Abstract
Galectin-9, a tandem-repeat-type β-galactoside-specific animal lectin with two carbohydrate recognition domains (CRDs) at the N- and C-terminal ends, is involved in chemoattraction, apoptosis, and the regulation of cell differentiation and has anti-allergic effects. Its ability to recognize carbohydrates is essential for its biological functions. Human galectin-9 (hG9) has high affinity for branched N-glycan-type oligosaccharides (dissociation constants of 0.16–0.70 μm) and linear β1–3-linked poly-N-acetyllactosamines (0.09–8.3 μm) and significant affinity for the α2–3-sialylated oligosaccharides (17–34 μm). Further, its N-terminal CRD (hG9N) and C-terminal CRD (hG9C) differ in specificity. To elucidate this unique feature of hG9, x-ray structures of hG9C in the free form and in complexes with N-acetyllactosamine, the biantennary pyridylaminated oligosaccharide, and α2–3-sialyllactose were determined. They are the first x-ray structural analysis of C-terminal CRD of the tandem-repeat-type galectin. The results clearly revealed the mechanism by which branched and α2–3-sialylated oligosaccharides are recognized and explained the difference in specificity between hG9N and hG9C. Based on structural comparisons with other galectins, we propose that the wide entrance for ligand binding and the shallow binding site of hG9C are favorable for branched oligosaccharides and that Arg221 is responsible for recognizing sialylated oligosaccharides.
Keywords: Carbohydrate-binding Protein, Carbohydrate Structure, Lectin, Oligosaccharide, X-ray Crystallography, Biantennary Oligosaccharide, Carbohydrate Recognition Domain, Galectin-9, Sialylated Oligosaccharide
Introduction
The galectins are a family of β-galactoside-specific animal lectins that contain conserved elements for carbohydrate recognition (1, 2) and have attracted much attention as novel regulators of the immune system (3). Recently, galectins have been shown to bind glycans on the surface of potentially pathogenic microorganisms and function as recognition and effect factors in innate immunity (4, 5). Currently, there are 14 members of the mammalian galectin family, classified into three subtypes on the basis of structure (6, 7). The prototype (galectin-1, -2, -5, -7, -10, -13, -14, and -15) has a single carbohydrate recognition domain (CRD)2 that usually forms a non-covalent homodimer. The chimera type (galectin-3) has a single CRD with an extended N-terminal non-lectin domain. The tandem-repeat-type galectins (galectin-4, -6, -8, -9, and -12) have two CRDs, one each in the N- and C-terminal regions, joined by a linker peptide.
Galectin-9 was independently identified as a tumor antigen in cells of patients with Hodgkin disease (8), as a new type of galectin from a kidney cDNA library (9), and as a novel eosinophil chemoattractant produced by T-lymphocytes (10). It is involved in various biological functions, including chemoattraction (11, 12), apoptosis (13–15), and the regulation of cell differentiation (16, 17), and has anti-allergic effects (18). The chemoattractant activity of galectin-9 depends on its carbohydrate binding activity and requires both CRDs (12). Zhu et al. (19) reported that galectin-9 is a ligand for T-cell immunoglobulin and mucin-containing-protein 3 (TIM3), which is a T-helper type 1 (Th1)-specific cell surface molecule. Galectin-9 binds to the N-glycan on TIM3 and stimulates cell death in TIM3+Th1 cells, leading to the termination of Th1-biased immunoreactions. Thus, it is likely that the ability of galectin-9 to recognize carbohydrates is essential to its biological functions.
A detailed analysis of the specificity of galectins in vitro was performed by frontal affinity chromatography, in which a dissociation constant Kd was calculated from the elution volume of an oligosaccharide sample applied to a galectin-immobilized column (20). According to the results, human galectin-9 (hG9) has high affinity for N-glycan-type oligosaccharides with branches (Kd values of 0.16–0.70 μm) and β1–3-linked poly-N-acetyllactosamines, namely oligolactosamines, with a linear structure (0.09–8.3 μm). Further, the hG9 N-terminal CRD (hG9N) and C-terminal CRD (hG9C) differed in specificity; hG9N had much higher affinity for oligolactosamines and glycolipid-type glycans, the Forssman pentasaccharide, and A-hexasaccharide than hG9C but similar affinity for branched oligosaccharides. Frontal affinity chromatography also revealed that hG9, as well as galectin-8 and galectin-3, have significant affinity for α2–3-sialylated oligosaccharides (17–34 μm). The unique specificity of hG9 would be closely related to its diverse biological functions.
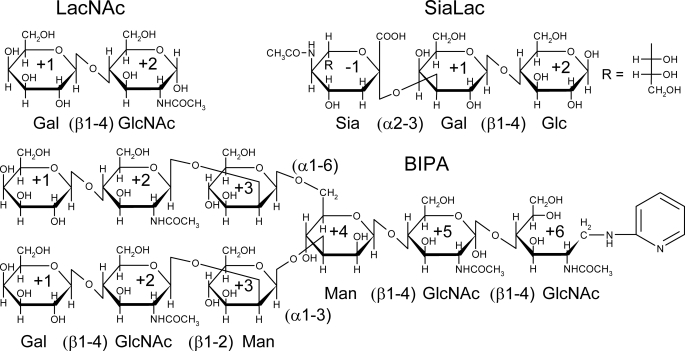
Structural studies on galectins alone and in complexes with oligosaccharides have provided important clues to their structure-function relationships (21–30). Nagae et al. (29, 30) reported the x-ray structures of hG9N in complexes with the Forssman pentasaccharide and oligolactosamines, clearly revealing the mechanism of recognition by hG9N. However, x-ray structures of hG9C have not been reported yet. The structure determinations of both CRDs should be important to understand the biological functions of hG9. Also, structural information on very few galectins in complexes with branched and/or sialylated oligosaccharides is available (23). We report here x-ray structures of hG9C in its free form and in complexes with N-acetyllactosamine (LacNAc), the biantennary pyridylaminated oligosaccharide (BIPA), and α2–3-sialyllactose (SiaLac), providing new insights into how hG9C recognizes these oligosaccharides (Fig. 1). The presented results are the first x-ray structural analysis of C-terminal CRD of the tandem-repeat-type galectin.
FIGURE 1.
The chemical structures of LacNAc, BIPA, and SiaLac. The names of sugar units with glucoside bonds and subsite numbers are indicated.
EXPERIMENTAL PROCEDURES
Purification and Crystallization
The expression and purification of hG9C have been reported previously (31). Briefly, hG9C was expressed as a glutathione S-transferase fusion protein in Escherichia coli BL21 cells and purified by affinity chromatography using glutathione-Sepharose 4B (GE Healthcare UK Ltd.). The bound protein (5 ml) was washed with 10 bed volumes of TBS buffer containing 0.03% CHAPS and 3 bed volumes of PBS in an Econo-Pac column (Bio-Rad), and incubated with 60 units of thrombin (GE Healthcare UK Ltd.) at 293 K overnight. The cleaved target hG9C was eluted from the gel with 2 bed volumes of PBS, and the protein solution was dialyzed against a buffer solution (10 mm Tris-HCl, pH 7.5, 100 mm NaCl) overnight. The purified protein solution was concentrated to 3.9 mg/ml using an Amicon Ultra-4 10-kDa Ultracel (Millipore).
Initial crystallization screening was performed using Crystal Screen kits 1 and 2 and PEG/Ion Screen (Hampton Research Corp.) and Emerald BioSystems Wizard I, II, and III (Emerald BioSystems, Inc.) by the sitting drop method with 96-well plates (Corning Inc.) at 293 K. Crystals of 0.1 × 0.1 × 0.1 mm using diffraction experiments were grown in a droplet containing 1.2 μl of protein solution (3.9 mg/ml in 10 mm Tris-HCl, pH 7.5, 100 mm NaCl) and 0.8 μl of reservoir solution (0.01 m NiCl2, 20% (w/v) PEG monomethyl ether 2000, and 0.1 m Tris-HCl, pH 8.5) against 500 μl of the reservoir solution by the hanging drop method using a 24-well plate (TPP AG).
X-ray Structure Determination
Data were collected at Photon Factory AR-NW12A and BL5A (Tsukuba, Japan). A crystal mounted in a loop was soaked in crystallization solution containing 20% (v/v) ethylene glycol and flash-cooled in a stream of evaporating nitrogen. The diffraction data were collected using an Area Detector Systems Corp. 210r or 315 CCD detector with a wavelength of 1.0 Å at 100 K. All data were processed using the HKL2000 system (32). Crystals of complexes with oligosaccharides were obtained by a soaking method. Aqueous solutions of LacNAc (1.0 m), SiaLac (400 mm), and BIPA (30 mm) were added to droplets of the crystals (0.2–0.4 μl) and incubated for 6 h (LacNAc), 2 days (SiaLac), and 4 weeks (BIPA). LacNAc and SiaLac were purchased from Sigma-Aldrich, and BIPA was prepared as described previously (33). The data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are of the high-resolution bin.
| Free hG9C | h9GC-LacNAc | hG9C-BIPA | hG9C-SiaLac | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | PF-AR NW12A | PF-AR NW12A | PF-BL5A | PF-AR NW12A |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Wavelength (Å) | 1.0 | 1.0 | 1.0 | 1.0 |
| Resolution range (Å) | 50.00–1.50 (1.55–1.50) | 50.00–2.34 (2.38–2.34) | 50.00–1.57 (1.60–1.57) | 50.00–1.99 (2.02–1.99) |
| No. of measured refinements | 105,029 | 68,565 | 96,753 | 55,160 |
| No. of unique refinements | 22,811 | 6,135 | 19,985 | 9,959 |
| Redundancy | 4.6 (4.5) | 11.2 (11.2) | 4.8 (3.9) | 5.5 (5.1) |
| Completeness (%) | 99.4 (99.5) | 100.0 (100.0) | 99.5 (99.9) | 99.7 (95.5) |
| Mean Io/σ(Io) | 10.3 (3.96) | 8.7 (7.4) | 16.6 (3.5) | 16.4 (7.9) |
| Rmergea (%) | 7.3 (39.7) | 7.5 (38.5) | 5.5 (38.7) | 4.7 (21.5) |
| Space group | P63 | P63 | P63 | P63 |
| Unit cell parameters (Å) | a = b = 70.6, c = 50.1, γ = 120.0 | a = b = 70.8, c = 50.2, γ = 120.0 | a = b = 71.0, c = 49.7, γ = 120.0 | a = b = 70.8, c = 50.5, γ = 120.0 |
| Refinement | ||||
| Resolution range (Å) | 26.10–1.50 (1.59–1.50) | 38.85–2.34 (2.49–2.34) | 35.50–1.57 (1.67–1.57) | 30.64–1.99 (2.11–1.99) |
| No. of refinementsb | 21,979 (3,441) | 5,989 (937) | 19,402 (3,046) | 9,744 (1,548) |
| Completeness (%) | 96.0 (90.9) | 97.5 (92.8) | 96.5 (91.7) | 97.8 (93.6) |
| Rfactor (%) | 18.4 (22.1) | 18.5 (18.0) | 19.1 (22.8) | 18.8 (19.0) |
| Rfree (%) | 20.5 (25.9) | 23.1 (25.6) | 21.2 (25.5) | 22.6 (22.9) |
| r.m.s.d.c bond lengths (Å) | 0.005 | 0.005 | 0.005 | 0.006 |
| r.m.s.d.c bond angles (°) | 1.5 | 1.5 | 1.5 | 1.5 |
| Ramachandran plot | ||||
| Most favored region (%) | 86.2 | 83.6 | 87.1 | 85.3 |
| Additional allowed region (%) | 13.8 | 15.5 | 12.9 | 13.8 |
| B-factor (Å2) | ||||
| Protein | 15.2 | 29.9 | 19.9 | 28.1 |
| Ligand | – | 30.6 | 30.2 | 41.9 |
| Solvent | 30.4 | 49.4 | 34.5 | 49.8 |
| PDB code | 3NV1 | 3NV2 | 3NV3 | 3NV4 |
a Rmerge = ΣΣ|Ii − 〈I〉|/Σ〈I〉.
b Reflections (Io > 0.0) were used for refinement calculations.
c r.m.s.d., root mean square deviation.
Molecular replacement was applied with the program MOLREP (34). The initial phase was determined using human galectin-3 CRD (Protein Data Bank (PDB) code: 1KJL), which has 42% sequence identity with hG9C. Further model building was performed with the programs Coot (35) and X-fit (36), and the structure was refined using the programs Refmac5 (37) and CNS (38). Water molecules were gradually introduced if the peaks above 3.5 σ in the (Fo − Fc) electron density map were in the range of a hydrogen bond. In a Ramachandran plot (39), the number of residues in the most favored regions was determined by the program PROCHECK (40). Refinement statistics are listed in Table 1. Figs. 2, 3, and 5 were drawn by the program PyMOL (41).
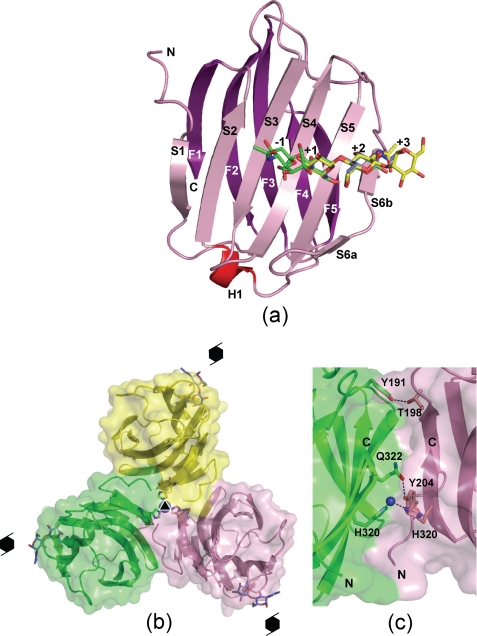
FIGURE 2.
Crystal structure of hG9C. a, the overall structure of hG9C is shown with the bound LacNAc (blue), BIPA (yellow), and SiaLac (green). Secondary elements and subsite numbers are also indicated. b, a trimer of hG9C with the three molecules (pink, yellow, and green) in a crystal is shown with a three-fold axis and 63 screw axes. Ni2+ on the three-fold axis, three coordinated His residues, and the bound LacNAc are also shown. c, area of contact between two molecules (pink and green) with intermolecular hydrogen bonds and metal coordination shown by dotted lines.
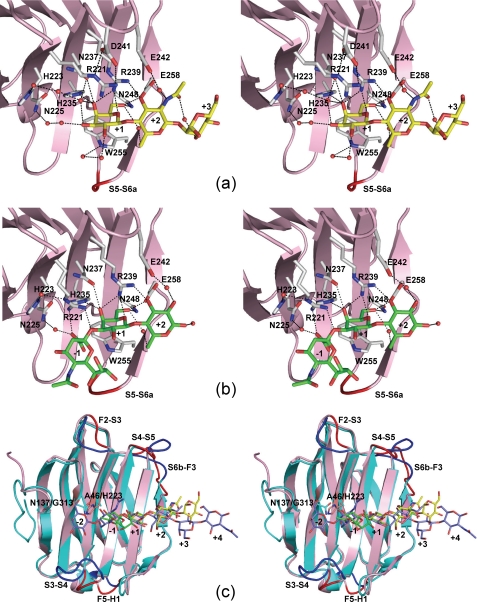
FIGURE 3.
Stereo views of the carbohydrate-binding sites of hG9C and a structural comparison between hG9C and hG9N. Selected hydrogen bonds with BIPA (a) and SiaLac (b) are shown by dotted lines. Although two conformers of Arg221 are found in hG9C-SiaLac, only conformer-2 is shown in b for clarity. c, superimposition of hG9C (pink) and hG9N (cyan) with the bound BIPA (yellow), SiaLac (green), and LN3 (blue). The loops with large deviations are indicated by dark colors.
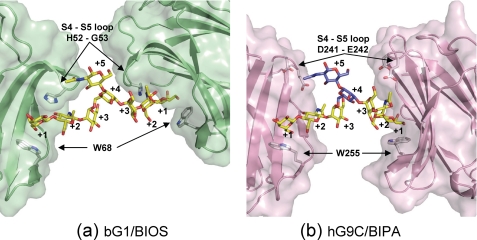
FIGURE 5.
The cross-linking of bG1 (PDB code: 1SLA, green) (a) and hG9C (pink) (b) by the biantennary oligosaccharides. Subsite numbers and selected amino acid residues are given. In b, the modeled sugar units at subsites +4 and +5 are shown in blue.
RESULTS
Overall Structure
We successfully determined the x-ray structures of free hG9C, hG9C-LacNAc, hG9C-BIPA, and hG9C-SiaLac complexes, all of which are isomorphous. There is one molecule of hG9C in an asymmetric unit. Almost all the atoms of the protein molecules, each of which comprises 138 amino acid residues, the bound oligosaccharides, and solvent molecules, fitted nicely to the final (2Fo − Fc) electron density map, except for the N-terminal residues (His188 and Pro189) and terminal sugar units of the oligosaccharides.
The hG9C adopts a β-sandwich structure formed by two anti-parallel β-sheets consisting of six (S1–S6) and five (F1–F5) β-strands, with a short α-helix, as shown in Fig. 2a. The nomenclature for β-strands follows that for hG9N (29). The β-strands align in the following sequence: S1, F2, S3, S4, S5, S6a/S6b, F3, F4, F5, S2, and F1, from the N to the C terminus, and an additional short α-helix is located between F5 and S2. Oligosaccharides bind to the concave surface formed by S3, S4, S5, and S6. In overall structure, free hG9C is almost identical to the hG9C in complexes with oligosaccharides, with the root mean square deviations for main chain atoms being 0.14–0.33 Å, suggesting that the binding of oligosaccharides was unlikely to induce a large structural change in hG9C. However, slight structural differences are found in the conformation of Arg221 and the loop region between S5 and S6a, depending on the bound oligosaccharides, as described later.
In a crystal, three molecules related by a crystallographic three-fold symmetry associate to give a trimer with an intermolecular contact area of 998 Å2 for each molecule (14% of the total surface area of the molecule) as calculated with the program AREAIMOL in the CCP4 program suite (42) (Fig. 2b). There is a Ni2+ on the three-fold axis, coordinated by three His320 residues from the three molecules, stabilizing the trimeric structure. Thr198 and Tyr204 form intermolecular hydrogen bonds with Tyr191 and Gln322 of one of the other molecules, respectively, and there are intermolecular hydrophobic interactions among the three molecules, also contributing to a stable trimer, as shown in Fig. 2c.
Structure of the Carbohydrate-binding Site
Oligosaccharides bind to hG9C via non-reducing and reducing ends located at S3 to S6. For clarity, sugar units are numbered −1, +1, +2, and +3, from the non-reducing end to reducing end. Because Gal residues of three oligosaccharides occupy the same positions in each complex, the position of Gal is defined as subsite +1 (Gal+1), as shown in Figs. 2a and 3. The bound LacNAc in hG9C-LacNAc has almost the same conformation as the LacNAc moiety in hG9C-BIPA, except that GlcNAc+2 in LacNAc adopts an α-anomer different from that in BIPA where the GlcNAc residue forms a β1–4-glucoside bond with Man+3 (supplemental Fig. S1a). Therefore, the description here concentrates on hG9C-BIPA and hG9C-SiaLac.
The structure of the carbohydrate-binding site with the bound BIPA is shown in Fig. 3a. Gal+1 nicely forms stacking interactions with Trp255 and six hydrogen bonds with the following amino acid residues: O4-His235, O4-Asn237, O4-Arg239, O5-Arg239, O6-Asn248, and O6-Glu258. The axial conformation of the O4 of Gal+1 is strictly recognized by three hydrogen bonds with His235, Asn237, and Arg239. The hydroxyl of the O2 and O3 of Gal+1 are directed toward the solvent-accessible surface and form hydrogen bonds with water molecules, which are linked to the amino acid residues, His223, Asn225, and Trp255, by a water-mediated hydrogen bond network. The O3 of GlcNAc+2 forms direct hydrogen bonds with the following amino acid residues: O3-Arg239 and O3-Glu258. Because Arg239 and Glu258 form bifurcated hydrogen bonds with both Gal+1 and GlcNAc+2, they could efficiently recognize the Gal (β1–4) GlcNAc moiety of BIPA. These hydrogen bonds by Arg and Glu residues are strictly conserved in galectin structures. The O6 of Gal+1 and O6 of GlcNAc+2 are also connected by a water-mediated hydrogen bond. An N-acetyl group of GlcNAc+2 forms a water-mediated hydrogen bond with Glu242. Although the electron density for Man+3 of BIPA is relatively poor when compared with that for Gal+1 and GlcNAc+2, the orientation of the pyranose ring of Man+3 could be determined (supplemental Fig. S1b). A water-mediated hydrogen bond between the O3 of Man+3 and the N-acetyl group of GlcNAc+2 was found, but the electron density for other water molecules in hydrogen bonds with Man+3 could not be determined. The electron density for additional sugar residues at the reducing end (Man+4, GlcNAc+5, and GlcNAc+6) could not be determined either, maybe due to the highly disordered structure.
The structure of the carbohydrate-binding site with the bound SiaLac is shown in Fig. 3b. As in hG9C-BIPA, Gal+1 forms stacking interactions with Trp255 and forms six hydrogen bonds with His235, Asn237, Arg239, Asn248, and Glu258. Gal+1 and Glc+2 are connected by bifurcated hydrogen bonds from Arg239, Glu258, and a water molecule, and the O2 of Glc+2 forms a water-mediated hydrogen bond with Glu242. The electron density for Sia−1 allowed us to determine the orientation of the pyranose ring and the conformation of the carboxyl group at the 2-position but not the conformation of an N-acetyl group at the 5-position or a glycerol part at the 6-position (supplemental Fig. S1c). The α2–3-glucoside bond between Sia−1 and Gal+1 is bent extensively, placing Sia−1 beyond the concave surface of the carbohydrate-binding site. Interestingly, Arg221 changes its side chain conformation to form hydrogen bonds with the carboxyl group and the glucoside oxygen atom of Sia−1. The Arg221 found in hG9C-BIPA is denoted as “conformer-1,” and that newly found in hG9C-SiaLac is denoted as “conformer-2.” The electron density of Arg221 in hG9C-SiaLac indicates both conformers (1 and 2), each with an occupancy of 0.5, suggesting that Arg221 could not completely fix the position and orientation of Sia−1 in motion with a large B-factor of 54.2 Å2 (supplemental Fig. S1d). Nevertheless, Arg221 is thought to be responsible for the recognition of sialylated oligosaccharides because two such conformers of Arg221 have not been observed in the structures of free hG9C and hG9C complexes with LacNAc and BIPA.
Depending on the binding of BIPA and SiaLac, a slight but significant structural difference is found in the loop region between S5 and S6a, as shown in Fig. 3, a and b in red. This loop contains a β-turn consisting of Ile251, Asp252, Asn253, and Ser254. The electron density showed that the β-turn is of type II in hG9C-BIPA and type I in hG9C-SiaLac with deviations of Cα atoms by 0.6, 2.6, 1.3, and 0.4 Å, for Ile251, Asp252, Asn253, and Ser254, respectively. The binding of a sugar unit to subsite −1 seems to affect the conformation of the β-turn because Sia−1 directs toward the β-turn. However, no direct interaction between Sia−1 and the β-turn could be found, and the correlation between the β-turn and the binding of a sugar unit to subsite −1 is still unclear.
Structural Comparison with hG9N
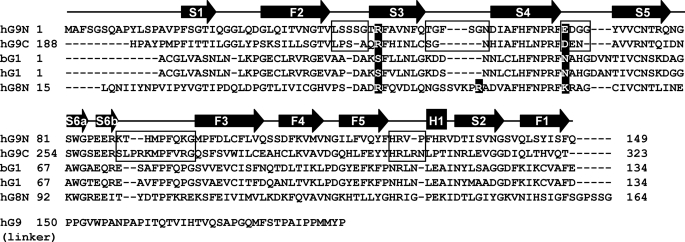
Fig. 3c shows the superimposition of hG9C (pink) and hG9N (cyan) with the bound oligosaccharides, BIPA (yellow) and SiaLac (green) in hG9C, and LacNAc trimer (LN3, blue), which is three LacNAc moieties linked by β1–3-glucoside bond, in hG9N (PDB code: 2ZHN). Structural differences between hG9C and hG9N are found in the loop regions. Apart from the N-terminal loop, the loop regions between F2 and S3 (F2–S3 loop), between S3 and S4, between S4 and S5, between S6b and F3, and between F5 and H1 deviate greatly. These structural differences are due to the insertion or deletion of amino acid residues (Fig. 4). The F2–S3 and S3–S4 loops of hG9N with additional residues protrude to cover the β-sheet of S2–S4, giving a favorable carbohydrate-binding site for LN3. The S4–S5 and S6b–F3 loops construct the entrance for ligand binding, which is relatively widely opened in hG9C, where sugar units at subsites +2 and +3 move to the backside; the deviations of the center of mass of pyranose rings at subsites +2 and +3 between BIPA and LN3 are 1.1 and 3.7 Å, respectively. The α-helix of hG9C also shifts to the outside of the protein by 2.8 Å due to an additional Arg302.
FIGURE 4.
Amino acid alignment among hG9N, hG9C, bG1, hG1, and hG8N, with the linker peptide sequence of hG9 short isoform (National Center for Biotechnology Information (NCBI) reference: NP_002299). Secondary elements are shown by arrows (β-strands) and a rectangle (α-helix). The loops with large deviations found between hG9N and hG9C are indicated by boxes. The positions of Arg221 and Asp241 in hG9C and of Arg66 in hG8N are highlighted in black.
The bound SiaLac in hG9C is bent at a glucoside bond between Sia−1 and Gal+1, placing Sia−1 out of the carbohydrate-binding site. On the other hand, the bound LN3 in hG9N has a linear structure that enables sugar units at subsite −1 and −2 to enter deeply into the carbohydrate-binding site, forming direct hydrogen bonds with Asn137, corresponding to Gly313 in hG9C. This is because His223 in hG9C causes steric hindrance with the sugar unit at subsite −1 of LN3. The corresponding residue is Ala46 in hG9N, and Nagae et al. (29, 30) proposed that Ala46 is one of the residues responsible for the striking affinity of hG9N for oligolactosamines, the Forssmann pentasaccharide, and A-hexasaccharide. This is also supported by the present results, and the protruding F2–S3 and S3–S4 loops of hG9N may contribute to the specificity of hG9N.
DISCUSSION
In a crystal, hG9C associates around a three-fold axis to give a trimer. Because Ni2+ from the crystallization solution is thought to be one of the major driving forces of molecular association, the trimer seems to be artificially constructed. However, 14% of the total surface area was buried by the formation of a trimer, and the significant intermolecular hydrogen bonds and hydrophobic interactions suggest another possibility. Also, the mouse galectin-9 N-terminal CRD has been reported to associate as a dimer in a crystal different from the dimer of the prototype galectin-1, -2, and -7 (29), and a tendency for hG9 to self-associate in solution was revealed by Ferguson plot analysis of polyacrylamide gel electrophoresis and MALDI-TOF mass spectroscopy (43). Thus, it is possible that hG9C partially associates as a trimer in solution, although it is unclear whether the formation of a trimer is related to the biological function of hG9C or not.
As indicated in Fig. 2b, the carbohydrate-binding site of hG9C is exposed to a 63-screw axis in a crystal. Three Man+3 residues in hG9C-BIPA around a three-fold symmetry resulting from the 63-screw axis approach one another with a distance of 4.9 Å between O1 atoms (supplemental Fig. S2). It is likely that two of the three Gal+1-GlcNAc+2-Man+3 moieties binding in hG9Cs belong to the same BIPA molecule, forming a cross-linking structure. If this is the case, Man+4 and GlcNAc+5 are almost on the 63-screw axis, and their electron density cannot be observed due to the highly disordered structure. In fact, there is a large solvent channel along the 63-screw axis, allowing enough space for the passing of BIPA molecules (supplemental Fig. S2).
The x-ray structure of bovine galectin-1 (bG1) in a complex with the biantennary octasaccharide (BIOS), which has exactly the same chemical structure as BIPA at subsites +1 to +5, has been reported in three crystal forms, hexagonal, trigonal, and monoclinic (23). Careful inspection revealed that BIPA in hG9C adopts a similar conformation to that in bG1-BIOS in the hexagonal form (PDB code: 1SLA), where the distance between O1 atoms of Man+3 is 4.9 Å equal to that in hG9C-BIPA (Fig. 5a). Using the structure of bG1-BIOS in the hexagonal form, a model of the cross-linking of hG9C-BIPA was built, as shown in Fig. 5b. The procedure of model building is given in supplemental Fig. S3. Although the model is possibly dependent on the protein-protein interactions in the crystals, the plausible cross-linking structure of hG9C was obtained. The deviation in the torsion angles of glucoside bonds of Man+3 to Man+4 is 2–17° between bG1-BIPA (crystal structure, PDB code: 1SLA) and hG9C-BIPA (model), showing that the model is chemically reliable. The α1–6-glucoside bond adopts a low energy gauche-gauche conformation in both structures. In bG1-BIOS, two Gal+1-GlcNAc+2-Man+3 moieties make an angle of 150°, and the cross-linked bG1 molecules are close together on the side of GlcNAc+5 with a distance of 8.3 Å (Gly53–Gly53) and far apart on the opposite side with a distance of 28.6 Å (Trp68–Trp68). The S4–S5 loop of bG1, including His52 and Gly53, overlaps BIOS, giving a deep carbohydrate-binding site. His52 and Trp68 sandwich Gal+1 and GlcNAc+2 to fix their positions, and Gly53 efficiently forms van der Waals contacts with Man+3 and Man+4. In hG9C-BIPA, two Gal+1-GlcNAc+2-Man+3 moieties make an angle of 118°, and the cross-linked hG9C molecules are located almost parallel to each other at a distance of 18.0 Å (Asp241–Asp241) and 19.5 Å (Trp255–Trp255). The S4–S5 loop of hG9C is not directed toward the Gal+1 and GlcNAc+2, giving a shallow carbohydrate-binding site. The entrance for the ligand binding of hG9C is widely opened, and Man+3 and Man+4 of BIPA are free from hG9C without any direct interactions.
According to the frontal affinity chromatography analysis, human galectin-1 (hG1) has significant affinity for branched oligosaccharides (Kd values of 4.5–7.7 μm) but less affinity than hG9N and/or hG9C. Because bG1 has 87% sequence identity to hG1 (Fig. 4), it should have comparable affinity. Supposing that a branched oligosaccharide binds to bG1 as a ligand, bG1, having a deep carbohydrate-binding site, can form a stable protein-ligand complex of low structural energy with many attractive interactions when compared with hG9C. However, the conformation of the bound branched oligosaccharide may be restricted by strong protein-ligand interactions. Gly53 of bG1 forming van der Waals contacts with Man+3 and Man+4 likely induces the conformational change of glucoside bonds for ligand binding. In the case of hG9C, protein-ligand interactions are limited to Gal+1 and GlcNAc+2, and other sugar units are free from the protein, meaning that hG9C can recognize the antennae of a branched oligosaccharide in various conformations. Because N-glycans are generally thought to be in dynamic motion in vitro and in vivo, the wide entrance and shallow binding site of hG9C seem favorable for binding. In hG9C, the less structural energy in the formation of a protein-ligand complex is probably compensated for by the ability to accept branched oligosaccharides in various conformations, without inducing any conformational changes. This is one of the reasons why hG9 has high affinity for N-glycan-type oligosaccharides. The subtle structural difference in the entrance for ligand binding found between hG9N and hG9C may explain the unidentified specificity of hG9N and/or hG9C toward the N-glycan type oligosaccharides.
The x-ray structure of hG9C-SiaLac revealed that Arg221 efficiently recognizes Sia−1 by changing its conformation. This Arg residue is conserved among the CRDs of galectins, which have significant affinity for sialylated oligosaccharides. As shown in Fig. 3a, Asp241 forms strong hydrogen bonds with Arg221 in hG9C-BIPA, partially preventing Arg221 from changing its conformation, and this may lead to the two conformers of Arg221 in hG9C-SiaLac. As well as the frontal affinity chromatography analysis, ELISA and the surface plasmon resonance assay showed that the human galectin-8 N-terminal CRD (hG8N) has great affinity for SiaLac (44). For the structural comparison of hG9C with hG8N, the x-ray structure of hG8N in a complex with lactose (PDB code: 2YXS) is available. Instead of Arg221 and Asp241, hG8N has Arg52 and Lys78, which are incapable of attractive interactions. In practice, the Arg52 of hG8N adopts conformer-2 to avoid short contacts with Lys78. There is no galectin CRD that has a positively charged amino acid residue at the position of Lys78 (hG8N) or Asp241 (hG9C), except for hG8N (Fig. 4). The superimposition of hG8N on hG9C-SiaLac also showed that Arg66 of hG8N, located in the long S3–S4 loop (9 amino acid residues, Fig. 4), particular to hG8N, directs its side chain toward the carboxyl group of Sia−1, possibly forming salt-bridge interactions. These results suggest that SiaLac binds to hG8N with the same conformation as found in hG9C-SiaLac and that Arg52, Lys78, and Arg66 of hG8N contribute to the striking affinity of hG8N for SiaLac.
The hG9 having two CRDs is divalent in glycan binding with different specificity. Structural comparison of hG9C with hG9N explained the difference in specificity between them. This structural feature of hG9 should be related to its various biological functions. When hG9 binds to ligands on different entities, it may mediate interactions between cells or cell and pathogen. When hG9 binds to ligands on a single cell, it may cross-link different target molecules. The presented structural information could give a clue to elucidate the molecular mechanism underlying the biological functions of hG9.
Supplementary Material
Acknowledgments
This research was performed with the approval of the Photon Factory Advisory Committee and the National Laboratory for High Energy Physics, Tsukuba, Japan.
This work was supported in part by Kagawa University Characteristic Prior Research Fund 2009-2010.
The atomic coordinates and structure factors (codes 3NV1, 3NV2, 3NV3, and 3NV4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CRD
- carbohydrate recognition domain
- TIM3
- T-cell immunoglobulin and mucin-containing-protein 3
- Th1
- T-helper type 1
- hG9
- human galectin-9
- hG9N
- hG9 N-terminal CRD
- hG9C
- hG9 C-terminal CRD
- bG1
- bovine galectin-1
- hG1
- human galectin-1
- hG8N
- human galectin-8 N-terminal CRD
- LacNAc
- N-acetyllactosamine
- LN3
- LacNAc trimer
- SiaLac
- α2–3-sialyllactose
- BIPA
- biantennary pyridylaminated oligosaccharide
- BIOS
- biantennary octasaccharide.
REFERENCES
- 1.Barondes S. H., Castronovo V., Cooper D. N., Cummings R. D., Drickamer K., Feizi T., Gitt M. A., Hirabayashi J., Hughes C., Kasai K., et al. (1994) Cell 76, 597–598 [DOI] [PubMed] [Google Scholar]
- 2.Kasai K., Hirabayashi J. (1996) J. Biochem. 119, 1–8 [DOI] [PubMed] [Google Scholar]
- 3.Rabinovich G. A., Baum L. G., Tinari N., Paganelli R., Natoli C., Liu F. T., Iacobelli S. (2002) Trends Immnol. 23, 313–320 [DOI] [PubMed] [Google Scholar]
- 4.Stowell S. R., Arthur C. M., Dias-Baruffi M., Rodrigues L. C., Gourdine J. P., Heimburg-Molinaro J., Ju T., Molinaro R. J., Rivera-Marrero C., Xia B., Smith D. F., Cummings R. D. (2010) Nat. Med. 16, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasta G. R. (2009) Nat. Rev. Microbiol. 7, 424–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper D. N. (2002) Biochim. Biophys. Acta. 1572, 209–231 [DOI] [PubMed] [Google Scholar]
- 7.Hirabayashi J., Kasai K. (1993) Glycobiology. 3, 297–304 [DOI] [PubMed] [Google Scholar]
- 8.Türeci O., Schmitt H., Fadle N., Pfreundschuh M., Sahin U. (1997) J. Biol. Chem. 272, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 9.Wada J., Kanwar Y. S. (1997) J. Biol. Chem. 272, 6078–6086 [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto R., Matsumoto H., Seki M., Hata M., Asano Y., Kanegasaki S., Stevens R. L., Hirashima M. (1998) J. Biol. Chem. 273, 16976–16984 [DOI] [PubMed] [Google Scholar]
- 11.Hirashima M. (1999) Int. Arch. Allergy. Immunol. 120, 7–10 [DOI] [PubMed] [Google Scholar]
- 12.Matsushita N., Nishi N., Seki M., Matsumoto R., Kuwabara I., Liu F. T., Hata Y., Nakamura T., Hirashima M. (2000) J. Biol. Chem. 275, 8355–8360 [DOI] [PubMed] [Google Scholar]
- 13.Kageshita T., Kashio Y., Yamauchi A., Seki M., Abedin M. J., Nishi N., Shoji H., Nakamura T., Ono T., Hirashima M. (2002) Int. J. Cancer. 99, 809–816 [DOI] [PubMed] [Google Scholar]
- 14.Kashio Y., Nakamura K., Abedin M. J., Seki M., Nishi N., Yoshida N., Nakamura T., Hirashima M. (2003) J. Immunol. 170, 3631–3636 [DOI] [PubMed] [Google Scholar]
- 15.Seki M., Sakata K. M., Oomizu S., Arikawa T., Sakata A., Ueno M., Nobumoto A., Niki T., Saita N., Ito K., Dai S. Y., Katoh S., Nishi N., Tsukano M., Ishikawa K., Yamauchi A., Kuchroo V., Hirashima M. (2007) Arthritis Rheum. 56, 3968–3976 [DOI] [PubMed] [Google Scholar]
- 16.Seki M., Oomizu S., Sakata K. M., Sakata A., Arikawa T., Watanabe K., Ito K., Takeshita K., Niki T., Saita N., Nishi N., Yamauchi A., Katoh S., Matsukawa A., Kuchroo V., Hirashima M. (2008) Clin. Immunol. 127, 78–88 [DOI] [PubMed] [Google Scholar]
- 17.Tanikawa R., Tanikawa T., Hirashima M., Yamauchi A., Tanaka Y. (2010) Biochem. Biophys. Res. Commun. 394, 317–322 [DOI] [PubMed] [Google Scholar]
- 18.Niki T., Tsutsui S., Hirose S., Aradono S., Sugimoto Y., Takeshita K., Nishi N., Hirashima M. (2009) J. Biol. Chem. 284, 32344–32352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu C., Anderson A. C., Schubart A., Xiong H., Imitola J., Khoury S. J., Zheng X. X., Strom T. B., Kuchroo V. K. (2005) Nat. Immunol. 6, 1245–1252 [DOI] [PubMed] [Google Scholar]
- 20.Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 21.Lobsanov Y. D., Gitt M. A., Leffler H., Barondes S. H., Rini J. M. (1993) J. Biol. Chem. 268, 27034–27038 [DOI] [PubMed] [Google Scholar]
- 22.Liao D. I., Kapadia G., Ahmed H., Vasta G. R., Herzberg O. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1428–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourne Y., Bolgiano B., Liao D. I., Strecker G., Cantau P., Herzberg O., Feizi T., Cambillau C. (1994) Nat. Struct. Biol. 1, 863–870 [DOI] [PubMed] [Google Scholar]
- 24.Seetharaman J., Kanigsberg A., Slaaby R., Leffler H., Barondes S. H., Rini J. M. (1998) J. Biol. Chem. 273, 13047–13052 [DOI] [PubMed] [Google Scholar]
- 25.Leonidas D. D., Vatzaki E. H., Vorum H., Celis J. E., Madsen P., Acharya K. R. (1998) Biochemistry 37, 13930–13940 [DOI] [PubMed] [Google Scholar]
- 26.López-Lucendo M. F., Solís D., André S., Hirabayashi J., Kasai K., Kaltner H., Gabius H. J., Romero A. (2004) J. Mol. Biol. 343, 957–970 [DOI] [PubMed] [Google Scholar]
- 27.Sörme P., Arnoux P., Kahl-Knutsson B., Leffler H., Rini J. M., Nilsson U. J. (2005) J. Am. Chem. Soc. 127, 1737–1743 [DOI] [PubMed] [Google Scholar]
- 28.Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. (2006) J. Biol. Chem. 281, 35884–35893 [DOI] [PubMed] [Google Scholar]
- 29.Nagae M., Nishi N., Nakamura-Tsuruta S., Hirabayashi J., Wakatsuki S., Kato R. (2008) J. Mol. Biol. 375, 119–135 [DOI] [PubMed] [Google Scholar]
- 30.Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. (2009) Glycobiology 19, 112–117 [DOI] [PubMed] [Google Scholar]
- 31.Nishi N., Itoh A., Fujiyama A., Yoshida N., Araya S., Hirashima M., Shoji H., Nakamura T. (2005) FEBS Lett. 579, 2058–2064 [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33.Nakakita S., Sumiyoshi W., Miyanishi N., Hirabayashi J. (2007) Biochem. Biophys. Res. Commun. 362, 639–645 [DOI] [PubMed] [Google Scholar]
- 34.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 35.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 37.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38.Brünger A. T. (1993) X-PLOR 3.1: A System for X-ray Crystallography and NMR, Yale University Press, New Haven, CT [Google Scholar]
- 39.Ramachandran G. N., Sasisekharan V. (1968) Advan. Protein. Chem. 23, 283–437 [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1992) PROCHECK Version 2: Programs to Check the Stereochemical Quality of Protein Structures, Oxford Molecular Ltd, Oxford [Google Scholar]
- 41.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 42.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 43.Miyanishi N., Nishi N., Abe H., Kashio Y., Shinonaga R., Nakakita S., Sumiyoshi W., Yamauchi A., Nakamura T., Hirashima M., Hirabayashi J. (2007) Glycobiology 17, 423–432 [DOI] [PubMed] [Google Scholar]
- 44.Ideo H., Seko A., Ishizuka I., Yamashita K. (2003) Glycobiology 13, 713–723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.