Abstract
Magic angle spinning solid-state NMR has been used to study the structural changes in the Pf1 filamentous bacteriophage, which occur near 10 °C. Comparisons of NMR spectra recorded above and below 10 °C reveal reversible perturbations in many NMR chemical shifts, most of which are assigned to atoms of hydrophobic side chains of the 46-residue subunit. The changes mainly involve groups located in patches on the interfaces between neighboring capsid subunits. The observations show that the transition adjusts the hydrophobic interfaces between fairly rigid subunits. The low temperature form has been generally more amenable to structure determination; spin diffusion experiments on this form revealed unambiguous contacts between side chains of neighboring subunits. These contacts are important constraints for structure modeling.
Keywords: Bacteriophage, NMR, Protein Assembly, Protein Structure, Spectroscopy, Hydrophobic Interactions, Magic Angle Spinning, Phase Transition
Introduction
The filamentous bacteriophage Pf1 consists of a closed circular, single-stranded DNA of 7349 nucleotides, encased by ∼7300 copies of a major capsid protein in a shell capped at the ends by a few copies of key proteins involved in virus assembly and host specificity (for reviews see Refs. 1–3). It is 1 of over 60 different filamentous phages that have been reported, about a dozen of which have been structurally characterized to various extents. Many of these phages integrate their genomes into the genomes of their bacterial hosts and some contribute to the virulence of pathogenic bacteria. The host of Pf1 is Pseudomonas aeruginosa, strain K (4). Pf1 has virtually the same structural genes as the much larger phage Pf4 (12,437 nucleotides), which occurs as a latent prophage integrated in the chromosome of P. aeruginosa PA01 that has been implicated as a virulence factor of this human pathogen (5, 6). Among the characterized filamentous phages, Pf1 is unique in having a unit chemical ratio of one major capsid subunit for each nucleotide in the packaged genome (1). Related to this stoichiometry, Pf1 does not show any evidence of long length scale gentle coiling characteristic of several other phages (7). Through many structural studies by a variety of techniques, Pf1 capsid structure has become a paradigm for the capsids of a number of filamentous phages (2, 3), even though key aspects of that structure are still puzzling.
An approach to the full determination and understanding of the Pf1 structure is through the use of a well known reversible structural phase transition near 10 °C between two ordered states that were first observed by x-ray fiber diffraction (8, 9). This transition temperature is well below the physiological temperature, and the structural response of Pf1 is associated with a change in the helical pitch and rise. A modification in salt concentration is also associated with a small change in subunit rise in both forms of the phage (10). Interestingly, a similar response to temperature has not been observed in any of Ff, Pf3, Xf, or PH75. One difference between Pf1 and the others that might account for the presence of a transition in Pf1 and not the others is its 1:1 stoichiometry. Diffraction studies of oriented phage in solution showed that the low and high temperature forms differ slightly in the orientation of the subunit with respect to the filament axis (10, 11). The two states have slightly different numbers of subunits per turn in the helical arrangement of their capsids, which produce very different diffraction patterns even though the structures are closely similar. The low temperature form (Pf1L)2 has an apparent structure repeat of about 21.9 nm containing 71 subunits in 13 turns, or 5.46 subunits/turn (2). To account for the unit stoichiometry, one can consider repeats of 71 dimers in 26 turns, where subunits in the dimer differ in structure slightly because of up-strand versus down-strand interactions (12). For Pf1H, the apparent repeat is about 7.8 nm, and the symmetry assigned is nominally 27 subunits per 5 turns (or 5.40 subunits/turn), but this symmetry does not account fully for features of some diffraction patterns and NMR data (13–15). The transition between the two symmetries is estimated to occur between 280 and 286 K depending on pH and salt concentration. The transition is clearly a property of a single virion particle, as shown by studies done at varying concentrations of 100 mg/ml by Raman optical activity (16), 50 mg/ml by NMR (17), 20 mg/ml by diffraction from oriented gels (10), and 0.5 mg/ml and lower by calorimetry and CD (18). However, the transition is apparently blocked at the very high concentrations of 400 mg/ml or more in fibers; dry fibers made at the high temperature exhibit the same diffraction pattern when the temperature is lowered to 4 °C and vice versa (9, 19). The electronic and vibrational spectroscopies revealed little if any change in protein conformation, although changes involving DNA have been observed by CD and Raman optical activity (16, 20).
Static solid-state NMR experiments on oriented samples at ∼50 mg/ml by Thiriot et al. (17) showed large changes in 15N chemical shift values in 15N-Ile enriched virus, revealing structural details for the two-state transition. A decrease of the tilt angle of the C terminus starting at residue alanine 29 was clearly observed in the dipolar wave analysis (see Fig. 5b in Ref. 17). This change results in a smaller overall diameter of the virion at low temperature, in agreement with measurements of the birefringence of aligned concentrated solutions of Pf1 in varying magnetic fields (21). Others had suggested that the N terminus is tilted 4° toward the filamentous axis in the low temperature form (10).
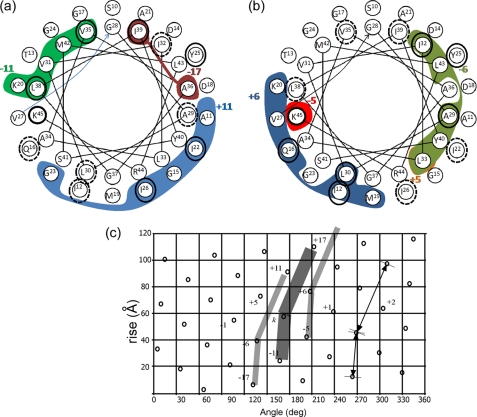
FIGURE 5.
Correlation of the measured shift perturbations Δδ and a 3.5-Å intersubunit interface in the low temperature form of Pf1, model 4IFM. A helical representation of residues Ser-10 to Lys-45 is shown. The internal circle represents the C-terminal residues Gly-28–Lys-45. The external circle is for Ser-10–Val-27. The colored areas enclose amino acids for which at least one carbon or nitrogen is within 3.5 Å of a neighboring subunit in the model, and the identity of this subunit is indicated by a colored number. a, contacts with subunits +11 (blue), −11 (green), and −17 (red) are shown (for +17 and only contacts with Ile-3 exist). b, contacts with subunits +6 (blue), −6 (green), and the two residues of subunit ±5 (orange and red) are shown. Amino acids circled in black show Δδ ≥0.4 ppm in our data. The dashed circles in a correspond to shifts in van der Waals contact areas of the subunits emphasized in b and vice versa. See text for further details. c, a lattice diagram corresponding to the low temperature symmetry of the Pf1 filamentous bacteriophage. A rotation of 65.9° and rise of 3.05 Å were used to generate the dots. A representative subunit k is sketched as a broad bent line, and the neighboring subunits ±6 are indicated by narrower lines. Subunits in contact with subunit k are numbered according to a right-handed sense of the Pf1 capsid helix.
This study of Pf1 and its phase transition is by means of magic angle spinning solid-state NMR on fully hydrated, infectious samples. Based on our prior assignments of the coat protein subunit (15), we have used 13C homonuclear correlation spectroscopy to map transition-induced perturbations in the assigned NMR chemical shifts. We report on a significant number of such perturbations. We also report the first intermolecular constraints on the virus; spin diffusion experiments on Pf1 have revealed internuclear contacts between neighboring subunits.
EXPERIMENTAL PROCEDURES
Sample Preparation
Pf1 phage was isolated and purified from infected cultures of P. aeruginosa, strain K, grown in minimal M9 media with 15NH4Cl and [U-13C]glucose as the sole nitrogen and carbon sources as described earlier (15). Following density gradient ultracentrifugation, the sample in high CsCl was precipitated with 4% w/v polyethylene glycol (PEG) 8000 (Fluka Biochemicals), pelleted, and redissolved to a concentration of ∼1 mg/ml in 10 mm Tris, pH 8.4. The cryoprotectant ethylene glycol was then added (22) to a final concentration of 30% v/v. A second precipitation was done by mixing MgCl to 5 mm and PEG8000 to 10% w/v. After several minutes of gentle mixing, the suspension was centrifuged for 10 min at 3700 × g. The loose pellet was transferred to a pipette tip sealed at the bottom and further pelleted at ∼20,000 × g for 2 h in an Eppendorf microcentrifuge to reduce excess supernatant. The concentration of Pf1 in the pellet was estimated to be ∼200 mg/ml based on the amount of Pf1 before precipitation (6 mg) and the approximate total volume of the resulting pellet (30 μl). The bottom seal of the pipette tip was then sliced off, and the pellet was sedimented from the tip into a 4-mm ZrO2 magic angle spinning NMR rotor. For Pf1H, all steps were carried out at room temperature. For Pf1L, the second precipitation was done in a cold room at ∼4 °C, and the pelleting into the pipette tips and the NMR rotor was done at 0 °C. The small NMR rotor was kept cold at all times and transferred to a pre-cooled NMR probe for the magic angle spinning experiments. Concentrations were based on UV spectra with an extinction coefficient ϵ = 2.1 mg−1 cm2 at 271 nm (7).
NMR Experiments
Two-dimensional homonuclear experiments were based on dipolar-assisted rotational resonance (DARR) (23) also known as RF-assisted spin diffusion (RAD mixing) (24), with heteronuclear scalar JCN decoupling; they were performed on a Varian Infinity Plus spectrometer operating at a 600-MHz proton frequency. Cross-polarization (25) was achieved by tangential ramp (26) on the 13C channel with the magic angle spinning Hartman-Hahn condition set to n = 1 (ν1H – ν1C = νr, ν1X being the RF power on channel X, and νr is the sample spinning rate) and a contact time of 1.25 ms. Two-pulse phase modulation (27) was used for proton decoupling with ν1H ∼75 kHz, phase 15°, and pulse segments of 6.5 μs. Relaxation delay was 3 s, and acquisition lengths were 20 ms (t2) and 11.5 ms (t1). 16–32 transients were collected for each FID using 32 dummy pulses to equilibrate the RF-produced sample heating. Experiments were performed with mixing times of 10, 50, and 200 ms. Temperature control was achieved using an FTS cooling unit operating at −20 °C for the low temperature samples and 15 °C for the high temperature samples. The low temperature of −20 °C ensures that friction and RF-induced heating do not elevate the temperature above the phage transition temperature.
Data Processing and Analyses
The data were processed using NMRPipe (28) and involved various apodization schemes, including Gauss-to-Lorentz transformation, cosine bell apodization, and exponential multiplication. All were used to verify the position and integrity of the cross-peaks. The resulting spectra were analyzed using the SPARKY program (29). The assignments of Pf1L were based on BMRB (30) accession number 15138 for the high temperature form of Pf1 and aided by cross-peaks from the long mixing time experiments. We have used the following strategy to assign the low temperature form. (i) For peaks with a similar location at Pf1L and Pf1H, we assumed an identical assignment. (ii) Side-chain resonances, which make up most of the perturbations, were tracked from the backbone peaks using a “side-chain walk” strategy. (iii) The existence of a peak shift between Pf1H and Pf1L, when other peaks of the same amino acid type overlap, allowed unambiguous determination of its identity. (iv) In cases of ambiguities, cross-peaks from long mixing time experiments were used to verify the identities of the peaks. In some cases, such peaks did not exist for all amino acids of the same type; however, by observing cross-peaks of other specific amino acids, we assigned others by elimination. For example, out of four leucine residues in the sequence, Leu-33 and Leu-38 have a similar Cα, but the identification of Leu-33 Cβ–Ala-34 Cα cross-peak in the DARR50 experiment (DARR with 50-ms mixing time) allowed us to assign unambiguously Leu-38 Cα and Leu-38 Cβ to 58.4 and 41.8 ppm, respectively. In our final shift perturbation table, the entire side chain of Leu-38 shows significant shift deviations between the two forms (0.4–1 ppm), whereas the shifts of Leu-33 are similar in both forms.
RESULTS
We performed a set of two-dimensional 13C-13C chemical shift correlation experiments (23, 24) on both Pf1H and Pf1L. An overlay of the aliphatic carbon regions from spectra of the two forms is shown in Fig. 1, a and b. The observed resonances in the DARR/RAD spectra are those of the coat protein, which contributes ∼94% of the total mass of the virion. Cross-peaks resulting from a 10-ms magnetization transfer period mainly correspond to correlations between carbons that are one or two bonds apart. Initial inspection of the Cα-Cβ region (Fig. 1b) suggests that the two spectra are similar, and therefore it is highly likely, as suggested by other studies, that significant backbone structural changes do not occur. However, several chemical shift perturbations induced by the temperature difference were observed, and they can be attributed to individual resonances. The most apparent ones are indicated by labeled arrows in Fig. 1, a and b, based on the prior assignment of Pf1H (15). The α-helical nature of Pf1 causes high congestion in some regions of the NMR spectra, and therefore verifications of many of the chemical shifts for Pf1L were sought through analyses of spectra obtained with longer mixing times (50 and 200 ms, discussed below). These spectra include cross-peaks of carbons that are further apart, up to 5–6 Å, providing more spectral dispersion for detection and assignment of the perturbations in the chemical shifts.
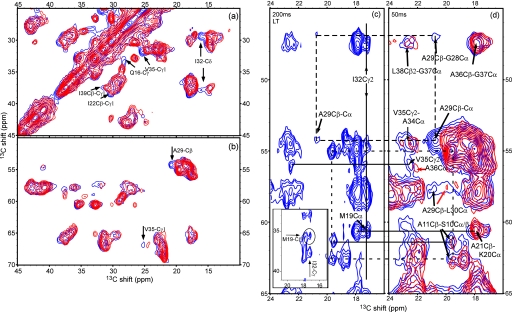
FIGURE 1.
a and b, overlay of spectra for the low and high temperature forms (blue, Pf1L; red, Pf1H). Both samples were separate aliquots of the same Pf1 preparation. Pf1L was precipitated at a temperature below the transition point and kept at that temperature, and Pf1H was prepared at high temperature throughout (see “Experimental Procedures”). The DARR spectra were measured on a 600-MHz Varian InfinityPlus spectrometer with a mixing time of 10 ms and identical experimental parameters other than the temperature. The signal-to-noise of the lowest contour is 15, and 12 contours are drawn at multiples of 1.3. a, side-chain cross-peaks region; b, mostly Cα-Cβ region; c and d, medium range correlations observed by long mixing time DARR experiments. d, typical region from the 50-ms DARR spectra of Pf1L (blue) and Pf1H (red) is shown. The black arrows indicate positions of intrasubunit contacts in Pf1L (red arrows point to similar cross-peaks in Pf1H). Especially important is the shift in Ala-29 Cβ between the two forms (Δδ = 1.1 ppm), which is verified by connectivities to Leu-30 Cα and Gly-28 Cα. The other peaks in this region indicate residues that are largely unchanged. c, a similar region is extracted from a 200-ms spectrum measured on Pf1L. Despite the decrease in intensity of many peaks, other peaks are well resolved and reinforce our assignments of Pf1L. The dashed and dotted rectangles link residues associated with the Cβ atoms of Ala-29 and Ala-11, respectively. Also indicated are the intersubunit contacts of the Cγ2 atom of Ile-32 with the Cα atom Met-19 belonging to a different subunit. In the inset, the correlation to Met-19 Cβ is shown, which resolves the ambiguity of Met-19 Cα with Lys-20 Cα.
In Fig. 1, c and d, we compare a portion of the 50-ms DARR spectra of both forms of Pf1 (Fig. 1d) and the 200-ms spectrum of Pf1L (Fig. 1c). The region shown in the spectra clearly indicates significant perturbation for residue Ala-29 by virtue of its correlations to its adjacent amino acids Gly-28 and Leu-30. Other typical peaks show mostly unchanged residues as indicated by the arrows in Fig. 1d. In general, the low temperature spectra of Pf1L show more intense peaks and more signals than Pf1H spectra, probably because of reduced dynamic effects. The same set of peaks is also detected in the 200-ms spectrum (Fig. 1c). In this spectrum, many of the peaks already lose intensity because of relaxation effects during the long longitudinal mixing period. The advantage is that some peaks, for example the alanine residues in the solvent-exposed region, become more resolved. The correlations of Ala-29 and Ala-11 are indicated in Fig. 1, c and d, by the dashed and dotted rectangles, respectively.
Significantly, the 200-ms DARR spectrum for Pf1L reveals three intersubunit contacts. One is between the well resolved Cγ2 atom of Ile-32 in one subunit (29.3 ppm) and the Cα (60.6) and Cβ (35.5) atoms of Met-19 in a neighboring subunit. This correlation is undoubtedly an intersubunit contact because the internuclear distances between atoms of residues 19 and 32 within a given α-helical subunit are too long to produce the observed data. By the same logic, the well resolved Ala-29 Cβ (20.7 ppm) with either of the overlapping Leu-38 Cδ (25.1) and Val-35 Cγ (25.2) is an intersubunit cross-peak, as is the well resolved Ile-12 Cβ (36.6 ppm) with either of the same two overlapping atoms, Leu-38 Cδ and Val-35 Cγ.
Based on our previous analysis of Pf1H spectra, we were able to assign many of the carbon atoms in Pf1L and hence establish the sites of the perturbations. A summary of the assigned chemical shifts for Pf1L appear in supplemental Table S1. The absolute values of the perturbations in chemical shifts observed in all our spectra appear in Fig. 2 and are tabulated in supplemental Table S2. Overall, 155/205 carbon atoms have been identified in both Pf1L and Pf1H (76% in 41 amino acids, missing are 4 glycines and Lys-45; 169 or 82% identified in Pf1L spectra). Of these, 23 atoms in 14 amino acids, all of which are hydrophobic, show shifts ≥0.4 ppm. In Fig. 2, backbone and side-chain carbons are plotted for each residue independently and are marked according to their positions in the chain. For a threshold of 0.4 ppm,3 it is immediately apparent that the most significant changes appear in the hydrophobic region of the capsid protein (residues 21–43) and are primarily observed for the side-chain atoms. The atoms that could not be resolved in the various two-dimensional spectra are indicated at the bottom of Fig. 2 (marked as NR).
FIGURE 2.
Absolute values of chemical shift perturbations Δδ = |δ(Pf1H) − δ(Pf1L)| between the low and high temperature forms of Pf1. The sequence of the coat protein of Pf1 and the corresponding residue numbers are shown on the horizontal axis. The emphasized broken line at the 0.4 ppm threshold (see footnote 3) is an indicator for the eye to recognize the significant shifts. The large majority of the perturbations we could resolve are lower than the threshold. Most of the significant changes appear at the C-terminal hydrophobic part of the protein. Data points at NR indicate nonresolved or undetected peaks. The following legends are used: diamonds, Cα; squares, Cβ; triangles, Cγ; ×, Cγ2; circles, Cδ and Cϵ, +, C′ (carbonyl).
After sufficient data had been collected for Pf1L, the sample temperature was increased to 15 °C to test whether the structural transition is reversible at the high concentration of ∼200 mg/ml used. Fig. 3 shows an overlay of the low (Pf1L) and low to high (Pf1L→H) forms, compared with an overlay of Pf1L→H and Pf1H forms. Arrows in Fig. 3 indicate chemical shift changes stemming from the transition, which have been fully reversed upon the temperature increase.
FIGURE 3.
Reversibility of the phase transition. a, spectrum of Pf1L→H (green) overlaid with Pf1L (blue), both measured on a 600-MHz spectrometer; b, Pf1L→H (green) overlaid with Pf1H (red). Pf1L→H is the original Pf1L sample measured at 15 °C. Pf1H is from a different preparation measured on a 750-MHz spectrometer. The concentrations of the precipitated, hydrated samples were ∼200 mg/ml. Solid arrows indicate significant chemical shift changes apparent in Pf1L of spectrum a, which move back to their high temperature shift location, indicated by the dashed arrows (green spectrum in a). The location of the perturbed resonances of Pf1L are indicated by an x in spectrum b. The resonances and their corresponding absolute shifts perturbations are as follows: 1, Gln-16 Cγ, 1.0 ppm; 2 and 7, Val-35 Cγ1, 0.7 ppm; 3, Leu-38 Cδ2, 0.4 ppm; 4, Ile-32 Cδ1, 1.8 ppm; 5, Leu-38 Cβ, 0.6 ppm; Cδ1, 1.0 ppm; 6, Ala-29 Cβ, 1.1 ppm; 8 Val-2 Cα/Cβ, very weak signal for Pf1L, intensity change (may stem from change in dynamics, not from the transition); 9, inset, Leu-38 Cβ, 0.6 ppm.
DISCUSSION
Characterization of the variations between the high and low temperature forms of Pf1 is of significant importance in obtaining a more accurate structural insight of Pf1H and in our understanding of the transition itself. In our data we observe chemical shift perturbations and some long range intersubunit contacts. The perturbations we observe are relatively small and are restricted mostly to side-chain atoms. Such changes are similar to those observed in “structure-activity relationship” studies by NMR of drug and ligand binding to proteins (31). In this way, our data are more compatible with changes in the intersubunit contact surfaces that occur upon the transition, rather than significant changes in the backbone structure of the coat protein.
Locations of Atoms with Perturbed Chemical Shifts on Hydrophobic Surfaces
According to the perturbation summary in Fig. 2, the largest chemical shift changes occur in well defined hydrophobic patches of side chains near the C terminus. Fig. 4, based on the capsid model 4IFM, illustrates the positions of these patches. Some minor perturbations also exist near the N terminus. A surface plot of the significant shifts (Δδ ≥ 0.4 ppm; Δδ = |δ(Pf1H) – δ(Pf1L)|) shows clearly the strong perturbations in the C-terminal patches. This indicates that reorganization of the packing of intermolecular hydrophobic contacts occurs in this structural transition, as was suggested earlier by Hinz et al. (18).
FIGURE 4.
a, a plot of the significant shifts (Δδ ≥0. 4 ppm) observed for the major coat protein in Pf1 phage. The residues are colored on a yellow-green scale (see color bar), and the surfaces of the residues exhibiting shifts ≥0.4 ppm are shown explicitly. The strongest and majority of shifts are located in the hydrophobic C-terminal region. The sequence of the 46-amino acid coat protein is shown below the monomer, and residues with significant shifts are indicated on the subunit (gray color marks unassigned atoms). The model was generated with the program PyMOL using the coordinates of the Pf1L model 4IFM. b, chemical shift perturbation surface of subunits 0, +6, and +11 from the same model. b1, view from the interior of the virion at the three subunits. Subunit 0 is indicated by a white-to-red color scale (0–2 ppm) according to the observed absolute shift perturbation Δδ, and a surface is plotted for atoms with Δδ ≥0.4 ppm. Subunits +6 and +11 are indicated by a white-blue and white-green color scales, respectively. The gray color indicates unassigned residues. b2, view in b1 zoomed into the main perturbation region showing the intersubunit patches involving residues Gln0-16, Ala6-29, Ile6-32, Val11-35, Leu11-38, and Ile11-39 at the bottom (superscripts indicate subunit) and residues Ile0-12 and Ile6-26 just above. b3, 180° rotation of the view in b2, showing the same region from the outside of the virion. c, axial and side views of a 60 subunit portion (∼0.8%) of the capsid of Pf1 showing locations of observed chemical shift perturbations Δδ colored according to their absolute values in ppm (Fig. 2). For Δδ ≥0.4 ppm, van der Waals surfaces are shown; otherwise stick models without surfaces are used in c1–c3. c1, axial view is from the N-terminal ends of the subunits on the outside toward their C-terminal ends near the center. c2, axial view is from the opposite direction as in c1. In the side view of c3, surfaces are only for Δδ values ≥0.4, and in c4, surfaces are for all residues regardless of the shift perturbation. The gray colors indicate unassigned atoms including all non-carbon atoms but not hydrogens.
A model of the interfaces within the capsid (Fig. 4b) locates the perturbations at the interaction surface between the subunits, based on Protein Data Bank code 4IFM. The assumption that the side chains are packed in the manner depicted agrees well with previous observations on residues that are not amenable to chemical modifications (32, 33). Fig. 4, b1, shows the three close-packed subunits 0, +6, and +11 (34) as viewed from the DNA axis toward the outside. Fig. 4, b2, is an enlargement of the principal contact region involving residue Ile-12 of subunit 0 (marked Ile0-12) and residue Ile6-26 located nearer the axis, as well as a bottom region involving residues Gln0-16, Ala6-29, Ile6-32, Val11-35, Leu11-38, and Ile11-39. A view from the outside in Fig. 4, b3 (a 180° rotation of Fig. 4, b2), shows that the largest chemical shift perturbations are not located in the external surface of the virion, but rather in the packing surfaces between the subunits.
To illustrate the location of the residues exhibiting significant shifts in the context of the entire virion, different views of an assembly of 60 subunits (<1% of the total virion length) are shown in Fig. 4c. Views in opposite directions along the axis of the virion are shown in Fig. 4, c1 and c2). The surfaces shown here emphasize where the regions of the largest chemical shift changes are concentrated. The side view of the virion (Fig. 4, c3) emphasizes the location of the main shift perturbations with respect to the exterior of the phage, and these perturbations become almost invisible in Fig. 4, c4, where the surface of the entire phage is drawn using a similar color scheme. It is apparent that the surface of the virion does not undergo any change, and the shifts we detect are not due to a different solvent exposure at the low temperature. Clearly these changes are buried between subunits.
It is important to note that interactions with the DNA could not be probed in these studies. Unfortunately, many of the atoms belonging to residues involved in interactions with the DNA (e.g. Met-42, Arg-44, and Lys-45) were assigned although broad in the high temperature form, and are missing or could not be unambiguously assigned in the present data for the low temperature form. Therefore, it is not clear if these residues are or are not shifted during the transition.
Correlation of Observed Perturbations with Capsid Models
To further explore correlations between the observed perturbations and intersubunit hydrophobic contacts at the atomic level, we first analyzed four models in the data base in terms of closest neighbors of adjacent subunits; Protein Data Bank codes 4IFM and 1PFI for the low temperature form and codes 2IFN and 1QLI for the high temperature form. We marked the amino acids belonging to an arbitrary subunit k = 0, for which at least one carbon or nitrogen atom is within 3.5 Å of a neighboring subunit, and binned those atoms to the contacted neighbor. The distance of 3.5 Å is approximately a van der Waals interaction surface of two carbon atoms. We then considered the models in low/high pairs to predict surface modifications that may induce chemical shift changes. One difficulty in the comparisons was that model 1PFI has DNA, although the others do not, and 1PFI was not fully refined. Also, there is no high temperature model that directly corresponds to 1PFI. Nevertheless, results are in agreement with the pair 1PFI/2IFN on 17 occasions in which a chemical shift change corresponds to a contact difference and with 4IFM/1QL1 (15 occasions). For both pairs, 12 unique shifts are explained by changes in the interface (out of 15 total), but then for the other two pairs we found 8 shifts (4IFM/2IFN, 8 unique) and 11 shifts (1PFI/1QL1, 9 unique).
Because the low temperature structure is inherently better defined, we focused on it and chose the empty capsid model 4IFM. In Fig. 5, a helical wheel representation of residues 10–45 is shown for the capsid. The surface lattice of the Pf1 capsid allows, for any subunit k, close contacts with neighboring subunits k ±5, k ±6, k ±11, and k ±17. Residues having atoms in contact with the particular subunit k are indicated by the colored areas in Fig. 5. The lattice diagram of the capsid arrangement for this symmetry is provided in Fig. 5c. In the helical wheels (Fig. 5, a and b), residues having atoms with Δδ ≥0.4 ppm are indicated by the black circles. They correlate well with the 3.5 Å interface. For the region shown, all shifts ≥0.4 ppm (with the exception of Tyr-25) can be attributed to one of the van der Waals interaction surfaces of the low temperature model, and many of the other residues involved in contacts are either unassigned in our data or show shifts just below 0.4 ppm. A similar analysis of both the high temperature models 2IFN and 1QL1 shows that Tyr-25 is in proximity (i.e. <3.5 Å) to subunit k −6, and therefore every shift perturbation we see in this region can be associated with a surface interaction in either Pf1L or Pf1H.
It is important to note that approximately one-half of the residues involved in the intersubunit surfaces exhibit chemical shift perturbations, whereas only a couple of residues not involved show substantial shift perturbations (Ala-7 and Thr-5). Among the residues belonging to subunit k = 0 in the 4IFM model of Pf1L that are within 3.5 Å of a neighboring subunit we found the following: for k +6, three fully assigned residues (Gln-16, Leu-30, and Ile-12) are perturbed, and Cϵ of Met-19 is unassigned; for subunit k −6, Ile-32 and Ala-29 (Leu-33 has two atoms shifted by ∼0.3 ppm and Cδ2 was detected only in Pf1L); for subunit k +11, Ile-26 and Ile-22 (Gly-15 and Gly-23 are unassigned); and for subunit k −11, Val-35 and Leu-38 and probably Met-42 (Val-31 is unassigned and Lys-20 contacts are at the very end of the chain, atoms Cϵ and Nζ. It was suggested that Lys-20 is in contact with the N terminus, potentially balancing the charge of the aspartic acid at position 4.). Overall, of the 26 residues involved in surface contacts, we could resolve resonances of 21, of which 11 or 13 show shift perturbations (for 0.4 or 0.3 ppm thresholds). Ignoring the first five N-terminal residues that may have sample- and condition-dependent structures and positions, we estimate a probability of 60–70% of observing at least one resonance shifted for a residue involved in the interfaces between subunits.
The perturbations also potentially provide a useful predictive tool to recognize interaction surfaces or changes in interaction surfaces. Among the 14 non-terminal residues in which chemical shift perturbations were detected (based on a 0.4 ppm cutoff), 11 or 78% are at an interaction surface of the low temperature model (8 or 57% for the high temperature model). Two additional residues (Tyr-25 and Val-8) are found on the interaction surface of the high temperature model 1QL1, increasing the number to 13, or 93%.
Intersubunit Contacts and Their Correlation to Capsid Models
Further characterization of the contact interfaces can be done via the identification of intersubunit cross-peaks. The helical nature of the coat protein causes significant ambiguity in the spectra of the fully enriched phage, and thus only very few contacts in our fully enriched sample can be expected to yield unambiguous correlations. In the spin diffusion experiments described above, we have identified three such intersubunit contacts. The cross-peak between Ala-29 Cβ and Leu-38 Cδ and/or Val-35 Cγ, despite the ambiguity in its assignment, is clearly not an intramolecular peak and therefore provides evidence regarding intersubunit contacts. A similar conclusion holds for Ile-12 Cβ and Leu-38 Cδ/Val-35 Cγ, and the Ile-32 Cγ–Met-19 Cα/Cβ contact (spectrum and inset shown in Fig. 1c). The latter was detected in the data for both Pf1H and Pf1L. Such quaternary contacts are useful for constraining the overall assembly of the coat protein. The contacts are generally compatible with existing models for the virion assembly, as shown in Fig. 6. For example, according to model 4IFM, the δ-carbons of Leu11-38 (but not Val11-35 Cγ) are 3.7–4.5 Å from the Cβ of Ala6-29 (located 5 positions down the virion axis). This proposed interpretation is depicted in the bottom portion of the binding pocket in Fig. 4b. For Ile0-12 Cβ, the γ-carbons of Val11-35 are within 4.5–5.5 Å but not Leu11-38. The contact between Ile-32 and Met-19 also appears at the interface linking subunits k = 0 (for Met-19) and k = 6 (Ile-32), and therefore reports on the pitch of the helix of subunit 0. (It is also expected that atoms from Tyr6-25 and Ile6-26 would also be in contact with Val11-35, and we identified a potential cross-peak between Val-35 Cγ and Tyr-25Cβ at 37.5–25.2 ppm; however, this contact is also ambiguous in that it is identified in relatively congested chemical shift regions of the spectra.) All three of these intersubunit interface contacts are important constraints for structure models of Pf1. Their presence in these spectra is encouraging regarding the feasibility of structural analysis.
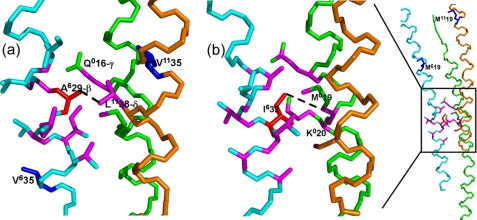
FIGURE 6.
13C-13C intersubunit contacts for Pf1L observed in the 200-ms mixing time experiments in relation to the Pf1L model 4IFM. The views are from the inside of the virion. Backbone carbons are green for subunit 0, cyan for +6, and orange for +11. The dashed lines connect atoms in the model that correspond to the assigned cross-peaks. In magenta are carbon atoms within 5Å of others in the model that show perturbations. a, contact between Ala6-29 Cβ (Ala residue in red) and Leu11-38 Cδ2. Although Gln0-16 Cγ is within 5 Å in the model, it does not show any cross-peak correlation for Pf1L, but its chemical shift is significantly perturbed during the transition (Fig. 2). Also, Val6-35 and Val11-35 (in blue), initially part of Leu-38/Val-35 ambiguity are far from Ala6-29- Cβ and hence are not involved in a contact. b, left, contact between Ile6-32 Cγ2 (Ile residue in red) and Met0-19 Cα/β. Despite their proximity to Ile6-32 in the 4IFM model, we could not detect any unique Lys0-20 atoms in correlation with Ile6-32 atoms. On the right, a zoom-out view of the same contact region showing the location of other Met residues (blue), demonstrating the clear assignment of this peak to an intersubunit contact.
Comparison with Other Studies Characterizing the Transition
On the basis of radial electron density distributions derived from x-ray diffraction by the virus both in fibers and in solution, Specthrie et al. (10) proposed that more changes are associated with the outer layer of the capsid than with the inner layer. Based on the calorimetry data, Hinz et al. (18) hypothesized that one or more hydrophobic side chains change positions to lower their surface contact with water. Welsh et al. (11), to account for their proposed conversion of a monomeric asymmetric unit in Pf1L to a trimeric asymmetric unit in Pf1H, suggested more extensive changes in subunit contacts and structure on the basis of proposed similarities to molecular crystals.
More in line with our observations, NMR measurements of oriented phage particles by static solid-state NMR by Thiriot et al. (17) led to the suggestion that the C-terminal region of the subunit α-helix has a slightly lower tilt angle at lower temperatures, producing a reduced radius of the virion and an overall slight extension. Dipolar wave analysis of these data show that the changes in the coat protein tilt angle occur at residues Gln-16 and Ala-29. In our data, we observe a significant Cβ chemical shift perturbation for Ala-29 (Δδ ∼1.1 ppm), that may be associated with a backbone shift. Torsion angle analysis of our data using either TALOS or PREDITOR of the fragment Gly-28–Ala-29–Leu-30 is not sufficiently accurate to verify that this perturbation is consistent with the proposed backbone tilt. We detect a small change for the Cβ of Gln-16 (0.3 ppm, below the threshold); however, a perturbation for Cγ of Gln-16 is large (1.0 ppm), in contrast with most other residues belonging to the N-terminal half of the protein. This change may be associated with an intersubunit interaction pocket discussed above. It was also observed in the same study of oriented phage solutions (17) that the changes of the 15N chemical shifts of all isoleucine residues occur simultaneously, suggesting that the whole protein is affected by the transition. Our data also indicate that chemical shift perturbations occur for all isoleucine residues (in addition to other residues) and that these perturbations occur mostly at the side-chain atoms. Nevertheless, the perturbations of the Ile chemical shifts in this case are not necessarily an indication of the whole protein, and our data show that most changes are located at the hydrophobic C-terminal part of the protein coat.
It is noteworthy that we observed a reversible transition in solutions of virus with concentrations as high as 200 mg/ml, suggesting that interactions between different virus particles even at this very high concentration are not strong enough to prevent the transition. In fact, for an approximate particle mass of ∼36 MDa, and assuming a rod-shaped particle of size 2000 × 7 nm, this concentration of 200 mg/ml results in an occupation of about one-third of the total volume, so that full hydration is maintained. This will leave enough solvent space between individual particles for the transition to occur.
Summary
We have used magic angle spinning solid-state NMR to study the intersubunit interface of the Pf1 filamentous phage, to better understand its structure via the structural phase transition that occurs at ∼10 °C, thus providing important insight toward understanding the assembly and morphogenesis of this bacteriophage. Our approach included re-assignment of the NMR resonances of the low temperature form of Pf1, identification of unambiguous intersubunit contacts, and detection of the chemical shift differences between its high and low temperature forms. These data provide evidence for the identities of side chains that interact at the intermolecular surfaces in the assembly and are most amenable to structural reorganization during the transition. We identify specific patches on the coat protein that show these interactions most clearly.
Supplementary Material
This work was supported by Public Health Research Institute (to L. A. D.), National Science Foundation MCB Grant 0316248 (to A. E. M.), and European Union, Marie Curie Action Grant IRG-FP7 224800 (to A. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
This threshold was chosen for two reasons. (i) This is the approximate line width for many of the peaks. (ii) The deviations between the reported assignments in BMRB accession number 15138 and the sample used for comparison (Pf1H) were up to 0.3 ppm except for rare cases (0.4 ppm for Ile-22 Cα, Lys-45 Cα, Ile-12 Cδ1, and 0.7 for Thr-5 Cγ2) and averaged 0.11 ± 0.1 ppm (1 S.D.).
- Pf1L
- Pf1 at the low temperature form
- DARR
- dipolar-assisted rotational resonance
- Pf1H
- Pf1 at the high temperature form.
REFERENCES
- 1.Day L. A., Marzec C. J., Reisberg S. A., Casadevall A. (1988) Annu. Rev. Biophys. Biophys. Chem. 17, 509–539 [DOI] [PubMed] [Google Scholar]
- 2.Marvin D. A. (1998) Curr. Opin. Struct. Biol. 8, 150–158 [DOI] [PubMed] [Google Scholar]
- 3.Mahy B. W. J., van Regenmortel M. H. V. (eds) (2008) Encyclopedia of Virology, 3rd Ed., pp. 117–124, Elsevier, Oxford [Google Scholar]
- 4.Takeya K., Amako K. (1966) Virology 28, 163–165 [DOI] [PubMed] [Google Scholar]
- 5.Webb J. S., Lau M., Kjelleberg S. (2004) J. Bacteriol. 186, 8066–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice S. A., Tan C. H., Mikkelsen P. J., Kung V., Woo J., Tay M., Hauser A., McDougald D., Webb J. S., Kjelleberg S. (2009) ISME J. 3, 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomar S., Green M. M., Day L. A. (2007) J. Am. Chem. Soc. 129, 3367–3375 [DOI] [PubMed] [Google Scholar]
- 8.Wachtel E. J., Marvin R. J., Marvin D. A. (1976) J. Mol. Biol. 107, 379–383 [DOI] [PubMed] [Google Scholar]
- 9.Nave C., Fowler A. G., Malsey S., Marvin D. A., Siegrist H., Wachtel E. J. (1979) Nature 281, 232–234 [DOI] [PubMed] [Google Scholar]
- 10.Specthrie L., Greenberg J., Glucksman M. J., Diaz J., Makowski L. (1987) Biophys. J. 52, 199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsh L. C., Symmons M. F., Marvin D. A. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56, 137–150 [DOI] [PubMed] [Google Scholar]
- 12.Liu D. J., Day L. A. (1994) Science 265, 671–674 [DOI] [PubMed] [Google Scholar]
- 13.Welsh L. C., Symmons M. F., Sturtevant J. M., Marvin D. A., Perham R. N. (1998) J. Mol. Biol. 283, 155–177 [DOI] [PubMed] [Google Scholar]
- 14.Thiriot D. S., Nevzorov A. A., Zagyanskiy L., Wu C. H., Opella S. J. (2004) J. Mol. Biol. 341, 869–879 [DOI] [PubMed] [Google Scholar]
- 15.Goldbourt A., Gross B. J., Day L. A., McDermott A. E. (2007) J. Am. Chem. Soc. 129, 2338–2344 [DOI] [PubMed] [Google Scholar]
- 16.Blanch E. W., Bell A. F., Hecht L., Day L. A., Barron L. D. (1999) J. Mol. Biol. 290, 1–7 [DOI] [PubMed] [Google Scholar]
- 17.Thiriot D. S., Nevzorov A. A., Opella S. J. (2005) Protein Sci. 14, 1064–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz H. J., Greulich K. O., Ludwig H., Marvin D. A. (1980) J. Mol. Biol. 144, 281–289 [DOI] [PubMed] [Google Scholar]
- 19.Marvin D. A., Nave C., Bansal M., Hale R. D., Salje E. K. (1992) Phase Transitions 39, 45–80 [Google Scholar]
- 20.Casadevall A., Day L. A. (1988) Biochemistry 27, 3599–3602 [DOI] [PubMed] [Google Scholar]
- 21.Torbet J., Maret G. (1981) Biopolymers 20, 2657–2669 [DOI] [PubMed] [Google Scholar]
- 22.Jakeman D. L., Mitchell D. J., Shuttleworth W. A., Evans J. N. (1998) J. Biomol. NMR 12, 417–421 [DOI] [PubMed] [Google Scholar]
- 23.Takegoshi K., Nakamura S., Terao T. (2001) Chem. Phys. Lett. 344, 631–637 [Google Scholar]
- 24.Morcombe C. R., Gaponenko V., Byrd R. A., Zilm K. W. (2004) J. Am. Chem. Soc. 126, 7196–7197 [DOI] [PubMed] [Google Scholar]
- 25.Hediger S., Meier B. H., Ernst R. R. (1995) Chem. Phys. Lett. 240, 449–456 [Google Scholar]
- 26.Baldus M., Geurts D. G., Hediger S., Meier B. H. (1996) J. Magn. Reson. A 118, 140–144 [Google Scholar]
- 27.Andrew E. B., Chad M. R., Michele A., Lakshmi K. V., Robert G. G. (1995) J. Chem. Phys. 103, 6951–6958 [Google Scholar]
- 28.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29.Goddard T. D., Kneller D. G. (2004) SPARKY, Version 3.1.1, University of California, San Francisco, CA [Google Scholar]
- 30.Ulrich E. L., Akutsu H., Doreleijers J. F., Harano Y., Ioannidis Y. E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C. F., Tolmie D. E., Kent Wenger R., Yao H., Markley J. L. (2008) Nucleic Acids Res. 36, D402–D408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. (1996) Science 274, 1531–1534 [DOI] [PubMed] [Google Scholar]
- 32.Nakashima Y., Konigsberg W. H. (1980) J. Mol. Biol. 138, 493–501 [DOI] [PubMed] [Google Scholar]
- 33.Marvin D. A. (1990) Int. J. Biol. Macromol. 12, 125–138 [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez A., Nave C., Marvin D. A. (1995) Acta Crystallogr. D Biol. Crystallogr. 51, 792–804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.