huxley layer (supplemental Fig. S1, H and H′). Conversely, IEM control studies using the desmoplakin antibody led to a significant and specific labeling of the desmosomal plaques directly beneath the cell membranes of cl and Huxley cells (Fig. 4D, cl-He) or between Henle cells (supplemental Fig. S1I, He-He).
Changes in Cellular Localization of K80 during Terminal Differentiation
Our studies have shown that in the course of K80 expression in the hair follicle, the cellular K80 label in the Henle layer underwent conspicuous changes. Although the lowermost Henle cells in the bulbar region remained unstained, the first cells to express K80 appeared at the level of the lower cortex and showed K80 staining at the cell margins (Fig. 4F, insert, and supplemental Fig. S2B, see also Fig. 2H, He). This staining abruptly changed in the last living cell and became strongly cytoplasmic (Fig. 4F and supplemental Fig. S2B, see also Fig. 2H, He#). CLS microscopy clearly confirmed this sudden appearance of the K80 label in the cytoplasm (Fig. 4, G and H, He#) as well as its disappearance in the first terminally differentiated cell (Fig. 4G, He*). Subsequent IEM studies revealed that the change of K80 from the cell margin to the cytoplasm (Fig. 4, F–H, He#) did not lead to visible alterations in cell shape and volume (Fig. 4F and supplemental Fig. S2A, He → He#). Also, the ultrastructural appearance of both trichohyaline granules and the IF network did not visibly differ in the respective cells (Fig. 4I and supplemental Fig. S2A, He→He#). In contrast, the step from the last living Henle cell with its cytoplasmic K80 localization (Fig. 4I and supplemental Fig. S2A′) to the terminally differentiated state led to the complete decomposition of the abundant trichohyaline granules concomitant with a strong compaction of now strictly vertically oriented IFs. Moreover, these processes entailed a dramatic reduction in cell diameter and volume (Fig. 4I and supplemental Fig. S2A, He# → He*) and the cells appeared as a completely amorphous mass of strongly contrasted electron microscopic density (Fig. 4I and supplemental Fig. S2A, He*). It should be mentioned that similar observations could not be made for the K80.1 variant, as compared with K80, the onset of K80.1 expression in the IRS occurred only after terminal differentiation of Henle cells (Fig. 2N).
A comparable intracellular displacement of K80 was also observed in the non-cornified, mucous-type of squamous epithelia such as vagina (Fig. 4J), esophagus, or gingiva (data not shown). Although the basal and lower suprabasal layers were K80 negative, in the upper suprabasal strata, K80 was clearly localized at the cell margins (Fig. 4J). In contrast, the uppermost living, still nucleated cells displayed a homogenous cytoplasmic staining (Fig. 4J and inset, #), interrupted only by cell nuclei which persisted in these cells.
Based on a detailed keratin expression study, we have recently shown that in the duct of eccrine sweat glands the differentiation processes in both the peripheral and, in particular, the luminal cell layer start very early, already within the lowermost, intraglandular portion of the duct (14). In line with this, Fig. 4K, and the higher magnification of the insert (Fig. 4K′), showed that shortly after the transition of the secretory gland (sg) into the intraglandular duct (igd), luminal cells began to exhibit the typical cell margin staining for K80 (Fig. 4K′, open arrows). Again, very rapidly and still within the intraglandular duct, this pattern changed from the marginal to a fully cytoplasmic staining of luminal cells (Fig. 4K′, closed arrows). This persisted in the entire slightly undulated intradermal duct (Fig. 4L), as well as in the intraepidermal duct of the acrosyringium in which it vanished in the uppermost cells toward the opening of the duct on the surface of the epidermis (Fig. 4M, triangle). It is worth mentioning that similar to cl cells in the hair follicle (see Fig. 4F), the K80 staining in luminal cells of the intradermal sweat duct became asymmetric and accumulated at the inner, luminal side of the cells, the so called duct cuticle (Fig. 4L, arrow). Throughout, peripheral duct cells were negative for K80 (Fig. 4, K and L).
K80 Expression in Non-stratified Epithelia and Cultured Epithelial Cells
We have shown that besides cornified, non-cornified, and hard-keratinizing epithelia, K80 was also demonstrable in the apical cytoplasm of most cells of non-stratified, simple epithelia such as the colon (see Fig. 2M) and, as shown in Fig. 5A, the duodenum. The apical K80 staining (red; merged yellow, respectively) was best seen following double staining with another simple epithelial keratin, e.g. K19 (green), which exhibited cytoplasmic staining throughout (Fig. 5, B and C). In contrast, the low differentiated duodenal crypt cells were widely K80 negative, (Fig. 5C, inset), or upon double staining with desmoplakin (green) showed, besides apical K80 co-localization (supplemental Fig. S3A, merged yellow), also staining of the lateral desmosomes (supplemental Fig. S3A, green). In liver tissue, K80 was seen in cells of the bile ducts, whereas parenchymal cells were largely unstained (supplemental Fig. S3B, red), whereas both cell types were positive for desmoplakin (supplemental Fig. S3A, dp, green). In ultrathin sections of duodenal epithelium using immunogold electron microscopy, K80 staining of apical cytoplasmic filaments could also clearly be demonstrated (supplemental Fig. S3, C and D).
FIGURE 5.
Localization of K80 in non-stratified, simple epithelia and cultured epithelial cells. A, IIF detects K80 in the apical cytoplasm of duodenum epithelium. Using double labeling, this apically concentrated K80 pattern (B and C, red) is well seen relative to the cytoplasmic staining of K19 (B and C, green). Note, less differentiated duodenum crypt cells are widely K80-negative (C), inset. In monolayer cultures of polarized epithelia, such as the mamma carcinoma line MCF-7 (D and inset), filamentous cytoplasmic K80 labeling is seen in most cells. In immortalized keratinocytes of line HaCaT (E and inset) and normal human epidermal keratinocytes (F), grown under conditions that induce local differentiation and stratification, differentiated suprabasal cells in the “domes” show cell margin staining of K80, whereas the single uppermost cells exhibit a cytoplasmic label. Note, monolayered and basal keratinocytes of the domes are K80-negative. Bars represent 25 (A, B, and D–F) and 10 μm (C).
Not surprising, a bright cytoplasmic K80 staining was also observed in cells of polarized epithelial cells in vitro, e.g. the mammary carcinoma line MCF-7 (Fig. 5D, inset) or the human liver carcinoma of line PLC (supplemental Fig. S3H).
Remarkably, the sequential marginal and cytoplasmic localization of K80 in the suprabasal epidermis was largely recapitulated in both immortalized keratinocytes of line HaCaT (Fig. 5E, inset) and primary human epidermal keratinocytes, normal human epidermal keratinocytes (Fig. 5F), grown under conditions that induced local differentiation and stratification. Although the monolayered areas and the basal cells of the stratified foci were not stained, suprabasal, differentiated cells in these structures mainly exhibited staining of their cell margins (Fig. 5, E, insert, and F). Few of them, however, clearly showed a cytoplasmic staining (Fig. 5, E and F). The occurrence of K80 in epithelial cell lines could also be confirmed by Western blots of the respective keratin extracts (Fig. 3B, lanes 5–7).
DISCUSSION
Initially, the present study was primarily aimed at providing a detailed description of the expression characteristics of the human type II keratin K80. However, we readily became aware that compared with other type II keratins, K80 possessed a number of unusual properties.
Genomic Localization and Sequence Characteristics
The KRT80 gene is located at the centromeric end of the type I keratin gene domain with 16 neighboring genes (Fig. 1A) being all expressed in the hair follicle, i.e. the hair forming compartment proper (KRT81–KRT86), the cl-IRS unit (KRT71–KRT75 and KRT6), and the ORS (KRT5, KRT6, and KRT7), whereas the remaining type II genes (3) are not expressed or their expression in this adnexal skin organ has not yet been determined (i.e. KRT78 and KRT79). The phylogenetic tree analysis of the type II keratins clearly indicates a structural vicinity of the K80 rod domain to that of hair keratins (Fig. 1B). This similarity is further corroborated by the strikingly high number of cysteine and proline residues in the non-α-helical head and tail domains of K80, which is clearly within the range of type II hair keratins (Fig. 1C and Table 1). In addition, the K80 non-α-helical regions completely lack GGG and GGX repeats (Fig. 1C), present in many type II epithelial keratins (15), but absent from hair keratins (16). From both the phylogenetic and structural point of view, K80 is closest to hair keratin K84 (Fig. 1C and Table 1). Recent studies have shown that the latter seems to represent the most primitive type II hair keratin as K84 homologues have been detected in the claws of lizards (17). Remarkably, but compatible with its position in the phylogenetic tree, the non-α-helical domains of the K80 neighbor K78 (Fig. 1C) reveals a hair keratin-like incidence of cysteine/proline residues (Table 1), in this case, however, concomitant with a low number of GGX repeats (not shown). Finally, also the incidence of cysteine residues in the rod domains of both K78 and K80 is distinctly higher than that of classical epithelial keratins and clearly approaches that of hair keratins (Table 1). Collectively, this would mean that the structural properties of K80 and to a lesser extent, K78, resemble those of hair keratins. In this respect, these two type II keratins are unique within the keratin family, considering that there are no keratin members in the type I family exhibiting structural properties intermediate between epithelial and hair keratins.
TABLE 1.
Cysteine and proline content in the head, tail, and rod domains of selected keratins
Based on this content (in % of the respective keratin domains), K78, K80, and K80.1 can be considered as keratins that are structurally intermediate between epithelial keratins and hair keratins.
| Keratin | Head |
Tail |
Rod | Keratin types | ||
|---|---|---|---|---|---|---|
| C | P | C | P | C | ||
| K7 | 0 | 3.37 | 0 | 0 | 0.32 | |
| K8 | 0 | 3.33 | 0 | 2.35 | 0 | Epithelial |
| K5 | 0 | 4.19 | 0.88 | 0.88 | 0.32 | |
| K1 | 1.11 | 2.79 | 0.64 | 0.64 | 0 | |
| K78 | 3.64 | 4.54 | 7.07 | 2.02 | 0.96 | |
| K80 | 6.10 | 9.76 | 1.61 | 4.87 | 0.97 | Intermediate |
| K80.1 | 6.10 | 9.76 | 6.45 | 12.90 | 0.97 | |
| K81 | 10.38 | 6.06 | 6.25 | 7.03 | 2.93 | |
| K82 | 5.83 | 7.50 | 5.81 | 6.89 | 3.91 | Hair |
| K84 | 3.64 | 6.06 | 6.25 | 7.03 | 1.62 | |
| K85 | 9.76 | 6.50 | 7.79 | 6.49 | 2.93 | |
Alternative Splice Variants
In the framework of investigations on sequence characteristics of human K80, database entries suggested the existence of two alternative K80 splice variants that differed in their carboxyl termini. By means of specific antisera, we were able to demonstrate the expression of both variants in skin and thus to prove their nature as genuine and functional keratins. Although transcripts from about 95% of the multiexon human genes are alternatively spliced (18), this represented the first demonstration of alternative keratin splicing. We therefore proposed a nomenclature system that distinguishes keratin splice variants from keratin gene isoforms (see “Results”). In this system, the designation K80 was preserved for the presently known large and evolutionary older variant (see below), whereas the novel and smaller variant was called K80.1.
It should be emphasized that the GenBank database contains multiple, as yet uncharacterized keratin sequences. In most cases the sequences do not encode functional keratins. Worth mentioning, however, is a postulated splice variant of the human type I keratin K13, which similar to K80.1 exhibited a truncated carboxyl terminus (Uniprot P13646-3). Interestingly, a corresponding cDNA had already been identified earlier and is suggested to encode “a truncated cytokeratin 13” resulting from an “alternative incorrect splicing” (19). It is, however, still today uncertain whether a smaller K13 protein variant is expressed in cells.3 Furthermore, it should also be recalled that a translated splice variant of the type I hair keratin K31 (formerly hHa1), which lacked the complete K31 carboxyl terminus had previously been reported by our group to occur along with K31 in about 5% of the human population. However, this distinctly smaller splice variant did not originate from an alternative splicing process sensu stricto, but was rather due to a polymorphism in the last donor splice site of the KRT31 gene leading to a premature carboxyl-terminal stop codon (20). Collectively, and in line with current knowledge on the involvement of keratin domains in the process of IF assembly, the existing data indicate that functional alternative splicing of keratins might mainly be restricted to variants exhibiting alterations of their carboxyl-terminal domain (see also Ref. 21). The extent and significance of functional alternative splicing in the keratin multigene family remains, however, to be explored.
Evolution of the Gene and Derived Splice Variants
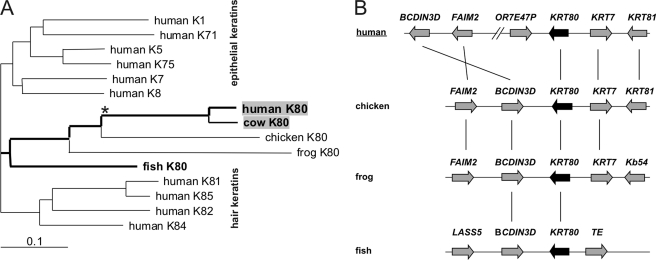
By screening the nucleotide/protein databases (GenBank, NCBI, and Uniprot), we found homologues of human K80 in a large number of vertebrate species that ranged from fish and frog to primates (supplemental Table S1). The homology of the KRT80 genes was shown by phylogenetic analysis of the encoded protein sequences (Fig. 6A) and demonstration of the largely conserved position of the respective genes relative to neighboring genes (Fig. 6B). The species distribution suggests that K80 evolved from a gene present in the last common ancestor of all modern vertebrates.
FIGURE 6.
Evolution of the KRT80 gene. A, phylogenetic analysis of K80 proteins of various species and representative human type II epithelial and hair keratins. Amino acid sequences of the rod domains of the keratins were aligned and a phylogenetic tree was built using the neighbor joining method. The branch lengths in the tree are proportional to the number of substitutions per site (scale: 0.1 substitution per site). B, comparison of the gene locus in vertebrates. Genes are represented by arrows pointing away from the 5′-end to the 3′-end of the respective gene. Homologous genes are connected by vertical lines. KRT, keratin; FAIM2, Fas apoptotic inhibitory molecule 2; BCDIN3D, bicoid-interacting 3 domain containing; OR7E47P, olfactory receptor family 7 subfamily E member 47 pseudogene; Kb54; Xenopus basic keratin 54; TE, transposable element derived 4, similar to piggyBac; LASS5, LAG1 homolog, ceramide synthetase 5.
At present, homologues of the human K80.1 variant could only be detected in mammals. This comprised mainly primates (i.e. chimpanzee, rhesus monkey, and common marmoset), but also the cow (supplemental Table S1). Considering the high sequence conservation of K80 in mammals (supplemental Table S1) as well as the strict association of the human K80.1 variant with the hair follicle, it may be speculated that generation of the latter might be generally confined to mammals. It remains to be seen from further studies on phylogenetically older mammals such as opposum and platypus whether the use of the alternative splice site leading to K80.1 might have coincided with a distinct step in the evolution of mammalian epithelia and in particular of hair.
Collectively, the present data indicate that K80 is encoded by an evolutionary ancient gene that apparently acquired the capacity of alternative, K80.1, splicing only in the mammalian line. Comparative studies involving the knockout of gene homologues in model species such as zebrafish and mouse are needed to identify and characterize ancient and derived functions of K80 in different phylogenetic clades.
Expression of Splice Variants
Another unexpected finding was the unprecedented broad expression spectrum of the large K80 variant. It not only comprised stratified keratinizing and non-keratinizing epithelia, including a large number of vertically differentiating single cell-layered epithelial structures such as the companion layer, the three IRS layers (Henle, Huxley, IRS cuticle) of the hair follicle, as well as the luminal cell layer of the duct of eccrine sweat glands. Remarkably, these special epithelial structures also encompassed the hard-keratinizing cuticle of the hair and, in line with this, the likewise hard-keratinizing posterior compartment of the filiform tongue papilla. In addition, keratin K80 was also detected in the cells of non-stratified, simple and polarized epithelia.
Independent of the type of stratified epithelia, including all single cell-layered vertical structures, K80 was absent from basal and parabasal cells, but occurred in suprabasal, differentiating cells. However, instead of being part of the conventional cytoplasmic IF network, K80 was only demonstrable along the cell margins as shown schematically in supplemental Fig. S5, A and B of keratinizing (foot sole epidermis, Henle layer) and non-keratinizing epithelia (vaginal mucosa, Huxley layer). Ultrastructurally, the K80-containing IF system emerged in the narrow area in which the cytoplasmic keratin IF bundles approaching the cell membrane underwent a fan-shaped spreading just in front of the desmosomes, into which the K80 filament system seemed to be interwoven, thus forming a compact IF meshwork internal to the desmosomal plaques. Shortly afterward, just before reaching the terminal differentiation stage and in general from one cell to the next, K80 suddenly appeared within the cytoplasmic IF network of the last living cells (supplemental Fig. S5, A and B, and Fig. 4, F and J). In non-keratinizing stratified epithelia, these cells represented the uppermost, still nucleated cells exposed to and then stepwise shed into the environment (supplemental Fig. S5B and Fig. 4, J and inset), whereas in keratinizing epithelia they underwent a similarly abrupt terminal differentiation concomitant with the loss of their nuclei and most of their keratin immunostainability (supplemental Fig. S5A and Fig. 2R).
This hitherto undescribed scenario of variable location of a distinct keratin IF network within differentiating cells of stratified epithelia raises several questions. Thus, differentiating cells of stratified epithelia exhibiting submembranous K80-containing IFs simultaneously display a prominent network of cytoplasmic IFs assembled from the respective differentiation-specific keratins of the tissues (see supplemental Fig. S5, A and B, and Table 2). The mechanism by which the entire bulk of K80-containing IFs is specifically targeted to the desmosomal plaques of these cells rather than, like K9 or K2, being incorporated into their existing cytoplasmic IF bundles is unknown at present. There is evidence that the latter join the desmosomal plaques through interaction of the head region of their type II keratins with the tail region of desmoplakin (11, 23–25). As the highly conserved desmoplakin binding sequence in the head domain of type II keratins of stratified epithelia is lacking in the head domain of the K80, a desmoplakin-mediated binding of this type II keratin to desmosomes is improbable. In the end, however, we do not exclude that the large body of partially conflicting data on desmosomal binding of keratins of stratified and simple epithelia (see Ref. 26) cannot indiscriminately be applied to K80 because of the strikingly different structures of its head and tail domains. As these strongly resemble the corresponding domains of hair keratins, and as K80 is able to form IFs with type I hair keratins, investigations in trichocytes may be key to the understanding of K80 binding to desmosomes.
TABLE 2.
Occurrence of K80 and K80.1 in the various epithelia and changes of the ratio of type I and type II keratins
The italics indicate the potential type I keratins able to form heteropolymers with K80 and K80.1, respectively.
| Epithelium | K80 | K80.1 | Type II keratin spectrum | Type I keratin spectrum | Type II:I - former- | Type II:I - present- | ||
|---|---|---|---|---|---|---|---|---|
| Scalp | Interfollicular epidermis | + | − | K5, K1, K2(K2e) | K80 | K14, K10 | 3:2 | 4:2 |
| Infundibulum (upper) | ++ | − | K5, K1, K2 | K80 | K14, K10 | 3:2 | 4:2 | |
| Infundibulum(upper→isthmus) | +++ | − | K5, K1 | K80 | K14, K10 | 2:2 | 3:2 | |
| Hair follicle | Companion layer(middle→upper) | +++ | ++ | K6, K75 | K80, K80.1 | K16, K17 | 2:2 | 4:2 |
| Inner root sheath (IRS) IRS-Henle layer(middle→upper) | +++ | ++ | K71 | K80 | K25, K27, K28 | 1:3 | 2:3 | |
| IRS-Huxley layer(middle→upper) | +++ | + | K71, K74 | K80, K80.1 | K25, K27, K28 | 2:3 | 4:3 | |
| IRS-cuticle(middle→upper) | +++ | ++ | K71, K72, K73 | K80, K80.1 | K25, K26 K27, K28 | 3:4 | 5:4 | |
| Hair fiber cuticle(middle→upper) | ++ | + | K82, K85 | K80, K80.1 | K32, K35, K39, K40 | 2:4 | 4:4 | |
| Hair fiber medulla(middle→upper) | +++ | ++ | K5, K6, K7, K75, K81, K83, K85, K86 | K80, K80.1 | K14, K16, K17, K19, K25, K27, K28, K31, K33a, K33b, K34, K36, K37, K38, K39 | 8:15 | 10:15 | |
| Glands | Eccrine sweat gland, secretory portion | + | − | K7, K8 | K80 | K18, K19, K15 | 2:3 | 2:3 |
| Eccrine sweat gland, luminal duct | ++ | K5, K6, K77(K1b) | K80 | K10, K14, K16, K17, K19 | 3:5 | 4:5 | ||
| Other epithelia | Stratified cornified epithelia (foot sole) | ++ | − | K5, K1, K2, K6 | K80 | K14, K10, K9, K16, K17 | 4:5 | 5:5 |
| Stratified non-cornified, epithelia | ++ | − | K5, K4, K6, K76(K2p) | K80 | K14, K13, K16, K17 | 3:5 | 4:5 | |
| Non-stratified “simple” epithelia | + | − | K8, K7 | K80 | K18, K19, K20 | 2:2 | 3:2 | |
Also the mechanism governing the sudden translocation, be it complete or partial, of the plaque-associated K80 IF system into the cytoplasm of the last living cell is as unknown as the concomitant concentration of cytoplasmic microtubules and microfilaments at cell junctions (27, 28). It is, however, evident that both are part of a plethora of highly orchestrated and interconnected processes that occur in these terminal cells to produce and correctly assemble the protein and lipid constituents required for the formation of the cornified envelope (29) before their programmed cell death and desquamation.
Regarding the function of the unique plaque association of K80 in differentiating cells, it should be recalled that a characteristic feature of stratified epithelia is the sequential expression of keratins exhibiting an increasing length of their head and tail regions, mainly by the accumulation of glycine-rich sequence motifs (Ref. 15, and references therein). Anatomical regions particularly exposed to wear and tear, such as the sole of the foot or the palms of the hand, express additional keratins of this type. Their glycine-rich sequences have been shown to form highly flexible loop configurations that are thought to be responsible for cellular plasticity and resilience of the epithelium (15, 30). As these glycine-rich loops are not present in the K80 molecule, the function of the keratin must lie elsewhere. There are a few examples where the cells of epithelial structures such as the companion layer (13), the cuticle of the hair follicle,3 or the luminal layer of the eccrine sweat gland duct (14) gain unilateral strengthening through the asymmetric accumulation of most of their keratin IFs. We believe that the way K80 IFs are tightly interwoven into the binding area of cytoplasmic keratin IFs at desmosomes along the entire cell membrane represents an ideal means to uniformly stabilize the latter to render it able to cope with the high cellular deformability provided by the glycine loops present in internal keratin IFs. In this respect, the submembraneous K80-containing keratin IF belt can be considered a harbinger of the shortly later formed cornified envelope, whose set-up may render it obsolete and entail the return of the K80 IF system into the cytoplasm.
In this context, it is important that due to the additional, locally identical expression of the small K80.1 variant in the cl-IRS unit and the hard-keratinizing hair cuticle and filiform tongue papilla, the membrane stabilizing effect in differentiating cells of these epithelia is further reinforced. This device is particularly insightful in the hair follicle, because the involved epithelial structures are all thought to participate in the molding and guiding of the forming hair shaft as well as in forming a solid coat of the ripe hair. In line with this, the K80.1 expression sites in the cl-IRS unit and the hair cuticle occur along the area of the hair forming compartment in which the upper bulb region narrows and is finally constricted to the width of the hair cortex.
Unlike cells of stratified epithelia, cells of simple epithelia are polarized into an apical and a basolateral domain, with the “primary” keratin pair K8/K18 of these cells, as well as further keratins such as K19, K20, and K7 (31), forming a thick layer of IFs within the terminal web region subjacent to the microvilli (32, 33). In contrast, only a few IF bundles occur along the lateral domains and can also be detected in the basal pole (Ref. 34 and references therein). As shown schematically in supplemental Fig. S5C for the small intestine, such a scenario is typical for both less differentiated duodenal cells in the crypts (35–37) and mature cells in the villous epithelium. In line with the expression of K80 in differentiating cells of stratified epithelia, the keratin is only found in mature cells of simple epithelia in which it co-localizes with the thick apical layer of K8/K18 or K8/K19 IF bundles (supplemental Fig. S5C, Table 2, and see also Fig. 5, B and C). Thus in both stratified and simple epithelia, K80 is strictly bound to the differentiation of the tissues. Although in the first, K80 generally contributes to the temporary stabilization of the plasma membrane of differentiating cells, in simple epithelial cells it is apparently involved in the reinforcement of the lowermost keratin IF zone of the apical region (38), and, as such, may contribute to the stabilization and maintenance of the polar nature of the cells.
Filament Formation and Promiscuity
The conspicuous formation in vivo of distinct keratin pairs in a given epithelium and a given stage of cellular differentiation strongly contrasts the striking lack of polymerization constraints on type I and type II keratins in vitro. Early studies have shown that obviously any type I keratin can form filaments in vitro with any type II keratin (22). The present study shows that the type II K80 exhibits a similarly bewildering promiscuity in vivo with regard to the choice of its type I partners. Based on the broad range of epithelia in which K80 is constitutively expressed, it can be calculated that K80 forms IFs with at least 21 different type I keratin partners (Table 2). We do not exclude that the actual number of potential K80 partners is even higher. Although the corneal keratin K12 might certainly be a candidate (5), the relevance of K15, K23, K24, K78, and K79 as partners for K80 remains to be seen. In this context, it should be mentioned that previously we provided further evidence for pronounced in vivo keratin promiscuity in the hair medulla. This central compartment of the human hair follicle contains a total of 9 type II and 15 type I randomly expressed soft and hard keratins that interact in a highly promiscuous manner (6).
Although the expression of the smaller splice variant K80.1 is restricted to only distinct subcompartments of the hair follicle and the filiform tongue papilla, the number of its identifiable type I partners still amounts to 16 keratins (Table 2). It should be emphasized that in both cases, the high number of putative K80 partners is mainly due to the complex keratin spectrum of the hair medulla of which 6 members can be partners for K80 or K80.1 in the upper portion of the medulla (Table 2).
Collectively, these data suggest that the in vivo assembly processes resulting from the numerous K80/KX or K80.1/KX combinations that like other in vivo keratin pairs have certainly been subject to a strong selective pressure during evolution, are not influenced by the high number of strongly varying head and tail domains of the individual keratins. It is also important to note that our study in the medulla has already provided evidence for multiple pairings of epithelial and hair keratins, including that of K80 with a type I hair keratin partner (6). In this study, however, this feature is unambiguously demonstrated by the expression of K80 in the hair cuticle and the filiform tongue papilla, which both leave no other choice for K80 than type I hair keratins for IF assembly.
Our study establishes K80 and its splice variant K80.1 as members of the keratin family exhibiting unique structural properties and expression characteristics. Both represent hitherto ignored ubiquitous constituents of epithelia. The elucidation of their ultimate significance and role in the process of epithelial differentiation is the subject of future work in our laboratories.
Supplementary Material
Acknowledgments
We thank Hans-Juergen Stark (German Cancer Research Center) for cultures of normal human epidermal keratinocyte and HaCaT cells, and Bernd Noecker (KAO Corporation, Tokyo, Japan), David A. Parry (Palmerston, New Zealand), and Walter Kittstein (formerly German Cancer Research Center) for fruitful discussions.
Addendum
Recently, Zhou et al. (39) reported on the identification of an alternative splice transcript of the intermediate filament protein vimentin.
This work was supported by Wilhelm Sander-Stiftung Munich Grant 2007.133.1 (to L. L.).
This article is dedicated with gratitude to Werner W. Franke on the occasion of his 70th birthday. His pioneering work on keratins has been pivotal to our own investigations in this field.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
L. Langbein, L. Eckhart, S. Praetzel-Wunder, and J. Schweizer, unpublished data.
- HaCaT
- immortalized human keratinocytes
- IFF
- indirect immunofluorescence
- IEM
- immunoelectron microscopy
- ISH
- in situ hybridization
- IRS
- inner root sheath
- cl
- companion layer.
REFERENCES
- 1.Rogers M. A., Winter H., Langbein L., Bleiler R., Schweizer J. (2004) Differentiation 72, 527–540 [DOI] [PubMed] [Google Scholar]
- 2.Rogers M. A., Edler L., Winter H., Langbein L., Beckmann I., Schweizer J. (2005) J. Invest. Dermatol. 124, 536–544 [DOI] [PubMed] [Google Scholar]
- 3.Schweizer J., Bowden P. E., Coulombe P. A., Langbein L., Lane E. B., Magin T. M., Maltais L., Omary M. B., Parry D. A., Rogers M. A., Wright M. W. (2006) J. Cell Biol. 174, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hesse M., Zimek A., Weber K., Magin T. M. (2004) Eur. J. Cell Biol. 83, 19–26 [DOI] [PubMed] [Google Scholar]
- 5.Lu H., Zimek A., Chen J., Hesse M., Büssow H., Weber K., Magin T. M. (2006) Eur. J. Cell Biol. 85, 803–811 [DOI] [PubMed] [Google Scholar]
- 6.Langbein L., Yoshida H., Praetzel-Wunder S., Parry D. A., Schweizer J. (2010) J. Invest. Dermatol. 130, 55–73 [DOI] [PubMed] [Google Scholar]
- 7.Langbein L., Spring H., Rogers M. A., Praetzel S., Schweizer J. (2004) Methods Cell Biol. 78, 413–451 [PubMed] [Google Scholar]
- 8.Langbein L., Rogers M. A., Winter H., Praetzel S., Beckhaus U., Rackwitz H. R., Schweizer J. (1999) J. Biol. Chem. 274, 19874–19884 [DOI] [PubMed] [Google Scholar]
- 9.Cowin P., Kapprell H. P., Franke W. W. (1985) J. Cell Biol. 101, 1442–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke W. W., Cowin P., Schmelz M., Kapprell H. P. (1987) Ciba Found. Symp. 125, 26–48 [DOI] [PubMed] [Google Scholar]
- 11.Smith E. A., Fuchs E. (1998) J. Cell Biol. 141, 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langbein L., Rogers M. A., Praetzel S., Winter H., Schweizer J. (2003) J. Invest. Dermatol. 120, 512–522 [DOI] [PubMed] [Google Scholar]
- 13.Langbein L., Rogers M. A., Praetzel-Wunder S., Helmke B., Schirmacher P., Schweizer J. (2006) J. Invest. Dermatol. 126, 2377–2386 [DOI] [PubMed] [Google Scholar]
- 14.Langbein L., Rogers M. A., Praetzel S., Cribier B., Peltre B., Gassler N., Schweizer J. (2005) J. Invest. Dermatol. 125, 428–444 [DOI] [PubMed] [Google Scholar]
- 15.Korge B. P., Gan S. Q., McBride O. W., Mischke D., Steinert P. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langbein L., Schweizer J. (2005) Int. Rev. Cytol. 243, 1–78 [DOI] [PubMed] [Google Scholar]
- 17.Eckhart L., Valle L. D., Jaeger K., Ballaun C., Szabo S., Nardi A., Buchberger M., Hermann M., Alibardi L., Tschachler E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18419–18423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barash Y., Calarco J. A., Gao W., Pan Q., Wang X., Shai O., Blencowe B. J., Frey B. J. (2010) Nature 465, 53–59 [DOI] [PubMed] [Google Scholar]
- 19.Kuruc N., Leube R. E., Moll I., Bader B. L., Franke W. W. (1989) Differentiation 42, 111–123 [DOI] [PubMed] [Google Scholar]
- 20.Winter H., Hofmann I., Langbein L., Rogers M. A., Schweizer J. (1997) J. Biol. Chem. 272, 32345–32352 [DOI] [PubMed] [Google Scholar]
- 21.Covaciu C., Castori M., De Luca N., Ghirri P., Nannipieri A., Ragone G., Zambruno G., Castiglia D. (2010) Br. J. Dermatol. 162, 1384–1387 [DOI] [PubMed] [Google Scholar]
- 22.Hatzfeld M., Franke W. W. (1985) J. Cell Biol. 101, 1826–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouklis P. D., Hutton E., Fuchs E. (1994) J. Cell Biol. 127, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalczyk A. P., Bornslaeger E. A., Norvell S. M., Palka H. L., Green K. J. (1999) Int. Rev. Cytol. 185, 237–302 [DOI] [PubMed] [Google Scholar]
- 25.Vasioukhin V., Bowers E., Bauer C., Degenstein L., Fuchs E. (2001) Nat. Cell Biol. 3, 1076–1085 [DOI] [PubMed] [Google Scholar]
- 26.Green K. J., Simpson C. L. (2007) J. Invest. Dermatol. 127, 2499–2515 [DOI] [PubMed] [Google Scholar]
- 27.Perez-Moreno M., Jamora C., Fuchs E. (2003) Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 28.Lechler T., Fuchs E. (2007) J. Cell Biol. 176, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candi E., Schmidt R., Melino G. (2005) Nat. Rev. Mol. Cell Biol. 6, 328–340 [DOI] [PubMed] [Google Scholar]
- 30.Hohl D., Mehrel T., Lichti U., Turner M. L., Roop D. R., Steinert P. M. (1991) J. Biol. Chem. 266, 6626–6636 [PubMed] [Google Scholar]
- 31.Owens D. W., Lane E. B. (2003) Bioessays 25, 748–758 [DOI] [PubMed] [Google Scholar]
- 32.Salas P. J., Rodriguez M. L., Viciana A. L., Vega-Salas D. E., Hauri H. P. (1997) J. Cell Biol. 137, 359–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellner J. C., Coulombe P. A. (2009) J. Cell Biol. 187, 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oriolo A. S., Wald F. A., Ramsauer V. P., Salas P. J. (2007) Exp. Cell Res. 313, 2255–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Es J. H., Jay P., Gregorieff A., van Gijn M. E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., Taketo M. M., Clevers H. (2005) Nat. Cell Biol. 7, 381–386 [DOI] [PubMed] [Google Scholar]
- 36.Barker N., van de Wetering M., Clevers H. (2008) Genes Dev. 22, 1856–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haegebarth A., Clevers H. (2009) Am. J. Pathol. 174, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hull B. E., Staehelin L. A. (1979) J. Cell Biol. 81, 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z., Kahn S., Nielsen A. L. (2010) Biol. Rep. 37, 2407–2413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.