Abstract
Androgen receptor (AR) plays a critical role in prostate cancer and male sexual differentiation. We have identified AR from a primitive vertebrate with a sex reversal characteristic, the rice field eel. AR of this species (eAR) is distinct from human AR, especially in the ligand binding domain (LBD), and its expression in gonads shows an increasing tendency during gonadal transformation from ovary via ovotestis to testis. eAR has a restricted androgen-dependent transactivation function after a nuclear translocation upon dihydrotestosterone exposure. A functional nuclear localization signal was further identified in the DNA binding domain and hinge region. Although nuclear export is CRM1-independent, eAR has a novel nuclear export signal, which is negatively charged, indicating that a nuclear export pathway may be mediated by electrostatic interaction. Further, our studies have identified critical sequences for ligand binding in the C terminus. A structure of three α-helices in the LBD has been conserved from eels to humans during vertebrate evolution, despite a distinct amino acid sequence. Mutation analysis confirmed that the LBD is essential for dihydrotestosterone-induced nuclear import of eAR and following transactivation function in the nucleus. In addition, eAR interacts with both Sox9a1 and Sox9a2, and their interaction regulates transactivation of eAR. Our data suggest that the primitive species conserves and especially acquires key novel domains, the nuclear export signal and LBD, for the eAR function in spite of a rapid sequence evolution.
Keywords: Cellular Regulation, Development, Evolution, Nuclear Translocation, Testosterone, Nucleocytoplasmic Shuttling, Prostate cancer, Sex Reversal, Steroid, Teleosts
Introduction
Androgen signaling is essential for male sexual differentiation, gonadal maturation, and maintenance of secondary male sexual characteristics, and, more importantly, it plays a critical role in development of prostate cancer. The action of androgens is generally mediated through their binding to and activating AR.3 The activated AR is translocated into the nucleus for a cascade of androgen signaling. AR mutations are frequent in metastatic prostate cancer, and more than 70 single-base substitutions have been described so far in prostate cancer (1, 2), which would be one of the main causes of androgen-independent prostate cancer. Although great progress has been made, development of prostate cancer is a complicated process, and its molecular mechanism remains elusive.
Androgen receptor is an X-linked intracellular receptor that belongs to a large family of nuclear receptors. The main action pathway of the AR is the AR-DNA interactions, which are mediated by androgen binding and activation. The activated AR is relocated into the nucleus and binds to androgen-responsive elements of androgen-regulated genes. At AR binding sites of target genes, a transcription complex is then assembled by recruiting coregulatory proteins, coactivators/corepressors, and general transcriptional factors, which ultimately stimulates or inhibits target gene transcription (3, 4). Meanwhile, nuclear AR can be exported to the cytoplasm by ligand withdrawal through an unknown mechanism (5–7). Recent findings in the DNA-independent AR actions (50, 51) add a layer of complexity to the AR signaling. AR may exert its functions in the cytoplasm, which does not need the nuclear import and DNA binding of AR (8–13). The androgen nongenomic signaling is involved in interactions between AR and relevant signaling proteins (e.g. phosphatidylinositol 3-OH kinase, Akt, Src/Shc/ERK/MAPK, JNK, and lipid rafts). Further complexity of the AR actions is the observation that the AR is likely to play a major role in androgen-independent prostate cancer (14, 15). These data indicate that the mechanisms by which AR acts and regulations of AR nucleocytoplasmic shuttling are not understood well.
Evidence is accumulating that endocrine-disrupting chemicals, such as androgen analog or AR antagonist, that impair androgen-mediated signaling pathways, are present in the environment, which disrupt fish reproductive processes and often result in reduced spawning and fecundity (16, 17). In addition, development of approaches to making a single-sex population or sex reversal in fish species using androgen or relevant steroid treatments are economically important for fish production. Androgen treatments have been used to explore approaches to making a sex reversal of genetic females into phenotypic males (18). Although AR has been identified in a number of teleosts, including zebrafish, medaka, tilapia, sea bass, rainbow trout, Western mosquitofish, Japanese eel, black porgy, and Spinibarbus denticulatus (19–28), further understanding of fish AR signaling and functions is needed.
The rice field eel, Monopterus albus, taxonomically belongs to teleosts, the family Synbranchidae of the order Synbranchiformes (Neoteleostei, Teleostei, Vertebrata); it is not only an economically important fish species but also has attractive biological features, including its primitive evolutionary status, relative small genome size, and natural sex reversal from female into male via intersex during its life cycle (29, 30), which is a perfect model for studies in genetics and sexual differentiation. Here we report identification of the AR gene from the species and show its up-regulation during gonadal transformation, its restricted androgen-dependent transactivation function, and a functional NLS in the DNA binding domain (DBD) and hinge region. We identified a novel NES and showed that nuclear export of eAR is CRM1-independent. We further determined critical sequences for ligand binding in the LBD, which show a similar three-dimensional structure of the LBD to human AR but distinct amino acid sequence. In addition, interaction of eAR with Sox9a1 or Sox9a2 was further determined.
EXPERIMENTAL PROCEDURES
Animals
The rice field eels (M. albus) were obtained from markets in the Wuhan area in China. Their sexes were conformed by microscopic analysis of gonad sections.
Degenerate PCR and RACE Cloning of AR Gene of the Rice Field Eel
SMART cDNAs were made from testis RNAs using the SMART cDNA library construction kit following a commercial protocol (Clontech). Based on a search of GenBankTM sequences, two conserved amino acid regions in the DNA binding domain of AR (ALTCGSC and LNELGER) were selected, and their degenerate oligonucleotides (sense, 5′-GCMCTCACYTGTGGMAGCTGC-3′; antisense, 5′-TCTCYCCSARCTCGTTRAGGC-3′) were synthesized and used as primers for degenerate PCR. PCR was performed in a 20-μl reaction mix containing 10 mm Tris-HCl, pH 8.3, 1.5 mm MgCl2, 50 mm KCl, 150 mm dNTP, 0.2 μm each primer, and 1 unit of TaqDNA polymerase. Amplification conditions were as follows: 94 °C for 30 s, 64 °C for 30 s, and 72 °C for 40 s for 35 cycles. PCR products was sequenced. Degenerate primer 5′-ARACAGCSAGGGAGCTSTGC-3′ and specific antisense primer 5′-GCCGACAAGACGGACAGTTC-3′ were used for a second PCR to obtain a longer 5′-sequence. 5′-RACE was performed using common SMARTIII primer 5′-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG-3′ and primer 5′-TTGCGGCACAAGGTGGGAG-3′. 3′-RACE was performed using common CDSIII primer 5′-ATTCTAGAGGCCGAGGCGGCCGACATGd(T)30N_1N-3′ (where N represents A, G, C, or T, and N_1 is A, G, or C) and primer 5′-CTGTCCGTCTTGTCGGCTAA-3′ and nested primer 5′-GGTTGCCCGGAGTATCTCTC-3′. RACE was performed under the condition of 35 cycles, each with 30 s at 94 °C, 30 s at 64 °C, and 1 min at 72 °C. All sequences were cloned and sequenced.
Phylogenetic Analysis and Molecular Modeling
AR protein sequences were aligned using the ClustalX 1.81 program. A phylogenetic tree was constructed using NJ (100 runs) (Phylip) and ML (100 runs) methods. The three-dimensional model of the ligand binding domain of the rice field eel AR was generated using an automated homology modeling server (see the Pôle BioInformatique Lyonnais Web site) running at Geno3D (Lyon, France), and human AR LBD was used as a control. Visualization of the three-dimensional structure was performed using Swiss-PDB ViewER 4.0.1 (available on the World Wide Web).
Real-time Fluorescent Quantitative RT-PCR
Real-time RT-PCR was used to quantify AR expression using the multichannel RotorGene 3000 (Corbett Research, Mortlake, Australia) according to the supplied protocol. Total RNA was prepared from heart, liver, brain, kidney, spleen, testis, ovotestis, and ovary of the rice field eel using TRIzol reagent (Invitrogen). All of the RNAs were digested by RNase-free DNase I and purified. 3 μg RNAs were used as template for reverse transcription using 0.5 μg of poly(T)20 primer and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). Amplification conditions were as follows: 95 °C for 30 s, 64 °C (AR) or 62 °C (HPRT) for 30 s, and 72 °C for 30 s for 32 cycles (AR) or 28 cycles (HPRT) in a 25-μl reaction mix containing 1× SYBR Green I. Primers were as follows: 5′-GGTTGCCCGGAGTATCTCTC-3′ and 5′-CTCCCATGTGCCCCAAAAATC-3′ for AR and 5′-AATCAAAGTAATCGGTGGCG-3′ and 5′-GACCTCGAATCCTACAAAGTCTG-3′ for HPRT. For robustness issues, each sample was performed in triplicate at least. Data were analyzed by the software Rotor-gene version 4.6 and then plotted in Microsoft Excel.
Northern Blot Hybridization
40 μg of total RNA from each tissue was denatured and separated by electrophoresis. Northern blots were performed as routine protocols, and hybridization at 42 °C was performed in ULTRAhyb solution (Ambion, Applied Biosystems Inc.) with [α-32P]dCTP-labeled probe. A fragment including the full coding sequence of eAR cDNA was used as the probe in the hybridizations.
In Situ Hybridization Analysis
For mRNA in situ hybridization onto gonadal sections, antisense and sense RNA probes were prepared separately from the transcriptional activation domain (TAD) and DBD of the AR gene of the rice field eel, labeled with digoxigenin-UTP, using SP6 or T7 RNA polymerase. Gonad tissues were cryosectioned. The sections were immediately hybridized at 42 °C, and hybridization signals were detected by the nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate system according to the manufacturer's instructions (Roche Applied Science).
Plasmid Constructs
Plasmids pSV-AR0, MMTV-Luc, and pRL-SV40-LUC were gifts from Professor Albert Brinkmann (Erasmus University, Rotterdam, The Netherlands). AR-GFP recombinant was constructed by cloning the encoding region of the AR gene of the rice field eel into HindIII and SacII sites of the pEGFP-N1 (Clontech, Mountain View, CA). Various deletion mutants of AR fused to GFP were subcloned into pEFGP-N1 using AR-GFP as a template. Site-directed mutagenesis was performed to generate the following AR-GFP mutants: AR-GFP(L576A, L579A,L581A), AR-GFP(K461A, K463A,K464A), and AR-GFP(L559A,K563A,K566A). The primers and PCR conditions for making these constructs are shown in Table 1. The full-length mouse AR was cloned using primers 5′-AGCCTCGAGATGGAGGTGCAGTTAGGGCT-3′ and 5′-ATAGTCGACTGTGTGTGGAAATAGATGGG-3′ and constructed into XhoI and SacII sites of the pEGFP-N1. The NES sequence of AR (residues 553–575) was amplified and cloned into AR-GFP(1–447) and AR-GFP(1–464) vectors, to generate expression vectors AR-GFP(1–447)+NES and AR-GFP(1–464)+NES. The nucleotide sequence for the polypeptides GSNELALKLAGLDINKTGGC, the NES of the heat-stable protein kinase inhibitor (PKI) (5, 6, 31, 32), was cloned into pEGFP-N1 to generate NESPKI-GFP. The two oligonucleotides for NESPKI (5′-AACAGCAATGAATTAGCCTTGAAATTAGCAGGTCTTGATA-3′ and 5′-TTCACCTTCTGTCTTGTTGATATCAAGACCTGCTAATTTC-3′), which are partially complementary, were used as PCR template after annealing. The primers for amplification for NESPKI are 5′-AGCCTCGAGAACAGCAATGAATTAGCC-3′ and 5′-AGCAAGCTTTTCACCTTCTGTCTTGTTG-3′. The pSV-eAR recombinant was constructed by replacement of human AR from pSV-AR0 (cut with SacII) with eAR coding sequence of the rice field eel. The primers for eAR are 5′-ATACCGCGGCATGAGCCAAACTAACCCACAG-3′ and 5′-ATACCGCGGCTCCCATGTGCCCCAAAAATC-3′. The full-length cDNAs of Sox9a1 and Sox9a2 were amplified and cloned into pEGFP-N1 and pDsRED-N1 to generate Sox9a1-GFP/RFP and Sox9a2-GFP/RFP. For a GST pull-down assay, various eel AR deletions were amplified and cloned into pGEX-4T-1 through EcoRI and XhoI sites for recombinant protein expression in bacteria, including GST-AR(340–468), GST-AR(384–500), GST-AR(384–483), GST-AR(384–460), and GST-AR(340–446). For the dihydrotestosterone (DHT) binding assay, pcHis-hAR-DBD-LBD was first subcloned into pET-32a through EcoRV and XhoI sites, and then a DNA fragment including the His tag and hAR DBD-LBD was amplified and cloned into pcDNA 3.0 by BamHI and ApaI sites to generate pcHis-hAR-DBD-LBD. Various mutations of eAR were first subcloned into reconstructed pET-32a (without the HindIII site) through EcoRI and XhoI sites, and then DNA fragments, including His tag and eel AR sequences, were amplified and cloned into pcDNA 3.0 by HindIII and ApaI sites to generate pcHis-DBD-LBD, pcHis-H6 deletion, pcHis-H5 mutation, pcHis-H4 mutation, pcHis-H3 mutation, pcHis-H3 deletion, and pcHis LBD deletion. All constructs were verified by sequencing.
TABLE 1.
Primers and PCR conditions for making AR mutant constructs
Sites for restriction enzymes are underlined. Sites for point mutations are in boldface type.
| Mutant name | Mutant primer | PCR |
|---|---|---|
| Deletion | ||
| WT AR | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATACCGCGGCTCCCATGTGCCCCAAAAATC-3′ | ||
| 1–588 | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATACCGCGGTAAAACAAAATGAGAAATGCC-3′ | ||
| 1–575 | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATACCGCGGATGAATAAATAACTCTGAG-3′ | ||
| 1–552 | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATACCGCGGACCTGGAAGTCCTTTCGCCC-3′ | ||
| 1–464 | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-ATACCGCGGTTTCTTTAGTTTACGTGCTCC-3′ | ||
| 1–447 | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-ATACCGCGGTAGCCGACAAGACGGACAG-3′ | ||
| 1–464+NES | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-ATACTCGAGTTTCTTTAGTTTACGTGCTCC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 30 s; 35 cycles | |
| 5′-AGCCTCGAGAGAACACCGGGTGAAGTTTTGG-3′ | ||
| 5′-ATACCGCGGATGAATAAATAACTCTGAG-3′ | ||
| 1–447+NES | 5′-AGCAAGCTTATGAGCCAAACTAACCCACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-ATACTCGAGTAGCCGACAAGACGGACAG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 30 s; 35 cycles | |
| 5′-AGCCTCGAGAGAACACCGGGTGAAGTTTTGG-3′ | ||
| 5′-ATACCGCGGATGAATAAATAACTCTGAG-3′ | ||
| Site-directed mutations | ||
| K461A,K463A,K464A | 5′-GGAGCACGTGCACTAGCGGCAATTGGACAA-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 7 min; 18 cycles |
| 5′-TTGTCCAATTGCCGCTAGTGCACGTGCTCC-3′ | ||
| L563A,L566A | 5′-GTAGATCAGGCGATTTTTGCATTTAATCAC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 7 min; 18 cycles |
| 5′-GTGATTAAATGCAAAAATCGCCTGATCTAC-3′ | ||
| L559A,L563A,L566A | 5′-TGAAGTTGCGGTAGATCAGGCGATTTTTGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 7 min; 18 cycles |
| 5′-GCAAAAATCGCCTGATCTACCGCAACTTCA-3′ | ||
| L576A,L579A,L581A | 5′-TTATTCATGCATGTATTGCCATGGCAGGCATTTC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 7 min; 18 cycles |
| 5′-GAAATGCCTGCCATGGCAATACATGCATGAATAA-3′ | ||
| GST tag mutants | ||
| 340–468 | 5′-ACAGAATTCTACCCCGGCGGGTTGCTG-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTGTTGTCCAATTTTCTTTAG-3′ | ||
| 384–500 | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCAGCTGGGAGCTGAAGTTC-3′ | ||
| 384–483 | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTCAGCAGGTTCCTGGACAG-3′ | ||
| 384–460 | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGTGCTCCAAGAGTCATTCCAG-3′ | ||
| 340–446 | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCCGACAAGACGGACAGTTC-3′ | ||
| His tag mutants | ||
| DBD-LBD | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTACTCCCATGTGCCCCAAAAATC-3′ | ||
| H5 mutation | ||
| H4 mutation | ||
| H6 deletion | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTATAAAACAAAATGAGAAATG-3′ | ||
| H3 mutation | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-GGCGCTGGTGGCCGCGGTGGCTGCTGATTC-3′ complementary to | ||
| 5′-GCGGCCACCTGCGCCAATGAGCTCGGAGAG-3′ | ||
| 5′-AGCCTCGAGCTACTCCCATGTGCCCCAAAAATC-3′ | ||
| H3 deletion | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-GTGGCTGCTGATTCTGGTTGG-3′ complementary to | ||
| 5′-GAATCAGCAGCCACCCTTCCAGGTAGAACACC-3′ | ||
| 5′-AGCCTCGAGCTACTCCCATGTGCCCCAAAAATC-3′ | ||
| LBD deletion | 5′-ACAGAATTCTTCACACCACAGAGAACATGC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTACAGCTGGGAGCTGAAG-3′ | ||
| pcHis tag mutants | ||
| DBD-LBD | 5′-AGAGGGCCCACCATGCACCATCATCATC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCAAGCTTCTACTCCCATGTGCCCCAAAAATC-3′ | ||
| H5 mutation | ||
| H4 mutation | ||
| H3 mutation | ||
| H3 deletion | ||
| H6 deletion | 5′-AGAGGGCCCACCATGCACCATCATCATC-3′ | 94 °C, 30 s; 64 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCAAGCTTCTATAAAACAAAATGAGAAATG-3′ | ||
| pcHis-hAR DBD-LBD | 5′-AGCGATATCGTTTTGCCCATTGAC 3′ | 94 °C, 30 s; 62 °C, 30 s; 72 °C, 1 min; 35 cycles |
| 5′-AGCCTCGAGCTGGGTGTGGAAATAGATGG-3′ | ||
| 5′-AGAGGGCCCACCATGCACCATCATCATC-3′ | ||
| 5′-ACAGGATCCTCACTGGGTGTGGAAATAG 3′ | ||
| Sox9a1/a2-GFP/RFP | ||
| Sox9a1 | 5′-AGCCTCGAGATGAATCTCCTCGACCCTTAC-3′ | 94 °C, 30 s; 62 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATAGGATCCGGCCTGGACAGCTGTGTG-3′ | ||
| Sox9a2 | 5′-AGCCTCGAGATGAATCTCCTCGACCCTTAC-3′ | 94 °C, 30 s; 62 °C, 30 s; 72 °C, 2 min; 35 cycles |
| 5′-ATAGGATCCGGTCTGGTGAGCTGGGTG-3′ | ||
Cell Preparation, Transfection, and DHT/leptomycin B (LMB) Treatments
COS-7 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (HyClone, Logan, UT), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) in the presence of 5% CO2 at 37 °C in a humidified incubator. We used COS-7 cells following methods described previously (33–35). Prior to transfection, cells were washed in PBS and grown in phenol red-free DMEM supplemented with 10% charcoal-stripped FBS (Biowest, Nuaillé, France) for 24 h. For transient transfections, cells growing on a coverslip were transfected with plasmids using Lipofectamine PLUSTM reagent as suggested by the supplier (Invitrogen).
To test the effect of androgens on inducing nuclear localization of AR, the cells were treated with DHT for 16 h. Mouse AR-GFP was used as a positive control in parallel experiments. The method for inhibition of CRM1-mediated nuclear export by leptomycin B was described previously (5, 6). Briefly, 4 h after transfection, cells were treated with 10−7 m DHT to induce nuclear localization of eAR-GFP. Nuclear export of eAR-GFP was induced by DHT withdrawal in the absence or presence of 15 ng/ml LMB. Localization of GFP fusion proteins was observed through fluorescence microscopy 8 h after LMB addition. NESPKI-GFP was used as a positive control in parallel experiments. Hoechst 33258 (Sigma) was used to visualize the nuclei, and the localization of GFP fusion proteins was observed using a DMLA fluorescence microscope (Leica, Bensheim, Germany).
Transcriptional Activity
COS-7 cells were grown in 48-well plates to 80% confluence. Prior to transfection, cells were washed and grown in phenol red-free DMEM with 10% charcoal-stripped FBS (Biowest) for 24 h. For the DHT dose-response experiments, COS-7 cells were co-transfected with 15 ng of pSV-eAR, 250 ng of MMTV-Luc, and 0.25 ng of pRL-SV40 (inner control, SV40 promoter with Renilla luciferase reporter) per well. Six hours after transfection, media were replaced with phenol red-free DMEM supplemented with charcoal-stripped FBS and an increasing concentration of DHT (from 10−10 to 10−6 m) or 0.1% ethanol (control). Expression plasmid pSV-AR0 (human AR) was used as a control in parallel experiments. Treated cells were harvested 24 h later. Luciferase activity was measured using the Dual-LuciferaseTM reporter assay system as suggested by the supplier (Promega).
To test the function of the ligand binding domain of AR, cells were transfected with 15 ng of GFP-tagged wild type or mutant AR constructs, 250 ng of MMTV-Luc, and 0.25 ng of pRL-SV40 per well. Six hours after transfection, medium was replaced with phenol red-free DMEM supplemented with charcoal-stripped FBS, in the presence or absence of 10−7 m DHT. Treated cells were harvested 24 h later. Luciferase activity was measured using the Dual-LuciferaseTM reporter assay system (Promega).
For the flutamide (Sigma) treatment assay, cells were transfected with 15 ng of pSV-eAR, 250 ng of MMTV-Luc, and 0.25 ng of pRL-SV40, and after 6 h, the medium was replaced with phenol red-free DMEM supplemented with charcoal-stripped FBS; meanwhile, an increasing concentration (from 10−6 to 10−4 m) of flutamide was added in the presence of 10−7 m DHT. After 24 h, luciferase activity was measured using the Dual-LuciferaseTM reporter assay system (Promega).
Antibodies
Antibodies used for the GST pull-down assay included anti-GST (GE Healthcare) and anti-GFP (Abcam Inc., Cambridge, CA). Anti-His antibody for detection of in vitro translated protein was purchased from Proteintech Group Inc. (Chicago, IL), and anti-mouse/goat HRP-conjugated secondary antibodies were from Thermo Fisher Scientific (San Jose, CA).
GST Pull-down
COS-7 cells transfected with Sox9a1-GFP or Sox9a2-GFP were lysed in radioimmune precipitation buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 50 mm NaF, 1% Nonidet P-40, 2 mm EDTA, 0.25% sodium deoxycholate, 1× mammalian protease inhibitor). After incubation with the glutathione-Sepharose 4B beads only (Amersham Biosciences), the supernatants of cell lysates were incubated with fresh glutathione-Sepharose 4B beads and various GST-fused AR proteins, including GST (negative control), GST-AR(340–468), GST-AR(384–500), GST-AR(384–483), GST-AR(384–460), or GST-AR(340–446), respectively, at 4 °C overnight. After washing, bound proteins were detected using anti-GFP antibody and anti-mouse HRP-conjugated secondary antibody, followed by Western blot analysis, and visualized using ImmobilonTM Western chemiluminescent HRP substrate (Millipore Corp., Billerica, MA).
DHT Binding Assay
In vitro translation of recombinant His-fused AR mutant proteins (His-hAR-DBD-LBD, His-eAR-DBD-LBD, His-H3/H4/H5 point mutations, and His-H3/H6/LBD deletions) was performed according to the methods supplied by the manufacturer's instructions (TNT® SP6 High-Yield Protein Expression System, Promega). Recombinant proteins were purified using nickel-Sepharose (Ni SepharoseTM 6 Fast Flow, Amersham Biosciences) and checked using anti-His antibody and anti-goat HRP-conjugated secondary antibody (Thermo Fisher Scientific). Equal amounts of proteins were prepared for the DHT binding assay. The purified proteins His-eAR-DBD-LBD, His-eAR-DBD (without LBD, negative control), and His-hAR-DBD-LBD (positive control) in the beads were incubated with an increasing concentration of DHT (from 10−9 to 10−7 m) in the absence or presence of a 500-fold concentration of flutamide in TEG buffer (10 mm Tris-HCl, pH 7.4, 1.5 mm EDTA, 10% glycerol) at 4 °C for 20 h (36). Unbound DHT was removed by washing nickel-Sepharose four times with TEG buffer, and then bound DHT was released to the supernatant after incubation with 120 μl of TEG buffer at 70 °C for 10 min. Quantitative determination of DHT in the supernatant was performed using a DHT ELISA kit, according to the methods supplied by the manufacturer (IBL international GmbH, Hamburg, Germany). The concentrations of bound DHT were calculated as follows. First, a standard curve with mean optical densities on the x axis and the standard calibrator concentrations on the y axis was generated, using DHT standards supplied by the manufacturer, and then an equation (y = 87.366x−1.6118) for the standard curve was obtained using an Excel worksheet. The concentrations of bound DHT were calculated using the above equation according to mean optical densities of the bound DHT. The relative DHT binding of various eAR mutant proteins was measured by incubating with 10−8 m DHT, and LBD deletion protein was used to determine the nonspecific binding amount. The specific DHT-binding amounts of various AR mutant proteins were determined by subtracting the nonspecific binding amounts from total binding amounts.
RESULTS
Characterization of AR in the Rice Field Eel
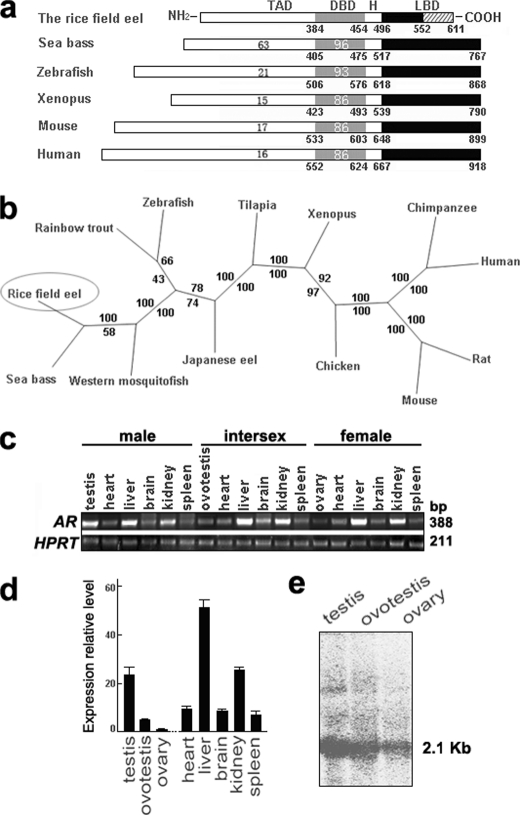
To investigate the role of AR in gonadal transformation of the rice field eel, we first cloned the eAR cDNA using degenerate PCR and RACE techniques. The full-length eAR cDNA (GenBankTM accession number FJ471538) contained an open reading frame encoding 611 amino acids, including the putative TAD, DBD, hinge domain (H), and LBD (Fig. 1a). Both sequence alignment and phylogenetic analysis showed that the AR gene of the rice field eel is close to those of fish, especially sea bass (Fig. 1, a and b). The DBD was highly conserved in vertebrates, including the rice field eel; however, the putative LBD of eAR was short and distinct in comparison with other vertebrate LBDs.
FIGURE 1.
Characterization of eAR gene of the rice field eel. a, structural comparison of AR protein of the rice field eel with ARs of other species. The TAD, DBD, hinge domain (H), and LBD are indicated. The numbers below each box refer to the positions of amino acids in the sequence. The numbers within each box indicate the percentage identity of TAD and DBD relative to those of the rice field eel. b, phylogenetic tree of the AR proteins. Phylogenetic analysis was performed with Phylip (Joseph Felsenstein, Washington University, St. Louis, MO). Numbers in the branches represent the boot strap values (percentage) from 100 replicates obtained using the maximum likelihood method (value above) and the neighbor-joining method (value below). GenBankTM accession numbers were as follows: human AR (M23263), chimpanzee AR (O97775), mouse AR (M37890), rat AR (M20133), chicken AR (P15143), Xenopus AR (U67129), Nile tilapia AR (AB045211), Japanese eel AR (AB023960), zebrafish AR (EF440290), rainbow trout AR (AB012095), Western mosquito fish AR (AB174849), sea bass AR (AY647256), and the rice field eel AR (FJ471538). c, semiquantitative RT-PCR analysis of AR gene expression in three kinds of sexes of the rice field eel (388 bp). HPRT (211 bp) was used as an internal control. d, real-time fluorescent quantitative RT-PCR analysis of AR gene expression in three kinds of gonads and other adult tissues of females. Error bars, S.E. e, Northern blot analysis of the AR gene expression in testis, ovotestis, and ovary of the rice field eel indicates a major band of 2.1 kb.
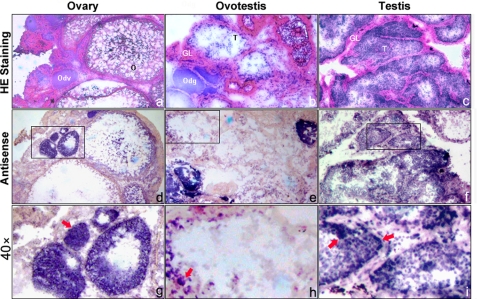
Both semi-quantitative RT-PCR and real-time fluorescent quantitative RT-PCR analysis showed that eAR was ubiquitously expressed highly in testis, liver, and kidney (Fig. 1, c and d). Interestingly, AR expression showed an increasing tendency during gonadal transformation of the rice field eel from ovary via ovotestis to testis (Fig. 1d). Northern blot analysis confirmed the expression trend, and a major band of 2.1 kb of the eAR was observed (Fig. 1e). We further analyzed eAR expression sites in three types of gonads by mRNA in situ hybridization. In testis, positive signals were mainly detected in the inner layer of the seminiferous tubule, including somatic and germ cells (Fig. 2, f and i), and in the ovotestis, signals were shown in the region of testis differentiation of the inner layer of gonadal lamella and degrading oocytes (Fig. 2, e and h), whereas in the ovary, positive signals appeared in the developing oocytes (Fig. 2, d and g), suggesting a potential role of eAR in gonadal transformation.
FIGURE 2.
Expression analysis of eAR gene by mRNA in situ hybridization to gonadal sections of female, intersex, and male. a–c, H&E staining of ovary, ovotestis, and testis, respectively. d–f, eAR expression in the relevant gonad samples is shown in the middle panel (magnification ×20). g–i, higher amplification (magnification ×40) from the squares in d–f, respectively. Red arrowheads indicate the positive signals. Positive signals were observed in developing oocytes of the ovary (female), in degrading oocytes and the gonadal lamella of the ovotestis (intersex), and in both somatic and germ cells of the testis (male). GL, gonadal lamella; Odv, developing oocytes; Odg, degrading oocytes; T, testis; O, ovary.
An Androgen-dependent Transactivation Function of eAR
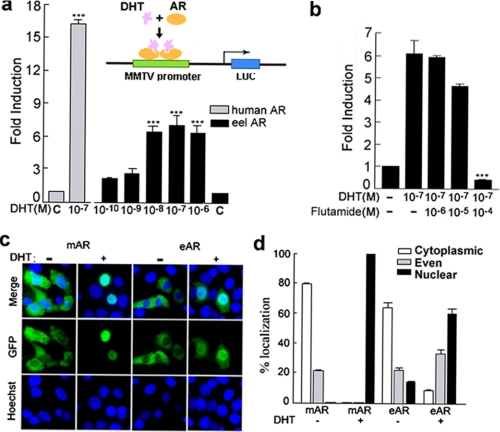
Because there is a special LBD of eAR, we investigated whether the LBD is functional and whether the eAR has an androgen-dependent transactivation role. Wild type eAR expression plasmid pSV-eAR was co-transfected with an androgen-regulated reporter, MMTV-Luc, into COS-7 cells (33) to test eAR transactivation capability (Fig. 3a). The eAR exhibited a remarkable transcriptional activity in a DHT dose-dependent manner, with the highest activity at a dose of 10−7 m DHT (p < 0.005) (Fig. 3a). However, the transactivation capability of eAR was markedly low, compared with that of human AR. The GFP-tagged eAR was further used to determine the intracellular localization of the eAR after DHT treatment. The GFP-tagged eAR was translocated into nuclei with a proportion of 60.8% upon DHT exposure, compared with a complete nuclear import of the mouse AR (Fig. 3, c and d). These results suggested an androgen-dependent transactivation function of eAR and a nuclear translocation upon DHT exposure in spite of a weak transactivation capability compared with mammalian AR. We further investigated whether anti-androgen flutamide could affect transactivation capability of eAR by competition with DHT for binding to eAR. As shown in Fig. 3b, the transactivation activity of eAR decreases remarkably at a dose of 10−4 m flutamide in combination with 10−7 m DHT.
FIGURE 3.
Subcellular localization and dose-dependent effect of DHT/flutamide on the transcriptional activation of eAR. a, plasmids pSV-eAR and MMTV-Luc were transiently co-transfected with plasmid pSL-SV40 into COS-7 cells. Cells were treated with increasing concentrations of DHT (from 10−10 to 10−6 m) or 0.1% ethanol (C, control). Plasmids pSV-AR0 (human AR) and MMTV-Luc were co-transfected with plasmid pSL-SV40 (inner control, SV40 promoter with Renilla luciferase reporter) and treated with 10−7 m DHT in parallel experiments. Data are expressed as -fold induction relative to control for three independent experiments. The asterisks indicate a significant difference between DHT treatment and ethanol treatment (control). ***, p < 0.005. Shown is a sketch model of MMTV-Luc transcription activated by AR and DHT. b, increasing concentration (from 10−6 to 10−4 m) of flutamide treatments in the presence of DHT (10−7 m) in COS-7 transfected with relevant constructs pSV-eAR, MMTV-Luc, and pSL-SV40. Data are expressed as -fold induction relative to nontreatment for three independent experiments. The asterisks indicate a significant difference between non-flutamide treatment and flutamide treatment. ***, p < 0.005. c, subcellular localization of mouse AR (mAR)-GFP and eAR-GFP fusion proteins in the transfected COS-7 cells in the presence or absence of 10−7 m DHT showed nuclear import following DHT exposure with high efficiency in mouse AR and relatively low efficiency in eel AR. Merge, the corresponding merged images of GFP and nuclear staining with Hoechst 33258. d, percentages of AR-GFP-expressing COS-7 cells in c that displayed cytoplasmic, even, or nuclear localization. The results were from three transfections, and at least 50 AR-GFP-expressing cells were counted each time. Error bars, S.E.
The eAR Contains a Functional NLS Sequence in the DBD and Hinge Region
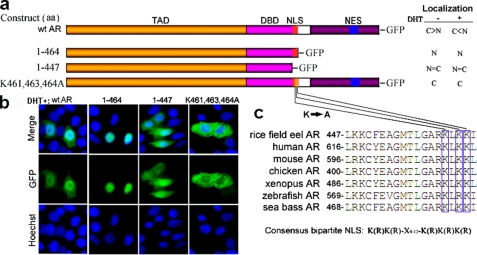
To explore the nuclear localization signal of eAR, sequence comparison of available DBDs and hinge regions from vertebrates, including mammals, birds, frogs, and fish, revealed two highly conserved basic clusters (KK and RKLKK) between amino acids 447 and 465 in eAR, which corresponded to bipartite NLS (RK and RKLKK) in the DBD and hinge region of human AR (32, 35, 37) (Fig. 4, a and c). To test the potential function of the NLS, GFP-tagged eAR and two deletion mutants were constructed (Fig. 4a) and transfected into COS-7 cells. Both wild type (WT) eAR and deletion mutant 1–464, which include the putative NLS sequence, could efficiently target the GFP to the nuclei upon DHT exposure. However, deletion mutant 1–447, which lacked the putative NLS sequence, showed 50% retention in the cytoplasm (Fig. 4, a and b). These results suggested that the NLS sequence between 447 and 465 is important for AR nuclear localization in the rice field eel. To further analyze critical amino acids of the nuclear localization signal, site-directed mutant eAR-GFP(K461A,K463A,K464A) was constructed and transfected into COS-7 cells. As shown in Fig. 4b, GFP fluorescence was completely localized in the cytoplasm, indicating that Lys461, Lys463, and Lys464 are pivotal sites for nuclear translocation of eAR.
FIGURE 4.
Identification of NLSAR by deletion and site-directed mutagenesis. a, scheme of WT AR, two deletion mutants, and one site-directed mutant fused to GFP. Nuclear (N)/cytoplasmic (C) localization ratios of eAR-GFP in the absence (−) or presence (+) of DHT after transfection into COS-7 are shown on the right. C > N, more cytoplasmic than nuclear localization; C < N, more nuclear than cytoplasmic localization; C = N equal in nuclear and cytoplasmic localization. b, subcellular localization of WT AR and various eAR mutants-GFP transiently transfected into COS-7 in the presence of DHT. Plasmids, including putative NLS (WT AR, aa 1–464) efficiently targeted the GFP to nuclei. A mutant without putative NLS (aa 1–447) showed even distribution. Site-directed mutant eAR-GFP(K461A,K463A,K464A) resulted in retention of GFP in the cytoplasm completely. c, sequence comparison of AR among different species showed a high degree of conservation around the putative NLS region. The consensus sequence of bipartite NLS is indicated below.
Nuclear Export of the AR is CRM1-independent
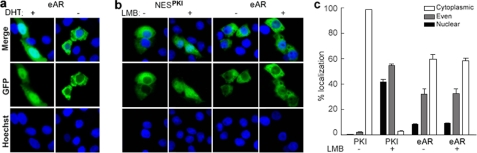
Because human AR translocates into the nuclei in the presence of androgens and can be exported to the cytoplasm by ligand withdrawal (5, 6), we investigated whether eAR may be also exported to the cytoplasm upon DHT withdrawal. The same nuclear export was observed in rice field eel as well as in human AR (Fig. 5a). To test whether the nuclear export of eAR-GFP depends upon CRM1/exportin 1, after transfection with eAR-GFP and following the addition of DHT for the induction of nuclear import, the cells were treated by DHT withdrawal in the presence or absence of LMB, an inhibitor of CRM1-dependent nuclear export (38, 39). eAR-GFP maintained cytoplasmic localization regardless of the presence or absence of LMB (Fig. 5, b and c), whereas in parallel control experiments, GFP-tagged NES of the heat-stable protein kinase inhibitor NESPKI, a CRM1-dependent nuclear export signal sensitive to LMB inhibition (5, 6, 31, 32), resulted in predominantly nuclear localization after LMB was added (Fig. 5, b and c). These results indicate that nuclear export of eAR-GFP is CRM1-independent, which is consistent with that of human AR (6).
FIGURE 5.
Characterization of nuclear export of eAR. a, DHT withdrawal experiment indicated nuclear export of eAR-GFP. COS-7 cells were transiently transfected with expression vector eAR-GFP. 10−7 m DHT was added to induce nuclear import of the eAR-GFP. After 16 h, medium was replaced by DHT-free medium (−) or there was no medium change (+) for culture of another 8 h. b, effect of LMB treatment on the nuclear export of the eAR-GFP. COS-7 cells were transfected with expression vector eAR-GFP or NESPKI-GFP. 10−7 m DHT was added for induction of nuclear import of eAR-GFP. After 16 h, export of nuclear eAR-GFP was induced by DHT withdrawal in the absence (−) or presence (+) of LMB for another 8 h. Cytoplasmic localization of eAR-GFP was observed regardless of addition of the LMB or not. NESPKI-GFP was used as a positive control for nuclear export (5, 6, 31, 32), which showed that the addition of LMB resulted in inhibition of nuclear export of the NESPKI-GFP. c, percentages of eAR-GFP- or NESPKI-GFP-expressing COS-7 cells in b that displayed cytoplasmic, even, or nuclear localization. The results were from three repeats of transfection, and at least 50 transfected cells were counted each time. Error bars, S.E.
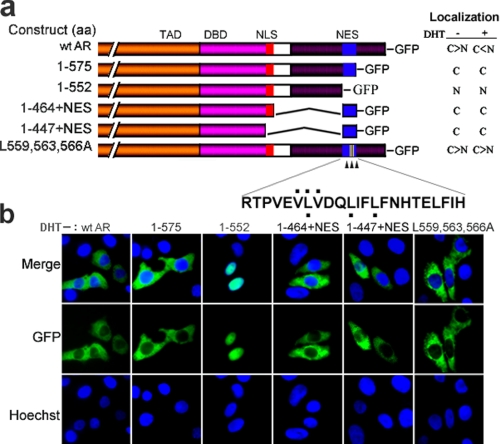
Identification of a Novel NES in the LBD of eAR
In an effort to understand nuclear export of eAR during DHT withdrawal, we further identified the NES sequence of eAR. Deletion mutagenesis was first performed to map the NES in the LBD of eAR. Deletion mutant eAR-GFP(1–575) showed completely cytoplasmic localization, whereas mutant eAR-GFP(1–552) exhibited completely nuclear localization (Fig. 6, a and b), indicating that the NES sequence of eAR was restricted within amino acids 553–575, a polypeptide of 23 aa. To further confirm the nuclear export function of putative NES, we generated GFP-fused deletion mutants AR-GFP(1–447)+NES and AR-GFP(1–464)+NES (1–447+NES and 1–464+NES in Fig. 6a). Transfection of both mutants exhibited completely cytoplasmic localization (Fig. 6, a and b), suggesting that the NES is necessary for eAR nuclear export and is dominant over the NLS in the DBD and hinge region in the absence of ligand DHT. Because the classical NES sequence is leucine-rich and the identified leucine-rich NES was generally accepted as a loose consensus, LX2,3(L/I/V/F/M)X2,3LX(L/I) (40), we compared the eAR NES with the consensus and observed similar leucine-rich residues within aa 553–575 in α-helix 4 of the LBD. However, GFP-tagged mutant eAR-GFP(L559A,L563A, L566A) had a slight influence on the cytoplasmic localization, whereas eAR-GFP(V558A,V560A,L559A) had no effect on the cytoplasmic localization, compared with wild type eAR in the absence of DHT (Fig. 6, a and b). Further, we observed that both mutants eAR-GFP(L559A,L563A,L566A) and eAR-GFP(V558A,V560A,L559A) remained in the cytoplasm upon DHT exposure, in comparison with nuclear translocation of wild type eAR. These results indicated that these mutation sites are critical for the DHT-induced nuclear import of eAR.
FIGURE 6.
Identification of NESAR by deletion mutagenesis of eAR. a, construction maps of WT AR and a series of mutants fused to GFP. Nuclear (N)/cytoplasmic (C) localization ratios of eAR-GFP in the absence (−) or presence (+) of DHT after transfection into COS-7 are shown on the right. The mutant sites of site-directed mutagenesis are indicated with a black dot above or below the sequence (L/V to A). b, subcellular localization of WT AR and various eAR mutants-GFP transiently transfected into COS-7 in the absence of DHT. Plasmids containing putative NES (WT AR, 1–575, 1–447+NES, and 1–464+NES) showed cytoplasmic localization, whereas mutant (aa 1–552) without putative NES exhibited nuclear localization. Mutant eAR-GFP(L559A,L563A,L566A) showed no significant influence on cytoplasmic localization of GFP-fused proteins.
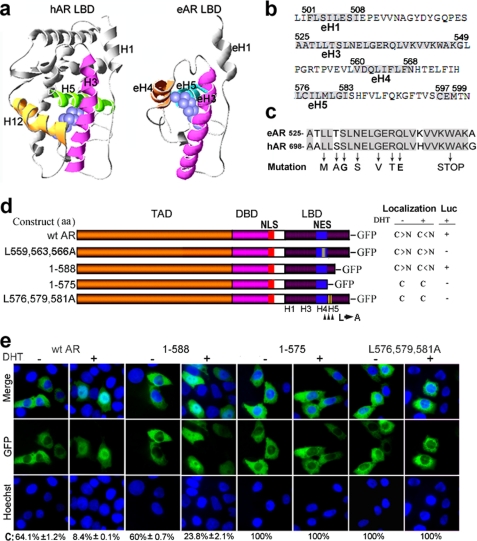
Identification of Critical Amino Acids in Helices for eAR Subcellular Localization and Transactication Function
Androgen-dependent transactivation function of eAR stimulates our interest in identifying critical amino acids in the LBD for subcellular localization and transactivation. Although the LBD sequence of eAR was different from those of other vertebrates, three α-helices (eH3, eH4, and eH5) were observed in the LBD of eAR, which were spatially similar to α-helices H3, H5, and H12 of human LBD (Fig. 7, a and b). The three α-helices in human AR LBD directly contacted with the bound ligand (41). Among the three α-helices, only the α-helix 3 was conserved between humans and rice field eels (Fig. 7c); its mutations resulted in human complete androgen insensitivity syndrome and partial androgen insensitivity syndrome diseases (1). Seven of eight key point mutations that led to low or no binding capacity to ligands (42–47) were identical between humans and rice field eels (Fig. 7c), suggesting a conserved androgen-binding ability of this α-helix 3. Mutations in the α-helix 4 of the eAR LBD (L559A,L563A,L566A and V558A,V560A,L559A) failed in DHT-induced nuclear import and transactivation function (Figs. 6 and 7). To further determine subcellular localization and transactivation function of α-helix 5, deletion and site-directed mutagenesis were performed for the helix. Wild type eAR and deletion mutant 1–588 exhibited nuclear translocation upon DHT exposure, whereas deletion mutant 1–575 (helix 5 absent) completely remained in the cytoplasm in transfected cells regardless of the absence or presence of DHT, indicating that amino acid residues 576–588 are critical for the nuclear localization of eAR (Fig. 7, d and e). Further, site-directed mutant eAR-GFP(L576A,L579A,L581A) showed that GFP fluorescence was completely in the cytoplasm in transfected cells regardless of the absence or presence of DHT. Furthermore, these point mutations and deletions severely affected the transactivation function of eAR (Fig. 7d).
FIGURE 7.
Identification of critical amino acids in helices for eAR subcellular localization and transactivation function by deletion and site-directed mutagenesis of eAR. a, simulation model of the three-dimensional structure of human AR LBD and eAR LBD. LBD region and potential location of DHT (blue spheres) are indicated. Key ligand binding α-helices 3, 5, and 12 of human AR LBD and relevant putative ligand binding α-helices 3, 4 and 5 of eAR LBD of the rice field eel are highlighted. b, eAR LBD amino acid sequence of the rice field eel. Numbers above the sequence indicate the positions of amino acids, and the residues of helices are shaded in gray. c, sequence comparison of α-helix 3 between human AR and eel AR. The conserved sites are shaded in gray. Mutation sites (arrows) were detected in human complete androgen insensitivity syndrome and partial androgen insensitivity syndrome. d, scheme of wt AR and a series of mutants fused to GFP. Nuclear (N)/cytoplasmic (C) localization ratios of eAR-GFP in the absence (−) or presence (+) of DHT after transfection into COS-7 and their relevant luciferase activities are showed on the right. e, subcellular localization of WT eAR-GFP and various eAR mutants-GFP transiently transfected into COS-7 in the absence (−) or presence (+) of DHT. The percentage of eAR-GFP-expressing COS-7 cells that displayed cytoplasmic localization is shown below. Mutant 1–588 efficiently targeted the GFP to nuclei upon DHT exposure, whereas mutant 1–575 resulted in the retention of GFP in the cytoplasm. Site-directed mutant eAR-GFP(L576A,L579A,L581A) showed completely cytoplasmic localization and a lack of luciferase activity.
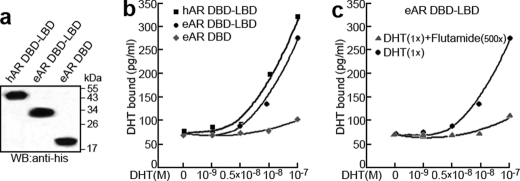
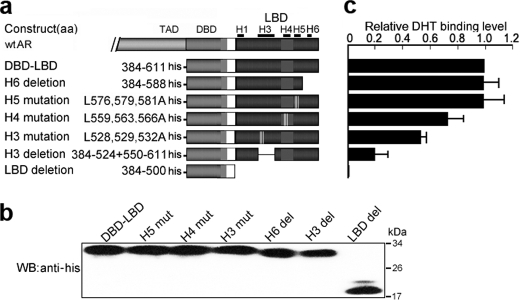
DHT Binding to eAR Directly
To further investigate whether eAR and DHT interact directly, receptor-ligand binding of eAR LBD with DHT was measured by a quantitative DHT-AR binding ELISA. Protein eAR DBD-LBD showed marked DHT binding at 10−8 or 10−7 m DHT in contrast with LBD deletion protein eAR DBD, although the DHT binding ability of the eAR DBD-LBD is weaker than that of human hAR DBD-LBD (Fig. 8). Further, flutamide competed with DHT for binding to eAR DBD-LBD and decreased DHT binding (Fig. 8). These results indicate that eAR binds directly to DHT. Further mutation analysis was used to confirm the conclusion. Although mutations of helices 5 and 6 of eAR LBD have DHT binding ability nearly equivalent to that of WT AR, mutations in the helices 3 and 4 of eAR LBD remarkably affected the eAR binding to DHT (Fig. 9), indicating that eAR LBD is able to bind DHT efficiently.
FIGURE 8.
DHT-binding analysis of eAR. a, detection of in vitro translation of His-hAR-DBD-LBD, His-eAR-DBD-LBD, and His-eAR-DBD proteins by SDS-PAGE and Western blot (WB) with anti-His antibody. b, DHT binding ability of hAR and eAR. An increasing concentration of DHT was incubated with an equal amount of translated proteins. The x axis indicates the concentrations of incubated DHT, whereas the amount of bound DHT is shown on the y axis, which was calculated using the equation, y = 87.366x−1.6118, obtained from the standard curve. c, DHT binding analysis of eAR with 500-fold flutamide treatment as a competitor of the DHT.
FIGURE 9.
DHT-binding analysis of eAR LBD helices through deletion and site-directed mutagenesis. a, construction sketch of a series of LBD mutants fused with His tags. b, detection of in vitro translation of the mutant proteins by SDS-PAGE and Western blot (WB) with anti-His antibody. c, DHT binding of various eAR LBD mutant proteins. Data are expressed relative to the DBD-LBD binding quantity for at least two independent experiments. Error bars, S.E.
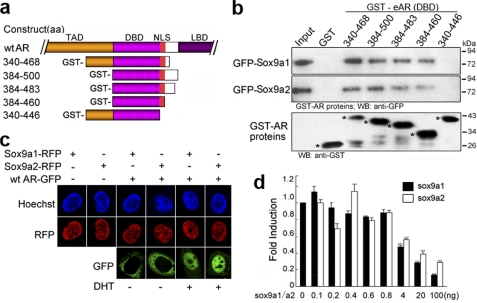
Sox9a1/Sox9a2 Interacts with eAR and Regulate eAR Transactivation
Previous work revealed that human SOX9 directly interacted with AR through its DNA binding domain (48). Because we previously identified two Sox9 genes in the genome of the rice field eel (49) instead of one SOX9 gene in humans, we wondered whether both Sox9 proteins could interact with eAR. Various GST-fused eAR deletions were generated and a GST pull-down assay was employed to detect the interaction between eAR and Sox9. Deletion analysis of eAR indicated that eAR could interact with both Sox9 proteins, and Sox9a1 interacted with eAR more efficiently than Sox9a2. A series of deletions of eAR indicated that the region aa 447–460 (corresponding to the NLS) of eAR was necessary for Sox9a1/a2 binding (Fig. 10b). Further subcellular localization analysis showed that eAR, Sox9a1, and Sox9a2 were co-localized in the nucleus in the presence of DHT (Fig. 10c), which is consistent with their interaction. We further investigated whether interaction of eAR with Sox9a1/a2 has an effect on eAR transactivation function by using the MMTV-Luc reporter system. Both Sox9a1 and Sox9a2 decreased eAR transactivation remarkably at a high level of Sox9a1/a2 transfection (from 4 to 100 ng), whereas they slightly increased eAR transactivation at a lower level of Sox9a1/Sox9a2 (Fig. 10d). These results are consistent with that for human SOX9 (48).
FIGURE 10.
Interaction of eAR with both Sox9a1 and Sox9a2. a, construction sketch of GST-fused eAR mutants. b, GST pull-down assay of GST-eAR with both Sox9a1-GFP and Sox9a2-GFP. COS-7 cells transfected with Sox9a1-GFP/Sox9a2-GFP were lysed and incubated with glutathione-Sepharose 4B beads coupled with GST-fused eAR proteins. Bound proteins were detected with anti-GFP antibody. The lower panel indicates the input amount of GST-fused proteins (marked by an asterisk). c, nuclear localization of eAR with both Sox9a1 and Sox9a2 in the presence of DHT. Plasmids Sox9a1-RFP and Sox9a2-RFP were transfected individually or co-transfected with WT eAR-GFP, respectively, into COS-7 cells in the absence or presence of DHT. Sox9a1-RFP and Sox9a2-RFP were localized in the nuclei, whereas WT eAR-GFP was localized in the cytoplasm in the absence of DHT. After DHT treatment, both GFP and RFP were observed mainly in the nuclei. d, both Sox9a1 and Sox9a2 modulate the transactivation function of eAR. COS-7 cells were transiently co-transfected with pSV-eAR (30 ng), MMTV-Luc (250 ng), pSL-SV40 (0.25 ng), and an increasing amount of Sox9a1-RFP or Sox9a2-RFP in the presence of DHT. DsRED-N1 was employed to balance the quantities of transfected plasmids. Luciferase activities are shown as -fold induction relative to Sox9a1/a2-non-transfected cells for three independent experiments. The asterisks indicate significant difference between Sox9a1/a2-transfected and non-transfected cells. ***, p < 0.005. Error bars, S.E.; WB, Western blot.
DISCUSSION
AR is composed of multiple domains or regions with distinct functions, including an N-terminal activating domain, a C-terminal ligand binding domain, and a DNA binding domain in the mid-region, in addition the NLS and NES. The interplay among these functional regions determines AR action, regardless of the presence or absence of the ligand. For eAR nucleocytoplasmic shuttling, we observed here that NES action is dominant over NLS in the absence of DHT, which results in the cytoplasmic retention of eAR. However, when eAR is activated upon DHT exposure, the NLS functions dominantly, which leads the activated eAR to the nucleus. We infer that the NES region of the activated eAR for nuclear export might be covered by the DHT, which would weaken the action of the NES and facilitate the NLS to exert nuclear import function.
We identified a novel NES sequence of eAR, which is a short polypeptide with only 23 amino acids. Its isoelectric point is pI = 5.27, and charge at pH 7.0 is −2.06, which shows that the nuclear export pathway may be mediated by electrostatic interaction of the negatively charged NES with positively charged residues, although eAR nuclear export is CRM1-independent, as is that of human AR (6). The NES sequence of eAR is distinct from human AR NES, which was observed in a region of 75 residues. These observations suggest that the novel NES must be generated despite a rapid sequence evolution of the gene during the vertebrate speciation, that ensures that AR exerts nuclear export functions. The cytoplasmic retention might be an essential step for AR functions through DHT-mediated regulation. The deficiency of the C terminus including NES will result in the nuclear retention of the AR because of the NLS action; consequently, the AR will play an androgen-independent role in the nucleus, a similar situation to androgen-independent prostate cancer (45).
Coherent action of NLS and the androgen-bound LBD is another essential step for AR nuclear import and functions in the nucleus. The NLS sequence is conserved from fish to frogs, birds, and mammals, and the consensus is bipartite NLS ((R/K)K and RKLKK), which suggests an essential role of the NLS in AR functions. Nevertheless, amino acid sequences of the LBD between the eel and other vertebrate ARs are different, which shows evolutionary divergence of the ARs. Although a restricted androgen-dependent transactivation function was observed in eAR, how does eAR exert its function in the primitive vertebrate? After analysis, we found that a three-dimensional structure with three α-helices (eH3, eH4, and eH5) in the LBD of eAR is spatially similar to α-helices H3, H5, and H12 of the human LBD. The three α-helices in the human AR LBD made direct contact with the bound ligand (41). Mutation analysis confirmed that the LBD is essential for DHT-induced nuclear import of eAR and following transactivation function in the nucleus. The LBD mutations were indeed observed in human complete androgen insensitivity syndrome and partial androgen insensitivity syndrome diseases and in prostate cancer (1, 2). Evolutionarily, three α-helices (eH3, eH4, and eH5) in the LBD are spatially essential for the AR function despite the different amino acid sequence; therefore, the structure of three α-helices has been conserved from eels to humans during vertebrate evolution. The α-helix structure in human AR has evolved into a precisely matched three-dimensional configuration of 12 helices with the smarter structure of three key α-helices. Overall, the conservation of the three-dimensional structure of the LBD seems more important than primary structure for the AR functions, which gives reasonable explanations for diverse androgen-binding capabilities among different point mutations in the LBD and heterogeneous phenotypes in human AR diseases. Given that the structure of three α-helices in the LBD is essential for androgen binding and following transactivation, it may helpful to design androgen analogs based on the structure for treatment of prostate cancer and for development of approaches for sexual control in fish.
This work was supported by the National Natural Science Foundation of China, National Key Basic Research Project Grant 2010CB126306, the Fundamental Research Funds for the Central Universities, the Program of Wuhan Subject Chief Scientist, and the 111 Project Grant B06018.
- AR
- androgen receptor
- eAR
- eel AR
- hAR
- human AR
- MMTV
- mouse mammary tumor virus
- NLS
- nuclear localization signal
- DBD
- DNA binding domain
- NES
- nuclear export signal
- RACE
- rapid amplification of cDNA ends
- LBD
- ligand binding domain
- TAD
- transcriptional activation domain
- PKI
- protein kinase inhibitor
- DHT
- dihydrotestosterone
- LMB
- leptomycin B
- aa
- amino acid.
REFERENCES
- 1.Gottlieb B., Beitel L. K., Wu J. H., Trifiro M. (2004) Hum. Mutat. 23, 527–533 [DOI] [PubMed] [Google Scholar]
- 2.Bergerat J. P., Céraline J. (2009) Hum. Mutat. 30, 145–157 [DOI] [PubMed] [Google Scholar]
- 3.Heemers H. V., Tindall D. J. (2007) Endocr. Rev. 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 4.Heemers H. V., Tindall D. J. (2009) Cancer Cell 15, 245–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi R. K., Lavrovsky Y., Ahn S. C., Song C. S., Chatterjee B., Roy A. K. (2000) Mol. Endocrinol. 14, 1162–1174 [DOI] [PubMed] [Google Scholar]
- 6.Saporita A. J., Zhang Q., Navai N., Dincer Z., Hahn J., Cai X., Wang Z. (2003) J. Biol. Chem. 278, 41998–42005 [DOI] [PubMed] [Google Scholar]
- 7.Nguyen M. M., Dincer Z., Wade J. R., Alur M., Michalak M., Defranco D. B., Wang Z. (2009) Mol. Cell. Endocrinol. 302, 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kousteni S., Bellido T., Plotkin L. I., O'Brien C. A., Bodenner D. L., Han L., Han K., DiGregorio G. B., Katzenellenbogen J. A., Katzenellenbogen B. S., Roberson P. K., Weinstein R. S., Jilka R. L., Manolagas S. C. (2001) Cell 104, 719–730 [PubMed] [Google Scholar]
- 9.Heinlein C. A., Chang C. (2002) Mol. Endocrinol. 16, 2181–2187 [DOI] [PubMed] [Google Scholar]
- 10.Kousteni S., Han L., Chen J. R., Almeida M., Plotkin L. I., Bellido T., Manolagas S. C. (2003) J. Clin. Invest. 111, 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron S., Manin M., Beaudoin C., Leotoing L., Communal Y., Veyssiere G., Morel L. (2004) J. Biol. Chem. 279, 14579–14586 [DOI] [PubMed] [Google Scholar]
- 12.Culig Z. (2004) Growth Factors 22, 179–184 [DOI] [PubMed] [Google Scholar]
- 13.Freeman M. R., Cinar B., Lu M. L. (2005) Trends Endocrinol Metab. 16, 273–279 [DOI] [PubMed] [Google Scholar]
- 14.Balk S. P. (2002) Urology 60, 132–138 [DOI] [PubMed] [Google Scholar]
- 15.Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., Jänne O. A., Balk S. P., Mehra R., Han B., Chinnaiyan A. M., Rubin M. A., True L., Fiorentino M., Fiore C., Loda M., Kantoff P. W., Liu X. S., Brown M. (2009) Cell 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ankley G. T., Jensen K. M., Makynen E. A., Kahl M. D., Korte J. J., Hornung M. W., Henry T. R., Denny J. S., Leino R. L., Wilson V. S., Cardon M. C., Hartig P. C., Gray L. E. (2003) Environ. Toxicol. Chem. 22, 1350–1360 [PubMed] [Google Scholar]
- 17.Martyniuk C. J., Alvarez S., McClung S., Villeneuve D. L., Ankley G. T., Denslow N. D. (2009) J. Proteome Res. 8, 2186–2200 [DOI] [PubMed] [Google Scholar]
- 18.Baron D., Houlgatte R., Fostier A., Guiguen Y. (2008) Gen. Comp. Endocrinol. 156, 369–378 [DOI] [PubMed] [Google Scholar]
- 19.Ikeuchi T., Todo T., Kobayashi T., Nagahama Y. (1999) J. Biol. Chem. 274, 25205–25209 [DOI] [PubMed] [Google Scholar]
- 20.Takeo J., Yamashita S. (1999) J. Biol. Chem. 274, 5674–5680 [DOI] [PubMed] [Google Scholar]
- 21.Todo T., Ikeuchi T., Kobayashi T., Nagahama Y. (1999) Biochem. Biophys. Res. Commun. 254, 378–383 [DOI] [PubMed] [Google Scholar]
- 22.He C. L., Du J. L., Lee Y. H., Huang Y. S., Nagahama Y., Chang C. F. (2003) Biol. Reprod. 69, 455–461 [DOI] [PubMed] [Google Scholar]
- 23.Ogino Y., Katoh H., Yamada G. (2004) FEBS Lett. 575, 119–126 [DOI] [PubMed] [Google Scholar]
- 24.Blázquez M., Piferrer F. (2005) Mol. Cell. Endocrinol. 237, 37–48 [DOI] [PubMed] [Google Scholar]
- 25.Park C. B., Takemura A., Aluru N., Park Y. J., Kim B. H., Lee C. H., Lee Y. D., Moon T. W., Vijayan M. M. (2007) Chemosphere 69, 32–40 [DOI] [PubMed] [Google Scholar]
- 26.Hossain M. S., Larsson A., Scherbak N., Olsson P. E., Orban L. (2008) Biol. Reprod. 78, 361–369 [DOI] [PubMed] [Google Scholar]
- 27.Cui J., Shen X., Yan Z., Zhao H., Nagahama Y. (2009) Biochem. Biophys. Res. Commun. 380, 115–121 [DOI] [PubMed] [Google Scholar]
- 28.Liu X., Su H., Zhu P., Zhang Y., Huang J., Lin H. (2009) Gen Comp. Endocrinol. 160, 93–101 [DOI] [PubMed] [Google Scholar]
- 29.Zhou R., Cheng H. H., Tiersch T. R. (2001) Rev. Fish Biol. Fisheries 11, 331–337 [Google Scholar]
- 30.Cheng H., Guo Y., Yu Q., Zhou R. (2003) Cytogenet. Genome Res. 101, 274–277 [DOI] [PubMed] [Google Scholar]
- 31.Wen W., Harootunian A. T., Adams S. R., Feramisco J., Tsien R. Y., Meinkoth J. L., Taylor S. S. (1994) J. Biol. Chem. 269, 32214–32220 [PubMed] [Google Scholar]
- 32.Zhou Z. X., Sar M., Simental J. A., Lane M. V., Wilson E. M. (1994) J. Biol. Chem. 269, 13115–13123 [PubMed] [Google Scholar]
- 33.Georget V., Lobaccaro J. M., Terouanne B., Mangeat P., Nicolas J. C., Sultan C. (1997) Mol. Cell. Endocrinol. 129, 17–26 [DOI] [PubMed] [Google Scholar]
- 34.Hofman K., Swinnen J. V., Claessens F., Verhoeven G., Heyns W. (2000) Mol. Cell. Endocrinol. 168, 21–29 [DOI] [PubMed] [Google Scholar]
- 35.Cutress M. L., Whitaker H. C., Mills I. G., Stewart M., Neal D. E. (2008) J. Cell Sci. 121, 957–968 [DOI] [PubMed] [Google Scholar]
- 36.Jørgensen A., Andersen O., Bjerregaard P., Rasmussen L. J. (2007) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 146, 561–568 [DOI] [PubMed] [Google Scholar]
- 37.Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. (1991) J. Biol. Chem. 266, 510–518 [PubMed] [Google Scholar]
- 38.Wolff B., Sanglier J. J., Wang Y. (1997) Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 39.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogerd H. P., Fridell R. A., Benson R. E., Hua J., Cullen B. R. (1996) Mol. Cell. Biol. 16, 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matias P. M., Donner P., Coelho R., Thomaz M., Peixoto C., Macedo S., Otto N., Joschko S., Scholz P., Wegg A., Bäsler S., Schäfer M., Egner U., Carrondo M. A. (2000) J. Biol. Chem. 275, 26164–26171 [DOI] [PubMed] [Google Scholar]
- 42.De Bellis A., Quigley C. A., Cariello N. F., el-Awady M. K., Sar M., Lane M. V., Wilson E. M., French F. S. (1992) Mol. Endocrinol. 6, 1909–1920 [DOI] [PubMed] [Google Scholar]
- 43.Pinsky L., Trifiro M., Kaufman M., Beitel L. K., Mhatre A., Kazemi-Esfarjani P., Sabbaghian N., Lumbroso R., Alvarado C., Vasiliou M. (1992) Clin. Invest. Med. 15, 456–472 [PubMed] [Google Scholar]
- 44.Radmayr C., Culig Z., Glatzl J., Neuschmid-Kaspar F., Bartsch G., Klocker H. (1997) J. Urol. 158, 1553–1556 [PubMed] [Google Scholar]
- 45.Melo K. F., Mendonca B. B., Billerbeck A. E., Costa E. M., Inácio M., Silva F. A., Leal A. M., Latronico A. C., Arnhold I. J. (2003) J. Clin. Endocrinol. Metab. 88, 3241–3250 [DOI] [PubMed] [Google Scholar]
- 46.Hannema S. E., Scott I. S., Hodapp J., Martin H., Coleman N., Schwabe J. W., Hughes I. A. (2004) J. Clin. Endocrinol. Metab. 89, 5815–5822 [DOI] [PubMed] [Google Scholar]
- 47.Melo K. F., Mendonça B. B., Billerbeck A. E., Costa E. M., Latronico A. C., Arnhold I. J. (2005) Arq. Bras. Endocrinol. Metabol. 49, 87–97 [DOI] [PubMed] [Google Scholar]
- 48.Wang H., McKnight N. C., Zhang T., Lu M. L., Balk S. P., Yuan X. (2007) Cancer Res. 67, 528–536 [DOI] [PubMed] [Google Scholar]
- 49.Zhou R., Liu L., Guo Y., Yu H., Cheng H., Huang X., Tiersch T. R., Berta P. (2003) Mol. Reprod. Dev. 66, 211–217 [DOI] [PubMed] [Google Scholar]
- 50.Heinlein C. A., Chang C. (2002) Mol. Endocrinol. 16, 2181–2187 [DOI] [PubMed] [Google Scholar]
- 51.Fix C., Jordon C., Cano P., Walker W. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10919–10924 [DOI] [PMC free article] [PubMed] [Google Scholar]