Abstract
Nitrogen starvation-mediated reduction of Ypk1 is suggested to suppress translational initiation, possibly in parallel with the target of rapamycin complex 1 (TORC1) signaling. However, the molecular mechanism that regulates Ypk1 in nitrogen-starved cells is poorly understood. Here we report that Ypk1 is a novel selective substrate for nitrogen starvation-triggered proteolysis requiring autophagy system. Among various nutrient starvation methods used to elicit autophagy, rapid Ypk1 degradation was specific to nitrogen starvation. In screening genes required for such nitrogen starvation-specific vacuolar proteolysis, we found that autophagy-related degradation of Ypk1 depended on the endosomal sorting complex required for transport (ESCRT) machinery, which is conventionally thought to function in endosomal trafficking. In microscopic analyses, the disruption of ESCRT subunits resulted in the accumulation of both Ypk1 and autophagosomal Atg8 at a perivacuolar site that was distinct from conventional endosomes. ESCRT machinery was not involved in autophagic flux induced by the TORC1 inhibitor rapamycin, thus suggesting that ESCRT represents an exclusive mechanism of nitrogen starvation-specific proteolysis of Ypk1. Overall, we propose a novel regulation of Ypk1 that is specific to nitrogen limitation.
Keywords: Autophagy, Protein Degradation, Protein Kinases, Sphingolipid, Yeast, Nitrogen Starvation, Ypk1
Introduction
Cells control their proliferation and growth by modulating anabolic and catabolic processes in response to extracellular nutrient availability. Nutrient starvation stops cellular anabolic processes by rapidly inhibiting ribosome biogenesis and protein translation to conserve limited nutrients. Concomitantly, starvation induces a cellular catabolic process, autophagy, to supply limited nutrients. The cellular response to starvation is regulated, at least in part, by the evolutionally and structurally well conserved target of rapamycin complex 1 (TORC1),3 which plays pivotal roles in the regulation of cellular proliferation and growth in nutrient-responsive signaling (1–4). Both nutrient limitation and blocking of TORC1 by rapamycin induce autophagy (5, 6); therefore, rapamycin-mediated autophagy is considered to mimic that of nutrient starvation, and a clear distinction between the two is not well documented.
YPK1 encodes a serine/threonine protein kinase belonging to the cyclic AMP-dependent protein kinase/protein kinase G/protein kinase C (AGC) kinase family. Although a direct substrate of this yeast protein kinase has not been identified, loss of Ypk1 causes several cellular deficiencies in actin cytoskeletal organization (7) and endocytosis (8) as well as resistance to ISP-1/myriocin, a potent inhibitor of sphingolipid biosynthesis (9). As a common feature of AGC kinases, Ypk1 kinase activity is regulated by three phosphorylations at the T-loop, turn motif, and hydrophobic motif. Phosphorylation of Ypk1 at the T-loop is regulated by Pkh1/2 kinases, yeast homologs of 3-phosphoinositide-dependent kinase (PDK1) (10), and those at the turn and hydrophobic motifs are mediated by rapamycin-insensitive TORC2 (11).
Although Ypk1 is considered a downstream target of TORC2 signaling (11), note that previous genetic studies imply a role for Ypk1 in nutrient-responsive signaling in parallel with the TORC1 pathway. Loss of Ypk1 leads to hypersensitivity to rapamycin, synthetic growth defects with TOR1 depletion (12, 13), and synthetic lethality with loss of its downstream effectors, 14-3-3 proteins (12, 13). These defects might be explained by the regulation of translational cap-dependent initiation of messenger RNA because the activity of both TORC1 (14) and Ypk1 (15) is required for stability of eIF4G. The regulatory mechanism of TORC1 under nutrient-limited conditions has been extensively studied (1, 3, 4). In contrast, study of Ypk1 in nutrient-starved cells is limited to the pioneering genetic analysis of Gelperin et al. (15) that showed that Ypk1 expression diminishes rapidly in response to nitrogen starvation but not glucose starvation or TORC1 inhibition by rapamycin.
Here we examined the regulatory mechanism of Ypk1 expression in nitrogen-starved yeast. Nitrogen starvation caused rapid vacuolar proteolysis of Ypk1 in as early as 1 h. This rapid Ypk1 degradation was selective to the limitation of nitrogen. Although rapamycin did not induce selective Ypk1 degradation, autophagy system was required for this proteolytic event. The autophagy-related selective proteolysis of Ypk1 utilized the endosomal sorting complex required for transport (ESCRT) in Ypk1 sorting to vacuoles. Overall, we demonstrated that nitrogen starvation triggers rapid and autophagy-related selective proteolysis of the translational regulator Ypk1 that precedes bulk degradation of cellular components by autophagy.
EXPERIMENTAL PROCEDURES
Yeast Strains, Culture Conditions, Reagents, and Plasmids
The yeast strains used in this study are listed in Table 1. Construction of the deletion strains was achieved through PCR-based homologous recombination as described previously (16, 17). The conditions for cell culture and plating assays were as reported previously (18). For nitrogen starvation, SD-N medium (1.7 g of DifcoTM yeast nitrogen base without amino acids and ammonium sulfate (BD Biosciences) and 20 g of glucose/liter) was used. SD-P and SD-S media were prepared as described in Ref. 19 except that 2% glucose was supplemented instead of 0.5%. Phenylmethylsulfonyl fluoride (PMSF; Nacalai Tesque, Kyoto, Japan) and rapamycin (Calbiochem) were dissolved in methanol. FM4-64 (Invitrogen) was dissolved in dimethyl sulfoxide and used at a final concentration of 20 μm. The polyclonal antibodies used against Ypk1 were the same as in Ref. 9. Monoclonal antibodies against GFP and Pgk1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and Invitrogen, respectively. The polyclonal antibodies against Ape1 were a kind gift from Dr. Daniel J. Klionsky (University of Michigan) (20). HRP-conjugated secondary antibodies were purchased from Zymed Laboratories Inc. (South San Francisco, CA). To construct the pRS415-GFP-YPK1, the GFP fragment from pFA6a-GFP(S65T)-HIS3MX6 was introduced into the MroI site between the promoter and the initiation codon of YPK1 of pRS415-YPK1. The BamHI-SacI fragment of GFP-YPK1, including promoter and polyadenylation signal of YPK1, was transferred into pRS413 to generate pRS413-GFP-YPK1. To construct the pRS415-TAT2-EGFP, the EGFP fragment was introduced into the SalI and XbaI sites prior to the termination codon of TAT2 of pRS415-TAT2, including the region from the promoter to the polyadenylation signal of TAT2. The pRS316-GFP-ATG8 plasmid was kindly provided by Dr. Yoshinori Ohsumi (Tokyo Institute of Technology) (21). The SacI-XhoI fragment of GFP-ATG8 was introduced into pRS415, generating pRS415-GFP-ATG8. The pRS414-RFP-ATG8 plasmid was kindly provided by Dr. Daniel J. Klionsky (22). The EcoRI-XhoI fragment of RFP-ATG8 was transferred into pRS413 to generate pRS413-RFP-ATG8, and the BamHI-XhoI fragment was transferred into BamHI-SalI sites of YEp351 to generate YEp351-RFP-ATG8.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| BY4741 | MATa his3-1 leu2-0 met15-0 ura3-0 | 16 |
| pep4Δ | BY4741 pep4::URA3 | This study |
| ypk1Δ | BY4741 ypk1::HIS3MX6 | This study |
| ypk1Δpep4Δ | BY4741 ypk1::URA3 pep4::KanMX4 | This study |
| pep4Δatg1Δ | BY4741 pep4::KanMX4 atg1::URA3 | This study |
| atg1Δ | BY4741 atg1::URA3 | This study |
| atg5Δ | BY4741 atg5::KanMX4 | 56 |
| atg7Δ | BY4741 atg7::KanMX4 | 56 |
| atg11Δ | BY4741 atg11::KanMX4 | 56 |
| ssa1Δ | BY4741 ssa1::KanMX4 | 56 |
| ssa2Δ | BY4741 ssa1::KanMX4 | 56 |
| ALD6-GFP | BY4741 ALD6-GFP::HIS3MX6 | 57 |
| vps27Δ | BY4741 vps27::KanMX4 | 56 |
| vps23Δ | BY4741 vps23::KanMX4 | 56 |
| vps22Δ | BY4741 vps22::KanMX4 | 56 |
| vps36Δ | BY4741 vps36::KanMX4 | 56 |
| vps2Δ | BY4741 vps2::KanMX4 | 56 |
| ubp3Δ | BY4741 ubp3::KanMX4 | 56 |
| bre5Δ | BY4741 bre5::KanMX4 | 56 |
| end3Δ | BY4741 end3::KanMX4 | 56 |
| rvs161Δ | BY4741 rvs161::KanMX4 | 56 |
| rvs167Δ | BY4741 rvs167::KanMX4 | 56 |
| sla1Δ | BY4741 sla1::KanMX4 | 56 |
| pep4Δvps27Δ | BY4741 pep4::URA3 vps27::KanMX4 | This study |
| pep4Δvps23Δ | BY4741 pep4::URA3 vps23::KanMX4 | This study |
Western Blotting Analysis
Harvested yeast cells were resuspended in lysis buffer (50 mm Tris-HCl buffer (pH 8.0) containing 5 mm EDTA, 150 mm NaCl, 50 mm sodium fluoride, 30 mm β-glycerophosphate, 1 mm PMSF, protease inhibitor mixture for general use (Nacalai Tesque), and 0.5% Triton X-100). The cell suspension was continuously vortexed for 30 min at 4 °C in a microtube mixer (model MT-360, Tomy, Tokyo, Japan) with a half-volume of glass beads to lyse the yeast cells. Unbroken cells and debris were removed by centrifugation at 500 × g for 10 min, and the supernatant containing 30 μg of protein was used for SDS-PAGE analysis. The signals were detected by a LAS-3000 imager (Fujifilm, Tokyo, Japan) with Chemilumi-One (Nacalai Tesque) as a chemiluminescent substrate.
Fluorescence Microscopy
Cells expressing GFP-Ypk1, Tat2-EGFP, or GFP-Atg8 were cultured to early log phase and stained with FM4-64 for 1 h. After removal of FM4-64, the cells were chased for 1 h in SD medium supplemented with auxotrophic nutrients. For nitrogen starvation, cells were washed with SD-N medium and resuspended in SD-N medium containing 1 mm PMSF. For colocalization analysis of GFP-Ypk1/Tat2-EGFP with RFP-Atg8 and confocal microscopic analysis of GFP-Ypk1 in FM4-64-stained pep4Δ cells, cell culture was terminated by the addition of 10 mm NaF and 10 mm NaN3 and 10-min incubation on ice. Cells were washed twice with ice-cold SD or SD-N medium and imaged immediately after these treatments. The fluorescent signals were observed using an inverted microscope (IX70, Olympus, Tokyo, Japan) equipped with a charge-coupled device camera (Cool SNAP HQ/OL, Photometrics, Tucson, AZ) or laser-scanning confocal microscope (FV1000D IX81, Olympus) using 488 and 559 nm lasers. For 3D imaging, fluorescent signals were collected along Z-sections at 0.2-μm intervals from the top to bottom of the GFP signal. All images were analyzed using Metamorph (Olympus) or ImageJ software. For quantification of the localization, frequencies of colocalization (GFP-Ykp1 versus FM4-64 or RFP-Atg8) or non-colocalization (Tat2-EGFP versus FM4-64 or RFP-Atg8) between two channels in the collected images from at least duplicated independent experiments were calculated.
RESULTS
Nitrogen Starvation Stimulates Rapid Vacuolar Proteolysis of Ypk1
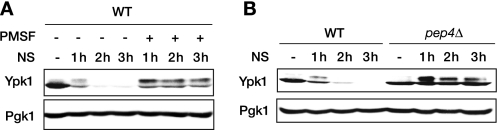
We were interested in the regulatory mechanism of Ypk1 in relation to nutrient signaling because ypk1Δ cells show a “slow growth” phenotype, and expression of Ypk1 is regulated in nitrogen-limited cells (15). Ypk1 and TORC1 may share an intracellular signaling pathway required for proper protein translation (14, 15, 23). In our semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis, YPK1 mRNA expression did not change in nitrogen-starved cells (supplemental Fig. S1). Therefore, we investigated whether rapid reduction of Ypk1 expression in nitrogen-starved cells was caused by proteolysis. In yeast cells, proteins can be degraded in either proteasomes or vacuoles. PMSF is a selective inhibitor of vacuolar serine proteases but does not affect proteasome function (24). Thus, we investigated Ypk1 expression in nitrogen-starved cells with this inhibitor. PMSF strongly blocked the nitrogen starvation-induced reduction of Ypk1 (Fig. 1A). In these assays, Ypk1 was detected as a doublet in which the slower migrating band represents the phosphorylated form of the protein (25). The nitrogen starvation-triggered decrease in Ypk1 was also inhibited in pep4Δ cells, which are deficient in mature vacuolar proteases (Fig. 1B). These observations indicate that vacuolar proteolysis is the main cause of the rapid reduction of Ypk1 upon nitrogen starvation.
FIGURE 1.
Nitrogen starvation induces vacuolar proteolysis of Ypk1. A, effects of a vacuolar protease inhibitor, PMSF, on nitrogen starvation-induced Ypk1 reduction. Logarithmically growing WT cells in SD medium supplemented with auxotrophic nutrients were starved of nitrogen (NS) in SD-N medium containing drug vehicle or 1 mm PMSF. Total cell lysates prepared at the indicated time points were resolved by SDS-PAGE and analyzed by Western blotting against Ypk1 (two bands) and a loading control, Pgk1. B, Ypk1 expression in nitrogen-starved WT and pep4Δ cells. Western blotting against Ypk1 and Pgk1 was performed as in A.
Ypk1 Is Sorted into Vacuoles and Localizes at Intravacuolar Exaggerated Structure in Vacuolar Protease-deficient Cells upon Nitrogen Starvation
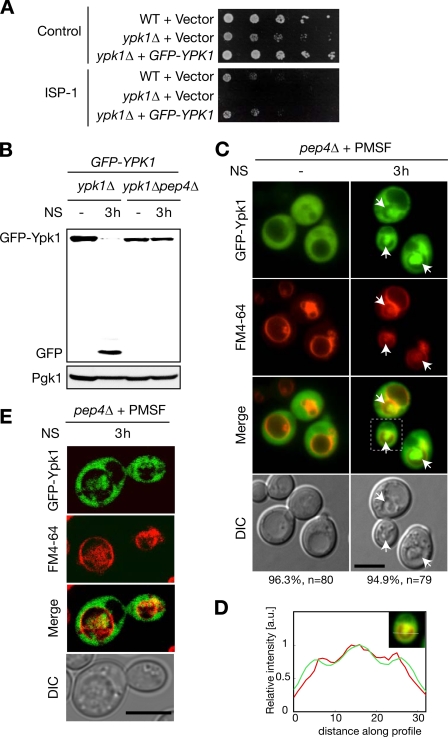
To further analyze vacuolar degradation of Ypk1, we investigated whether Ypk1 was sorted into vacuoles in response to nitrogen starvation. For microscopic analysis, we constructed N-terminal GFP fusion Ypk1 (GFP-Ypk1) because C-terminal fusion Ypk1-EGFP is not functional (25). Expression of N-terminal fusion GFP-Ypk1 from a single copy vector complemented ypk1Δ phenotypes both in terms of cellular proliferation and resistance to ISP-1/myriocin (Fig. 2A). Consistently, Western blotting against GFP showed a simultaneous decrease in GFP-Ypk1 and increase in a smaller fragment around 27 kDa in nitrogen-starved control cells (ypk1Δ cells expressing GFP-Ypk1) but not in pep4Δ cells (Fig. 2B). Because GFP is relatively resistant to vacuolar proteolysis in yeast (26, 27), this fragment could be a cleaved form of GFP inferring in situ vacuolar proteolysis of GFP-Ypk1. These results indicate that the N-terminal fusion GFP-Ypk1 behaves as WT Ypk1. We treated pep4Δ cells with PMSF to visualize vacuolar structure in differential interference contrast images upon nitrogen starvation because pep4Δ cells exhibited less defined vacuoles under such conditions without PMSF as reported previously (28). In live cell imaging, FM4-64 lipophilic dye staining was performed to visualize vacuolar membranes prior to nitrogen starvation. Unlike our previous report in which overexpressing Ypk1-EGFP from a multicopy vector resulted in significant plasma membrane association (25), GFP-Ypk1 primarily localized in the cytoplasm of both WT and pep4Δ cells under nutrient-supplied conditions (supplemental Fig. S2 and Fig. 2C). When nitrogen was removed for 3 h, prominent accumulation of GFP-Ypk1 in the vacuolar space was observed in PMSF-treated pep4Δ cells (Fig. 2, C and D). This was consistent with the vacuolar protease-dependent nitrogen starvation-mediated proteolysis of both Ypk1 and GFP-Ypk1 (Figs. 1 and 2B). To completely rule out a possible protrusion of GFP-Ypk1 signal from the cytoplasmic space, we used a confocal microscope and three-dimensional reconstruction. Accumulated Ypk1 signal was surrounded by the FM4-64-positive vacuolar membrane (supplemental Movie 1 and Fig. 2E). Therefore, we hereafter refer to this Ypk1 accumulation as “intravacuolar.” The intravacuolar GFP-Ypk1 colocalized with exaggerated structures in differential interference contrast images. These abnormal structures were composed of membrane as indicated by the fact that FM4-64 was also incorporated (supplemental Movie 1 and Fig. 2C, arrows). These structures resembled previously reported nitrogen starvation-triggered autophagic bodies (28, 29).
FIGURE 2.
GFP-Ypk1 is sorted into vacuoles upon nitrogen starvation. A, complementation assays on GFP-YPK1 in ypk1Δ background. WT or ypk1Δ cells transformed with vector (pRS415) control or pRS415-GFP-YPK1-harboring ypk1Δ cells were plated on leucine-deficient SD plates containing methanol or 250 ng/ml ISP-1 to examine complementation of the proliferative defect or ISP-1 resistance, respectively. The plates were incubated at 30 °C for 2 days. B, effect of N-terminal GFP fusion on nitrogen starvation-mediated vacuolar proteolysis of Ypk1. Expression of GFP-Ypk1 and cleaved GFP were analyzed by Western blotting with an antibody against GFP in ypk1Δ or ypk1Δpep4Δ cells transformed with pRS415-GFP-YPK1 upon nitrogen starvation (NS). C and D, fluorescence microscopic analyses of GFP-Ypk1 in nitrogen-starved pep4Δ cells. Logarithmically growing pep4Δ cells harboring pRS415-GFP-YPK1 were pulsed with FM4-64 lipophilic dye for 1 h and chased for 1 h. The FM4-64-labeled cells were starved of nitrogen in the presence of PMSF. Localization of GFP-Ypk1 fluorescence was examined 3 h after nitrogen starvation. Differential interference contrast (DIC) images were utilized to locate exaggerated structures inside vacuoles (arrows). Percentages noted below the images represent frequencies of the indicated phenotypes. Fluorescence intensity collected along a line was normalized with the maximum intensity in each channel. Relative fluorescence intensities of GFP-Ypk1 (green) and FM4-64 (red) were plotted (D). E, confocal microscopic analyses of GFP-Ypk1 in nitrogen-starved pep4Δ cells. Logarithmically growing pep4Δ cells harboring pRS415-GFP-YPK1 were pulse-chased with FM4-64 and starved of nitrogen as described in C. Cell culture was terminated by NaF and NaN3 treatment on ice as described under “Experimental Procedures,” and the fluorescence was immediately imaged by confocal microscope after this treatment. See also supplemental Movie 1. Bars, 5 μm. a.u., arbitrary units.
Vacuolar Sorting and Proteolysis of Ypk1 Are under Control of Autophagy System upon Nitrogen Starvation
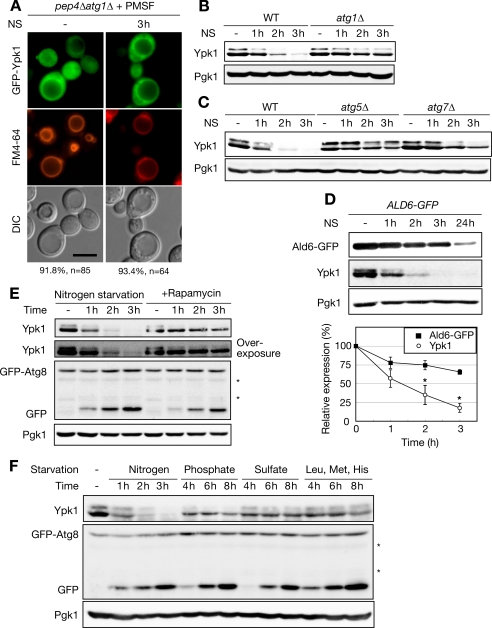
Autophagy may sort cytoplasmic Ypk1 into vacuoles. However, expression of Ypk1 was not altered in cells with limited glucose or treated with rapamycin (15), both of which induce autophagy (5, 28). Nevertheless, because nitrogen starvation is known to trigger autophagic proteolysis, we assessed the possible involvement of autophagy in Ypk1 degradation. In general, autophagy accounts for bulk proteolysis in nutrient-starved cells to supply amino acids using a specific membrane-trafficking system composed of Atg (autophagy-related) proteins (30). ATG1 encodes a serine/threonine protein kinase required for the induction of autophagy (6, 31). The null mutant of this gene is defective in initiating the formation of autophagosomes, a unique double membrane structure that sequesters cytoplasmic proteins or organelles from the cytosol (32). To investigate whether the vacuolar sorting of Ypk1 and the intravacuolar structures observed in PMSF-treated pep4Δ cells were regulated by autophagy, we analyzed the localization of GFP-Ypk1 in PMSF-treated pep4Δatg1Δ cells. We did not detect the accumulation of intravacuolar GFP-Ypk1 or the exaggerated structures, although a portion of GFP-Ypk1 localized proximally to vacuolar membranes (Fig. 3A). In addition to the defective vacuolar sorting, proteolysis of Ypk1 was also inhibited in atg1Δ cells (Fig. 3B) and other autophagy mutants such as atg5Δ and atg7Δ cells (Fig. 3C), both of which show defective autophagosome formation (Ref. 30 and references therein). Atg1, Atg5, and Atg7 are also involved in the cytoplasm-to-vacuolar targeting (Cvt) pathway, a constitutive vacuolar sorting pathway (30, 33), and cargo for this pathway resides in the cytoplasm; thus, we investigated the involvement of the Cvt pathway in nitrogen starvation-triggered Ypk1 proteolysis. Deletion of ATG11, which is required for the maturation of Ape1 in the Cvt pathway but not in autophagic transport (34), did not block the degradation of Ypk1 (supplemental Fig. S3A). In addition, nitrogen starvation-activated Hsp70-mediated vacuolar delivery, another system to sort cytoplasmic proteins to vacuoles (35), was not involved in Ypk1 proteolysis because deletion of SSA1, which also affects the maturation of Ape1 in the Cvt pathway (36), did not affect Ypk1 instability in nitrogen-starved cells (supplemental Fig. S3, B and C, and supplemental text). These results indicate that autophagy system, but neither the Cvt pathway nor Hsp70-mediated delivery, is involved in nitrogen starvation-triggered vacuolar sorting of Ypk1 for degradation and that the exaggerated intravacuolar structures observed in vacuolar protease-deficient cells (Fig. 2C) could be related to autophagic bodies.
FIGURE 3.
Ypk1 is a novel and selective substrate for nitrogen starvation-specific proteolysis requiring autophagy system. A, fluorescence microscopic analyses of GFP-Ypk1 in PMSF-treated pep4Δatg1Δ cells. Logarithmically growing pep4Δatg1Δ cells harboring pRS415-GFP-YPK1 were pulse-chased with FM4-64, starved of nitrogen, and examined by fluorescence microscopy for GFP-Ypk1 and FM4-64 as described in Fig. 2C. Percentages noted below the images represent frequencies of the indicated phenotypes. Bar, 5 μm. B and C, time course analysis of Ypk1 expression in nitrogen-starved WT, atg1Δ (B), atg5Δ (C), and atg7Δ (C) cells. Logarithmically growing cells were starved of nitrogen (NS) for the times indicated. Western blot analysis was performed as described in Fig. 1A. D, comparison of the degradation kinetics between Ypk1 and Ald6-GFP. Total cell lysates were prepared from ALD6-GFP cells starved of nitrogen for the indicated times and analyzed by Western blotting. The expression of Ald6-GFP was detected using anti-GFP antibody. Relative expressions of Ald6-GFP and Ypk1 were normalized by Pgk1 signal intensities and plotted on a line graph as means of three independent experiments. Error bars indicate S.E. Asterisks show statistical significance at p < 0.05, determined using Student's t test. E, comparison of degradation kinetics Ypk1 between nitrogen-starved or rapamycin-treated WT cells harboring pRS316-GFP-ATG8. Logarithmically growing cells in SD medium supplemented with auxotrophic nutrients were shifted to SD-N medium or treated with 200 ng/ml rapamycin. Cells were harvested at the indicated times, and Western blots against Ypk1 and Pgk1 were performed as described in Fig. 1A. Autophagic progression was monitored by GFP-Atg8 processing by Western blotting using an antibody against GFP. Asterisks indicate nonspecific signals. F, effect of various types of starvation on Ypk1 expression. Logarithmically growing cells harboring pRS316-GFP-ATG8 in SD medium supplemented with auxotrophic nutrients were shifted to SD-N medium, SD-P/SD-S medium with auxotrophic nutrients, or SD medium lacking auxotrophic nutrients. Cells were harvested at the indicated time point, and total cell lysates were analyzed by Western blotting as described in E. DIC, differential interference contrast.
Characterization of Autophagy-related Degradation of Ypk1
Although Atg proteins were required for Ypk1 degradation, the proteolytic properties of Ypk1 did not fit well with conventional autophagic protein degradation; autophagy-related proteolysis of Ypk1 occurred within 2 h of nitrogen starvation (Fig. 3, B and C), whereas bulk autophagy takes days to deplete long lived proteins. Therefore, we further characterized Ypk1 degradation. As Ald6 is a preferred substrate for rapid and selective autophagic degradation prior to bulk autophagy (37), we compared the kinetics of Ypk1 degradation with those of Ald6 and found that Ypk1 degradation was even faster than that of Ald6 (Fig. 3D). This indicates that Ypk1 can be a novel selective substrate for nitrogen starvation-induced proteolysis requiring autophagy system. Another conflict with conventional autophagic proteolysis is that Ypk1 seemed to diminish only in response to nitrogen starvation, not to glucose starvation or rapamycin treatment (15) also shown in Fig. 3E, whereas bulk autophagy is induced by various starvation conditions and the TORC1 inhibitor rapamycin (5, 28). Therefore, we tested whether these autophagy-inducible conditions could elicit Ypk1 degradation. Autophagy progression was monitored by GFP-Atg8 processing (27). When autophagy is induced, Atg8 is conjugated to phosphatidylethanolamine and regulates formation of autophagosomes. As a result, GFP-Atg8 is transported to vacuoles within autophagosomes and subsequently processed. This event can be monitored by a proteolysis-resistant cleaved, and thus size-shifted, GFP tag in vacuoles. Monitoring this cleaved GFP from GFP-Atg8 allows us to examine the total flux of autophagy. Ypk1 was rather stable in rapamycin-treated cells (Fig. 3E) compared with nitrogen-starved cells. In contrast, rapamycin induced GFP-Atg8 processing as much as nitrogen starvation. Although a subtle decrease in the phosphorylated form of Ypk1 was detected, overexposure of the blot clearly showed this phosphorylated species of Ypk1, indicating that Ypk1 phosphorylation is not an event downstream of TORC1 (Fig. 3E, overexposed blot for Ypk1). Only gradual reduction of Ypk1 was observed in cells starved of natural nutrients (phosphate and sulfate) or auxotrophic amino acids (leucine, methionine, and histidine; Fig. 3F and Table 2), although starvation of these nutrients caused accumulation of cleaved GFP comparable with that of nitrogen starvation. These results indicate that rapid degradation of Ypk1 is exclusively induced by extracellular nitrogen limitation. To the best of our knowledge, Ypk1 is the first reported cargo protein exhibiting such nitrogen starvation selectivity for autophagy-related degradation. In addition, inhibition of TORC1 by rapamycin did not mimic nitrogen starvation as far as Ypk1 degradation is concerned.
TABLE 2.
Comparison of autophagy and Ypk1 proteolysis under various culture conditions
ESCRT Machinery Facilitates Autophagy-related Proteolysis of Ypk1 upon Nitrogen Starvation
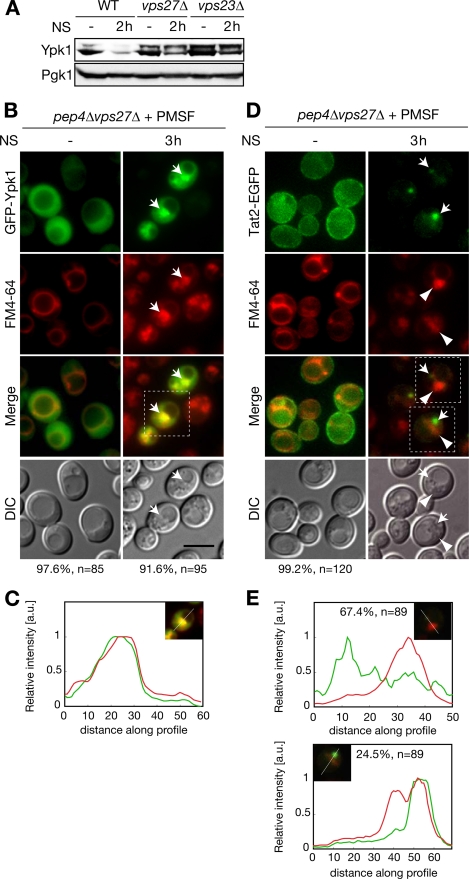
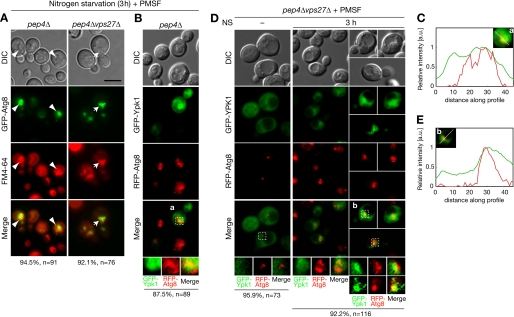
Because Ypk1 degradation exhibited distinct characteristics as a conventional autophagy cargo protein, the mechanistic basis for such a distinction should be present in the pathway in which Ypk1 is degraded in vacuoles upon nitrogen starvation but not after other autophagic stimuli (Table 2). Therefore, we target-screened for genes enabling this pathway. We examined changes in Ypk1 expression upon nitrogen starvation by Western blotting to determine the responsible vacuolar sorting mechanism(s). We surveyed cells from the Saccharomyces Genome Deletion Project (Invitrogen) related to vacuolar sorting events such as the vacuolar protein sorting (Vps) pathway, ribophagy, and endocytosis. Screening results are summarized in supplemental Table S2. We found that nitrogen starvation-specific Ypk1 degradation was inhibited in cells lacking Vps27 and Vps23 (Fig. 4A). To analyze whether this inhibition of Ypk1 proteolysis in these mutant cells is due to a defect in the vacuolar sorting of Ypk1, localization of GFP-Ypk1 was examined. In a vps27Δ background, nitrogen starvation-triggered vacuolar sorting of Ypk1 was inhibited, and the intravacuolar exaggerated structures were not observed (Fig. 4, B and C). Instead, in nitrogen-starved pep4Δvps27Δ cells, GFP-Ypk1 accumulated at an enlarged perivacuolar structure where FM4-64 was strongly incorporated (Fig. 4, B, arrows, and C). A similar distribution of GFP-Ypk1 and FM4-64 was observed in PMSF-treated pep4Δvps23Δ cells (supplemental Fig. S4). These results suggest that Vps27 and Vps23 are involved in the vacuolar sorting process of Ypk1 in nitrogen-starved cells. In addition, we also found that some endocytosis mutants (end3Δ, rvs161Δ, and rvs167Δ) inhibited Ypk1 proteolysis to varying degrees (supplemental Table S2). However, GFP-Ypk1 was transported to vacuoles in PMSF-treated pep4Δend3Δ cells (supplemental Fig. S5, arrows), indicating that endocytosis functions in Ypk1 proteolysis via an undetermined mechanism that may be unrelated to the autophagy-related vacuolar sorting observed in Fig. 3, A–C. Therefore, hereafter, we focused on the intracellular localization of Ypk1 in vps27Δ and vps23Δ cells to identify the specific vacuolar sorting mechanisms of Ypk1 that seemed to account for the selectivity of Ypk1 degradation.
FIGURE 4.
Involvement of ESCRT machinery in autophagy-related vacuolar sorting of Ypk1 that is distinct from conventional endosomal sorting. A, Ypk1 expression in nitrogen-starved WT, vps27Δ, and vps23Δ cells. Logarithmically growing cells were starved of nitrogen (NS) for the times indicated. Expressions of Ypk1 and Pgk1 were analyzed by Western blotting as described in Fig. 1A. B and C, fluorescence microscopic analyses of GFP-Ypk1 localization in pep4Δvps27Δ cells. FM4-64 labeling and nitrogen starvation were performed as described in Fig. 2C. Arrows indicate accumulated GFP-Ypk1 at the perivacuolar structures where FM4-64 was strongly incorporated. Relative fluorescence intensities of GFP-Ypk1 (green) and FM4-64 (red) were plotted (C) as in Fig. 2D. Percentages noted below the images represent frequencies of the indicated phenotypes. D and E, fluorescence microscopic analyses of Tat2-EGFP localization in pep4Δvps27Δ cells. Logarithmically growing cells harboring pRS415-TAT2-EGFP were labeled with FM4-64 and starved of nitrogen as described in Fig. 2C. The arrows indicate focus localization of Tat2-EGFP, and the arrowheads point to FM4-64-incorporated structures. Relative fluorescence intensities of Tat2-EGFP (green) and FM4-64 (red) in D were plotted (E) as in C. Bar, 5 μm. a.u., arbitrary units; DIC, differential interference contrast.
Distinctive Functions of ESCRT Machinery in Autophagy-related Vacuolar Sorting of Ypk1 from Endocytosis
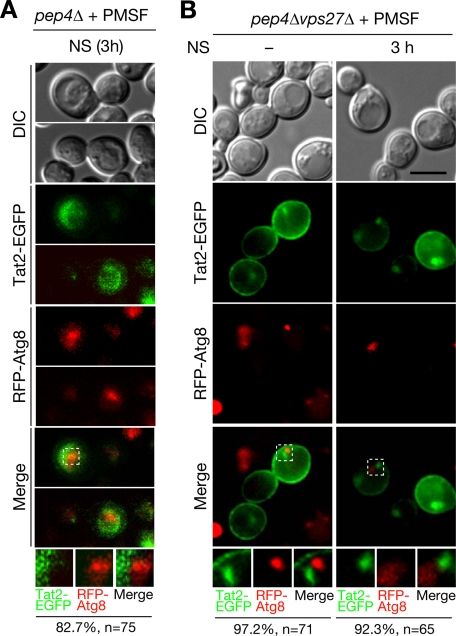
Vps27 and Vps23, which are class E Vps proteins, are the subunits of ESCRT-0 and ESCRT-I machinery, respectively, which function in the formation of multivesicular bodies on endosomal membranes (38–40). This process is required for endosome maturation, which is followed by fusion with vacuoles and subsequent degradation of endocytosed or sorted cargos from the trans-Golgi network. The deletion of class E VPS genes, including VPS27 and VPS23, leads to the formation of morphologically abnormal FM4-64-labeled foci, class E compartments, which are considered to be enlarged late endosomes (41, 42). However, the FM4-64-incorporated perivacuolar structures observed in nitrogen-starved pep4Δvps27Δ cells seemed morphologically distinct from the reported class E compartment. To confirm this, we compared this structure with endocytosed cargo. Because the class E compartment and the Ypk1-incorporated perivacuolar structure were indistinguishable by FM4-64 staining, we monitored the cellular distribution of Tat2, a high affinity tryptophan permease. Tat2 is also degraded in vacuoles upon nitrogen starvation, and this vacuolar degradation of Tat2 is independent of autophagy but dependent on endosomal transport, including endocytosis or sorting from the trans-Golgi network (43). The strains used here were derived from BY4741, which is prototrophic to tryptophan; therefore, Tat2-EGFP was mainly found at the plasma membrane in SD medium (44). Upon nitrogen starvation, Tat2-EGFP was internalized to foci that were morphologically identical to the class E compartment but not sorted into vacuoles in PMSF-treated pep4Δvps27Δ cells (Fig. 4D). This Tat2-EGFP-localized class E compartment was intermediate to endocytic vacuolar sorting because neither internalization nor accumulation of Tat2-EGFP at the foci was detected in PMSF-treated pep4Δend3Δ cells (supplemental Fig. S6A). In addition, the distribution of Tat2-EGFP was seldom or only partially colocalized with the FM4-64-stained perivacuolar structures in PMSF-treated pep4Δvps27Δ cells (Fig. 4, D and E), which is in sharp contrast to the case of Ypk1 (Fig. 4, B and C). Similar results were observed in pep4Δvps23Δ cells expressing Tat2-EGFP (supplemental Fig. S6, B and C). These results indicate that GFP-Ypk1 localizes in membrane structures that are distinct from the class E compartment in nitrogen-starved ESCRT mutant cells. Furthermore, localization of Tat2-EGFP was mutually exclusive with FM4-64-enriched exaggerated intravacuolar structures in which GFP-Ypk1 accumulated in nitrogen-starved PMSF-treated pep4Δ cells (supplemental Fig. S7, A and B). These results demonstrate that autophagy-related vacuolar sorting of Ypk1 and endocytic vacuolar transport of Tat2 do not share the same compartment during vacuolar sorting, although both trafficking events require ESCRT machinery.
GFP-Ypk1 Closely Associates with RFP-Atg8-containing Autophagic Bodies
The intravacuolar exaggerated structures in vacuolar protease-deficient cells (Fig. 2C) appeared to be reported autophagic bodies (28). Moreover, GFP-Ypk1-localized perivacuolar structures in nitrogen-starved ESCRT mutant cells (Fig. 4B and supplemental Fig. S4) would be expected to be intermediates of autophagy-related vacuolar sorting of Ypk1 in response to nitrogen starvation. If this were the case, the autophagic marker Atg8 would also be expected to associate with these structures. To investigate this, we examined the distribution of GFP-Atg8 with FM4-64 dye. GFP-Atg8 fluorescence accumulated in FM4-64-incorporated intravacuolar structures of PMSF-treated pep4Δ cells (Fig. 5A, arrowheads) and perivacuolar structures in PMSF-treated pep4Δvps27Δ cells (Fig. 5A, arrows). Because GFP-Ypk1 was also enriched in FM4-64-incorporated membrane structures (Figs. 2C and 4B and supplemental Fig. S4), these structures seemed to be related to autophagosomes. Intravacuolar GFP-Ypk1 was closely associated with RFP-Atg8 at the exaggerated structure(s) in PMSF-treated pep4Δ cells (Fig. 5, B and C). Similar close association of GFP-Ypk1 and RFP-Atg8 was also observed by confocal microscope analysis to exclude the possibility of fluorescent superimposition (supplemental Fig. S8). In PMSF-treated pep4Δvps27Δ cells, GFP-Ypk1 was exclusively localized with RFP-Atg8 in nitrogen-containing medium, whereas these proteins closely associated at the perivacuolar structures upon nitrogen starvation (Fig. 5, D and E). These results indicate that GFP-Ypk1 is transported by the autophagosome-related membrane structures. In contrast, the endocytic cargo marker Tat2-EGFP neither colocalized nor closely associated with RFP-Atg8 both inside the vacuoles of pep4Δ cells (Fig. 6A) and at the perivacuolar sites of pep4Δvps27Δ cells (Fig. 6B). These results further demonstrate that vacuolar transport of Ypk1 is clearly distinct from endocytic cargo trafficking even though both events require ESCRT machinery.
FIGURE 5.
Ypk1 closely associates with autophagy-related structures in both pep4Δ and ESCRT mutant cells. A, fluorescence microscopic analyses of GFP-Atg8 localization in the nitrogen-starved pep4Δ and pep4Δvps27Δ cells. Images were obtained for cells harboring pRS415-GFP-ATG8 as described in Fig. 2C. Arrowheads point to intravacuolar localization in pep4Δ cells, whereas arrows indicate perivacuolar sites in pep4Δvps27Δ cells where GFP-Atg8 localized within FM4-64-incorporated structures. B–E, fluorescence microscopic analyses of GFP-Ypk1 and RFP-Atg8 localization in pep4Δ (B and C) and pep4Δvps27Δ (D and E) cells. Logarithmically growing cells harboring pRS415-GFP-YPK1 and pRS413-RFP-ATG8 were starved of nitrogen (NS) for 3 h in the presence of PMSF. Cell culture was terminated by NaF and NaN3 treatment on ice as described under “Experimental Procedures,” and localization of GFP-Ypk1 and RFP-Atg8 fluorescence was immediately imaged after this treatment. Relative fluorescence intensities of GFP-Ypk1 (green) and RFP-Atg8 (red) in a and b were plotted (C and E, respectively) as in Fig. 2D. Percentages noted below the images represent frequencies of the indicated phenotypes. Bar, 5 μm. a.u., arbitrary units; DIC, differential interference contrast.
FIGURE 6.
Endocytic cargo protein Tat2-EGFP did not colocalize with autophagy-related structures in both pep4Δ and ESCRT mutant cells. A and B, fluorescence microscopic analyses of Tat2-EGFP and RFP-Atg8 localization in pep4Δ (A) and pep4Δvps27Δ (B) cells. Localization of Tat2-EGFP and RFP-Atg8 fluorescence was examined as in Fig. 5, B and D. Percentages noted below the images represent frequencies of the indicated phenotypes. Bar, 5 μm. DIC, differential interference contrast; NS, nitrogen starvation.
ESCRT Machinery Plays Roles in Autophagic Flux Induced by Nitrogen Starvation but Not by Rapamycin
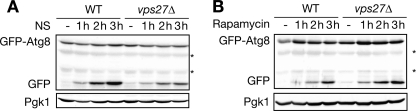
We showed that nitrogen starvation-triggered vacuolar sorting of Ypk1 utilizes ESCRT machinery (Fig. 4, B and C, and supplemental Fig. S4), which conventionally functions in endosomal trafficking. Although cytoplasmic autophagosome accumulation occurs in ESCRT subunit knockdown mammalian cells (45–47), the direct roles of ESCRT machinery in autophagy have not been well characterized. Moreover, deficiency of autophagy in an ESCRT mutant has not been reported in yeast cells despite intensive autophagy mutant screening in this organism. Thus, we investigated whether the loss of ESCRT machinery would reduce the nitrogen starvation-induced autophagic flux as a cause of reduced Ypk1 proteolysis. We analyzed autophagic flux by monitoring GFP-Atg8 processing, which requires the E1-like enzyme Atg7 (30). Ypk1 proteolysis utilized autophagy requiring the Atg8 conjugation system because deletion of the ATG7 gene inhibited nitrogen starvation-induced proteolysis of Ypk1 (Fig. 3C). Cleaved GFP was induced upon nitrogen starvation in WT cells but significantly reduced in nitrogen-starved vps27Δ cells (Fig. 7A). These data indicate that the dysfunctional ESCRT machinery reduced nitrogen starvation-triggered autophagic flux. Therefore, we further investigated whether ESCRT machinery is involved in autophagy induced by other stimuli. Rapamycin equally induced GFP-Atg8 processing both in WT and vps27Δ cells (Fig. 7B), indicating that ESCRT machinery is not involved in rapamycin-induced autophagic flux. Taken together, these data suggest that ESCRT machinery is required in nitrogen starvation- but not rapamycin-induced autophagic flux.
FIGURE 7.
ESCRT machinery is required for nitrogen starvation-triggered but not rapamycin-mediated autophagy. A, effect of ESCRT deficiency on nitrogen starvation-triggered GFP-Atg8 processing. Logarithmically growing WT and vps27Δ cells harboring pRS316-GFP-ATG8 were starved of nitrogen (NS). Total cell lysates were prepared at the indicated times and analyzed by Western blotting as in Fig. 3E. Asterisks indicate nonspecific signals. B, no effect due to ESCRT deficiency in rapamycin-triggered GFP-Atg8 processing. Logarithmically growing WT and vps27Δ cells harboring pRS316-GFP-ATG8 were treated with 20 ng/ml rapamycin. Expressions of GFP-Atg8 and cleaved GFP were analyzed by Western blotting as described in Fig. 3E.
DISCUSSION
Nitrogen Starvation-specific Ypk1 Proteolysis
Protein translation, the most energy-consuming process for cells, is regulated at multiple levels by modulation of translational initiation, ribosome biogenesis, mRNA turnover, and uptake of amino acids. TORC1 positively regulates these cellular processes by sensing extracellular nutrients (2). Lines of genetic evidence suggest that Ypk1 also plays an important role in protein translation by modulating translational initiation, possibly in parallel to TORC1 signaling, because both pathways are required for eIF4G stability (14, 15). However, the molecular mechanism of Ypk1 regulation in response to nutrients has not been characterized. In the present study, we showed that Ypk1 undergoes rapid autophagy-related degradation in response to nitrogen starvation. This finding was unexpected because Ypk1 repression was not induced by rapamycin treatment, which is known to induce autophagy-related protein degradation (5). To the best of our knowledge, Ypk1 is the first reported selective autophagic cargo specifically induced upon nitrogen starvation. It was reported that TORC1 activity toward Sch9, a proposed yeast counterpart of S6K, was inhibited by depletion of a number of nutrients, including glucose, phosphate, and nitrogen (48). Our result indicates that TORC1 and Ypk1 are differentially controlled by extracellular nutrient(s), although a downstream effecter for both of these signaling molecules involves eIF4G (14, 15). Currently, the biological role of rapid nitrogen starvation-specific Ypk1 clearance is not clear. Nitrogen is stored as proteins within cells; thus, limited amounts of this nutrient should trigger simultaneous cellular responses to supply nitrogen (autophagic protein breakdown) and to save nitrogen (translational arrest). This cellular storage of nitrogen is in sharp contrast to the storage of carbohydrate, which is stored as glycogen, and is thus readily usable upon activation of glycogen conversion (49). Rapid Ypk1 degradation using an autophagy system may ensure that yeast cells manage translational arrest and possibly autophagic protein breakdown in cellular nitrogen utilization. In addition to protein translation, Ypk1 is also involved in various cellular signaling pathways such as actin regulation and endocytosis (7, 8). Cellular nitrogen utilization may control these cellular events. Thus, additional studies are clearly required to elucidate the significance and relation of these cellular events as a whole to nitrogen starvation.
Distinction of Autophagy-related Proteolysis between Nitrogen Starvation and Rapamycin Treatment
TORC1 inhibition by rapamycin down-regulates anabolic processes and up-regulates catabolic processes similarly to nitrogen and carbon starvation (2). Therefore, rapamycin treatment has been utilized to mimic starved conditions (5, 14, 50). However, our results show that autophagy-related proteolysis of Ypk1 is specific to nitrogen starvation and not responsive to rapamycin treatment. It was previously reported that nitrogen starvation-induced autophagy in osmotically stressed cells was inhibited by deleting the HOG1 gene (51), which encodes a yeast ortholog of p38 MAPK required for osmotic stress responses (52). However, this osmosensitivity of autophagy in hog1Δ cells was not observed in rapamycin-induced autophagy (51). These results support the idea that rapamycin treatment is not identical to nitrogen-starved conditions in the induction of autophagy. Moreover, autophagic degradation induced by rapamycin was additively enhanced by inhibition of cyclic AMP-dependent protein kinase A (53, 54). Therefore, it is possible that several different intracellular signaling pathways may induce autophagy, and some may not be mimicked by rapamycin.
Involvement of ESCRT Machinery in Nitrogen Starvation-induced Ypk1 Proteolysis
In candidate-based screening of the genes required for nitrogen starvation-triggered autophagy-related proteolysis of Ypk1, we found that ESCRT-0 and ESCRT-I are required for Ypk1 proteolysis upon nitrogen starvation. Because rapamycin-induced autophagic flow was not affected by disruption of VPS27, involvement of ESCRT machinery is specific to nitrogen starvation-triggered autophagy. Recent reports have shown that knockdown of ESCRT subunits in mammalian cells causes a striking increase in the number of autophagosomes (45–47), suggesting three possibilities: (i) deficiency in fusion of autophagosomes with lysosomes, (ii) failure of autophagosome closure, and/or (iii) cellular stress caused by ESCRT dysfunction. In the case of our nitrogen-starved ESCRT mutants, dysfunction of ESCRT in (iii) is least likely to explain the role of ESCRT in autophagy because we found that ESCRT deficiency itself was insufficient to induce autophagic vacuolar sorting of Ypk1 and Atg8, and did not accumulate these proteins at the perivacuolar structure (Fig. 4B, supplemental Fig. S4, and Fig. 5D). Deficiency of autophagosome-vacuole fusion in (i) could lead to accumulation of cargo at perivacuolar spaces, although it is unknown whether ESCRT machinery mediates membrane fusion. Alternatively, because Ypk1 undergoes autophagy-related proteolysis, it is likely associated with the isolation membrane prior to autophagosome maturation. Therefore, a defect in the autophagosome closure step in (ii) could explain the accumulation of Ypk1 at the perivacuolar structure in nitrogen-starved ESCRT mutants (Fig. 4B and supplemental Fig. S4). In any case, we believe that this report is the first to show ESCRT-dependent autophagy in yeast cells. It is currently unknown whether ESCRT machinery functions in substrate selectivity in autophagy-related Ypk1 proteolysis. This may be the case because autophagic vacuolar transport of immature cytoplasmic Ape1 was not affected in nitrogen-starved ESCRT mutants (55).
Supplementary Material
Acknowledgments
We thank Dr. Daniel J. Klionsky (University of Michigan) for kindly providing Ape1 antibodies and the pRS414-RFP-ATG8 plasmid. We thank Dr. Yoshinori Ohsumi (Tokyo Institute of Technology) for kindly providing the pRS316-GFP-ATG8 plasmid. We also thank Drs. Hye-Won Shin and Hiroyuki Takatsu (Kyoto University) for help with confocal microscopic analysis and helpful discussion.
This work was supported by Grant-in-aid for Scientific Research 19590065 (to Y. K.) from the Japan Society for the Promotion of Science and CREST and by Grant-in-aid for Creative Scientific Research 16GS0313 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S8, Movie 1, and Tables S1 and S2.
- TORC1
- target of rapamycin complex
- AGC
- cyclic AMP-dependent protein kinase/protein kinase G/protein kinase C
- Ape1
- aminopeptidase I
- Cvt
- cytoplasm-to-vacuolar targeting
- ESCRT
- endosomal sorting complex required for transport
- Vps
- vacuolar protein sorting
- SD
- synthetic defined
- FM4-64
- N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide
- Atg
- autophagy-related.
REFERENCES
- 1.Rohde J., Heitman J., Cardenas M. E. (2001) J. Biol. Chem. 276, 9583–9586 [DOI] [PubMed] [Google Scholar]
- 2.De Virgilio C., Loewith R. (2006) Oncogene 25, 6392–6415 [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 4.Rohde J. R., Bastidas R., Puria R., Cardenas M. E. (2008) Curr. Opin. Microbiol. 11, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda T., Ohsumi Y. (1998) J. Biol. Chem. 273, 3963–3966 [DOI] [PubMed] [Google Scholar]
- 6.Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmelzle T., Helliwell S. B., Hall M. N. (2002) Mol. Cell. Biol. 22, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.deHart A. K., Schnell J. D., Allen D. A., Hicke L. (2002) J. Cell Biol. 156, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanoue D., Kobayashi T., Sun Y., Fujita T., Takematsu H., Kozutsumi Y. (2005) Arch. Biochem. Biophys. 437, 29–41 [DOI] [PubMed] [Google Scholar]
- 10.Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. (1999) Curr. Biol. 9, 186–197 [DOI] [PubMed] [Google Scholar]
- 11.Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. (2005) Mol. Cell. Biol. 25, 7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertram P. G., Zeng C., Thorson J., Shaw A. S., Zheng X. F. (1998) Curr. Biol. 8, 1259–1267 [DOI] [PubMed] [Google Scholar]
- 13.Beck T., Hall M. N. (1999) Nature 402, 689–692 [DOI] [PubMed] [Google Scholar]
- 14.Berset C., Trachsel H., Altmann M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelperin D., Horton L., DeChant A., Hensold J., Lemmon S. K. (2002) Genetics 161, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 17.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi T., Takematsu H., Yamaji T., Hiramoto S., Kozutsumi Y. (2005) J. Biol. Chem. 280, 18087–18094 [DOI] [PubMed] [Google Scholar]
- 19.Brauer M. J., Huttenhower C., Airoldi E. M., Rosenstein R., Matese J. C., Gresham D., Boer V. M., Troyanskaya O. G., Botstein D. (2008) Mol. Biol. Cell 19, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky D. J., Cueva R., Yaver D. S. (1992) J. Cell Biol. 119, 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen W. L., Legakis J. E., Nair U., Klionsky D. J. (2007) Mol. Biol. Cell 18, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronova S., Wedaman K., Anderson S., Yates J., 3rd, Powers T. (2007) Mol. Biol. Cell 18, 2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D. H., Goldberg A. L. (1996) J. Biol. Chem. 271, 27280–27284 [DOI] [PubMed] [Google Scholar]
- 25.Sun Y., Taniguchi R., Tanoue D., Yamaji T., Takematsu H., Mori K., Fujita T., Kawasaki T., Kozutsumi Y. (2000) Mol. Cell. Biol. 20, 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reggiori F., Black M. W., Pelham H. R. (2000) Mol. Biol. Cell 11, 3737–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shintani T., Klionsky D. J. (2004) J. Biol. Chem. 279, 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992) J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Huang W. P., Klionsky D. J. (2001) J. Cell Biol. 152, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 31.Matsuura A., Tsukada M., Wada Y., Ohsumi Y. (1997) Gene 192, 245–250 [DOI] [PubMed] [Google Scholar]
- 32.Tsukada M., Ohsumi Y. (1993) FEBS Lett. 333, 169–174 [DOI] [PubMed] [Google Scholar]
- 33.Klionsky D. J., Ohsumi Y. (1999) Annu. Rev. Cell Dev. Biol. 15, 1–32 [DOI] [PubMed] [Google Scholar]
- 34.Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr., Klionsky D. J. (2001) J. Cell Biol. 153, 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horst M., Knecht E. C., Schu P. V. (1999) Mol. Biol. Cell 10, 2879–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silles E., Mazón M. J., Gevaert K., Goethals M., Vandekerckhove J., Leber R., Sandoval I. V. (2000) J. Biol. Chem. 275, 34054–34059 [DOI] [PubMed] [Google Scholar]
- 37.Onodera J., Ohsumi Y. (2004) J. Biol. Chem. 279, 16071–16076 [DOI] [PubMed] [Google Scholar]
- 38.Katzmann D. J., Odorizzi G., Emr S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893–905 [DOI] [PubMed] [Google Scholar]
- 39.Williams R. L., Urbé S. (2007) Nat. Rev. Mol. Cell Biol. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 40.Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 41.Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. (1992) Mol. Biol. Cell 3, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck T., Schmidt A., Hall M. N. (1999) J. Cell Biol. 146, 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umebayashi K., Nakano A. (2003) J. Cell Biol. 161, 1117–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J. A., Beigneux A., Ahmad S. T., Young S. G., Gao F. B. (2007) Curr. Biol. 17, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 47.Rusten T. E., Stenmark H. (2009) J. Cell Sci. 122, 2179–2183 [DOI] [PubMed] [Google Scholar]
- 48.Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., Loewith R. (2007) Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 49.Jørgensen H., Olsson L., Rønnow B., Palmqvist E. A. (2002) Appl. Microbiol. Biotechnol. 59, 310–317 [DOI] [PubMed] [Google Scholar]
- 50.Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. (1996) Mol. Biol. Cell 7, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prick T., Thumm M., Häussinger D., Vom Dahl S. (2006) Autophagy 2, 241–243 [DOI] [PubMed] [Google Scholar]
- 52.Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C. (1993) Science 259, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 53.Yorimitsu T., Zaman S., Broach J. R., Klionsky D. J. (2007) Mol. Biol. Cell 18, 4180–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reggiori F., Wang C. W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. (2004) Mol. Biol. Cell 15, 2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 57.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.