Abstract
The emerging concept of generating cancer stem cells from epithelial-mesenchymal transition has attracted great interest; however, the factors and molecular mechanisms that govern this putative tumor-initiating process remain largely elusive. We report here that miR-200a not only regulates epithelial-mesenchymal transition but also stem-like transition in nasopharyngeal carcinoma cells. We first showed that stable knockdown of miR-200a promotes the transition of epithelium-like CNE-1 cells to the mesenchymal phenotype. More importantly, it also induced several stem cell-like traits, including CD133+ side population, sphere formation capacity, in vivo tumorigenicity in nude mice, and stem cell marker expression. Consistently, stable overexpression of miR-200a switched mesenchyme-like C666-1 cells to the epithelial state, accompanied by a significant reduction of stem-like cell features. Furthermore, in vitro differentiation of the C666-1 tumor sphere resulted in diminished stem-like cell population and miR-200a induction. To investigate the molecular mechanism, we demonstrated that miR-200a controls epithelial-mesenchymal transition by targeting ZEB2, although it regulates the stem-like transition differentially and specifically by β-catenin signaling. Our findings reveal for the first time the function of miR-200a in shifting nasopharyngeal carcinoma cell states via a reversible process coined as epithelial-mesenchymal to stem-like transition through differential and specific mechanisms.
Keywords: beta-Catenin, Cell Differentiation, Epithelial Cell, Gene Regulation, MicroRNA, ZEB2, Cancer Stem Cells, Epithelial-Mesenchymal Transition, Nasopharyngeal Carcinoma
Introduction
Epithelial-mesenchymal transition (EMT)3 is a crucial developmental program in which immotile epithelial cells acquire mesenchymal traits. Activation of EMT triggers tumor cell invasion and dissemination and is thus considered as the initiating step of cancer metastasis (1, 2). Recently, several studies have demonstrated that EMT is associated with cells with stem cell properties. For example, induction of EMT by potent EMT inducers, such as Twist, Snail, and TGFβ1, in human mammary epithelial cells produces CD44+CD24−/low breast cancer stem cells (CSCs) (3, 4). Similarly, breast CSCs can also be generated from CD8 T cell-induced EMT in epithelial breast cancer cells (5). These phenomena have been suggested to be mediated by the aberrant activation of Ras/MAPK signaling and suppression of stemness-inhibiting miR-203 by ZEB1 (4, 6).
MicroRNAs (miRNAs) are known to regulate tumor progression by suppressing target gene expression (7–9). Ample studies have implicated the important role of miRNAs in regulating EMT (10, 11). In particular, the miR-200 family has been documented to induce mesenchymal-epithelial transition (MET) by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2 (also know as SIP1) in murine mammary carcinoma cells (12), Madin Darby canine kidney epithelial cells (13), human bladder (14), prostate (15), and several other cancer cell lines (16, 17). Notably, the expressions of miR-200b-200a-429 and miR-200c-141 clusters, presumably encoded in form of polycistronic transcripts, are reciprocally repressed by their downstream targets ZEB1 and ZEB2 (18, 19). Among all five members of the miR-200 family, we are particularly interested in miR-200a, which has been identified as a prognostic marker for advanced ovarian (20) and cervical cancers (21).
Here, we investigated the function and mechanism of miR-200a on EMT-associated CSC generation in nasopharyngeal carcinoma (NPC). NPC is a head and neck tumor originating in the nasopharynx. With a distinct ethnic predilection, this malignancy is particularly common in Southeast Asia and Northern Africa. NPC patients tend to present at an advanced stage of disease, as the primary tumor is often located in a silent area. Moreover, NPC exhibits high invasive and metastatic potential, and therefore the prognosis of patients with metastatic NPC is poor (22). In addition, both epithelium-like CNE-1 and mesenchyme-like C666-1 NPC cell lines were available for this study. Using these two cell lines, we have recently reported that miR-200a differentially inhibits NPC cell growth via β-catenin signaling, and it suppresses NPC cell migration and invasion by ZEB2 down-regulation (23). In this study, we focused on investigating the potential role and mechanisms of miR-200a on EMT and in particular the generation of stem-like cells.
By loss-of-function studies, we first demonstrated that suppression of miR-200a promotes the transition of epithelium-like CNE-1 cells to mesenchymal phenotype. Importantly, we found that it also induces stem cell-like traits, including the CD133+ side population, sphere formation ability, and in vivo tumorigenicity. Consistently, overexpression of miR-200a not only promoted the transition of mesenchyme-like C666-1 cells to the epithelial state but also caused significant reduction of stem-like cell features. Finally, we showed that miR-200a controls EMT by targeting ZEB2 and regulates stemness transition differentially and specifically via β-catenin signaling. Results from our study suggest for the first time the effect and mechanism of miR-200a on shifting NPC cells through a reversible process coined as epithelial-mesenchymal to stem-like transition (EMST) and the potential associations with cell migration and proliferation. This knowledge will shed light on the origin and characteristics of CSCs and open up new avenues for developing novel cancer therapeutic strategies aimed at targeting EMT and CSCs.
EXPERIMENTAL PROCEDURES
Establishment of NPC Cell Lines Stably Lacking or Overexpressing miR-200a
CNE-1 and C666-1 NPC cell lines were obtained and cultured as described previously (23). One day before transfection, cells were seeded onto 24-well plates at 60% confluence. CNE-1 and C666-1 cells were, respectively, transfected with anti-miR-200a construct (miRZip-200a; System Biosciences, Mountain View, CA) and pLL3.7-miR-200a precursor plasmid using FuGENE HD transfection reagent (Roche Diagnostics) in the absence of antibiotics. After 48 h, cells were subcultured to 10% confluence in medium containing 1 μg/ml (for CNE-1 cells) or 5 μg/ml (for C666-1 cells) of puromycin (Sigma). When all cells in the nontransfected control culture were killed, antibiotic-resistant cells were pooled and passaged in medium containing a half-concentration of puromycin as in the first round of selection. For re-expression of CTNNB1, C666-1-pLL3.7-miR-200a cells (5 × 104) were transfected with pCI-neo-β-catenin WT plasmid (Addgene, Cambridge, MA). Cells were selected in medium containing 500 μg/ml (for first round) or 300 μg/ml (for second round) of G418 antibiotic (Sigma).
RNA Extraction, Quantitative RealTime PCR and Semi-quantitative Reverse Transcription-PCR (RT-PCR)
Total RNA was extracted from stable NPC cell lines, tumor spheres, or tumor tissues with the use of TRIzol reagent (Invitrogen). miR-200a, CTNNB1, ZEB2, Oct-4 (24), and aldehyde dehydrogenase 1 (ALDH1) (25) mRNA expressions were detected by quantitative real time PCR or semi-quantitative RT-PCR according to our previously described protocols (23, 26).
Immunoblotting
For the detection of epithelial and mesenchymal marker expressions, stable NPC cells were harvested for immunoblotting analysis according to our previously described protocol (26). Rabbit E-cadherin (1:400 dilution), mouse vimentin (1:300 dilution), and rabbit fibronectin (1:300 dilution) antibodies were products of Boster Biological Technology (Wuhan, China). Mouse β-catenin monoclonal antibody (1:400 dilution) and rabbit ZEB2 polyclonal antibodies (1:500 dilution) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Flow Cytometry
Established CNE-1 and C666-1 stable cell lines (1 × 106) were detached by treatment with 0.25% trypsin/EDTA (Invitrogen) and washed once with phosphate-buffered saline. The cells were then resuspended in 100 μl of Staining Buffer (eBioscience, San Diego) containing 1% fetal bovine serum (FBS; Invitrogen) and place on ice for 20 min to block Fc receptors. After incubating with primary phycoerythrin anti-human CD133 or Alexa Fluor 488 anti-human Oct-4 antibodies (eBioscience) for another 45 min on ice in the dark, the cells were washed twice with 1 ml of ice-cold Staining Buffer and centrifuged (400 × g) for 5 min at 4 °C. Cells resuspended in 0.5 ml of 2% formaldehyde fixation buffer were analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences). All flow cytometry results were obtained from two independent experiments performed in triplicate.
Sphere Formation Assay
Single cells (1 × 103) were plated onto a 24-well ultra-low attachment plate (Corning Glass) in serum-free DMEM/F-12, supplemented with 10 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor, and 0.4% bovine serum albumin and B-27 supplement (1:50 dilution; Invitrogen). After 14 days of culture, the number of tumor spheres formed (diameter >40 μm) was counted under an inverted microscope.
Sphere Differentiation Assay
C666-1 cells were subjected to sphere formation assay as described above. After 14 days of sphere formation culture, spheres were harvested using 40-μm cell strainers (BD Biosciences), seeded onto 6-well cell culture plates, and cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS in the absence of the aforementioned growth factors. Sphere adherence and differentiation could be observed after culturing in serum-containing medium for 24 h. At 36 and 72 h post-seeding, the differentiated cells (1 × 106) were collected for flow cytometry analysis of CD133 or Oct-4-positive cell population. Total RNA was also isolated from the differentiated cells for the detection of miR-200a, CTNNB1, and ZEB2 expressions by real time quantitative PCR. Both sphere formation assay and sphere differentiation assay were performed twice in triplicate with consistent results.
Transient siRNA Transfection
Transient transfection of C666-1 cells with control siRNA (siControl) or siRNAs targeting CTNNB1 and ZEB2 mRNAs (siCTTNB1 and siZEB2) was performed as described previously (23). The siCTTNB1 and siZEB2 were synthesized by GenePharma Co. (Shanghai, China) in accordance with the sequences reported previously (13, 27). The effects of siCTTNB1 and siZEB2 were confirmed by three independent transfection experiments.
Tumor Xenografts in Nude Mice
Female BALB/c athymic mice (nu/nu, 5–8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA), housed under aseptic conditions, and cared for in accordance with the guidelines of the Laboratory Animal Unit of the University of Hong Kong. Following our previous protocol (28), NPC tumor xenografts were established by subcutaneous injection of stable CNE-1-miRZip-200a cells (5 × 106), C666-1-pLL3.7-miR-200a cells (1 × 106), or respective control cells into the back flanks of mice (n = 6 mice per group). Tumor size was measured every 3 days for 15 days with the use of a caliper. The tumor volume (V) was calculated according to the formula V = ab2/2, where a and b are major and minor axes of the tumor foci, respectively. This animal study was approved by the Department of Health of the Government of Hong Kong and by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong.
Immunohistochemistry (IHC)
Tissue sections were deparaffinized in xylene and rehydrated in graded alcohols and distilled water. Slides were processed for antigen retrieval by a standard microwave heating technique. Specimens were incubated with anti-β-catenin or anti-ZEB2 antibodies (1:50 dilution). The immunoreactions were visualized with diaminobenzidine as a chromogen and counterstained with hematoxylin. The slides were washed in tap water, dehydrated in alcohol, and mounted.
Statistical Analysis
Experimental data were presented as mean ± S.D. All statistical analyses were performed using a two-tailed Student's t test (SPSS 12.0; SPSS Inc., Chicago, IL). Differences were considered to be statistically significant at p < 0.05.
RESULTS
Stable Knockdown of miR-200a Promotes Epithelial-Mesenchymal to Stem-like Transition in NPC Cells
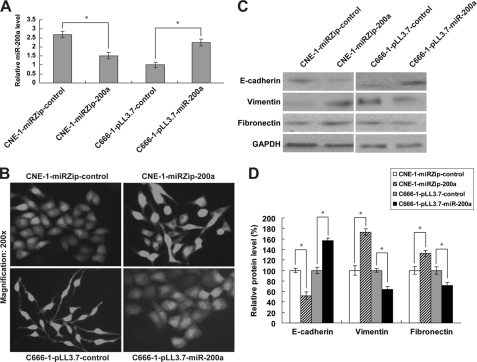
Morphologically, CNE-1 cells highly expressing miR-200a are tightly bound cells with epithelial phenotype. We first tested the ability of miR-200a to regulate EMT in CNE-1 cells. To this end, we established CNE-1 cells stably suppressing miR-200a (CNE-1-miRZip-200a). The down-regulation of miR-200a was confirmed by quantitative real time PCR (Fig. 1A). Dramatic morphological change, which is indicative of EMT, was observed in CNE-1-miRZip-200a cells (Fig. 1B, upper panel). We further examined the expressions of epithelial (E-cadherin) and mesenchymal (vimentin and fibronectin) markers. As anticipated, reduced E-cadherin expression and induction of vimentin and fibronectin were detected in CNE-1-miRZip-200a cells (Fig. 1, C and D), indicating that stable knockdown of miR-200a induces EMT in CNE-1 cells.
FIGURE 1.
Effect of miR-200a on EMT in NPC cells. A, quantitative real time PCR analysis of miR-200a expression in CNE-1 cells lacking miR-200a (CNE-1-miRZip-200a) and C666-1 cells overexpressing miR-200a (C666-1-pLL3.7-miR-200a). B, morphological changes in established NPC stable cell lines. C, Western blotting analysis of epithelial (E-cadherin) and mesenchymal (vimentin and fibronectin) markers. GAPDH was shown as an internal control. D, GAPDH-normalized protein levels of EMT markers relative to their respective controls. *, p < 0.05, as compared with respective controls.
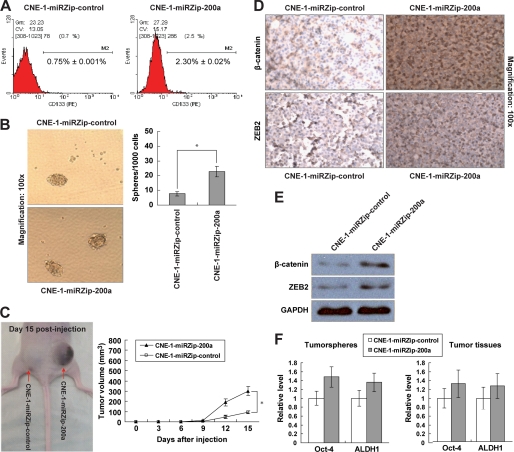
To test the hypothesis that miR-200a knockdown could further affect NPC stem cell population, CNE-1-miRZip-200a cells were assayed for the presence of the CD133 marker, a conserved cell surface molecule that has been associated with cancer stem-like cells in various human cancers, including brain, colon, liver, lung, pancreatic, prostate, skin, thyroid, and head and neck cancers (29–31). Flow cytometry analysis indicated that the CD133+ cell population was significantly enriched in CNE-1-miRZip-200a cells (Fig. 2A). To affirm the stemness of the CD133+ stem-like population, we investigated the self-renewal potential of CNE-1-miRZip-200a cells by sphere formation assay. With miR-200a suppression, CNE-1-miRZip-200a cells generated 3-fold more spheres than CNE-1-miRZip-control cells (Fig. 2B, 2.27 ± 0.35% versus 0.77 ± 0.15%, p < 0.05). In addition, we also assessed the in vivo tumorigenicity of CNE-1-miRZip-200a cells in subcutaneous xenograft nude mouse model. At day 15 post-injection, the mean volume of tumors generated from CNE-1-miRZip-200a cells was significantly larger than that originated from CNE-1-miRZip-control cells (Fig. 2C). IHC staining in NPC xenografts revealed that the staining intensities of β-catenin and ZEB2, both of which are known targets of miR-200a (23, 32), were remarkably higher in tumor tissues derived from CNE-1-miRZip-200a cells (Fig. 2D). The increase in β-catenin and ZEB2 protein levels in CNE-1-miRZip-200a tumor tissues was also confirmed by Western blotting analysis (Fig. 2E). We further ascertained the induction of stem-like cell traits by detection of two undifferentiated stem cell markers Oct-4 (33) and ALDH1 (34, 35). In both tumor spheres and tumor tissues originated from CNE-1-miRZip-200a cells, Oct-4 and ALDH1 expressions were up-regulated (Fig. 2F), suggesting that the increase in sphere number and tumor size by miR-200a knockdown is due, at least in part, to the acquisition of stem-like cell traits. These data reveal that reduced expression of miR-200a not only triggers EMT but also, in turn, transits CNE-1 cells to the stem-like state. Therefore, we propose a term epithelial-mesenchymal to stem-like transition (EMST) to describe this combined reprogramming process.
FIGURE 2.
Induction of stem-like cell phenotypes by stable knockdown of miR-200a in CNE-1 cells. A, flow cytometry analysis of CD133+ cell distribution in CNE-1-miRZip-control and CNE-1-miRZip-200a cells (1 × 106 cells per sample). The numbers represented the mean percentage of CD133+ cells from triplicate data. PE, phycoerythrin. B, representative images of tumor spheres generated after 14 days of single cell cultures. The bar graph indicated the number of spheres with diameter of at least 40 μm per 1 × 103 single cell seeded. C, subcutaneous NPC tumor in nude mouse (n = 6) at 15 day post-injection of CNE-1-miRZip-control and CNE-1-miRZip-200a cells (5 × 106 cells). The graph indicated the tumor volume measured every 3 days for 15 days. D, IHC staining; E, Western blotting analysis of β-catenin and ZEB2 protein expressions in tumor tissues. F, quantitative PCR analysis of Oct-4 and ALDH1 mRNA expressions in tumor spheres (left) and tumor tissues (right). *, p < 0.05, as compared with CNE-1-miRZip-control group.
Overexpression of miR-200a Switches Mesenchyme-like C666-1 Cells to Epithelial State, Accompanied by the Significant Reduction of Stem-like Cell Features
C666-1 cells that have a low level of miR-200a are spindle-shaped similar to mesenchymal cells (Fig. 1B, lower left panel). To test the reversible nature of EMST and the role of miR-200a in regulating EMST, we established stable C666-1 cells overexpressing miR-200a (C666-1-pLL3.7-miR200a) (Fig. 1A). In contrast to C666-1-pLL3.7-control cells, C666-1-pLL3.7-miR-200a cells were present with an epithelium-like phenotype (Fig. 1B, lower right panel). Consistently, the stimulated E-cadherin, but suppressed vimentin and fibronectin expressions, support that miR-200a-overexpressing C666-1 cells had undergone an MET (Fig. 1, C and D).
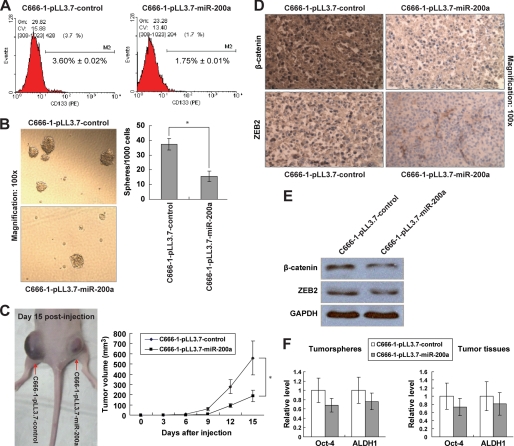
The stable miR-200a expression also diminished several stem-like traits in C666-1-pLL3.7-miR-200a cells, including significantly decreased CD133+ cell population (Fig. 3A) and the reduced ability to form tumor spheres than C666-1-pLL3.7 control cells (Fig. 3B, 1.57 ± 0.35% versus 3.73 ± 0.41%, p < 0.05). Moreover, in subcutaneous nude mice xenografts, C666-1-pLL3.7-miR-200a cells produced tumors with smaller sizes (Fig. 3C) and lower β-catenin and ZEB2 protein expressions (Fig. 3, D and E). The expressions of Oct-4 and ALDH1 stem cell markers were also decreased in tumor spheres and tumor tissues derived from C666-1-pLL3.7-miR-200a cells (Fig. 3F). These results suggest that miR-200a is capable of reversing EMST in favor of the epithelial state.
FIGURE 3.
Loss of stem cell traits in C666-1 cells with stable miR-200a expression. Experiments were performed using C666-1-pLL3.7-miR-control and C666-1-pLL3.7-miR-200a cells in the same way as described in Fig. 2. A, CD133+ cell population. B, sphere formation assay. C, in vivo tumorigenicity and tumor volume of C666-1-pLL3.7-miR-200a cells (1 × 106 cells; n = 6). D, IHC staining; E, Western blotting analysis of β-catenin and ZEB2 protein expressions in tumor tissues. F, mRNA levels of Oct-4 and ALDH1 in tumor spheres (left) and xenograft tumor tissues (right). *, p < 0.05, as compared with C666-1-pLL3.7-miR-control group.
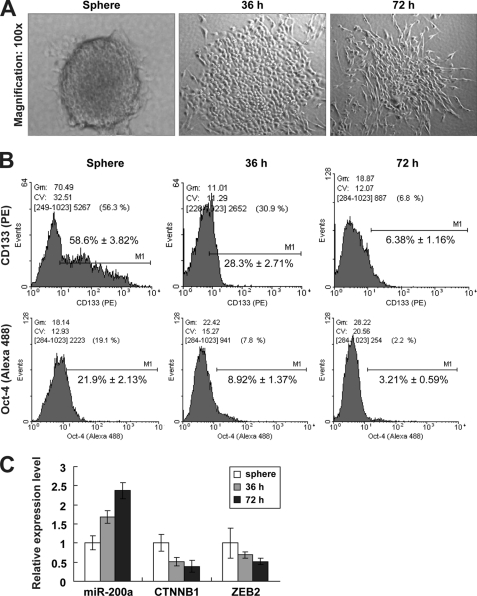
Spheric C666-1 Cell Differentiation Results in Reduced Stem-like Cell Population and miR-200a Induction
To determine whether the tumor spheres are enriched with stem-like cells that are capable of redifferentiating into parental phenotypes, we analyzed the in vitro differentiation of a sphere derived from miR-200a-low C666-1 cells. When culturing in serum-containing medium in the absence of growth factors, C666-1 spheres adhered to the plate and eventually acquired a typical differentiated morphological feature after 72 h (Fig. 4A). By flow cytometry analysis, we found that the CD133+ cell population was gradually decreasing over the course of sphere differentiation (Fig. 4B). This result was seconded by the decrease of an additional Oct-4 stem cell marker (Fig. 4B), confirming a reduction of stem-like population upon differentiation.
FIGURE 4.
In vitro differentiation of C666-1 spheres. A, microscopic observation of C666-1 cells cultured as nonadherent undifferentiated spheres and adherent cells under differentiation conditions for 36 and 72 h. B, flow cytometry analysis of CD133+ (upper) and Oct-4-positive (lower) cell populations in C666-1 spheres and sphere-derived adherent progeny at 36 and 72 h. C, quantitative real time PCR analysis of miR-200a, CTNNB1, and ZEB2 expressions. mRNA levels in sphere were normalized as 1. PE, phycoerythrin.
To further investigate the association between miR-200a and NPC stem-like properties, we also monitored the expressions of miR-200a and its direct downstream targets at different time points of differentiation. At 36 and 72 h post-differentiation, miR-200a expression was significantly increased, accompanied by the down-regulation of its targets CTNNB1 and ZEB1 during spheric cell differentiation (Fig. 4C). These results strongly support that miR-200a regulates the stem-like state.
miR-200a-mediated EMST Is Differentially Modulated by ZEB2 and CTNNB1 Down-regulation
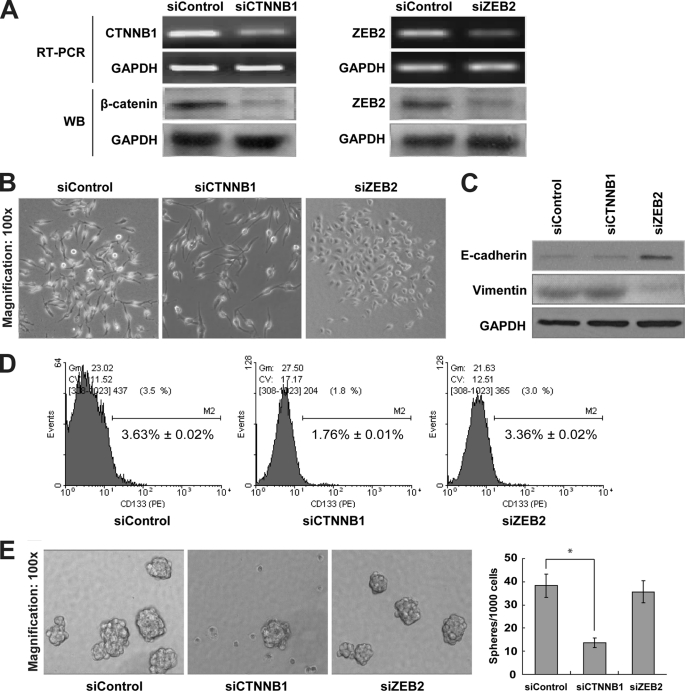
To understand how miR-200a regulates the two-step process of EMST, we tested the effects of silencing CTNNB1 and ZEB2 on EMT and stem-like characteristics (CD133+ cell population and sphere formation) in C666-1 cells. CTNNB1 and ZEB2 were successfully knocked down at 48 h post-transfection of siCTNNB1 and siZEB2, respectively (Fig. 5A). Interestingly, morphological change from mesenchymal to epithelial phenotype was observed in siZEB2-transfected but not in siCTNNB1-transfected C666-1 cells (Fig. 5B). This change was concurrent with E-cadherin induction and vimentin suppression (Fig. 5C), suggesting that miR-200a regulates MET by targeting ZEB2 rather than CTNNB1. Concerning the effects of miR-200a on stem-like traits, we demonstrated in C666-1 cells that the CD133+ cell population and the number of tumor spheres were solely significantly reduced by CTTNB1 but not ZEB2 down-regulation (Fig. 5, D and E). These results reveal that during EMST, miR-200a specifically controls EMT and stem-like properties via ZEB2 and CTNNB1, respectively.
FIGURE 5.
Differential regulation of EMST by ZEB and CTTNB1 down-regulation in C666-1 cells. A, RT-PCR and Western blotting (WB) analysis of CTNNB1 and ZEB2 levels in siControl-, siCTNNB1-, and siZEB2-transfected C666-1 cells at 48 h post-transfection. GAPDH was shown as an internal control. B, phase contrast images of C666-1 cells after transient knockdown of CTNNB1 and ZEB2. C, Western blot analysis of epithelial (E-cadherin) and mesenchymal (vimentin) markers. D, flow cytometry analysis of CD133+ cell population; E, sphere formation assay in siControl-, siCTNNB1-, and siZEB2-transfected C666-1 cells. *, p < 0.05, as compared with siControl group.
Re-expression of CTNNB1 Restores Stem-like Cell Phenotypes in miR-200a-overexpressing Cells
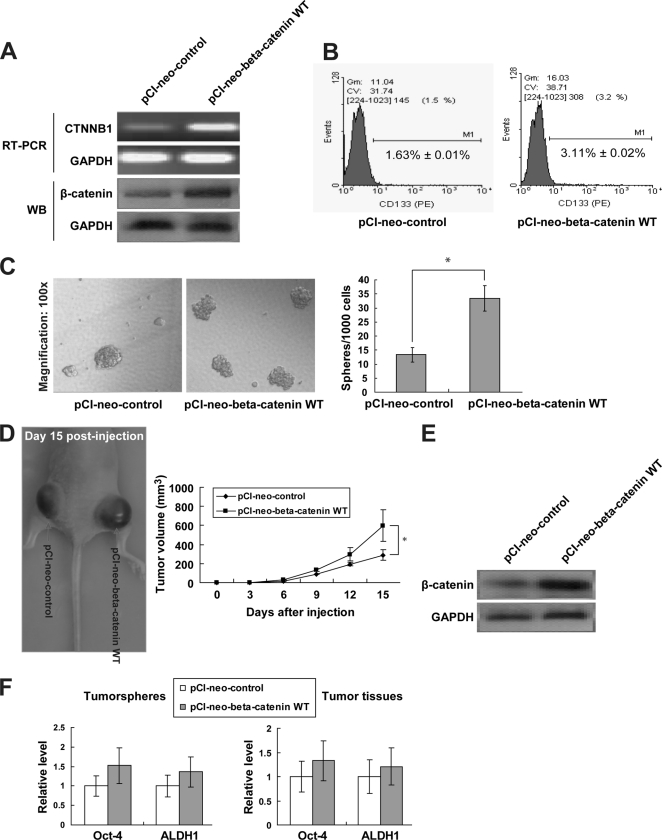
To corroborate the role of CTNNB1 in miR-200a-regulated stem-like transition, we also rescued miR-200a-targeted CTNNB1 suppression in C666-1-pLL3.7-miR-200a cells by transfection of a plasmid carrying the wild-type CTNNB1 gene (pCI-neo-β-catenin WT). The re-expression of CTNNB1 was validated by semi-quantitative RT-PCR (Fig. 6A). The CD133+ cell population and sphere generation ability were partially, if not totally, restored upon CTNNB1 re-expression (Fig. 6, B and C). In addition, inoculation of C666-1-pLL3.7-miR-200a cells with CTNNB1 re-expression generated tumors with larger volume in vivo (Fig. 6D). Such re-expression of CTNNB1 in tumor tissues was clearly detected (Fig. 6E). The restoration of stem-like cell phenotypes was also evident by the up-regulation of Oct-4 and ALDH1 markers in tumor spheres and tumor tissues (Fig. 6F), confirming that miR-200a regulates stem-like phenotypes by targeting CTNNB1.
FIGURE 6.
Restoration of stem-like traits by re-expression of CTNNB1. A, RT-PCR and Western blotting (WB) analysis showing CTNNB1 re-expression in C666-1-pLL3.7-miR-200a cells transfected with wild-type β-catenin plasmid (pCI-neo-beta-catenin WT). B, CD133+ cell distribution, C, tumor sphere formation; D, in vivo tumorigenicity and tumor volume of C666-1-pLL3.7-miR-200a cells expressing pCI-neo-control or pCI-neo-β-catenin WT. E, WB confirmation of CTNNB1 re-expression in xenograft NPC tissues. F, Oct-4 and ALDH1 mRNA levels in tumor spheres (left) and xenograft tumor tissues (right). *, p < 0.05, as compared with pCI-neo-control group.
DISCUSSION
In this study, we demonstrated for the first time that miR-200a not only controls EMT, but also mediates the acquisition of stem-like cell phenotypes in NPC cells. We found that epithelium-like CNE-1 cells stably lacking miR-200a underwent an EMT and exhibited an increased CD133+ cell population whose stem-like properties are well supported by the enhanced sphere-forming capacity, in vivo tumorigenicity, and stem cell marker expression. Reversely, stable overexpression of miR-200a reduced stem-like cell population and concurrently promoted MET in mesenchyme-like C666-1 cells, suggesting that these cell state transitions are reversible. To extend our current concept about EMT, we propose the new term epithelial-mesenchymal to stem-like transition (EMST) to describe such putative transition of cells between epithelial, mesenchymal, and stem cell states.
CSCs, also known as tumor-initiating cells, are rare subpopulation of cells within a tumor that possess the capacity for self-renewal and differentiation into heterogeneous lineages of cancer cells (36). These CSCs have been isolated from leukemia (37), brain (38), breast (39), colon (40), liver (41), lung (42), pancreatic (43), and prostate cancers (44), as well as from thyroid carcinoma (31) and NPC cell lines (45). Given the fact that differentiated tumor cells lacking self-renewal capability are in principle unable to establish macroscopic metastases at distant sites, it has been proposed that the migrating CSCs are required for the metastatic spread of tumors (46).
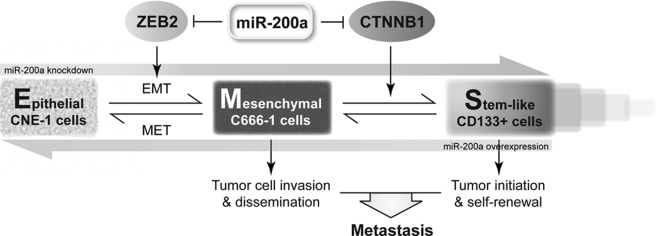
With this mentioned, our discovery of miR-200a-mediated EMST has important implications in the context of cancer metastasis. Based on our presented data, we hypothesize that, in primary tumor, down-regulation of miR-200a could activate the EMT program to promote tumor cell invasion. Furthermore, migrating CSCs would also be generated via EMST for dissemination through the circulatory system. By doing so, CSCs capable of self-renewal could initiate metastatic colonies in distant organs, where some of the CSCs might redifferentiate into the epithelial state via reverse EMST and some might maintain stemness for supporting the continuous growth of tumor (Fig. 7).
FIGURE 7.
Proposed model of the role of miR-200a in EMST.
Regarding the molecular mechanisms underlying miR-200a-mediated EMST, we specifically identified ZEB2 and CTNNB1 as the effectors of miR-200a during EMT and stem-like cell generation, respectively. ZEB2 is a member of the Zfh1 family of two-handed zinc finger/homeodomain proteins. It interacts with activated SMADs and represses E-cadherin transcription by direct binding to paired E-box sites of the proximal E-cadherin promoter (47). Because the 3′-untranslated region of ZEB2 has target sites for all five miR-200 family members, it is not surprising that miR-200a also targets ZEB2 for EMT regulation. On the other hand, CTNNB1 encodes β-catenin, which is a key mediator of canonical Wnt/β-catenin cascade important for NPC progression. Activation of this signaling is required for maintaining the stemness of CSCs in tumors (48–50). We also showed an association of stem-like transition with NPC cell growth and EMT with NPC cell migration and invasion. Our results are in line with the essential role of Wnt/β-catenin pathway and suggest the requirement of cell growth in sustaining CSC phenotypes.
We manifested that miR-200a, as a single factor, could regulate EMT and stem-like cell population in NPC cells, highlighting the potency of miRNAs in generating CSCs via EMST. Indeed, an increasing number of genes have been shown to orchestrate EMT during embryogenesis and tumor progression. Among these EMT regulators are, to name a few, cytokines (TGF-β), growth factors (FGF, EGF, HGF, and PDGF), transcription factors (Twist, Snail1, Snail2, ZEB1, and ZEB2), miRNAs (miR-200 family and miR-205), and members of the BMP, Wnt, and Notch signaling pathways (51). Our results raise the possibility that these genes are also involved in EMT-induced CSC generation. Revisiting their effects on EMST may reveal novel mechanisms for CSC generation. Furthermore, in addition to metastasis, CSCs can also account for the recurrence of cancers after chemotherapy. Thus, developing strategies aimed at targeting miR-200a or other EMST regulators offer great therapeutic opportunities for eradicating cancer at its root.
Apart from cytokine 19 that has been associated with CSC-like side population cells in a poorly differentiated CNE-2 cell line (45), report on NPC CSC marker is lacking. In our study, we could barely detect the presence or any significant change in cytokine 19 (data not shown). This could be due to the very small population of side population cells in CNE-1 (0.7%) and C666-1 (0.1%), as compared with CNE-2 (2.6%) cells. In addition, different culture conditions may also contribute to our failure to detect cytokine 19 expression. Therefore, we used CD133 as a NPC CSC marker because in our study CD133 expression is associated with other CSC characteristics, including tumor sphere formation, in vivo tumorigenicity in nude mice xenografts, and Oct-4 and ALDH1 stem cell marker expression.
Our characterization of stem-like cell traits are largely based on sphere formation and in vivo tumorigenicity assays, both of which are subjected to the influence of cell growth. We and others have previously reported the inhibitory effect of miR-200a on NPC and meningioma cell growth (23, 32). Taking this into consideration, one might argue that the effects of miR-200a on sphere formation and tumor volume are indeed due to altered growth rate of NPC cells rather than their self-renewal ability. To resolve this, we detected two well characterized stem cell markers in tumor spheres and tumor tissues whose expressions are strongly associated with the sphere number and tumor volume. These results clearly indicate that the self-renewal capability of putative stem-like cells is in action. As a matter of fact, upon miR-200a manipulation, the number of tumor spheres is changed by more than 3-fold, whereas the change in NPC cell growth is less than 1-fold only (data not shown). Therefore, the changes in sphere number and tumor volume should be dominantly governed by the self-renewing stem-like cells, with partial contribution from NPC cell growth.
In conclusion, this is the first study demonstrating the regulation of epithelial-mesenchymal to stem-like transition by miR-200a in NPC, as well as the differential mechanisms by which miR-200a modulates EMT and stem-like cell generation through targeting ZEB2 and β-catenin signaling, respectively. These results underscore the importance of miRNA in governing the stem-like properties of cancer cells. Developing miR-200a-based therapy may be beneficial for the clinical treatment of cancers, including NPC.
Acknowledgment
We thank Dr. Samuel S. Ng for constructive comments.
This work was supported by Research Grants Council of Hong Kong Grant 467109 and National Basic Research Program of China (973 Program) Grants 2010CB529400 and 2010CB912800.
- EMT
- epithelial-mesenchymal transition
- CSC
- cancer stem cell
- miRNA
- microRNA
- MET
- mesenchymal-epithelial transition
- NPC
- nasopharyngeal carcinoma
- EMST
- epithelial-mesenchymal to stem-like transition
- IHC
- immunohistochemistry.
REFERENCES
- 1.Tsuji T., Ibaragi S., Hu G. F. (2009) Cancer Res. 69, 7135–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavert N., Ben-Ze'ev A. (2008) Trends Mol. Med. 14, 199–209 [DOI] [PubMed] [Google Scholar]
- 3.Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel A. P., Lièvre M., Thomas C., Hinkal G., Ansieau S., Puisieux A. (2008) PLoS One 3, e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santisteban M., Reiman J. M., Asiedu M. K., Behrens M. D., Nassar A., Kalli K. R., Haluska P., Ingle J. N., Hartmann L. C., Manjili M. H., Radisky D. C., Ferrone S., Knutson K. L. (2009) Cancer Res. 69, 2887–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V. G., Morton J., Sansom O., Schüler J., Stemmler M. P., Herzberger C., Hopt U., Keck T., Brabletz S., Brabletz T. (2009) Nat. Cell Biol. 11, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 7.Medina P. P., Slack F. J. (2008) Cell Cycle 7, 2485–2492 [DOI] [PubMed] [Google Scholar]
- 8.Kent O. A., Mendell J. T. (2006) Oncogene 25, 6188–6196 [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 10.Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. (2008) Cell Cycle 7, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 11.Cano A., Nieto M. A. (2008) Trends Cell Biol. 18, 357–359 [DOI] [PubMed] [Google Scholar]
- 12.Korpal M., Lee E. S., Hu G., Kang Y. (2008) J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 14.Adam L., Zhong M., Choi W., Qi W., Nicoloso M., Arora A., Calin G., Wang H., Siefker-Radtke A., McConkey D., Bar-Eli M., Dinney C. (2009) Clin. Cancer Res. 15, 5060–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong D., Li Y., Wang Z., Banerjee S., Ahmad A., Kim H. R., Sarkar F. H. (2009) Stem Cells 27, 1712–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korpal M., Kang Y. (2008) RNA Biol. 5, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008) Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008) Cancer Res. 68, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 19.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X., Macdonald D. M., Huettner P. C., Feng Z., El Naqa I. M., Schwarz J. K., Mutch D. G., Grigsby P. W., Powell S. N., Wang X. (2009) Gynecol. Oncol. 114, 457–464 [DOI] [PubMed] [Google Scholar]
- 21.Hu X., Schwarz J. K., Lewis J. S., Jr., Huettner P. C., Rader J. S., Deasy J. O., Grigsby P. W., Wang X. (2010) Cancer Res. 70, 1441–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou J., Lin Y. C., Kim J., You L., Xu Z., He B., Jablons D. M. (2008) Head Neck 30, 946–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia H., Ng S. S., Jiang S., Cheung W. K., Sze J., Bian X. W., Kung H. F., Lin M. C. (2010) Biochem. Biophys. Res. Commun. 391, 535–541 [DOI] [PubMed] [Google Scholar]
- 24.Hart A. H., Hartley L., Parker K., Ibrahim M., Looijenga L. H., Pauchnik M., Chow C. W., Robb L. (2005) Cancer 104, 2092–2098 [DOI] [PubMed] [Google Scholar]
- 25.Visus C., Ito D., Amoscato A., Maciejewska-Franczak M., Abdelsalem A., Dhir R., Shin D. M., Donnenberg V. S., Whiteside T. L., DeLeo A. B. (2007) Cancer Res. 67, 10538–10545 [DOI] [PubMed] [Google Scholar]
- 26.Xia H., Qi Y., Ng S. S., Chen X., Chen S., Fang M., Li D., Zhao Y., Ge R., Li G., Chen Y., He M. L., Kung H. F., Lai L., Lin M. C. (2009) Biochem. Biophys. Res. Commun. 380, 205–210 [DOI] [PubMed] [Google Scholar]
- 27.Simpson J. C., Cetin C., Erfle H., Joggerst B., Liebel U., Ellenberg J., Pepperkok R. (2007) J. Biotechnol. 129, 352–365 [DOI] [PubMed] [Google Scholar]
- 28.Ng S. S., Cheung Y. T., An X. M., Chen Y. C., Li M., Li G. H., Cheung W., Sze J., Lai L., Peng Y., Xia H. H., Wong B. C., Leung S. Y., Xie D., He M. L., Kung H. F., Lin M. C. (2007) J. Natl. Cancer Inst. 99, 936–948 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q., Shi S., Yen Y., Brown J., Ta J. Q., Le A. D. (2010) Cancer Lett. 289, 151–160 [DOI] [PubMed] [Google Scholar]
- 30.Wu Y., Wu P. Y. (2009) Stem Cells Dev. 18, 1127–1134 [DOI] [PubMed] [Google Scholar]
- 31.Zito G., Richiusa P., Bommarito A., Carissimi E., Russo L., Coppola A., Zerilli M., Rodolico V., Criscimanna A., Amato M., Pizzolanti G., Galluzzo A., Giordano C. (2008) PLoS One 3, e3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saydam O., Shen Y., Würdinger T., Senol O., Boke E., James M. F., Tannous B. A., Stemmer-Rachamimov A. O., Yi M., Stephens R. M., Fraefel C., Gusella J. F., Krichevsky A. M., Breakefield X. O. (2009) Mol. Cell. Biol. 29, 5923–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lengner C. J., Welstead G. G., Jaenisch R. (2008) Cell Cycle 7, 725–728 [DOI] [PubMed] [Google Scholar]
- 34.Jiang F., Qiu Q., Khanna A., Todd N. W., Deepak J., Xing L., Wang H., Liu Z., Su Y., Stass S. A., Katz R. L. (2009) Mol. Cancer Res. 7, 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginestier C., Hur M. H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C. G., Liu S., Schott A., Hayes D., Birnbaum D., Wicha M. S., Dontu G. (2007) Cell Stem Cell 1, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001) Nature 414, 105–111 [DOI] [PubMed] [Google Scholar]
- 37.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M. A., Dick J. E. (1994) Nature 367, 645–648 [DOI] [PubMed] [Google Scholar]
- 38.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 39.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. (2007) Nature 445, 111–115 [DOI] [PubMed] [Google Scholar]
- 41.Ma S., Chan K. W., Hu L., Lee T. K., Wo J. Y., Ng I. O., Zheng B. J., Guan X. Y. (2007) Gastroenterology 132, 2542–2556 [DOI] [PubMed] [Google Scholar]
- 42.Kim C. F., Jackson E. L., Woolfenden A. E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R. T., Jacks T. (2005) Cell 121, 823–835 [DOI] [PubMed] [Google Scholar]
- 43.Li C., Heidt D. G., Dalerba P., Burant C. F., Zhang L., Adsay V., Wicha M., Clarke M. F., Simeone D. M. (2007) Cancer Res. 67, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 44.Miki J., Furusato B., Li H., Gu Y., Takahashi H., Egawa S., Sesterhenn I. A., McLeod D. G., Srivastava S., Rhim J. S. (2007) Cancer Res. 67, 3153–3161 [DOI] [PubMed] [Google Scholar]
- 45.Wang J., Guo L. P., Chen L. Z., Zeng Y. X., Lu S. H. (2007) Cancer Res. 67, 3716–3724 [DOI] [PubMed] [Google Scholar]
- 46.Brabletz T., Jung A., Spaderna S., Hlubek F., Kirchner T. (2005) Nat. Rev. Cancer 5, 744–749 [DOI] [PubMed] [Google Scholar]
- 47.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. (2001) Mol. Cell 7, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 48.Malanchi I., Peinado H., Kassen D., Hussenet T., Metzger D., Chambon P., Huber M., Hohl D., Cano A., Birchmeier W., Huelsken J. (2008) Nature 452, 650–653 [DOI] [PubMed] [Google Scholar]
- 49.Yamashita T., Budhu A., Forgues M., Wang X. W. (2007) Cancer Res. 67, 10831–10839 [DOI] [PubMed] [Google Scholar]
- 50.Reya T., Clevers H. (2005) Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 51.Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]