Abstract
Protein-disulfide isomerase-associated 3 (Pdia3) is a multifunctional protein hypothesized to be a membrane receptor for 1,25(OH)2D3. In intestinal epithelium and chondrocytes, 1,25(OH)2D3 stimulates rapid membrane responses that are different from genomic effects via the vitamin D receptor (VDR). In this study, we show that 1,25(OH)2D3 stimulates phospholipase A2 (PLA2)-dependent rapid release of prostaglandin E2 (PGE2), activation of protein kinase C (PKC), and regulation of bone-related gene transcription and mineralization in osteoblast-like MC3T3-E1 cells (WT) via a mechanism involving Pdia3. Pdia3 was present in caveolae based on co-localization with lipid rafts and caveolin-1. In Pdia3-silenced (Sh-Pdia3) cells, 1,25(OH)2D3 failed to stimulate PKC and PGE2 responses; in Pdia3-overexpressing cells (Ov-Pdia3), responses to 1,25(OH)2D3 were augmented. Downstream mediators of Pdia3, PLA2-activating protein (PLAA) and arachidonic acid, stimulated similar PKC activation in wild-type, Sh-Pdia3, and Ov-Pdia3 cells supporting the hypothesis that Pdia3 mediates the membrane action of 1,25(OH)2D3. Treatment of MC3T3-E1 cells with 1,25(OH)2D3 for 9 min stimulated rapid phosphorylation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) and increased expression of alkaline phosphatase, MMP-13, and osteopontin but decreased expression of osteocalcin, osteoprotegerin (mRNA and protein), and smad2. These effects were attenuated in Sh-Pdia3 cells. Sh-Pdia3 cells produced higher numbers of von Kossa-positive nodules and alizarin red-positive nodules compared with WT cells with or without 1,25(OH)2D3 treatment whereas Ov-Pdia3 did not show any mineralization. Our data suggest Pdia3 is an important initiator of 1,25(OH)2D3-stimulated membrane signaling pathways, which have both genomic and non genomic effects during osteoblast maturation.
Keywords: Calcification, Caveolae, Phospholipase A, Plasma Membrane, Protein Kinase C (PKC), shRNA, Steroid Hormone Receptor, Vitamin D, Osteoblast, Pdia3
Introduction
The secosteroid 1,25-dihydroxyvitamin D3 (1α,25(OH)2D3)2 regulates osteoblasts through both the classic vitamin D receptor (VDR)-mediated genomic pathway and through membrane receptor-mediated rapid responses (1–3). In osteoblasts, the VDR acts by binding with vitamin D response elements (VDRE) to modulate gene transcription (4, 5). Rapid membrane signaling has been shown to regulate calcium, phosphate, and chloride transport through ion channels (6–8). The VDR has been implicated in these rapid responses to 1α,25(OH)2D3 (9, 10). However, studies using analogues to the secosteroid with low VDR binding affinities indicate that the structural features of 1α,25(OH)2D3 are important in stimulating the membrane response, suggesting the existence of a specific membrane receptor (8).
Protein-disulfide isomerase-associated 3 (Pdia3, also known as ERp60, ERp57, Grp58, and 1,25-MARRS) has been identified as a potential candidate as an alternate membrane-associated receptor for 1α,25(OH)2D3 (11). It has been intensively studied as a chaperone protein in glycoprotein folding (12) and major histocompatibility complex I loading (13). Unlike other protein-disulfide isomerase family members, Pdia3 exists not only in the endoplasmic reticulum but also in the nucleus, extracellular matrix, and plasma membrane, suggesting additional functions (3). Pdia3 purified from chick intestinal epithelium (14) exhibits saturable binding to 1α,25(OH)2D3. More recently, antibody blocking and ribosome knock down experiments have further linked this protein to rapid responses to 1α,25(OH)2D3 both in rat chondrocytes and chick intestinal epithelial cells (11, 15). Taken together, these observations support a role for Pdia3 as a membrane receptor for the secosteroid.
Multiple cell models have been used to elucidate the role of membrane signaling by 1α,25(OH)2D3. In growth plate chondrocytes, 1α,25(OH)2D3 regulates phospholipase A2 (PLA2), phospholipase C (PLC), intracellular Ca2+, and protein kinase C (PKC) (16–18). In osteoblasts, 1α,25(OH)2D3 has also been shown to activate PLC and PKC as well as regulating voltage-gated ion channels and increasing PLA2 activity and prostaglandin production (PGE1 and PGE2) (10, 19–21). These data suggest that the rapid membrane signaling pathway discovered in chondrocytes may also function in osteoblasts.
One function of membrane signaling is to modulate gene transcription. 1α,25(OH)2D3 regulates rat growth zone chondrocytes via PKC and extracellular signal-regulated kinases 1 and 2 (ERK1/2) in a Pdia3-dependent pathway (22). Microarray analysis of rat osteoblastic ROS 17/2.8 cells treated with a 1,25(OH)2D3 analog with low binding affinity to VDR also demonstrated regulation of a large number of genes through an intracellular calcium-dependent mechanism (23, 24). Whether Pdia3 mediates 1α,25(OH)2D3-dependent gene expression in osteoblasts is not known, nor are the overall physiological consequences of rapid responses to the secosteroid clear.
In this study, we first examined 1α,25(OH)2D3 stimulated rapid membrane signaling in osteoblasts. Second, we showed Pdia3 is required to initiate this rapid signaling. Third, we showed that 1α,25(OH)2D3-stimulated Pdia3-dependent non-genomic effects eventually control genomic changes affecting mineralization.
EXPERIMENTAL PROCEDURES
Pdia3 Knock Down and Overexpression
Pdia3 shRNA probes were designed to target the mouse Pdia3 mRNA (NM_007952). Five different sequences were generated and incorporated into lentivirus particles (SHCLNV-NM 007592, Sigma-Aldrich). To select the optimal shRNA, mouse MC3T3-E1 osteoblasts (CRL-2593, ATCC, Manassas, VA) were plated at a density of 20,000 cells/cm2 in a 24-well plate in α-MEM supplemented with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (P/S). After 24 h, media were changed to α-MEM supplemented with 10% FBS, 1% P/S, and 8 μg/ml hexadimethrine bromide and transduced with lentivirus particles at a multiplicity of infection (MOI) of 7.5. Cells containing shRNAs or empty vectors were selected by culturing the cells for 2 weeks in medium containing 2.0 μg/ml puromycin. Loss of Pdia3 expression was quantified by real-time PCR and verified by Western blot using anti-Pida3 antibody (Alpha Diagnostic International Inc., San Antonio, TX). A stable transduced cell line with 80% knock down of Pdia3 was chosen. The shRNA targeted the 3′-UTR region of Pdia3 mRNA (ggaccagtttatgtttgtggttt, N0000011436, Sigma-Aldrich).
To overexpress Pdia3, MC3T3-E1 cells were plated at a density of 20,000 cells/cm2 in a 24-well plate in α-MEM supplied with 10% FBS. After 24 h, 100 μl of Opti-MEM (Invitrogen, Carlsbad, CA) containing 2.0 μl of Lipofectamine 2000 (Invitrogen) and 0.8 μg of pCMV6-Kan/Neo empty vector (OriGene, Rockville, MD) or overexpression plasmid (MC200134, OriGene) that contained the full-length of mouse Pdia3 cDNA (BC003285.1) was added into each well. After 48 h, cells were selected in medium containing 550 μg/ml G418 for 2 weeks. mRNA and protein were quantified by real-time PCR and Western blot as described above. A stable transfected cell line with 100% overexpression of Pdia3 was chosen.
Cell Culture
Wild-type MC3T3-E1 cells, MC3T3-E1 cells silenced for Pdia3 (Sh-Pdia3), and MC3T3-E1 cells overexpressing Pdia3 (Ov-Pdia3) were plated in T75 flasks (10,000 cells/cm2) and cultured in α-MEM containing 10% FBS and 1% P/S. Puromycin (2 μg/ml) was included in the medium of Sh-Pdia3 cells and G418 (550 μg/ml) was included in the medium of Ov-Pdia3 cells. At confluence, cells were subcultured at the same plating density. 48 h after plating, the media were replaced with α-MEM supplemented with 5% FBS, 1% P/S, and 1% vitamin C. After 12 days in culture, the cells were treated with medium containing either the 1α,25(OH)2D3 vehicle (ethanol) alone or with the appropriate dose of 1α,25(OH)2D3.
Plasma Membranes and Caveolae
A detergent-free method of plasma membrane and caveolae isolation was used as described previously (25). MC3T3-E1 cells were cultured for 12 days and harvested by scraping in isolation buffer (0.25 m sucrose, 1 mm EDTA, 20 mm Tricine, pH 7.8). Samples were homogenized using a tissue grinder for twenty strokes. Homogenates were centrifuged at 20,000 × g for 10 min to pellet cell debris, including nucleus, mitochondria, and endoplasmic reticulum. The supernatant was collected, placed on top of 30% Percoll (GE Healthcare, Piscataway, NJ) in isolation buffer, and then centrifuged for 30 min at 84,000 × g. The plasma membranes formed a visible band and were collected by aspiration. Plasma membranes were layered on a 10–20% OptiPrep gradient (Sigma-Aldrich) and centrifuged at 52,000 × g for 90 min. The top layer was collected, overlaid with 5% OptiPrep, and centrifuged at 52,000 × g for another 90 min. Fractions were collected from the top to the bottom in thirteen fractions. The caveolae existed as an opaque band, which was collected in Fraction 3, based on the presence of caveolin-1 as described below.
Western Blot
Gel electrophoresis was performed using NuSep 4–20% LongLife Gels (NuSep, Lawrenceville, GA). Proteins were transferred from gels to nitrocellulose membrane using an iBlot Dry Blotting System (Invitrogen). The membrane was subsequently blotted in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h. Next, samples were incubated overnight with antibodies against Pdia3 (Alpha Diagnostic International); caveolin-1 (Sc-894, Santa Cruz Biotechnology, Santa Cruz, CA), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (MAB374, Millipore, Billerica, MA). After washing three times with PBS containing 0.05% Tween-20, the membrane was incubated with goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) in PBS containing 5% dry milk and 0.05% Tween-20 for 1 h. After three washes, the membrane was developed using SuperSignal West Pico Chemiluminescent System (Thermo Fisher Scientific, Rockford, IL) and imaged with VersaDoc imaging system (Bio-Rad).
Immunofluorescence
Immunofluorescence was used to assess Pdia3 protein in intact cells. Wild type, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1 cells were plated at 10,000 cells/cm2 on a glass chamber slide for 24 h. The cells were fixed in 4% paraformaldehyde for 20 min and permeabilized with 0.01% Triton-X 100 for 10 min. Cells were then incubated with 1:100 dilution of Hoechst 33342 (Invitrogen), Pdia3 primary antibody, and 1:40 dilution of Alexa Fluor 555 phalloidin (Invitrogen) in a PBS containing 1% BSA. After washing, cells were incubated with goat anti-rabbit Alexa 488 (Invitrogen) in 1% BSA-PBS.
To determine if Pdia3 was associated with a specific plasma membrane compartment, co-localization experiments were performed. Cells in suspension were labeled with Vybrant Lipid Rafts Labeling Kits-Alexa Fluor 594 (Invitrogen). After labeling, cells were fixed in 4% paraformaldehyde for 20 min. Then, cells were further stained with Pdia3 primary antibody and 1:100 (v/v) Hoechst 33342 in 1%BSA-PBS. After washing, cells were incubated with goat anti-rabbit Alexa 488 in 1% BSA-PBS, fixed with GEL/MOUNT (Biomeda Corp, Foster City, CA), and imaged using a Zeiss LSM 510 confocal microscope.
Signaling by 1α,25(OH)2D3
To study the effect of 1α,25(OH)2D3 on PKC activity, cells were treated with vehicle (ethanol) or 10−10, 10−9, or 10−8 m 1α,25(OH)2D3 for 9 min, based on the observation that 1α,25(OH)2D3 activates PKC in chondrocytes at this time point (26). The effect of 1α,25(OH)2D3 on PKC is via a PLA2-dependent pathway (16). To determine if this is also the case for MC3T3-E1 cells, one-half of the cultures were treated with 10−5 m of the PLA2 inhibitor quinacrine (Calbiochem). Quinacrine was added to media 30mins before and maintained during 1α,25(OH)2D3 treatment. Cells were also treated for 9 min with 0, 10−8, 10−7, 10−6 m PLA2-activating protein (PLAA) (Enzo Life Sciences International, Inc, Plymouth Meeting, PA) as well as with 0, 10−6, 10−5, or 10−4 m arachidonic acid (AA) (Calbiochem), which is the product of PLA2 action. Media were collected, and cell layers were washed twice with PBS and lysed in 300 μl RIPA buffer (20 mm Tris-HCl, 150 mm NaCl, 5 mm disodium EDTA, 1%Nonidet P-40). PKC activity was measured using a commercial kit (RPN77, GE Healthcare) and normalized by total protein. To measure rapid PGE2 release into the media, cells were treated with vehicle (ethanol), 10−10, 10−9, or 10−8 m 1α,25(OH)2D3. After 30 min treatment, the media were acidified and PGE2 was measured using a commercial kit (Perkin Elmer, Waltham, MA) and normalized by cell number.
ERK1/2
To determine if the rapid responses to 1α,25(OH)2D3 result in ERK1/2 activation, phosphorylation of ERK1/2 was determined using an ELISA assay. After 12 days of culture, cells were treated for 9 min with medium containing vehicle (ethanol) or 10−8 m 1α,25(OH)2D3. Media were replaced, and cells were harvested 0, 30, and 90 min later. Phospho-ERK1/2 was measured in cell layer lysate using a mouse phospho-ERK1/2 ELISA kit (R&D system, Minneapolis, MN).
Gene Expression
After 12 days of culture, cells were treated with medium containing vehicle (ethanol) or 10−8 m 1α,25(OH)2D3. After 9 min, media were replaced with fresh media for another 8 h. RNA was extracted using TRIzol (Invitrogen) and reverse-transcribed into cDNA using the Omniscript RT kit (Qiagen, Valencia, CA) according to the manufacturer's directions. Real-time PCR was performed using SYBR Green SuperMix 170–8882 (Bio-Rad) for osteocalcin (OCN, NM_001032298), alkaline phosphatase (ALP, NM_007431), bone sialoprotein (BSP, NM_008318), matrix metalloproteinase 13 (MMP-13, NM_008607), osteopontin (OPN, NM_009263), osteoprotegerin (OPG, NM_011613), Pdia3 (NM_007952), Runx2 (NM_009820), inositol 1,4,5-trisphosphate 3-kinase A (ITPKA, NM_146125), osteonectin (OTN, NM_009242), Smad2 (NM_010754), Vitamin D receptor (VDR, NM_009504), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, NM_008084). Oligonucleotide primers were designed using Beacon Designer 7.0 software (supplemental Table S1). A homology blast search was performed within the mammalian genome to exclude the possibility of sequence similarity. Real-time PCR was performed using an iCycler PCR machine (Bio-Rad) with iCycler software. Data were normalized to the endogenous reference gene GAPDH.
Osteopontin
Changes in osteopontin secreted into the conditioned media were used as an outcome measure of the rapid signaling pathway. The role of Ca2+ ions was examined by incubating the cells with 10−4 m Ca2+ chelator BAPTA-AM (Sigma-Aldrich). The role of PLA2 was assessed by treating the cells with 10−5 m quinacrine. For these experiments, cultures were treated with media containing the inhibitor vehicle (ddH2O) or inhibitor for 30 min. Fresh media containing the 1,25(OH)2D3 vehicle (ethanol) or the inhibitor plus 10−8 m 1α,25(OH)2D3 were added. 9 min later, the media were replaced by fresh media, and these media were collected 24 h later. Osteopontin in the conditioned media was determined by ELISA using a mouse osteopontin ELISA kit (R&D Systems).
Calcification
To determine if Pdia3 modulates terminal osteoblast differentiation, we assayed the ability of the cells to mineralize their extracellular matrix. Cells were plated in 24-well plates in full media and cultured as described above. At confluence, full media were changed for osteogenic media (α-MEM supplemented with 5% FBS, 1% P/S, 100 μg/ml ascorbic acid, 10 mm β-glycerol phosphate and 10−7 m dexamethasone) or growth media (α-MEM supplemented with 5% FBS, 1%P/S). Every 48 h, the osteogenic media were replaced with growth media containing vehicle (ethanol) or 10−8 m 1α,25(OH)2D3 for 9 min; these media were then removed and replaced with osteogenic media. Growth media were replaced every 2 days without additional 1α,25(OH)2D3. On day 28 after plating, media were replaced with osteogenic media containing 10% alamar blue for 40 min to assess viability of the cells. In healthy cells, alamar blue is reduced by components of the electron transport chain, resulting in red fluorescence. The cultures were then fixed in 10% formalin and alizarin red and von Kossa staining were performed.
Statistical Analysis
For each experiment, each data point represents the means ± S.E. for six individual cultures. Each experiment was repeated two or more times to ensure the validity of the data. The data presented are from a single representative experiment. Significance was determined by analysis of variance and post hoc testing performed using Bonferroni's modification of Student's t test for multiple comparisons. p ≤ 0.05 was considered to be significant.
RESULTS
Rapid Membrane Response in MC3T3-E1 Cells
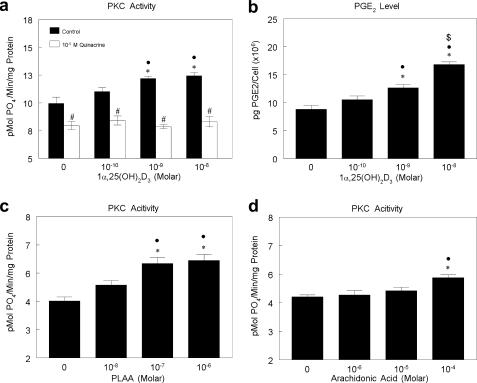
PKC activity in MC3T3-E1 cells was stimulated by 1α,25(OH)2D3 in a dose-dependent manner (Fig. 1a). This effect was rapid, occurring within 9 min of treatment. 1α,25(OH)2D3 also caused a dose-dependent increase in PGE2 content of the conditioned media at 30 min (Fig. 1b), suggesting that PLA2 had been activated. Inhibition of PLA2 with quinacrine blocked the stimulating effect of 1α,25(OH)2D3 on PKC at all concentrations treated. Moreover, the PLA2 activator, PLAA, and the product of PLA2 action, arachidonic acid both stimulated PKC activity to a similar extent. PLAA caused a dose-dependent increase in PKC activity at 10−7 m to 10−6 m (Fig. 1c); arachidonic acid increased PKC at 10−4 m (Fig. 1d). These results indicate that 1α,25(OH)2D3 regulated PKC via a mechanism that required PLA2.
FIGURE 1.
Effect of 1α,25(OH) 2D3 on PGE2 production and PKC activity and role of PLA2 in PKC activation in MC3T3-E1 cells. a, MC3T3-E1 cells were treated with vehicle (ethanol) or 10−10, 10−9, or 10−8 m 1α,25(OH)2D3 with or without 10−5 m quinacrine (PLA2 inhibitor) for 9 min. PKC activity was normalized to total protein. b, MC3T3-E1 cells were treated with vehicle (ethanol) or 10−10, 10−9, or 10−8 m 1α,25(OH)2D3 for 30 min. Conditioned media were collected, and PGE2 was measured and normalized to cell number. c, MC3T3-E1 cells were treated with vehicle (ddH2O) or 10−8, 10−7, or 10−6 m PLAA for 9 min. PKC activity was normalized to total protein level. d, MC3T3-E1 cells were treated with vehicle (media) or 10−6, 10−5, or 10−4 m AA for 9 min. PKC activity was normalized to total protein level. *, p < 0.05, treatment versus control; ●, p < 0.05, 10−8 m and 10−9 m versus 10−10 m; $, p < 0.05, 10−8 m versus 10−9 m; #, p < 0.05 quinacrine versus control.
Subcellular Location of Pdia3
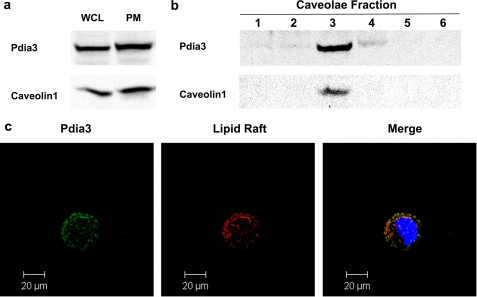
Western blots of whole cell lysates and isolated plasma membranes indicated that both Pdia3 and caveolin-1 were present (Fig. 2a). Pdia3 was present in factions 1, 2, 3, and 4 of the plasma membrane whereas caveolin-1 was present only in faction 3 (Fig. 2b). These observations were supported by confocal microscopy. Both Pdia3 and lipid rafts were distributed over the surface of non-permeabilized MC3T3-E1 cells. When the immunofluorescent images were merged, part of staining overlapped. However, a small portion of fluorescently labeled Pdia3 was not associated with lipid rafts.
FIGURE 2.
Western blot and confocal microscope image of MC3T3-E1 cells. MC3T3-E1 cells were cultured as previously described. Whole cell lysates, plasma membrane fractions, and caveolae fractions were collected separately. Western blots against caveolin-1 and Pdia3 were performed. a, Western blot of whole cell lysates and membrane fractions. Thirteen fractions were collected; fractions one to six are shown. Caveolae exist in fraction three (b). c, confocal image of non-permeabilized MC3T3-E1 cells. Green: Pdia3; red: lipid rafts; yellow: merge of Pdia3 and lipid rafts.
Pdia3 Expression in MC3T3-E1 Cells
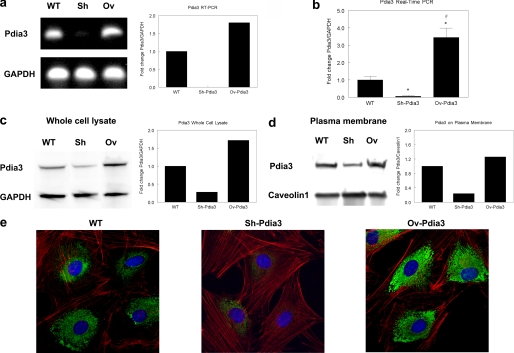
Two MC3T3-E1 cell lines were established that were silenced for expression of Pdia3 or that overexpressed this protein. RT-PCR indicated that mRNA for Pdia3 was reduced in Sh-Pdia3 cells compared with wild-type cells whereas expression was increased in Ov-Pdia3 cells (Fig. 3a). Real-time PCR analysis of six independent cultures showed Pdia3 expression was decreased by 80% in Sh-Pdia3 cells; it increased by 200% in Ov-Pdia3 cells (Fig. 3b). Western blots of whole cell lysates confirmed that Pida3 protein was affected in a comparable manner. Pdia3 protein was reduced by 75% in the Sh-Pdia3 cells and increased by 70% in Ov-Pdia3 cells (Fig. 3c). Western blots of plasma membranes showed similar results. Pdia3 protein was decreased by 80% in plasma membranes from Sh-Pdia3 cells and increased by 30% in plasma membranes from Ov-Pdia3 cells (Fig. 3d). Similarly, MC3T3-E1 cells exhibited intense immunofluorescence for Pdia3 surrounding the nucleus, but this staining was largely decreased in Sh-Pdia3 cells; it was augmented in Ov-Pdia3 cells (Fig. 3e).
FIGURE 3.
Silencing and overexpression of Pdia3 in MC3T3-E1 cells: RT-PCR, real-time PCR, Western blot and confocal microscopy. a, RT-PCR. Left: gel electrophoresis of RT-PCR product. Right: quantitative RT-PCR for the fold change of Pdia3 levels relative to GAPDH control. b, real-time PCR. Fold change of Pdia3 levels relative to GAPDH control. c, Western blot of whole cell lysate. Left: blotting image. Right: quantitative Western blot for the fold change of Pdia3 levels relative to GAPDH control. d, Western blot of plasma membranes. Left: blotting image. Right: quantitative Western blot for the fold change of Pdia3 levels relative to GAPDH control. e, confocal microscopy of permeabilized cells. Red: actin; green: Pdia3; blue: nucleus. Cells were permeabilized before staining.
Role of Pdia3 in the Rapid Response to 1α,25(OH)2D3
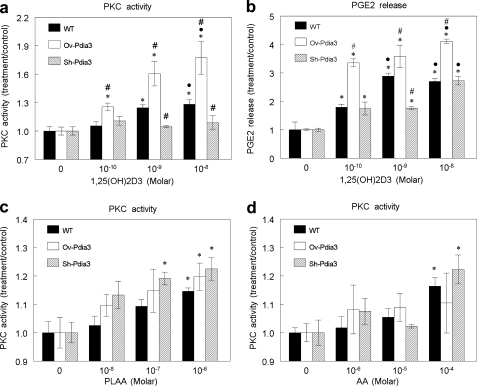
Pdia3 mediated the rapid response of MC3T3-E1 cells to 1α,25(OH)2D3. Whereas 1α,25(OH)2D3 increased PKC in wild-type cells, it had no effect on PKC activity in Sh-Pdia3 cells (Fig. 4a). In contrast, in Ov-Pdia3 cells, 1α,25(OH)2D3 increased PKC activity over the stimulatory effect of the secosteroid in wild-type cells. 10−8 m 1α,25(OH)2D3 caused a 1.8-fold increase of PKC activity in Ov-Pdia3 compared with a 1.3-fold increase in wild-type cells (Fig. 4a). These effects were specific to Pdia3 since the cells transfected with empty vectors responded to 1α,25(OH)2D3 as the wild-type MC3T3-E1 cells (supplemental Fig. S1).
FIGURE 4.
Effect of 1α,25(OH)2D3, PLAA and AA on PKC activity and effect of 1α,25(OH) 2D3 on PGE2 release in wild type, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1 cells. a, the effect of 1α,25(OH)2D3 on PKC activity of WT, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1 cells. MC3T3-E1 cells were treated with vehicle (ethanol) or 10−10, 10−9, or 10−8 m 1α,25(OH)2D3 for 9 min. PKC activity was normalized to total protein. b, 1α,25(OH)2D3 effect on PGE2 release of WT, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1. MC3T3-E1 cells were treated with vehicle (ethanol) or 10−10, 10−9, or 10−8 m 1α,25(OH)2D3 for 30 min. PGE2 in conditioned media was measured and normalized to cell number. c, PLAA effect on PKC activity of WT, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1 cells. MC3T3-E1 cells were treated with vehicle (ddH2O) or 10−6, 10−7, or 10−8 m PLAA for 9 min. PKC activity was measured and normalized to total protein. d, AA effect on PKC activity of WT, Sh-Pdia3, and Ov-Pdia3 MC3T3-E1 cells. MC3T3-E1 cells were treated with vehicle (media) or 10−4, 10−5, or 10−6 m AA for 9 min. PKC activity was measured and normalized by total protein. *, p < 0.05, treatment versus control; ●, p < 0.05, 10−8 and 10−9 versus 10−10; #, p < 0.05 Sh-Pdia3 and Ov-Pdia3 versus WT for the same treatment.
Pdia3 also mediated the 1α,25(OH)2D3-dependent PLA2 signaling pathway resulting in PKC activation. Production of PGE2, a downstream metabolite of PLA2 action, was increased by 1α,25(OH)2D3 in MC3T3-E1 cells, and this effect was enhanced in Ov-Pdia3 cells at all concentrations tested (Fig. 4b). In contrast, the stimulatory effect of 1α,25(OH)2D3 was reduced in Sh-Pdia3 cells. The PLA2 pathway was functional in all three cell lines, however. Treatment of the cells with PLAA caused a dose-dependent increase in PKC that was comparable in all three cell types (Fig. 4c). Similarly arachidonic acid activated PKC to a comparable extent as 1α,25(OH)2D3 in all three cell lines (Fig. 4d).
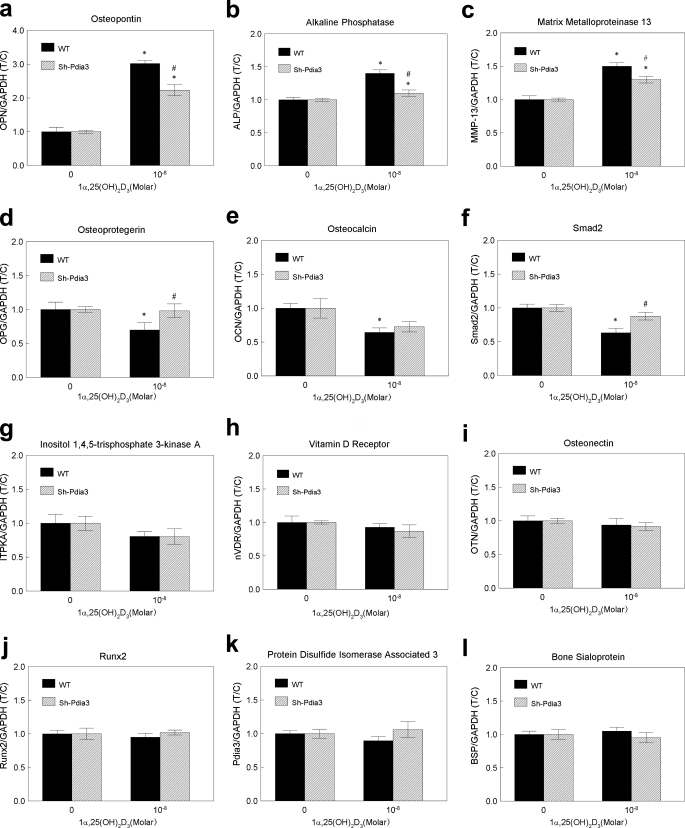
Pdia3-dependent Rapid Signaling Regulates Gene Expression
Gene expression was regulated, at least in part, by Pdia3-mediated signaling (Fig. 5). Treatment of MC3T3-E1 cells with 1α,25(OH)2D3 for 9 min resulted in changes in mRNA levels for bone related genes. OPN, ALP, and MMP-13 were up-regulated, while OPG, Smad2, and OCN were down-regulated. BSP, ITPKA, Runx2, OTN, VDR, and Pdia3 showed no significant differences as a function of 1α,25(OH)2D3 treatment. 1α,25(OH)2D3 did not affect mRNA for BSP, IPTKA, Runx2, OTN VDR or Pdia3 in Sh-Pdia3 cells. However, genes that were down regulated in wild-type cells (OPG, OCN, and Smad2) were unaffected by 1α,25(OH)2D3 in Sh-Pdia3 cells. For the three up-regulated genes (OPN, ALK, and MMP-13), the effects of 1α,25(OH)2D3 were reduced in the Sh-Pdia3 cells.
FIGURE 5.
Effect of 1α,25(OH)2D3 on gene transcription in wild-type and Sh-Pdia3 MC3T3-E1 cells. Wild-type and Sh-Pdia3 cells were treated with vehicle (ethanol) or 10−8 m 1α,25(OH)2D3. After 9 min, media were replaced with fresh media for another 8 h before harvest. Real-time PCR was performed against twelve bone-related genes: (a) osteopontin; (b) alkaline phosphatase; (c) matrix metalloproteinase 13; (d) osteoprotegerin; (e) osteocalcin; (f) Smad2; (g); inositol 1,4,5-trisphosphate 3-kinase A; (h) vitamin D receptor; (i) osteonectin; (j) Runx2; (k) Pdia3; and (l) bone sialoprotein. T/C represents the treatment over control ratio. Absolute values of targeting genes were first normalized by GAPDH. Then the normalized values from treatment groups were further divided by the normalized values from vehicle (ethanol) control groups. *, p < 0.05, treatment versus control; #, p < 0.05, Sh-Pdia3 versus WT for the same treatment.
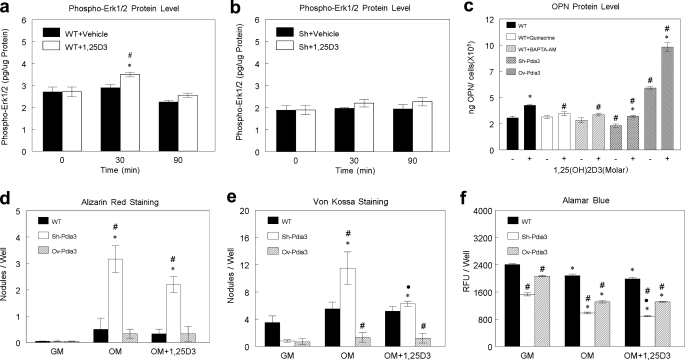
Pdia3 was required for 1α,25(OH)2D3-dependent activation of ERK1/2. 1α,25(OH)2D3 treatment caused phosphorylation of ERK1/2 by 30mins; this effect was lost by 90mins (Fig. 6a). In contrast, 1α,25(OH)2D3 had no effect on ERK1/2 phosphorylation in Sh-Pdia3 cells (Fig. 6b).
FIGURE 6.
Effect of 1α,25(OH)2D3 on ERK1/2 phosphorylation, OPN secretion, and in vitro mineralization in wild-type, Sh-Pdia3, and Ov-Pdia3 cells. a and b, wild-type and Sh-Pdia3 MC3T3-E1 cells were treated with vehicle (ethanol) or 10−8 m 1α,25(OH)2D3 for 9 min. The media were replaced, and cells were harvested at 0 min (no treatment), 30 min, and 90 min after treatment. Intracellular phospho-ERK1/2 was measured by ELISA and normalized to total protein. c, wild-type, Ov-Pdia3, and Sh-Pdia3 MC3T3-E1 cells were treated with vehicle (ethanol) or 10−8 m 1α,25(OH)2D3 for 9 min with or without 10−5 m quinacrine or 10−6 m BAPTA-AM. After 9 min, the media were replaced and after 24 h, OPN was measured in the conditioned media using an ELISA assay. OPN levels were normalized to cell number. d—f, wild-type, Ov-Pdia3, and Sh-Pdai3 cells were cultured in growth media or osteogenic media with or without pulse treatments (9 min) with 10−8 m 1α,25(OH)2D3 every 48 h. Four weeks after seeding, cultures were examined by alamar blue, alizarin red, and van Kossa staining. d, number of alizarin red-positive nodules; e, number of von Kossa-positive nodules. f, relative fluorescence units of alamar blue stain. Numbers indicate the cell viability. Each data point represents mean ± S.E. for n = 6 independent cultures. *, p < 0.05, 30 min, and 90 min versus 0 min for a and b or 1α,25(OH)2D3 versus vehicle (ethanol) for C or OM± 1α,25(OH)2D3 versus GM for d, e, and f; ●, p < 0.05, OM+1α,25(OH)2D3 versus OM-1α,25(OH)2D3 for d, e, and f; #, p < 0.05, 1α,25(OH)2D3 versus vehicle (ethanol) for A and B or Sh-Pdia3, Ov-Pdia3, and WT with inhibitors versus WT for the same conditions for c, d, e, and f.
Changes in protein production correlated with changes in gene expression. Treatment of wild-type MC3T3-E1 cells with 1α,25(OH)2D3 for 9 min caused a 200% increase in OPN mRNA at 8 h (Fig. 5a) and a marked increase in OPN protein at 24 h (Fig. 6c). This effect was attenuated in Sh-Pdia3 cells while Ov-Pdia3 cells showed an augmented increase. The basal level of OPN protein was also lower in Sh-Pdia3 cells and higher in Ov-Pdia3 cells. Inhibition of rapid membrane signaling using quinacrine or bapta-AM also blocked the 1α,25(OH)2D3-dependent increase in OPN protein in the conditioned media in cultures of wild-type cells. Neither inhibitor affected the basal level of OPN, indicating they did not block the normal transcription and translation activity of this protein.
Pdia3 Regulates Mineralization
Wild-type MC3T3-E1 cells, Sh-Pdia3 cells, and Ov-Pdia3 cells all formed von Kossa-positive nodules. The number of alizarin-red/von Kossa-positive nodules formed by wild-type cells were comparable in cultures grown in growth media, osteogenic media, or osteogenic media plus 1α,25(OH)2D3 (Fig. 6, d and e). In growth media, von Kossa-positive nodule formation was reduced in Sh-Pdia3 and Ov-Pdia3 cultures compared with wild-type cells although differences in Pdia3 expression did not affect the number of alizarin-red-positive nodules. When Sh-Pdia3 cells were cultured in osteogenic media, the number of alizarin-red/von Kossa-positive nodules was markedly increased in comparison to wild-type cells. In contrast, von Kossa-positive nodule formation was reduced in cultures of Ov-Pdia3 cells. Treatment with 1α,25(OH)2D3 blocked the stimulatory effect of the osteogenic media on von Kossa-positive nodule formation in cultures of Sh-Pdia3 cells but had no effect on alizarin-red-positive nodules. Moreover, treatment did not affect wild-type or Ov-Pdia3 cultures. For all cell types, growth in osteogenic media resulted in decreased alamar blue staining compared with growth media (Fig. 6f). Treatment with 1α,25(OH)2D3 caused a further decrease only in Sh-Pdia3 cells.
DISCUSSION
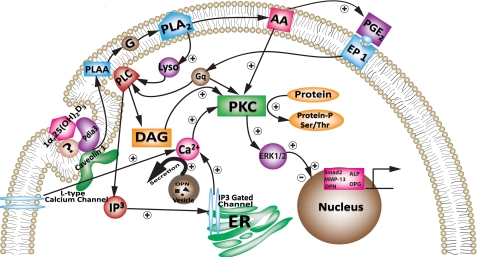
Our data show that Pdia3 mediates effects of 1α,25(OH)2D3 on osteoblasts, including rapid production of PGE2 and activation of PKC. Importantly, the results demonstrate that Pdia3-dependent signaling results in changes in gene expression, via phosphorylation of transcription factors such as ERK1/2. Taking these observations together with our previous observations using growth plate chondrocytes (16, 27, 28) and osteoblasts (19, 21), we propose a mechanism initiated at the plasma membrane and culminating in downstream genomic regulation (Fig. 7). In this signaling pathway, 1α,25(OH)2D3 first binds with Pdia3 or a Pdia3-membrane receptor complex in caveolae, activating PLA2. This results in arachidonic acid release; the arachidonic acid is processed further to PGE2. In growth plate chondrocytes (1, 16, 18). PGE2 binds its G-protein coupled receptor and activates PLC. Activated PLC acts on phosphatidylinositol, releasing inositol-trisphosphate (IP3) and diacylglycerol (DAG). Increased intracellular Ca2+ due to IP3 works with DAG to translocate and activate PKC on the plasma membrane. This pathway also results in activation of ERK1/2 (22). The fact that key components of the signaling pathway are also present in osteoblasts (19, 21) and that PKC and PLC were also reported to be rapidly activated by 1α,25(OH)2D3 in other cell types (29–32), suggests that the proposed pathway is likely to be conserved in 1α,25(OH)2D3-responsive cells.
FIGURE 7.
Proposed mechanism of 1α,25(OH)2D3 stimulated rapid responses in osteoblasts.
Pdia3 has been reported to be present in endoplasmic reticulum, cytosol, nucleus, plasma membranes, extracellular matrix, and matrix vesicles (33). Our results show that Pdia3 is present in the cytosol and plasma membranes of MC3T3-E1 osteoblasts as well. Part of the Pdia3 is in specialized compartments of the plasma membranes, co-localizing with lipid rafts by fluorescence microscopy. Moreover, Western blot shows that Pdia3 is in the caveolar fraction of the plasma membrane based on the presence of caveolin-1 (34). We previously reported caveolae are required for rapid 1α,25(OH)2D3-dependent PKC signaling in chondrocytes, and caveolin-1 must be present based on studies using chondrocytes from Cav-1−/− mice (27). This suggests that caveolae provide a microenvironment that permits the interaction of Pdia3 with 1α,25(OH)2D3 together with other components of the signaling complex. Pdia3 was also present in plasma membrane fractions not associated with caveolin-1 and immunofluorescence demonstrated that not all Pdia3 was associated with lipid rafts. What role this Pdia3 may play with respect to 1α,25(OH)2D3, if any, is not clear.
In the present study, we were able to construct stably transfected cell lines that exhibited reduced or over expression of Pdia3. The changes in expression were confirmed by real-time PCR, confocal microscopy, and Western blots. These changes in Pdia3 correlated with changes in cell response to 1α,25(OH)2D3, including activation of PKC, phosphorylation of ERK1/2, downstream regulation of gene expression and protein production, and ultimately, in von Kossa-positive nodule formation. These results strongly implicate Pdia3 as a membrane receptor for 1α,25(OH)2D3. Moreover, we showed that by regulating expression of Pdia3, signaling could be either blocked or augmented. These data suggest that Pdia3 could be a determining factor, the abundance of which directly correlates with the magnitude of the membrane response.
Previous studies using antibodies to Pdia3 demonstrated a role for this protein in Ca2+ ion uptake in chicken intestinal epithelium (35) and as a regulator of PKC signaling (36), but they did not demonstrate in a conclusive manner that Pdia3 is upstream of the earliest steps in the signaling pathway. Here we showed that the PLA2 pathway was intact in the Sh-Pdia3 and Ov-Pdia3 cells. If Pdia3 had been a mediator downstream of PLA2, either PLAA, the activator of PLA2, or arachidonic acid, the product of PLA2 action would stimulate a different response among the three-cell lines. However, we observed a similar effect of PLAA and arachidonic acid on PKC activation in all three-cell lines, indicating that Pdia3 functions in the very first step of this signaling cascade, potentially as the membrane receptor.
It has been unclear whether the Pdia3-dependent rapid membrane response to 1α,25(OH)2D3 also has a genomic function. By silencing and overexpressing Pdia3, we were able to assess the potential effect of rapid signaling on gene transcription in a new perspective. In growth plate chondrocytes, the rapid translocation and activation of PKC leads to the phosphorylation of ERK1/2 (22). ERK1/2 has been shown to regulate osteoblast differentiation in multiple signaling pathways (37–39). Therefore, it is very likely for the rapid response to have a role in gene transcription through the activation of ERK1/2. In wild-type cells, among the 12 bone related genes we studied, six were regulated by a 9 min treatment with 1α,25(OH)2D3. Among the six genes, only OCN and OPN have been reported to have VDR response elements (VDRE) (40). All of the six genes have been previously reported to be regulated through a VDR-independent membrane pathway (24). Here we showed in Sh-Pdia3 cells, both rapid activation of ERK1/2 and regulation of these six genes were either completely blocked or significantly attenuated. Our result confirms the previous report and further, shows that this VDR-independent genomic effect is mediated by Pdia3.
It should be noted that three out of the six 1α,25(OH)2D3-regulated genes did not completely lose their response to 1α,25(OH)2D3 in Sh-Pdia3 cells. There are two possible explanations for this. First, in Sh-Pdia3 cells, 30% of Pdia3 were still on the plasma membrane to mediate the effect. Second, Pdia3-dependent rapid membrane signaling participates in these mechanisms by using kinase cascades to change activity of the transcription factor complex. Therefore, this kind of regulation may cross-talk with other pathways. For example, VDR could be phosphorylated and activated by PKC (41).
In wild-type MC3T3-E1 cells, three genes were up-regulated in response to1α,25(OH)2D3: ALP is an early stage osteoblast differentiation marker (42); MMP-13 is an extracellular matrix remodeling proteinase (43); and OPN has been shown to be a mineralization inhibitor (44). Three genes were down-regulated. OCN is a late stage osteoblast differentiation marker (45), and OPG inhibits osteoclast differentiation (46). This suggests that the effect of pulse treatment with 1α,25(OH)2D3 promotes early osteoblast differentiation and extracellular matrix remodeling but prevents late stage osteoblast differentiation and mineralization. Others have shown that OCN is increased in response to 1α,25(OH)2D3 via VDR-mediated transcription (47), however, our study suggests that it is regulated via Pdia3 as well, and in the experimental design used for this study, the Pdia3 pathway was dominant. Taken together, this suggests that Pdia3 and VDR act at different points in osteoblast differentiation and that the relative contributions of the mechanisms are dose-dependent as well as time-dependent.
If Pdia3 action modulates terminal differentiation of MC3T3-E1 cells, we would expect that 1α,25(OH)2D3 would cause an increase in mineralization in Sh-Pdia3 cells but a decrease in Ov-Pdia3 cells. Both of the transfected cell lines exhibited fewer von Kossa-positive nodules when cultured in growth media than were seen in wild type cells, related at least in part to the reduced levels of viable cells in these cultures. In osteogenic media, however, we observed more von Kossa-positive nodules in Sh-Pdia3 cultures and fewer nodules in Ov-Pdia3 cultures compared with wild-type MC3T3-E1 cells and this was matched by corresponding differences in alizarin red-positive nodules. Pulse treatment with 1α,25(OH)2D3 had no effect on nodule number in cultures of wild-type or Ov-Pdia3 cells, whether determined as a function of calcium (alizarin red) or phosphate (von Kossa). In contrast, pulse treatment of the Sh-Pdia3 cultures with osteogenic media containing 1α,25(OH)2D3 reduced the number of von Kossa-positive nodules without affecting the number of alizarin red positive nodules. von Kossa staining detects phosphate, which is produced by the action of alkaline phosphatase. Both wild-type and Sh-Pdia3 cells exhibited increased ALP expression when pulse treated with 1α,25(OH)2D3, but only the Sh-Pdia3 cultures had increased von Kossa nodule formulation when grown in osteogenic media containing 1α,25(OH)2D3. These results support the hypothesis that Pdia3 acts in a dose-dependent manner and its actions vary with cell differentiation. The 1α,25(OH)2D3 content of the osteogenic media was determined by the content of the secosteroid in FBS, ∼10−12 m (48). When differentiated osteoblasts were pulsed with exogenous 1α,25(OH)2D3, thereby increasing the concentration, the effect of the secosteroid was to reduce phosphate content, but without impacting Ca2+. This was more pronounced in the Sh-Pdia3 cultures, in part due to the reduction in viable cells. In conclusion, our data suggest that in MC3T3-E1 cells, Pdia3 and its rapid membrane signaling decrease mineralization and by silencing Pdia3 mineralization could be largely augmented.
Considering the multiple cellular functions of Pdia3 when studying the consequence of silencing Pdia3, the effects of other Pdia3-dependent mechanisms must be considered. For example, Pdia3 is a chaperone protein that participates in the folding of N-glycosylated integral membrane proteins (12). Pdia3 also exists in the nucleus and binds with DNA, but the effect of this binding on transcription is still unclear (49). Thus, changing Pdia3 expression levels could result in changing various cellular processes and how much of these side effects contribute to our observation is not known. The observation that inhibitors targeting mediators of the rapid response successfully blocked 1α,25(OH)2D3-stimulated OPN protein, supports the idea that the observed effects in Sh-Pdia3 and Ov-Pdia3 are contributed by the specific role of Pdia3 in the 1α,25(OH)2D3 pathway, rather than side effects.
In conclusion, we mapped out a detailed mechanism of 1α,25(OH)2D3 stimulated rapid response in osteoblasts. The importance of this protein to the mechanism was shown by silencing and overexpressing Pdia3. The function of Pdia3 in the first step of our pathway was demonstrated. By establishing the role of Pdia3 in 1α,25(OH)2D3-dependent gene transcription, protein secretion and mineralization, we showed that the proposed Pdia3 signaling pathway has significant physiological relevance. This conclusion is supported by studies showing that mice with reduced levels of Pdia3 (Pdia3+/−) have a defective bone phenotype (50).
Supplementary Material
Acknowledgments
We thank Sharon L. Hyzy, Ashley L. Grozier, and Reyhaan A. Chaudhri for technical assistance.
This work was supported by the Price Gilbert, Jr. Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- 1α,25(OH)2D3
- 1,25-dihydroxyvitamin D3
- VDR
- vitamin D receptor
- Pdia3
- protein-disulfide isomerase-associated 3
- Sh-Pdia3
- Pdia3-silenced cells
- Ov-Pdia3
- Pdia3-overexpressing cells
- PLA2
- phospholipase A2
- PLC
- phospholipase C
- PLAA
- PLA2-activating protein
- AA
- arachidonic acid
- PGE2
- prostaglandin E2.
REFERENCES
- 1.Boyan B. D., Wang L., Wong K. L., Jo H., Schwartz Z. (2006) Steroids 71, 286–290 [DOI] [PubMed] [Google Scholar]
- 2.Zanello L. P., Norman A. (2006) Steroids 71, 291–297 [DOI] [PubMed] [Google Scholar]
- 3.Khanal R. C., Nemere I. (2007) Curr. Med. Chem. 14, 1087–1093 [DOI] [PubMed] [Google Scholar]
- 4.Breen E. C., van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraichely D. M., MacDonald P. N. (1998) Front Biosci. 3, d821–833 [DOI] [PubMed] [Google Scholar]
- 6.Liu R., Li W., Karin N. J., Bergh J. J., Adler-Storthz K., Farach-Carson M. C. (2000) J. Biol. Chem. 275, 8711–8718 [DOI] [PubMed] [Google Scholar]
- 7.Veldman C. M., Schläpfer I., Schmid C. (1997) Bone 21, 41–47 [DOI] [PubMed] [Google Scholar]
- 8.Zanello L. P., Norman A. W. (1997) J. Biol. Chem. 272, 22617–22622 [DOI] [PubMed] [Google Scholar]
- 9.Zanello L. P., Norman A. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanello L. P., Norman A. W. (2004) Steroids 69, 561–565 [DOI] [PubMed] [Google Scholar]
- 11.Nemere I., Farach-Carson M. C., Rohe B., Sterling T. M., Norman A. W., Boyan B. D., Safford S. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7392–7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jessop C. E., Chakravarthi S., Garbi N., Hämmerling G. J., Lovell S., Bulleid N. J. (2007) EMBO J. 26, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peaper D. R., Wearsch P. A., Cresswell P. (2005) EMBO J. 24, 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemere I., Dormanen M. C., Hammond M. W., Okamura W. H., Norman A. W. (1994) J. Biol. Chem. 269, 23750–23756 [PubMed] [Google Scholar]
- 15.Boyan B. D., Bonewald L. F., Sylvia V. L., Nemere I., Larsson D., Norman A. W., Rosser J., Dean D. D., Schwartz Z. (2002) Steroids 67, 235–246 [DOI] [PubMed] [Google Scholar]
- 16.Schwartz Z., Graham E. J., Wang L., Lossdörfer S., Gay I., Johnson-Pais T. L., Carnes D. L., Sylvia V. L., Boyan B. D. (2005) J. Cell. Physiol. 203, 54–70 [DOI] [PubMed] [Google Scholar]
- 17.Schwartz Z., Sylvia V. L., Larsson D., Nemere I., Casasola D., Dean D. D., Boyan B. D. (2002) J. Biol. Chem. 277, 11828–11837 [DOI] [PubMed] [Google Scholar]
- 18.Schwartz Z., Gilley R. M., Sylvia V. L., Dean D. D., Boyan B. D. (1999) Bone 24, 475–484 [DOI] [PubMed] [Google Scholar]
- 19.Baran D. T. (1994) J. Cell. Biochem. 56, 303–306 [DOI] [PubMed] [Google Scholar]
- 20.Wali R. K., Kong J., Sitrin M. D., Bissonnette M., Li Y. C. (2003) J. Cell. Biochem. 88, 794–801 [DOI] [PubMed] [Google Scholar]
- 21.Schwartz Z., Dennis R., Bonewald L., Swain L., Gomez R., Boyan B. D. (1992) Bone 13, 51–58 [DOI] [PubMed] [Google Scholar]
- 22.Schwartz Z., Ehland H., Sylvia V. L., Larsson D., Hardin R. R., Bingham V., Lopez D., Dean D. D., Boyan B. D. (2002) Endocrinology 143, 2775–2786 [DOI] [PubMed] [Google Scholar]
- 23.Farach-Carson M. C., Bergh J. J., Xu Y. (2004) Steroids 69, 543–547 [DOI] [PubMed] [Google Scholar]
- 24.Farach-Carson M. C., Xu Y. (2002) Steroids 67, 467–470 [DOI] [PubMed] [Google Scholar]
- 25.Huhtakangas J. A., Olivera C. J., Bishop J. E., Zanello L. P., Norman A. W. (2004) Mol. Endocrinol. 18, 2660–2671 [DOI] [PubMed] [Google Scholar]
- 26.Sylvia V. L., Schwartz Z., Schuman L., Morgan R. T., Mackey S., Gomez R., Boyan B. D. (1993) J. Cell. Physiol. 157, 271–278 [DOI] [PubMed] [Google Scholar]
- 27.Boyan B. D., Wong K. L., Wang L., Yao H., Guldberg R. E., Drab M., Jo H., Schwartz Z. (2006) J. Bone. Miner Res. 21, 1637–1647 [DOI] [PubMed] [Google Scholar]
- 28.Schwartz Z., Shaked D., Hardin R. R., Gruwell S., Dean D. D., Sylvia V. L., Boyan B. D. (2003) Steroids 68, 423–437 [DOI] [PubMed] [Google Scholar]
- 29.Buitrago C. G., Pardo V. G., de Boland A. R., Boland R. (2003) J. Biol. Chem. 278, 2199–2205 [DOI] [PubMed] [Google Scholar]
- 30.Yarram S. J., Tasman C., Gidley J., Clare M., Sandy J. R., Mansell J. P. (2004) Mol. Cell. Endocrinol. 220, 9–20 [DOI] [PubMed] [Google Scholar]
- 31.Li J., Fleet J. C., Teegarden D. (2009) J. Cell. Biochem. 107, 1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boland R., De Boland A. R., Buitrago C., Morelli S., Santillán G., Vazquez G., Capiati D., Baldi C. (2002) Steroids 67, 477–482 [DOI] [PubMed] [Google Scholar]
- 33.Turano C., Coppari S., Altieri F., Ferraro A. (2002) J. Cell. Physiol. 193, 154–163 [DOI] [PubMed] [Google Scholar]
- 34.Smart E. J., Ying Y. S., Mineo C., Anderson R. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10104–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemere I., Campbell K. (2000) Steroids 65, 451–457 [DOI] [PubMed] [Google Scholar]
- 36.Boyan B. D., Sylvia V. L., McKinney N., Schwartz Z. (2003) J. Cell. Biochem. 90, 1207–1223 [DOI] [PubMed] [Google Scholar]
- 37.Dai Z., Li Y., Quarles L. D., Song T., Pan W., Zhou H., Xiao Z. (2007) Phytomedicine. 14, 806–814 [DOI] [PubMed] [Google Scholar]
- 38.Jadlowiec J., Koch H., Zhang X., Campbell P. G., Seyedain M., Sfeir C. (2004) J. Biol. Chem. 279, 53323–53330 [DOI] [PubMed] [Google Scholar]
- 39.Kanno T., Takahashi T., Tsujisawa T., Ariyoshi W., Nishihara T. (2007) J. Cell. Biochem. 101, 1266–1277 [DOI] [PubMed] [Google Scholar]
- 40.Haussler M. R., Whitfield G. K., Haussler C. A., Hsieh J. C., Thompson P. D., Selznick S. H., Dominguez C. E., Jurutka P. W. (1998) J. Bone Miner Res. 13, 325–349 [DOI] [PubMed] [Google Scholar]
- 41.Hsieh J. C., Jurutka P. W., Galligan M. A., Terpening C. M., Haussler C. A., Samuels D. S., Shimizu Y., Shimizu N., Haussler M. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zernik J., Twarog K., Upholt W. B. (1990) Differentiation 44, 207–215 [DOI] [PubMed] [Google Scholar]
- 43.Kusano K., Miyaura C., Inada M., Tamura T., Ito A., Nagase H., Kamoi K., Suda T. (1998) Endocrinology 139, 1338–1345 [DOI] [PubMed] [Google Scholar]
- 44.Ono N., Nakashima K., Rittling S. R., Schipani E., Hayata T., Soma K., Denhardt D. T., Kronenberg H. M., Ezura Y., Noda M. (2008) J. Biol. Chem. 283, 19400–19409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bronckers A. L., Gay S., Finkelman R. D., Butler W. T. (1987) Bone Miner 2, 361–373 [PubMed] [Google Scholar]
- 46.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., Tsuda E., Morinaga T., Higashio K., Udagawa N., Takahashi N., Suda T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lian J. B., Coutts M., Canalis E. (1985) J. Biol. Chem. 260, 8706–8710 [PubMed] [Google Scholar]
- 48.Schwartz Z., Schlader D. L., Ramirez V., Kennedy M. B., Boyan B. D. (1989) J Bone Miner Res 4, 199–207 [DOI] [PubMed] [Google Scholar]
- 49.Coppari S., Altieri F., Ferraro A., Chichiarelli S., Eufemi M., Turano C. (2002) J. Cell. Biochem. 85, 325–333 [DOI] [PubMed] [Google Scholar]
- 50.Wang Y., Chen J., Lee C. S., Nizkorodov A., Riemenschneider K., Martin D., Hyzy S., Schwartz Z., Boyan B. D. (2010) J Steroid Biochem. Mol. Biol. 121, 257–260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.