Abstract
Eukaryotic mRNAs are generally considered monocistronic and encode only one protein. Although dicistronic mRNAs encoding two proteins were found in fungi, plants, and animals, polycistronic mRNAs encoding more than two proteins have remained elusive so far in any eukaryote. Here we demonstrate that a single mRNA from silkworm encodes the precursor of an insect cytokine paralytic peptide (PP) and two new cytokine precursor-like proteins, uENF1 and uENF2. RT-PCR analysis showed that this mRNA is widely conserved in moths. Western blot analyses and reporter assays using its modified mRNAs, created by replacing each one of the three ORFs with the firefly luciferase ORF, showed that all three proteins were translated from this mRNA in cell lines, larval tissues, and cell-free systems. Insertion experiments using the Renilla luciferase ORF or a stem loop ruled out the possible involvement of internal ribosome entry site in the three protein translation. On the other hand, systematic mutation analysis of the translation initiation sequence of the 5′-proximal uENF1 ORF suggested that the context-dependent leaky-scanning mechanism is involved in translation of the downstream uENF2 and PP ORFs. In vitro, a synthetic peptide corresponding to the putative mature form of uENF1 stimulated spreading of hemocytes as did the synthetic PP, whereas that of uENF2 antagonized the stimulating activities of PP and the uENF1 peptide, suggesting that the three proteins control cellular immunity interactively. Thus, eukaryotes have a cellular tricistronic mRNA that encodes three functionally related proteins as in an operon.
Keywords: Cytokine, Innate Immunity, Insect, MRNA, Translation, Bombyx mori, Leaky Scanning, Operon, Paralytic Peptide, Polycistronic mRNA
Introduction
In prokaryotes, protein translation from an mRNA begins by direct recruitment of the 30 S small ribosomal subunit to the Shine-Dalgarno sequence located in the vicinity of the initiation methionine codon (1, 2). As a result, proteins could be translated from multiple sites in a single mRNA. Indeed, such polycistronic mRNAs are common in prokaryotes, and often they encode functionally related proteins as in an operon. In contrast, for translation of proteins in eukaryotes, the eukaryotic 40 S small ribosomal subunit first makes contact with the 5′ m7G cap structure of the mRNA through the mediation of the initiation factor proteins and then migrates downstream to scan the initiation codon (1, 3). After the translation of a protein, the 80 S ribosome complex, which is assembled at the initiation codon, is dissociated from the mRNA. Thus, in eukaryotes, the 5′-proximal open reading frame (ORF) is translated preferentially, and most cellular mRNAs are monocistronic and encode only one protein.
However, dicistronic mRNAs with two relatively long ORFs have also been found in fungi, plants, and animals (4–15). Annotation of the genome sequence of the fruit fly Drosophila melanogaster predicted that more than 200 mRNA species are dicistronic (16). Although it was believed that two proteins would be translated from each one of these dicistronic mRNAs, evidence supporting this notion is small in number. Notably, the D. melanogaster stoned and Alcohol dehydrogenase (Adh)-Adh related (Adhr) mRNAs are some rare examples of such cases (7, 8). In the stoned dicistronic mRNA, the stoned B protein encoded by the downstream ORF can be translated via the context-dependent leaky-scanning mechanism in which a population of ribosomes failed to start translation at the initiation codon of the first ORF and continuously scanned the mRNA to initiate the translation of the second ORF (17). In the Adh-Adhr dicistronic mRNA, a distinct mechanism enables translation of the ADHR protein encoded by the downstream ORF; in this case ribosomes are directly recruited to an internal ribosome entry site, located in the intercistronic region between the two ORFs, without prior binding to the 5′ m7G cap structure and translate the ADHR protein (18). The internal ribosome entry site is commonly used in viruses to translate proteins from viral mRNAs lacking the 5′ m7G cap structure (19). Such 5′ m7G cap -independent translation of the downstream ORF has also been postulated in the case of some other eukaryotic cellular dicistronic mRNAs (5, 11, 15). In addition to those dicistronic mRNAs, a polycistronic mRNA encoding several short peptides has been recently reported in insects (20–22). So far, however, the presence of any polycistronic cellular mRNA encoding three or more proteins of >50 amino acid residues, commonly found in prokaryotes, has remained elusive in eukaryotes.
Paralytic peptide (PP)4 is an insect cytokine belonging to the ENF peptide family (23, 24). In the silkworm, PP plays multiple roles on innate immunity and development (25–27). PP normally exists in the hemolymph as a part of the C-terminal end of its inactive precursor (proPP) protein, which is subsequently cleaved to form the active peptide immediately after bleeding. We have previously shown that there are two different sizes of PP mRNAs in the silkworm and already reported the isolation of the shorter 0.6-kb monocistronic PP mRNA encoding a 131-amino acid-long PP precursor (24). In this study we have characterized the longer 1.5-kb PP mRNA. Surprisingly, this mRNA encoded not only the PP precursor but also encoded two cytokine precursor-like proteins. Thus, it appears to be a tricistronic mRNA. We have shown that the three proteins were indeed translated from the single mRNA in all of the cell lines, silkworm larval tissues, and cell-free systems and have also demonstrated that the proteins encoded by the downstream ORFs were translated via the context-dependent leaky scanning, as has been found in the case of the D. stoned dicistronic mRNA. Physiological studies using the synthetic PP and putative mature form of the two cytokine precursor-like proteins suggested that they regulate cellular immunity interactively. Thus, this report demonstrates that the eukaryotes also have a cellular tricistronic mRNA, which like an operon, encodes three functionally related proteins.
EXPERIMENTAL PROCEDURES
Insects
An F1 hybrid strain C145 x N140 and a non-diapausing strain pnd-w1 of Bombyx mori were reared as described previously (24). To break the embryonic diapause of the C145 x N140 strain, eggs were dipped in 20% hydrochloric acid for 1 h at a room temperature 1 day after oviposition. Samia cynthia pryeri was maintained on Ailanthus altissima leaves. Mamestra brassicae eggs were gifts from Dr. Ken Tateishi in NIAS. Larvae of Neogurelca himachala sangaica and Theretra japonica were collected in Tukuba, Japan.
cDNA Cloning
The full-length cDNA corresponding to the uENF1-uENF2-PP tricistronic mRNA of B. mori was cloned from a cDNA pool synthesized from diapause-broken C145 x N140 eggs 3 days after oviposition using the RACE techniques. The procedure and primers used for this purpose were the same as those to clone the 0.6-kb monocistronic PP mRNA (24). The nucleotide sequence of the cDNA was deposited in the GenBankTM/EMBL/DDBJ databases (accession number AB511032). Total RNA was isolated from M. brassicae eggs 2–3 days after oviposition and from the heads of feeding last instar larvae of S. cynthia pryeri, N. himachala sangaica, and T. japonica following the acid guanidinium-phenol-chloroform method using TRIzol (Invitrogen). Partial cDNA sequences spanning from uENF2 to PP ORFs of these four moths were amplified by RT-PCR using degenerated primers that were designed based on the corresponding cDNA sequences of B. mori and Pseudaletia separata (28). First, PCR was carried out using the uENF1-F1 (5′-ATGATHGAMGTNCCNCCNAA-3′) and ENF-R1 (5′-CCRTCNGCNGTNCKYTTRAA-3′) primers. The second nested PCR was carried out using the uENF2-F2 (5′-GTNGTNTTYAAYTTYMRNGA-3′) and ENF-R2 (5′-CANCCNCCNACRAARTTYTC-3′) primers. Complete cDNAs corresponding to the uENF1-uENF2-PP (ENF peptide) tricistronic mRNA of these moth species were obtained by combining 5′-RACE and 3′-RACE using primers that were designed based on the sequences of the respective RT-PCR products. Sequences of cDNAs were deposited in the GenBankTM/EMBL/DDBJ databases (accession numbers AB511033, AB511034, AB511035, and AB511037).
Genomic Cloning
The genomic clone 14D7L containing the silkworm PP gene was isolated from a BAC (bacterial artificial chromosome) library (29). The genomic DNA region covering from uENF1 to PP genes was completely sequenced using internal primers, and the obtained sequence was deposited in the GenBankTM/EMBL/DDBJ databases (accession number AB511038). Genomic DNAs were isolated from the larval midguts of S. cynthia pryeri, N. himachala sangaica, and T. japonica using DNAzol (Invitrogen), and each genomic DNA sample was used to amplify the uENF2 to PP genomic region by PCR. Primer sets used were the same as those used in the RT-PCR analysis. The obtained genomic sequences were deposited in the GenBankTM/EMBL/DDBJ databases (accession numbers AB511039, AB511041, and AB511042). The ENF peptide gene sequences of the P. separata (number AB012294) (28) and M. brassicae (AB126696) (30) were also used for analyzing the genomic structure of the uENF1-uENF2-PP (ENF peptide) mRNA of the two moths.
Sequence Analysis
Multiple alignments of amino acid sequences were performed using ClustalX (eBiotools 3.0, Mac OS X) or GENETYX-MAC (Genetyx, Tokyo, Japan). N-terminal signal peptides were predicted by using the Signal P 3.0 program.
Northern Blot Analysis
RNA sample was separated on a guanidine thiocyanate 1% agarose gel (31) and transferred to a Hybond NX nylon membrane (GE Healthcare). The membranes were hybridized with a 32P-labeled DNA probe corresponding to the PP ORF as described previously (24).
Plasmid Construction
The full-length cDNA corresponding to the 1.5-kb uENF1-uENF2-PP mRNA was amplified by RT-PCR from the B. mori egg cDNA pool and cloned into the pIZT/V5-His expression vector (Invitrogen), and the resultant plasmid was called pIZT-1-2-PP. The insert of this plasmid was deleted from the 3′ direction to generate a series of plasmids containing ORFs for all the three proteins (pIZT-1-2-PP-V5), ORFs of uENF1, and uENF2 (pIZT-1-2-V5) and the ORF of only uENF1 (pIZT-1-V5). In each plasmid the V5 epitope and histidine tag encoding sequences, which were already present in the pIZT/V5-HIS vector, were fused in-frame to the 3′-end of the most downstream ORF.
The firefly luciferase ORF without the initiation methionine codon was amplified by PCR from the pGL3-basic luciferase reporter plasmid (Promega). Linearized pIZT-1-2-PP lacking most of the uENF1 ORF (corresponding to 103–357 bp of the full-length cDNA sequence), uENF2 ORF (corresponding to 481–630 bp of the full-length cDNA), or PP ORF (corresponding to 944–1306 bp of the full-length cDNA) was amplified by PCR, and each amplified fragment was then ligated with the luciferase ORF to generate pIZT-1[fluc]-2-PP, pIZT-1-2[fluc]-PP, or pIZT-1-2-PP[fluc] reporter plasmid, respectively. Each plasmid encoded a luciferase protein fused with the first 10–30 amino acids of uENF1, uENF2, or PP precursor.
The translation initiation sequence (AAAATGA) of the uENF1 ORF in pIZT-1[fluc]-2-PP, pIZT-1-2[fluc]-PP, and pIZT-1-2-PP[fluc] was changed to ACCATGG by PCR to generate pIZT-1opt[fluc]-2-PP, pIZT-1opt-2[fluc]-PP, and pIZT-1opt-2-PP[fluc], respectively. The uENF1 translation initiation sequence was similarly changed to TTTATGT to generate pIZT-1weak[fluc]-2-PP, pIZT-1weak-2[fluc]-PP, and pIZT-1weak-2-PP[fluc].
The Renilla luciferase ORF was amplified by PCR from the pRL-null luciferase reporter plasmid (Promega), the amplified fragment was cloned into the pIZT/V5-HIS, and the resultant plasmid was called pIZT-rluc. The same Renilla luciferase ORF was inserted in front of the uENF1 ORF of pIZT-1[fluc]-2-PP, pIZT-1-2[fluc]-PP, and pIZT-1-2-PP[fluc] to generate pIZT-rluc-1[fluc]-2-PP, pIZT-rluc-1-2[fluc]-PP, and pIZT-rluc-1-2-PP[fluc], respectively. Linearized pIZT-1-2[fluc]-PP and pIZT-1-2-PP[fluc] lacking the entire uENF1 ORF was amplified by PCR. Each amplified fragment was then ligated with the Renilla luciferase ORF to generate pIZT-1[rluc]-2[fluc]-PP and pIZT-1[rluc]-2-PP[fluc], respectively. Likewise, the uENF2 ORF of pIZT-1-2-PP[fluc] was replaced with the Renilla luciferase ORF to generate pIZT-1-2[rluc]-PP[fluc].
To add a 22-bp stable stem loop structure at the 5′-end of each transcript, a BglII site was inserted into just downstream of the transcription initiation site of pIZT-1[fluc]-2-PP, pIZT-1-2[fluc]-PP, and pIZT-1-2-PP[fluc] by PCR. A 5′ overhang double-stranded fragment of the stem loop region was generated by self-annealing the oligonucleotide 5′-GATCGGGGCGCGTGGTGGCGGCTGCAGCCGCCACCACGCGCCCC-3′ (32). The double-stranded oligonucleotide (SL) was cloned into the BglII site of each plasmid to generate pIZT-SL-1[fluc]-2-PP, pIZT-SL-1-2[fluc]-PP, and pIZT-SL-1-2-PP[fluc], respectively. The ORF of the green fluorescent protein ZsGreen1 was amplified by PCR from the pZsGreen1-N1 plasmid (Clontech), the amplified fragment was cloned into the pIB/V5-His-DESTTM expression vector (Invitrogen), and the resultant plasmid was called pIB-ZsGreen.
Western Blot Analysis
The moth Spodoptera frugiperda Sf9 cells (Invitrogen) were cultured at 25 ± 1 °C in IPL-41 medium (Invitrogen) with 10% (v/v) fetal bovine serum. Cells were seeded at 1 × 105/well in 24-well plates and transfected with 1 μg of pIZT-1-2-PP using a transfection regent FuGENE HD (Roche Diagnostics). Two weeks later the conditioned medium was collected, boiled, and then centrifuged to remove cell debris. The cleared supernatant was then used for Western blot analysis using an anti-PP antibody as described previously (24). Similarly, after lipofection with pIZT-1-2-PP-V5, pIZT-1-2-V5, or pIZT-1-V5, the V5 epitope-tagged protein was detected using a primary anti-V5 mouse antibody (Invitrogen) and a secondary goat anti-mouse IgG conjugated with horseradish peroxidase (Zymed Laboratories Inc.). In this experiment, protein samples were concentrated by TCA/acetone precipitation after boiling the conditioned medium.
Luciferase Reporter Assay Using Cell Lines
Sf9 cells or B. mori NIAS-Bm-aff3 (aff3) cells were seeded at 1 × 104/well in 96-well plates and transfected with 200 ng of one of the firefly luciferase expressing plasmid and 50 ng of pIZT-rluc using FuGENE HD. Three days later, firefly and Renilla luciferase activities were measured using a Dual-luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activities were normalized by Renilla luciferase activities. In the case where the plasmid contained both firefly and Renilla luciferase ORFs, 200 ng of only this plasmid was used for lipofection, and firefly luciferase activity-only was measured using a Luciferase assay system (Promega) according to the manufacturer's instructions.
In Vivo Lipofection into Silkworm Larval Tissues
One microgram of pIB-ZsGreen was mixed with 2.5 μl of the lipofection reagent Transfast (Promega) in 10 μl of 10 mm Tris-HCl buffer (pH 8.0), and the mixture was incubated for 10 min at room temperature and then injected into the hemocoel of newly molted third instar silkworm larvae. Three days later, ZsGreen expression in each tissue was observed under a fluorescence microscope (IX70, Olympus) with a specific optical filter cube for GFP detection (excitation, 460–490 nm; barrier, 510–550 nm; U-MWIBA/GFP, Olympus).
One microgram of pIZT-1[fluc]-2-PP, pIZT-1-2[fluc]-PP, or pIZT-1-2-PP[fluc] was mixed with 500 ng of pIZT-rluc and 4 μl of Transfast in 10 μl of Tris-HCl buffer, and each mixture was incubated for 10 min and then injected into the hemocoel of newly molted third instar silkworm larvae. Three days later the hemolymph was individually collected from their incised prolegs into chilled Pringle's buffer (154.1 mm NaCl, 27 mm KCl, 14 mm CaCl2, 22.2 mm dextrose) with a small amount of phenylthiourea. The hemolymph was centrifuged at 500 × g for 5 min, and the resultant pellet was re-suspended in the Passive Lysis Buffer supplied in the Dual-luciferase Reporter Assay kit with a small amount of phenylthiourea. After the hemolymph sampling, each larva was dissected, and the body wall of the abdominal segments, mainly composed of epidermis and fat body, was collected. Each of them was homogenated in the phenylthiourea-supplemented Passive Lysis Buffer. Both firefly and Renilla luciferase activities in these tissues were measured using the Dual-luciferase Reporter Assay kit, and the firefly luciferase activities were normalized with respect to the Renilla luciferase activities. In this in vivo reporter assay, gene introduction efficiency varied considerably from one larva to another, and both luciferase genes were expressed very weakly in some larvae. Therefore, only the larvae in which the Renilla luciferase activities both in the hemocyte and in the body wall was 10-times or much higher than those of the not lipofected controls were used for the data analysis.
In Vitro Transcription and Translation Reactions
Templates used for in vitro transcription reactions were amplified by PCR from the reporter plasmids using the pIZT/V5-HIS vector primers, T7-OpIE2-F (GGATCCTAATACGACTCACTATAGGCGCAACGATCTGGTAAACAC) and polyT-OpIE2-R (TTTTTTTTTTTTTTTTTTTTTTTGACAATACAAACTAAGATTTAGTCAG), which contained the T7 promoter sequence and oligo(dT) sequence, respectively.
RNAs were transcribed in vitro from the PCR fragments using RiboMAX large scale RNA production systems (Promega) and purified with Sephadex G-50 gel filtration columns (GE Healthcare). After confirming the purity of the RNAs by electrophoresis on a guanidine thiocyanate-containing denaturing agarose gel (31), equal amounts of the RNAs were used for in vitro translation reaction using cellular lysates prepared from the moth S. frugiperda Sf21 cells (Transdirect insect cell, Shimadzu), rabbit reticulocytes (Rabbit Reticulocyte Systems nuclease-treated, Promega), or wheat germs (Wheat Germ Extract, Promega) according to the manufacturers' instructions.
Peptide Synthesis and Bioassay
Peptides corresponding to the C-terminal portions of uENF1 (C-uENF1, NH2-VPPNTAGCQEQGTYLDKSGVCRRPW-COOH) and uENF2 (C-uENF2, NH2-VPELECPLGQRRDALGNCRQRF-COOH) were synthesized by Operon Biotechnologies. The mature PP peptide (NH2-ENFVGGCATGFKRTADGRCKPTF-COOH) was synthesized by the Peptide Institute (Osaka, Japan) as was reported previously (24). The underlined two cysteine residues in each peptide formed a disulfide bridge. Effects of the synthetic C-uENF1 and C-uENF2 peptides, and synthetic mature PP on P. separata plasmatocytes were examined as described in a previous report (23, 25). For testing the combined effects of C-uENF2 and PP or C-uENF2 and C-uENF1, various concentrations of C-uENF2 was first added to the hemocytes, and 3 min later 100 nm PP or 30 μm C-uENF1 was added. Dunnett's test was employed to detect statistically significant differences between the BSA-treated control and the peptide-treated groups using JMP7 software (SAS Institute Inc.).
RESULTS
cDNA Cloning of a Tricistronic uENF1-uENF2-PP mRNA from Various Moths
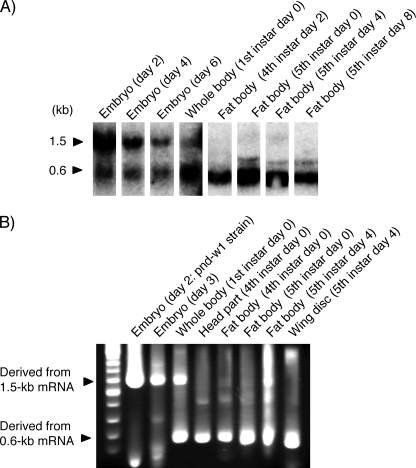
Northern blot analysis using a portion of the PP ORF as a probe detected two different sizes of mRNAs (Fig. 1A). The longer 1.5-kp mRNA was highly expressed in the embryo, particularly in the early embryonic stage. In contrast, the shorter 0.6-kb mRNA, which has been identified to be a PP-encoding monocistronic mRNA (Fig. 2A) (24), was highly expressed in the larval fat body. 5′-RACE analysis using multiple embryonic and larval cDNA pools as templates amplified two major fragments (Fig. 1B). The amplification patterns of the two fragments correlated well with expression profiles of the two mRNA species as found by the Northern blot analysis. The longer fragment was amplified from the embryonic cDNAs, and the shorter fragment was amplified from the larval cDNAs, indicating that the longer PCR fragment was amplified from the 1.5-kb PP mRNA. The longer fragment was amplified from the larval fat body cDNA of the fifth instar day 4 (Fig. 1B) and could be amplified from the other larval cDNAs by increasing cycle numbers of PCR (data not shown), indicating that the 1.5-kb PP mRNA was expressed weakly also at the larval stages.
FIGURE 1.
The uENF1-uENF2-PP tricistronic mRNA is highly expressed during embryogenesis. A, Northern blot analysis of embryonic and larval mRNAs using a portion of the PP ORF as the DNA probe is shown. Embryonic and first larval mRNAs were prepared from the pnd-w1 strain, and fourth and fifth larval mRNAs were prepared from the C145 x N140 strain. B, 5′-RACE analysis of embryonic and larval mRNAs using a reverse primer that was designed based on the nucleotide sequence of the PP ORF is shown. All mRNAs, except that from day 2 embryo, was prepared from the C145 x N140 strain. Cloning and sequence analysis confirmed that the longer fragment derived from the 1.5-kb uENF1-uENF2-PP tricistronic mRNA, and the shorter fragment was derived from the 0.6-kb PP monocistronic mRNA.
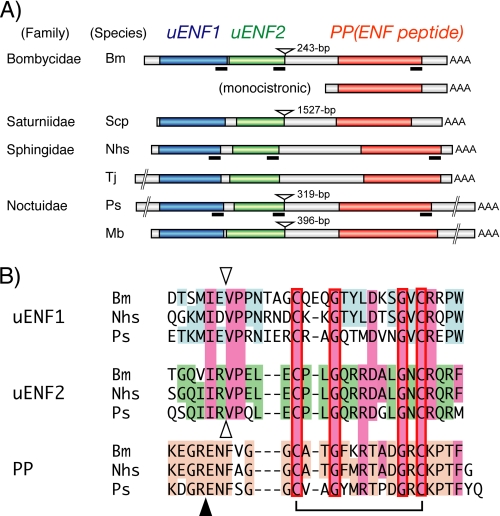
FIGURE 2.
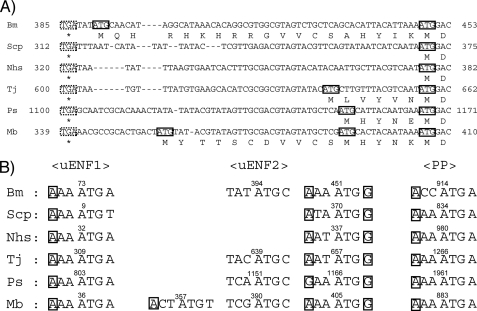
The uENF1-uENF2-PP tricistronic mRNA is expressed in a variety of moths. A, structural organization of the tricistronic mRNA identified from six moths is shown. The uENF1, uENF2, and PP (ENF peptide) ORFs are indicated using blue, green, and orange boxes, respectively. The structure of the B. mori PP monocistronic mRNA is also shown. Deduced amino acid sequences corresponding to the underlined portions of the mRNA are compared in panel B. The inverted triangles show the positions where the introns were found in the respective genes. The length of each intron is indicated on the right side of each inverted triangle. Bm, B. mori; Scp, S. cynthia pryeri; Nhs, N. himachala sangaica; Tj, T. japonica; Ps, P. separata; Mb, M. brassicae. B, alignment of the C termini of uENF1, uENF2, and PP (ENF peptide) precursor of three moths is shown. Conserved amino acid sequences are colored. The closed triangle represents the site where the PP precursor is cleaved to form the mature PP, and the inverted open triangles indicate the corresponding putative cleavage sites for uENF1 and uENF2. A disulfide bond formed between the conserved cysteine residues of PP (ENF peptide) is also indicated. The mature PP and putative mature peptides of uENF1 and uENF2 were synthesized and used in the bioassay shown in Fig. 9.
We cloned the full-length cDNA corresponding to the 1.5-kb PP mRNA from an embryonic cDNA pool by combining 5′-RACE and 3′-RACE. DNA sequence analysis showed that the cloned cDNA contained two additional long ORFs upstream of the PP ORF (Fig. 2A). Predicted proteins encoded by these two newly identified ORFs, named here as uENF1 (upstream of ENF peptide 1) and uENF2 (upstream of ENF peptide 2), consisted of 105 and 89 amino acids, respectively. The N-terminal ends of both proteins contained putative signal peptides, whereas their C-terminal ends showed sequence similarities to the mature PP (Figs. 2B and 3, A and B). Particularly, two cysteine residues, which are involved in forming a disulfide bridge in the ENF peptides (25), and two glycine residues were conserved in the C termini of both uENF1 and uENF2. Sequencing of a B. mori genomic BAC clone revealed that the genomic region corresponding to this mRNA contained an intron (Fig. 2A), thus suggesting that evolutionally it was not horizontally transferred from a virus or bacterium.
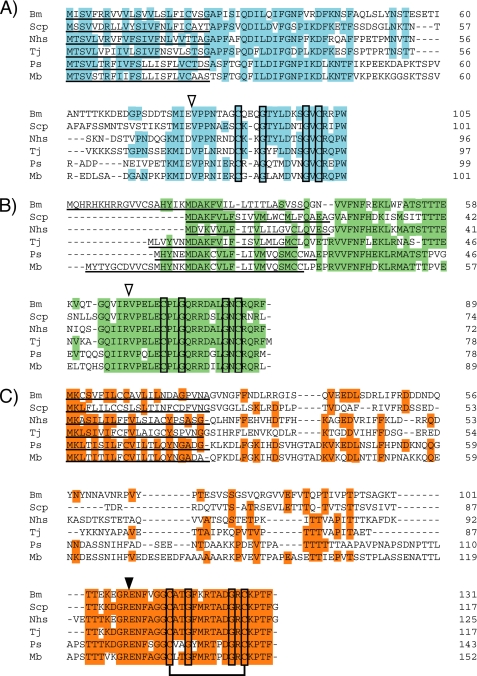
FIGURE 3.
Comparison of the deduced amino acid sequences of uENF1 (A), uENF2 (B), and PP (ENF peptide) (C) precursor proteins identified from six moths. Conserved amino acid sequences are colored. The two cysteine and two glycine residues that are conserved among all the proteins are boxed. The disulfide bond formed between the two cysteine residues of PP (ENF peptide) is also indicated. Putative secretion signals are underlined. The closed triangle in panel C represents the site where the PP precursor is cleaved to form the mature PP, and the open triangles in panels A and B indicate the corresponding putative cleavage sites for uENF1 and uENF2. Bm, B. mori; Scp, S. cynthia pryeri; Nhs, N. himachala sangaica; Tj, T. japonica; Ps, P. separata; Mb, M. brassicae.
Reevaluation of a previously identified 2.5-kb mRNA encoding the moth P. separata ENF peptide gene (28) showed that this mRNA also contained the uENF1 and uENF2 ORFs upstream of the ENF peptide ORF (Fig. 2A). RT-PCR analysis of four additional moth species from three families showed that they all expressed a homologous mRNA that contained the three ORFs (Fig. 2A). Deduced amino acid sequences of uENF1, uENF2, and PP precursor encoded by these homologous mRNAs are highly conserved among the six moths, especially in the C-terminal region (Fig. 3). Furthermore, the C-terminal regions showed sequence similarity to each other (Fig. 2B). The two cysteine and two glycine residues were perfectly conserved among them.
PCR analysis of the genomic DNA revealed that the genomic region corresponding to the intercistronic region between the uENF2 and PP of two Sphingidae moths lacked an intron, whereas the genome of the other three moths had an intron immediately after the uENF2 ORF, as did the Bombyx genome (Fig. 2A). We searched databases for homologous cDNAs or genes and found corresponding ESTs only from several moths, suggesting that this mRNA is present only in Lepidoptera (the family of moths and butterflies).
Translation of All Three Proteins from the Tricistronic uENF1-uENF2-PP mRNA in Cell Lines
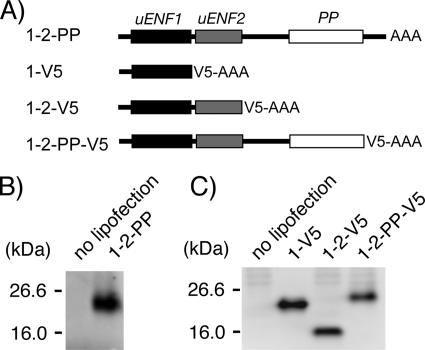
We examined whether all three ORFs in the Bombyx uENF1-uENF2-PP mRNA are indeed translated into proteins. When the entire mRNA was expressed in a moth Sf9 cell line, the PP precursor (proPP) was detected in the medium by Western blot analysis using an anti-PP antibody (Fig. 4B). When the plasmid constructs, which were created by sequentially deleting the 3′-UTR, PP ORF, and uENF2 ORF from the 3′-end of the uENF1-uENF2-PP cDNA and fusing a V5 epitope tag encoding sequence to the 3′-end of the most downstream ORF, were expressed in the Sf9 cells, each one of V5-epitope-tagged uENF1, uENF2, and PP precursor proteins was detected in the medium by Western blot analysis using an anti-V5 antibody (Fig. 4C). These results strongly suggest that all three proteins are, in fact, translated from the uENF1-uENF2-PP mRNA and that they all are processed and secreted from cells.
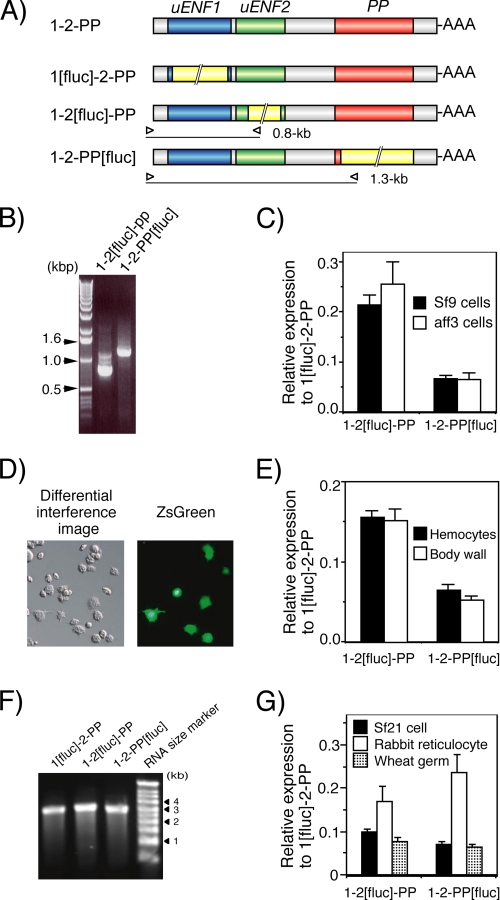
FIGURE 4.
The uENF1, uENF2, and PP precursor proteins are translated from the uENF1-uENF2-PP tricistronic mRNA in Sf9 cells. A, shown is a schematic representation of mRNAs used in experiments. B, detection of the proPP protein in conditioned medium of Sf9 cells expressing the intact tricistronic mRNA (1-2-PP) by Western blot using an anti-PP antibody is shown. C, detection of V5 epitope-tagged uENF1, uENF2, and proPP proteins in the conditioned medium of Sf9 cells expressing the 1-V5, 1-2-V5, or 1-2-PP-V5 mRNA, respectively, by Western blot using an anti-V5 antibody is shown. To achieve uniform expression levels, a one-fifth volume of the 1-V5 protein solution was loaded on the gel as compared with the others.
We next compared the translation efficiencies of the three ORFs using a luciferase reporter assay. When the reporter plasmids, created by replacing each one of the three ORFs in the uENF1-uENF2-PP mRNA expression plasmid, one at a time, with the luciferase ORF, were lipofected into Sf9 cells, all treated cells exhibited strong luciferase activity, and based on these results the relative expression levels of uENF2 and PP were calculated to be about 22 and 7%, respectively, that of the uENF1 (Fig. 5C). A 5′-RACE analysis showed that the expected size mRNA was transcribed from each plasmid, confirming that the luciferase was translated from the modified tricistronic mRNA and not from the processed monocistronic mRNA (Fig. 5B). When the same sets of luciferase reporter plasmids were lipofected individually into the NIAS-Bm-aff3 silkworm cell line, expression levels of uENF1, uENF2, and PP were very similar to those in Sf9 cell line (Fig. 5C). Thus, in both moth cell lines, significant amounts of uENF2 and PP precursor proteins were translated along with the uENF1 protein from the uENF1-uENF2-PP mRNA.
FIGURE 5.
Significant amounts of uENF2 and PP precursor proteins are translated from the uENF1-uENF2-PP tricistronic mRNA, indicated by luciferase reporter assays using cell lines, larval tissues, and cell-free systems. A, shown is a schematic representation of mRNAs used in the reporter assays. The firefly luciferase ORF (1.7 kb), indicated by a yellow box with a double diagonal line, was inserted in-frame in each ORF. Open triangles indicate positions of primers used for 5′-RACE analysis in panel B. The calculated size of 5′-RACE product amplified from each mRNA is indicated on the right side of the triangle, which represents the reverse primer that was designed based on the nucleotide sequence of the luciferase ORF. B, 5′-RACE analysis of mRNAs expressed in Sf9 cells after lipofection of the reporter plasmids is shown. As shown, a DNA fragment of the expected size indicated in panel A was amplified from the cells that were transfected with the plasmid expressing the indicated mRNA, confirming that luciferase was truly translated from the luciferase ORF-containing tricistronic mRNAs and not from the processed monocistronic mRNAs. C, shown are relative levels of expression from uENF2 and PP ORFs compared with that from the uENF1 ORF in two cell lines as determined by the luciferase reporter assay. Error bars represents S.E. (n = 42 for Sf9 cells and n = 10 for aff3 cells; each replicate represents average of data from 3–4 wells). Experiments were repeated using a total of 4 different lots of reporter plasmids on 11 and 3 different days for Sf9 and aff3 cells, respectively. D, expression of ZsGreen1 protein in hemocytes 3 days after in vivo lipofection of pIB-ZsGreen into newly molted third instar larvae is shown. Strong green fluorescence was observed in some hemocytes, particularly in the granulocytes. E, shown are relative levels of expression from the uENF2 and PP ORFs compared with that from the uENF1 ORF in the hemocytes and body wall after in vivo lipofection of the reporter plasmids into newly molted third instar larvae as determined by the luciferase reporter assay. Error bars represents S.E. (n = 20 for both tissues). Each datum was obtained from one individual. Experiments were repeated on two different days. F, ethidium bromide staining of the tricistronic RNAs synthesized in vitro and separated on a denaturing agarose gel show that a single band of RNA of an expected size (2.7–2.9 kb) was present on each lane. G, relative levels of expression from the uENF2 and PP ORFs compared with that from the uENF1 ORF in cell-free systems was determined by the luciferase reporter assay. After confirming the purity of RNAs synthesized in vitro by electrophoresis as in panel F, equal amounts of the RNAs were used for in vitro translation reactions using the cellular lysates prepared from three organisms in parallel. Error bars represents S.E. (n = 4). Four different sets of RNAs were synthesized independently and used for the in vitro translation reactions.
Translation of All Three Proteins from the Tricistronic uENF1-uENF2-PP mRNA in Silkworm Larval Tissues
We then examined whether all three proteins are translated from the uENF1-uENF2-PP tricistronic mRNA in vivo. To individually introduce reporter plasmids into larval tissues, we used an in vivo lipofection technique in which the luciferase reporter plasmids were mixed with a lipofection reagent and injected into the hemocoel of the silkworm larvae. This very simple method resulted in introducing the reporter plasmids into many tissues, including the fat body, epidermis, midgut, hemocytes, and silk glands (data not shown). The highest transfer efficiency was observed in the hemocytes, particularly in the granulocytes, in which green fluorescence was detected after lipofection with a green fluorescence protein expression plasmid (Fig. 5D). A similar efficient foreign gene expression in hemocytes was previously obtained using a combination of lipofection and sonoporation (33).
When the luciferase reporter plasmids used in the in vitro reporter assay were lipofected into silkworm larvae, significant luciferase activities were detected in the hemocytes with any one of the reporter plasmids 3 days later. The relative expression levels of uENF2 and PP were calculated to be 16 and 7%, respectively, that of the uENF1 (Fig. 5E). Similarly, strong luciferase activities were detected from the body wall, composing mainly of epidermis and fat body, and the relative expression ratios of uENF1, uENF2, and PP were found to be almost same as those observed in the hemocytes (Fig. 5E). These results suggest that the three proteins are also translated from the tricistronic mRNA in vivo.
Translation of All Three Proteins from the Tricistronic uENF1-uENF2-PP mRNA in Cell-free Systems
When the tricistronic mRNAs, in which either of the three ORFs was replaced with the firefly luciferase ORF, were synthesized in vitro and used as templates for in vitro translation reactions using the lysate of the S. frugiperda Sf21 cells, significant amounts of firefly luciferase were translated from any of the RNAs. The relative expression levels of the uENF2 and PP ORFs were calculated to be 10 and 7%, respectively, that of the uENF1 ORF (Fig. 5G). Similarly, translation of all the three ORFs was detected when the rabbit reticulocyte or wheat germ lysates were used for in vitro translation reaction. The relative translation levels of uENF2 and PP in the wheat germ lysates were similar to those observed in the Sf21 cellular lysates, and those ratios in the rabbit reticulocyte lysates were higher than those observed in the two lysates (Fig. 5G). Particularly, the expression level of the PP ORF in the rabbit reticulocyte lysate was above 20% that of the uENF1 ORF, which was even higher than those obtained in the live cells and larval tissues (Fig. 5, C and E). Thus, the three proteins were translated from the tricistronic mRNA in the cellular lysates prepared not only from insects but also from mammals and plants. These results suggest that basic molecular mechanisms shared by the eukaryotes enable translation of the three proteins.
Translational Mechanism That Enables Expression of All Three Proteins
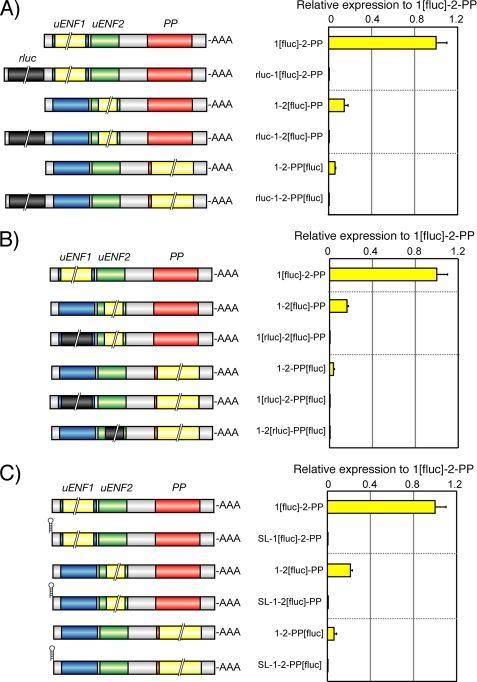
To delineate the translational mechanism involved in the expression of these three proteins, we constructed systematically mutated reporter plasmids and evaluated the translational efficiencies of each ORF in Sf9 or aff3 cells. First, we tested the possibility that the uENF2 or PP ORF is translated via an internal ribosome entry site that is located in the intercistronic region between the uENF1 and uENF2 ORFs or between the uENF2 and PP ORFs, respectively. Insertion of an ORF encoding the Renilla luciferase in front of the uENF1 ORF very strongly blocked the luciferase expression from the uENF1, uENF2, and PP ORFs (Fig. 6A). Replacement of the uENF1 or uENF2 ORF with the Renilla luciferase ORF also inhibited the luciferase expression from both uENF2 and PP ORFs (Fig. 6B). In contrast, the Renilla luciferase expression was not affected by the presence or absence of the downstream firefly ORF, indicating that the downstream ORFs does not affect the translational efficiency of the upstream ORFs in the tricistronic mRNA (data not shown). We then tested the effects of insertion of a 22-bp stem loop near the 5′-end of the tricistronic mRNA. This stem loop is known to create a rigid secondary structure and thereby prohibits translation after it (32). Our results showed that insertion of this 22-bp stem loop also decreased the luciferase expression strongly from all three ORFs (Fig. 6C). Taken together, these results strongly suggest that translation of proteins from the uENF2 and PP ORFs depends on the association of the translation apparatus with the 5′-end of the mRNA and the translation of their upstream ORFs, and therefore, our results rule out the possibility that the uENF2 or PP ORF is translated via internal initiation.
FIGURE 6.
Internal ribosome entry site is not used to translate uENF2 or PP ORF from the uENF1-uENF2-PP tricistronic mRNA. A, shown are the effects of insertion of the Renilla luciferase ORF (0.9 kb), indicated by a dark gray box with a double diagonal line, in front of the uENF1 ORF on the translation of proteins from the uENF1, uENF2, and PP ORFs. B, shown are the effects of replacement of the uENF1 or uENF2 ORF with the Renilla luciferase ORF on the translation of proteins from the downstream ORFs. C, shown are the effects of insertion of a 22-bp stem loop at the 5′-end of the mRNA on the translation of proteins from the uENF1, uENF2, and PP ORFs. Schematic structures of mRNAs expressed in Sf9 cell lines were shown on the left side of each graph. Error bars represent S.E. (n = 3–4 for each experiment. Each replicate is an average of results from 3–4 wells). Experiments were repeated on two to four different days.
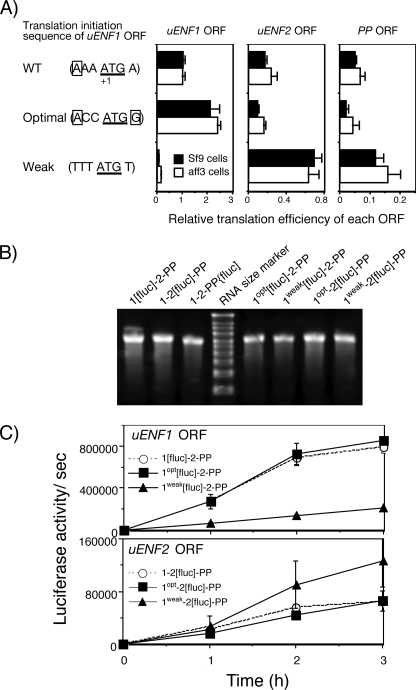
Next, we tested the possibility that the context-dependent leaky-scanning mechanism enables translation of uENF2 or PP ORF. The optimal translation initiation sequence in eukaryotes is (A/G)CCATGG, in which presence G at the +4 position and the presence of A or G at the −3 position are important (34). The translation initiation site of the uENF1 ORF has an A at the −3 position and an A instead of a G at the +4 position (Fig. 7B). Therefore, the translational activity of the uENF1 ORF is expected to be high but not optimal. We first changed this translation initiation site to the optimal sequence (ACCATGG) by mutagenesis and then measured the luciferase expression from each ORF (Fig. 8A). This change doubled the luciferase expression from the uENF1 ORF as was expected. In contrast, those from the uENF2 and PP ORFs were almost halved. Next, we changed the translation initiation site into a weaker translation initiation site (TTTATGT), which lacked both A/G at the −3 position and G at the +4 position. This change strongly reduced the luciferase expression level from the uENF1 ORF below one-tenth of the wild type but increased those from both uENF2 and PP ORFs by 2–4-fold (Fig. 8A). Thus, the luciferase expression levels of the uENF2 and PP ORFs were negatively affected by changes in those of the uENF1 ORF.
FIGURE 7.
Comparison of potential translation initiation sequences of uENF1, uENF2, and PP ORFs of six moths. A, alignment of cDNA sequences spanning from the end of the uENF1 ORF to the beginning of the uENF2 ORF is shown. The translation termination codons of the uENF1 ORFs and potential translation initiation codons of uENF2 ORFs are boxed with dotted and solid lines, respectively. B, comparison of potential translation initiation sequences of the three ORFs is shown. Nucleotides matching the deduced optimal translation initiation sequence of the eukaryote ((A/G)CCATGG) (34) at the most important −3 or +4 positions are boxed. Bm, B. mori; Scp, S. cynthia pryeri; Nhs, N. himachala sangaica; Tj, T. japonica; Ps, P. separata; Mb, M. brassicae.
FIGURE 8.
Leaky scanning of the uENF1 ORF enables translation from the uENF2 and PP ORFs. A, the translation efficiency of each ORF in the uENF1-uENF2-PP mRNA was measured by luciferase reporter assay using two cell lines expressing 1[fluc]-2-PP, 1-2[fluc]-PP, 1-2-PP[fluc], or their modified mRNAs in which the translation initiation sequence of the uENF1 ORF was changed to an optimal or a weak sequence. In both WT and mutated uENF1 translation initiation sequences, shown in the left side, nucleotides matching the deduced optimal translation initiation sequence of the eukaryote ((A/G)CCATGG) (34) at the most important −3 or +4 positions are boxed. Error bars represent S.E. (n = 6–12 for both cell lines; each replicate represents average of data from 3–4 wells). Experiments were repeated on three-to-six different days. B, ethidium bromide staining of the tricistronic RNAs synthesized in vitro and separated on a denaturing agarose gel show that a single band of RNA of an expected size (2.7–2.9 kb) was present on each lane. The RNA size marker used was the same as that in Fig. 5F. C, shown is the time course of the luciferase translation from the 1[fluc]-2-PP, 1-2[fluc]-PP, or their modified mRNAs in the Sf21 cellular lysate. After confirming the purity of RNAs synthesized in vitro by electrophoresis as in panel B, equal amounts of the RNAs were used for in vitro translation reactions. Error bars represent S.E. (n = 2). Two different sets of RNAs were synthesized independently and used for translation reactions.
We also compared the translation of the uENF1 and uENF2 ORFs after changing the uENF1 translation initiation sequence in the cell-free system using the Sf21 cellular lysates. Increasing the translation of the uENF1 ORF by the use of the optimal translation initiation sequence decreased the translation of the uENF2 ORF (Fig. 8C). Conversely, decreasing the translation of the uENF1 ORF by the use of the weak translation initiation sequence increased the translation of the uENF2 ORF. Thus, the negative relationships between the translational levels of the uENF1 and uENF2 ORFs were also observed in the cell-free system.
These results support the context-dependent leaky scanning hypothesis in which the ribosomes that fail to initiate translation of the uENF1 ORF are used for the translation of the uENF2 or PP ORFs. In addition, these results contradict the possibility that the uENF2 or PP ORF is translated via the re-initiation mechanism, in which ribosomes, after competing the translation of the upstream ORF, could again initiate translation from a downstream ORF without being dissociated from the mRNA (4), mainly because in this mechanism the translation efficiencies of the upstream and downstream ORFs should be positively correlated.
Biological Functions of uENF1 and uENF2 in Cellular Immunity
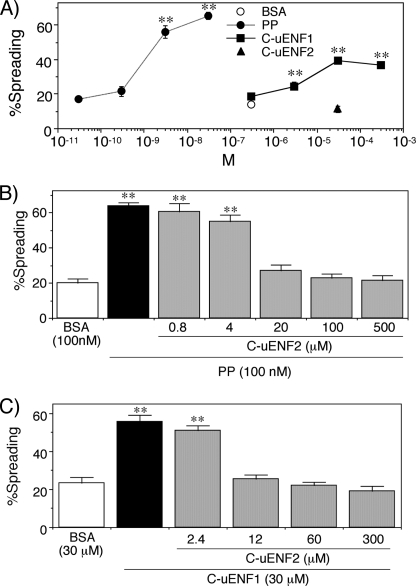
Sequence similarities of the C-terminal ends of uENF1 and uENF2 proteins to PP (Fig. 2B) suggest the possibility that the respective peptides, formed as a result of limited hydrolysis, might function as cytokines. Therefore, we synthesized putative mature uENF1 and uENF2 peptides, corresponding to their respective C-terminal portions (C-uENF1 and C-uENF2) (Fig. 2B), and assessed their biological activities. Because PP strongly stimulates spreading behavior of the plasmatocyte, a key hemocyte subtype in cellular defensive reactions (25, 35), we tested the effects of the synthetic peptides on purified plasmatocytes. As shown, 3 μm or more of the C-uENF1 peptide triggered spreading of the plasmatocytes in vitro (Fig. 9A). The threshold concentration of C-uENF1 necessary for triggering the plasmatocyte spreading was, however, a thousand times higher than that of the synthetic PP. In contrast, the C-uENF2 peptide had no plasmatocyte-stimulating activity even at 30 μm (Fig. 9A).
FIGURE 9.
Effects of synthetic PP and synthetic C termini peptides of uENF1 and uENF2 (labeled as C-uENF1 and C-uENF2) on P. separata plasmatocytes in vitro. A, shown is dose-dependent effects of PP and C-uENF1 on the spreading behavior of plasmatocytes. n = 7–12. B and C, shown are the antagonizing effects of C-uENF-2 on the plasmatocyte spreading activity of PP (B) and C-uENF1 (C). Indicated concentrations of C-uENF2 were first supplemented to cultured plasmatocytes, and 3 min later 100 nm PP or 30 μm C-uENF1 was added. Asterisks (**) indicate that the values are significantly different from that of the BSA control at p < 0.01 by Dunnett's test (n = 7–15).
Next, we tested whether the C-uENF2 could affect the stimulating activities of PP or C-uENF1 on plasmatocytes. The plasmatocytes were pretreated with different concentrations of C-uENF2 and then treated with PP or C-uENF1. The spreading of plasmatocytes by 100 nm PP was inhibited by supplementation of 20 μm or more of C-uENF2 (Fig. 9B). Similarly, the spreading of plasmatocytes by 30 μm of C-uENF1 was inhibited by supplementation of 12 μm or more C-uENF2 (Fig. 9C). Thus, C-uENF2 antagonized the plasmatocyte spreading activity both of the PP and C-uENF1. These results suggest that the C-terminal ends of both uENF1 and uENF2, like PP, function as cytokines, and these three cytokines regulate cellular immunity interactively.
DISCUSSION
Results described in this study clearly documents that the silkworm uENF1-uENF2-PP mRNA is a very unusual example of a eukaryotic cellular tricistronic mRNA. Indeed, three proteins were translated from this mRNA in cultured cells, silkworm larval tissues, and cell-free translation systems (Figs. 4 and 5). The uENF1-uENF2-PP mRNAs were stable and not further processed into monocistronic mRNAs (Fig. 4C), unlike the case of polycistronic mRNAs of Caenorhabditis elegans (5).
Many prokaryotic genomes contain operons that code for polycistronic mRNAs, which are translated into multiple functionally related proteins (2). Similar to these operons, the uENF1, uNNF2, and PP genes, all of which could regulate hemocyte behaviors (Fig. 9), are arranged tandemly in a small region of the genome, transcribed into the tricistronic mRNA, and translated all together. In Drosophila, two meiotic recombination genes, mei-217 and mei-218, and several pairs of odorant receptor genes are located nearby in the genome and transcribed as dicistronic mRNAs (11, 15). Two zebrafish Wint8 proteins with redundant functions in mesoderm and neurectoderm patterning and two tomato enzymes involved in proline synthesis, γ-glutamyl kinase, and γ-glutamyl phosphate reductase, are also encoded in dicistronic mRNAs (9, 12). Thus, operon-like organization of genes and their coexpression as polycistronic, mostly dicistronic, mRNAs have been found rarely but are widely seen in eukaryotes (5).
Multiple mechanisms have been proposed for the translation of the downstream ORFs in eukaryotic dicistronic mRNAs (4, 5, 17, 18). Systematic mutation analyses indicated that it is neither the internal initiation nor the re-initiation mechanism, both of which are postulated to be involved in translation of some eukaryotic dicistronic mRNAs (5, 11, 15), but the context-dependent leaky-scanning mechanism that is involved in the translation of uENF2 and PP from the Bombyx uENF1-uENF2-PP tricistronic mRNA (Figs. 6 and 8). According to this mechanism, the translation initiation site of the upstream ORF should be suboptimal, and that of the downstream ORF be optimal (4). The translation initiation sequence of uENF1 ORF is indeed suboptimal (Fig. 7B). However, the translational initiation sequence of the uENF2 ORF lacks A/G at −3 or G at +4 positions, predicting that it has only a weak translational activity. Incidentally, the surrounding sequence of the second methionine codon coincides with the consensus optimal translation initiation sequence (Fig. 7B). Because this latter sequence is conserved in all six moths, we presume that this might be the actual translation initiation site of uENF2. No matter which methionine codon is used as the translation initiation methionine, an identical uENF2 protein is secreted out of the cells because the putative signal peptide cleavage site is located downstream of the second methionine codon (Fig. 3B). Thus, there is a good correlation between our experimental results and the translation initiation sequences of uENF1 and uENF2 ORFs.
The translation efficiency of the PP ORF in cells and silkworm larval tissues was 25–40% that of the uENF2 ORF (Fig. 5, C and E). The ratio of translation efficiencies between PP and uENF2 was found to be higher than that between uENF1 and uENF2 (translation efficiency of the uENF2 ORF was 15–25% that of the uENF1). This result, however, is inconsistent, as the PP translation initiation sequence, like the uENF1 translation initiation sequence, is also suboptimal (Fig. 7B), thus, predicting that ribosomes that failed to translate uENF1 would also translate much less amount of protein from the PP ORF if the same leaky-scanning mechanism were to work for both uENF2 and PP ORFs. Therefore, an alternative mechanism should exist that would stimulate the translation of the PP ORF. The result obtained using the rabbit reticulocyte lysates, in which translation efficiency from the PP ORF is higher than that from the uENF2 ORF (Fig. 5G), supports this possibility.
The downstream ORFs of the Drosophila stoned and Snapin dicistronic mRNAs were also translated via the context-dependent leaky-scanning mechanism (17). The upstream ORFs of the stoned and Snapin dicistronic mRNAs do not have an in-frame internal methionine codon, which is necessary for the translation of the downstream ORFs. In contrast, the uENF1 ORF has one conserved internal methionine codon just upstream of the putative cleavage site (Fig. 3A). Therefore, it appears that in the leaky-scanning mechanism, the absence of an in-frame internal methionine codon in the upstream ORF is not a prerequisite for the translation of the downstream ORFs. This result implies that the context-dependent leaky-scanning mechanism could be used for translating the second ORF in more eukaryotic dicistronic mRNAs than assumed.
Both of uENF1 and uENF2 proteins have signal peptides at their N termini (Fig. 3, A and B) and are likely secreted from cells like the PP precursor (Figs. 4C). They also have conserved potential cleavage sites for proteases; for example, I(E/D)VP residues of uENF1 for the glutamyl endopeptidase (36) and IRVP residues of uENF2 for DESC1 peptidase and hepsin (37) (Fig. 3, A and B). Because synthetic peptides containing the C-terminal portions after these putative cleavage sites (C-uENF1 and C-uENF2) regulated the spreading behaviors of plasmatocytes (Fig. 9), it is very likely that both of uENF1 and uENF2 are cleaved at this site or at a proximal site, and the resultant C-terminal peptide, like the mature PP, functions as cytokines. However, concentrations of C-uENF1 and C-uENF2 necessary to regulate the spreading behavior of plasmatocytes were much higher than that of PP, suggesting that they may be processed at different sites or that they have other main biological functions in vivo.
Why are uENF1, uENF2, and PP genes encoded in a single mRNA? This is an essential question to answer for understanding the biological significance of this tricistronic mRNA. So far, we have no experimental data to directly answer this question. Our results showed that the uENF1, uENF2, and PP genes were expressed virtually at similar ratios (100:15–25:5–7) in moth cell lines and in silkworm larval tissues (Fig. 5, C and E). Thus, they are seemingly expressed in a fixed proportion from this mRNA independent of the cell type or condition. Synthetic C-terminal peptides of all three proteins regulated hemocyte behavior interactively in vitro (Fig. 9). Based on these results, we speculate that interaction between these three proteins at this stoichiometric ratio is essential for some yet to be determined biological processes, and they are being encoded in the tricistronic mRNA to ensure this possibility. Besides cellular immunity, embryogenesis is another candidate event that needs co-expression of the three proteins because the tricistronic mRNA is expressed strongly in embryo (Fig. 1). Indeed, we have already shown that the ENF peptide gene of M. brassicae plays an essential role on the morphogenesis of the embryonic head portion (30). Additionally, when a higher proportion of PP protein is required for any purpose, it can be achieved by co-expressing the protein from the monocistronic PP mRNA, which was detected throughout the embryogenesis and larval development (Fig. 1). Further molecular genetic and physiological analysis using the silkworm embryo will hopefully clarify the biological role of the tricistronic mRNA.
Our results suggest that the structural organization of eukaryotic mRNAs is more diverse than anticipated. Although we have found the uENF1-uENF2-PP tricistronic mRNA only in the moth lineage, tricistronic mRNAs and polycistronic mRNAs encoding more than three proteins may also exist in other eukaryotes. Indeed, it has been suggested that six sugar receptors are encoded in a single polycistronic mRNA in Drosophila (38). Recently, Calvo et al. (39) showed that around half of human and mouse transcripts have at least one upstream open reading frame (uORFs), and the uORFs reduce protein expression of the downstream main ORFs, depending on the strength of the uORF translation initiation sequence and the uORF number. The lengths of the uORFs vary from <20 bp to >300 bp, and around 10% of the uORFs are longer than 150 bp, encoding proteins of >50 amino acid residues. Thus, dicistronic mRNAs and probably polycistronic mRNAs encoding three or more proteins are much more abundant than anticipated, and the uENF1-uENF2-PP mRNA could be a pioneering example of such polycistronic mRNAs. We suggest that this information might necessitate modification of the genome annotation process, as large amounts of genomic data are now being produced from a wide variety of organisms.
Acknowledgments
We thank Dr. Ken Tateishi for providing M. brassicae eggs, Dr. Shigeo Imanishi for providing NIAS-Bm-aff3 cell line, Dr. Katsura Kojima and Dr. Takahiro Kusakabe for providing information on in vivo lipofection, Dr. Hideki Sezutsu and Dr. Keiro Uchino for assisting in carrying out preliminary experiments, Dr. Nobuhiko Nakashima for providing valuable comments on the manuscript, and Chihiro Ueno for assisting in cell culture and lipofection experiments.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and Grants-in-aid for Scientific Research 22380039 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
- PP
- paralytic peptide
- uENF1
- upstream of ENF peptide 1
- uENF2
- upstream of ENF peptide 2
- uORF
- upstream open reading frames.
REFERENCES
- 1.Kozak M. (1999) Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 2.Adhya S. (2003) Sci. STKE 2003, pe22. [DOI] [PubMed] [Google Scholar]
- 3.Merrick W. C. (1992) Microbiol. Rev. 56, 291–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. (2002) Gene 299, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal T. (2004) Brief. Funct. Genomic. Proteomic. 3, 199–211 [DOI] [PubMed] [Google Scholar]
- 6.Lee S. J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4250–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews J., Smith M., Merakovsky J., Coulson M., Hannan F., Kelly L. E. (1996) Genetics 143, 1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brogna S., Ashburner M. (1997) EMBO J. 16, 2023–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Ríos M., Fujita T., LaRosa P. C., Locy R. D., Clithero J. M., Bressan R. A., Csonka L. N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8249–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray T. A., Saitoh S., Nicholls R. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Jang J. K., Graham J., Nycz K., McKim K. S. (2000) Genetics 154, 1735–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lekven A. C., Thorpe C. J., Waxman J. S., Moon R. T. (2001) Dev. Cell 1, 103–114 [DOI] [PubMed] [Google Scholar]
- 13.Chung K. R., Daub M. E., Ehrenshaft M. (2003) Curr. Genet. 43, 415–424 [DOI] [PubMed] [Google Scholar]
- 14.Pavlik P., Konduri V., Massa E., Simonette R., Beckingham K. M. (2006) Genomics 88, 347–359 [DOI] [PubMed] [Google Scholar]
- 15.Ray A., van Naters W. G., Shiraiwa T., Carlson J. R. (2007) Neuron 53, 353–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin M. F., Carlson J. W., Crosby M. A., Matthews B. B., Yu C., Park S., Wan K. H., Schroeder A. J., Gramates L. S., St Pierre S. E., Roark M., Wiley K. L., Jr., Kulathinal R. J., Zhang P., Myrick K. V., Antone J. V., Celniker S. E., Gelbart W. M., Kellis M. (2007) Genome Res. 17, 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wall A. A., Phillips A. M., Kelly L. E. (2005) J. Biol. Chem. 280, 27670–27678 [DOI] [PubMed] [Google Scholar]
- 18.Ramanathan P., Guo J., Whitehead R. N., Brogna S. (2008) RNA Biol. 5, 149–156 [DOI] [PubMed] [Google Scholar]
- 19.Balvay L., Soto Rifo R., Ricci E. P., Decimo D., Ohlmann T. (2009) Biochim. Biophys. Acta 1789, 542–557 [DOI] [PubMed] [Google Scholar]
- 20.Savard J., Marques-Souza H., Aranda M., Tautz D. (2006) Cell 126, 559–569 [DOI] [PubMed] [Google Scholar]
- 21.Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y. (2007) Nat. Cell Biol. 9, 660–665 [DOI] [PubMed] [Google Scholar]
- 22.Galindo M. I., Pueyo J. I., Fouix S., Bishop S. A., Couso J. P. (2007) PLoS Biol. 5, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strand M. R., Hayakawa Y., Clark K. D. (2000) J. Insect Physiol. 46, 817–824 [DOI] [PubMed] [Google Scholar]
- 24.Kamimura M., Nakahara Y., Kanamori Y., Tsuzuki S., Hayakawa Y., Kiuchi M. (2001) Biochem. Biophys. Res. Commun. 286, 67–73 [DOI] [PubMed] [Google Scholar]
- 25.Miura K., Kamimura M., Aizawa T., Kiuchi M., Hayakawa Y., Mizuguchi M., Kawano K., Yasui H., Hayasaka S. (2002) Peptides 23, 2111–2116 [DOI] [PubMed] [Google Scholar]
- 26.Ishii K., Hamamoto H., Kamimura M., Sekimizu K. (2008) J. Biol. Chem. 283, 2185–2191 [DOI] [PubMed] [Google Scholar]
- 27.Ishii K., Hamamoto H., Kamimura M., Nakamura Y., Noda H., Imamura K., Mita K., Sekimizu K. (2010) J. Biol. Chem. 285, 28635–28642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa Y., Noguchi H. (1998) Eur. J. Biochem. 253, 810–816 [DOI] [PubMed] [Google Scholar]
- 29.Wu C., Asakawa S., Shimizu N., Kawasaki S., Yasukochi Y. (1999) Mol. Gen. Genet. 261, 698–706 [DOI] [PubMed] [Google Scholar]
- 30.Tsuzuki S., Sekiguchi S., Kamimura M., Kiuchi M., Hayakawa Y. (2005) Mech. Dev. 122, 189–197 [DOI] [PubMed] [Google Scholar]
- 31.Goda S. K., Minton N. P. (1995) Nucleic Acids Res. 23, 3357–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. (1989) Mol. Cell. Biol. 9, 5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J. M., Takahashi M., Mon H., Koga K., Kawaguchi Y., Kusakabe T. (2005) Cell Biol. Int. 29, 976–979 [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. (1981) Curr. Top. Microbiol. Immunol. 93, 81–123 [DOI] [PubMed] [Google Scholar]
- 35.Nakahara Y., Kanamori Y., Kiuchi M., Kamimura M. (2003) Arch Insect. Biochem. Physiol. 52, 163–174 [DOI] [PubMed] [Google Scholar]
- 36.Mil'gotina E. I., Voyushina T. L., Chestukhina G. G. (2003) Russ. J. Bioorg. Chem. 29, 511–522 [Google Scholar]
- 37.Béliveau F., Désilets A., Leduc R. (2009) FEBS J. 276, 2213–2226 [DOI] [PubMed] [Google Scholar]
- 38.Slone J., Daniels J., Amrein H. (2007) Curr. Biol. 17, 1809–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo S. E., Pagliarini D. J., Mootha V. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]