Abstract
In Saccharomyces cerevisiae, the forkhead transcription factor Hcm1 is involved in chromosome segregation, spindle pole dynamics, and budding. We found that Hcm1 interacts with the histone deacetylase Sir2 and shifts from cytoplasm to the nucleus in the G1/S phase or in response to oxidative stress stimuli. The nuclear localization of Hcm1 depends on the activity of Sir2 as revealed by activators and inhibitors of the sirtuins and the Δsir2 mutant. Hcm1-overexpressing cells display more mitochondria that can be attributed to increased amounts of Abf2, a protein involved in mitochondrial biogenesis. These cells also show higher rates of oxygen consumption and improved resistance to oxidative stress that would be explained by increased catalase and Sod2 activities and molecular chaperones such as Hsp26, Hsp30, and members of Hsp70 family. Microarray analyses also reveal increased expression of genes involved in mitochondrial energy pathways and those allowing the transition from the exponential to the stationary phase. Taken together, these results describe a new and relevant role of Hcm1 for mitochondrial functions, suggesting that this transcription factor would participate in the adaptation of cells from fermentative to respiratory metabolism.
Keywords: Mitochondria, Oxidative Stress, Sirtuins, Transcription Factors, Yeast, Forkhead Transcription Factors, Hcm1, Sir2
Introduction
Mammalian FoxO transcription factors (FoxO1, 3, 4, and 6), a subfamily of forkhead transcription factors (FKH-TFs),3 control various biological functions, including stress resistance, DNA repair, metabolism, cell cycle arrest, and apoptosis (reviewed in Refs. 1–3). According to this variety of functions, FoxO transcription factors are regulated in these diverse functions by a wide range of external stimuli, such as insulin, insulin-like growth factor, other growth factors, neurotrophins, nutrients, cytokines, and oxidative stress stimuli. These stimuli control FoxO protein levels, subcellular localization, DNA binding, and transcriptional activity. FoxO are subject to several post-translational modifications including phosphorylation, acetylation, and ubiquitination (1, 4).
Saccharomyces cerevisiae has four genes (FKH1, FKH2, FHL1, and HCM1) homologous to the forkhead family of eukaryotic transcription factors, classified on the basis of a conserved DNA-binding domain (5). Fkh1 (Forkhead homolog 1) and its closest homolog, Fkh2, are involved in transcriptional silencing, cell morphology and cell cycle (6–8). Fhl1 has been described as regulating transcription of ribosome-associated proteins (9, 10). Hcm1 was first identified as a high copy suppressor of a defect in spindle pole assembly (11). This cell cycle-specific transcription factor regulates the transcription of genes involved in chromosome segregation (7). HCM1 is periodically transcribed and expressed in late G1 and early S phase. Genes regulated by Hcm1 peak primarily during the late S phase, and some of them are involved in chromosome organization, spindle dynamics, and budding (12). Although chromosome segregation is impaired in the absence of Hcm1, budding kinetics under optimal growth conditions in Δhcm1 cells is quite similar to wild-type cells (12).
In mammals, oxidative stress (H2O2 treatment) increases both FoxO phosphorylation and nuclear localization (13, 14), although decreased nuclear localization has also been reported (15). In addition to phosphorylation, FoxO acetylation/deacetylation in response to oxidative stress stimuli also affects FoxO subcellular localization. Deacetylation is mainly catalyzed by Sirt1 (the ortholog of yeast Sir2), although other histone deacetylases may regulate FoxO deacetylation and localization (14). At present, most studies indicate that FoxO acetylation functions to repress FoxO activity. In Caenorhabditis elegans, extension of lifespan by Sir2 requires the presence of DAF16, which is the only homolog of the FOXO family of FKH-TF (16). This suggests that acetylation directly or indirectly regulates Daf16.
Yeast Sir2 is a NAD+-dependent histone deacetylase involved in silencing telomeres, mating locus and rDNA, maintaining genome integrity, and aging (17). Deletion of SIR2 shortens life span, whereas yeast strains with an extra copy of the SIR2 extend lifespan (18). Lifespan extension by Sir2 also was observed in C. elegans (19), suggesting that the role of this sirtuin is extended through evolution. Although it is established that FOXO genes are highly conserved in various organisms, the connection between oxidative stress, yeast FKH-TF, and Sir2 is poorly known. Oxidative stress can damage proteins, lipids, and nucleic acids and thereby compromise cell viability. The effects of external oxidative stress on yeast cell cycle depend on the stress-exerting agent. At low concentrations, H2O2 results in S phase delay followed by G2/M arrest; menadione (MD) arrests cells at G1 (20). Through a transcription regulatory complex, yeast FKH-TF (Fkh1 and especially Fkh2) is involved in H2O2-induced cell cycle arrest and explains the difference between H2O2 and MD effects on cell cycle arrest (21). However, there is no experimental evidence of a stress response directly orchestrated by the forkhead factor.
In this study we describe a new role of Hcm1 as a transcription factor involved in oxidative stress resistance that includes Sir2 activity. Hcm1 interacts with Sir2, and Sir2 activity increases nuclear Hcm1 localization. The results also uncover a novel role of Hcm1 in mitochondrial metabolism and biogenesis that contributes to stress resistance and an adapting role in an early response to nutrient scarcity.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
The S. cerevisiae strains employed in this work are described in Table 1. Standard protocols were used for DNA manipulations and cell transformations (22). Plasmid pCYC106 (centromeric; geneticin resistance) (23) contains a 3HA-tagged HCM1 sequence with its own promoter region. Plasmid pCYC86 contains GFP-tagged HCM1 (centromeric, geneticine resistance). HA and GFP were C-terminally labeled. Null mutants were obtained by using the short flanking homology approach after PCR amplification of the natMX4 cassette in the case of HCM1 and SIR2 (24). Disruption was confirmed by PCR analysis. Overexpression of Hcm1 protein was obtained by replacing the endogenous promoter with a tetO7 promoter using the short flanking homology approach after PCR amplification of the pCM225 plasmid (25). The strain carrying the integrated pCM225 plasmid was labeled MJRC07. The addition of the antibiotic repressed expression of the tet construction, resulting in cells with no detectable levels of Hcm1. The cells were grown at 30 °C by incubation in a rotary shaker using YPD medium (1% yeast extract, 2% peptone, 2% glucose) or SC medium (0.67% yeast nitrogen base, 2% glucose plus dropout mixture, and auxotrophic requirements) (26). Specific supplements were omitted for selection of the corresponding plasmid-carrying cells.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Comment(s) | Source |
|---|---|---|---|
| CML128 | MATaura3-52 his4 leu2-3,112 trp1 | Wild type | Ref. 23 |
| MJRC05 | CML128 hcm1::natMX4 | HCM1 disruption with natMX4 cassete | This study |
| GRB2405 | CML128 HCM1-3HA::natMX4 | Chromosomal HCM1 tagged with 3HA using the natMX4 cassette | This study |
| GRB2409 | CML128 FKH1-3HA::natMX4 | Chromosomal FKH1 tagged with 3HA using the natMX4 cassette | This study |
| GRB2411 | CML128 FKH2-3HA::natMX4 | Chromosomal FKH2 tagged with 3HA using the natMX4 cassette | This study |
| GRB2408 | CML128 FHL1-3HA::natMX4 | Chromosomal FHL1 tagged with 3HA using the natMX4 cassette | This study |
| MJRC07 | GRB2405 tetO7-HCM1-3HA::kanMX4 | Integration of tetO7-regulatable HCM1-HA in GRB2405 | This study |
| MJRC08 | CML128 HCM1-GFP:: kanMX4 | Chromosomal HCM1 tagged with GFP using the sGFP-ADH1t-kanMX4 cassette | This study |
| MJRC01 | CML128 FKH1-GFP:: kanMX4 | Chromosomal FKH1 tagged with GFP using the sGFP-ADH1t-kanMX4 cassette | This study |
| MJRC02 | CML128 FKH2-GFP:: kanMX4 | Chromosomal FKH2 tagged with GFP using the sGFP-ADH1t-kanMX4 cassette | This study |
| MJRC04 | CML128 FHL1-GFP:: kanMX4 | Chromosomal FHL1 tagged with GFP using the sGFP-ADH1t-kanMX4 cassette | This study |
| MJRC11 | MJRC08 sir2::natMX4 | SIR2 disruption with natMX4 cassette in MJRC08 (HCM1 tagged with GFP) | This study |
Enzyme Activities
Cell extracts were obtained using glass beads. Catalase activity was measured at 25 °C as described (27). Alcohol dehydrogenase was measured spectrophotometrically by NADH formation (28). Citrate synthase was measured in a coupled assay to reduce 5,5′-dithiobis-(2-nitrobenzoic acid) (29). Superoxide dismutase (SOD) activities were assayed in a native gel as described (30), with modifications. See the supplemental materials for details.
Western Blot Analysis
The cell extracts were obtained as described (31), separated in SDS-polyacrylamide gels, and transferred to polyvinylidene difluoride membranes. Immunodetection was performed using the SNAP i.d.TM system (Millipore). The antibodies used and the working dilutions are described in the supplemental materials. In vivo protein-protein cross-linking was performed using p-formaldehyde (PFA) (32, 33). Also see the supplemental materials for details on cross-linking. For immunoprecipitation, 10 μg of anti-Sir2 was added to 4 mg of cell extract. When anti-HA was used to immunoprecipitate, 5 μg of anti-HA was added to 1.5 mg of cell extract. In any case, the mixture was rocked for 2 h at 4 °C, and 100 μl of protein G-agarose (Roche Applied Science; reference number 11719416001) was added. After rocking for 2–3 h more at 4 °C, lysates were loaded in spin columns (Pierce; reference number 69705), and washed six times with lysis buffer. Bound material was eluted with 50 mm Tris-HCl, pH 7.5, 10 mm EDTA, 1% SDS for 10 min at 65 °C. SDS-PAGE loading buffer was added to eluted material, heated at 95 °C for 3 min, and loaded to SDS-PAGE gels. In the case of cells treated with PFA, SDS-PAGE loading buffer was added to eluted material and either loaded directly to SDS-PAGE gels or heated at 95 °C for 20 min for cross-link reverse prior to electrophoresis.
Yeast Cell Synchronization
The cells were synchronized either by centrifugal elutriation or using a block-and-release protocol. Centrifugal elutriation separates cells on the basis of their size, mass, and shape using centrifugal force to sediment cells in the presence of counterflowing media. Small G1 phase daughter cells were obtained from HCM1-GFP and HCM1-GFP Δsir2 cultures grown exponentially in YPD. Elutriation was performed in a Beckman centrifuge using a JE-5.0 rotor as described (34). α-Factor-based synchronization was performed in HCM1-GFP cells. The cultures were grown to an A600 = 0.5 in YPD, an asynchronous sample was taken, and α-factor was added to a concentration of 5 μg/ml. After 120 min the α-factor was removed by pelleting the cells for 5 min at 3,000 rpm and decanting the supernatant. The cells were washed twice, and the G1-arrested cells were resuspended in fresh YPD. In both cases, the samples were taken at different times, and Hcm1-GFP fluorescence was observed in a fluorescent microscope. A minimum of 100 cells were measured at each time point.
Microscopy Studies
To analyze Hcm1 cellular localization upon oxidative stress, HCM1-GFP-labeled cells grown in YPD medium were treated with 0.25, 0.5, or 1 mm H2O2 and analyzed every 5 min for 30 min by fluorescence microscopy (Olympus DP30 BW) using 488-nm laser excitation for GFP. The pictures were taken every 5 min, and the number of cells with an absence of nuclear stain was counted with Image J 1.42b software and represented as a percentage of the total. The cells showing either exclusively nuclear and both nuclear and cytoplasm localization were grouped and indicated as nuclear. Approximately 5% of the cells showed very low fluorescent levels and were removed from the statistics. All of the quantitative values represent averages from at least 300 cells from three independent experiments (∼100 cells/experiment/time point). To analyze mitochondrial content, the cells were grown in YPD medium and stained with 50 nm MitoTracker Red CM-H2XRos (Invitrogen; reference number M-7513) for 10 min in the dark. The cells were analyzed by fluorescence microscopy using 579-nm laser excitation. For electron microscopy studies, the cells were grown in YPD medium at 30 °C and fixed for 1 h at 4 °C in 0.1 m phosphate buffer, pH 7.4, containing 2.5% glutaraldehyde. After rinsing the pellet twice with phosphate buffer at 4 °C, the cells were postfixed in buffered OsO4, dehydrated in ethanol, and embedded in a EMbed-812 kit (Electron Microscopy Sciences). The ultrathin sections mounted on copper grids were counterstained with uranyl acetate and lead citrate. The images were collected on a transmission electron microscope (Zeiss EM 910).
Gene Expression Analysis
Microarray analysis was performed as described (35) in the genomic facilities at the Universitat Autònoma de Barcelona. All of the quantitative real time PCRs were performed using the TaqMan System (Applied Biosystems). See the supplemental materials for details on RNA preparation and the microchip and RT-PCR analysis.
Analysis
Resistance to oxidative stress was tested on cells grown in YPD at the middle of exponential phase (A600 = 0.5). We then added 2 mm H2O2 or 6 mm MD to the culture, and after a 45-min treatment we measured viability by plating serial dilutions (1:5) on YPD plates. Viability after heat shock resistance (48 °C for 15 and 30 min) was measured by plating serial dilutions (1:10) on YPD plates. The dihydroethidium (Fluka) oxidation rate was performed as described (36). Oxygen consumption was measured in a Clark detector as described (37). The results shown represent the means ± S.D. of three experimental trials. Growth curves were determined in a Microplate Spectrophotometer PowerWave XS (Biotek) apparatus, and the values were analyzed with Gen5 1.06 software. The cells (500 μl) were grown at 30 °C with shaking; absorbance at 600 nm was measured every 30 min for 40 h. Glucose concentration in the culture medium was measured using the BioSystem glucose kit (reference number 11803) following the glucose oxidase/peroxidase method. Statistical analysis between WT and mutant cells was performed using Student's t test.
RESULTS
Sir2 Activity and Oxidative Stress Modulate Cellular Localization of Hcm1
The yeast histone deacetylase Sir2 and its mammalian orthologous Sirt1 have been related with calorie restriction, oxidative stress, and aging (17). In this context, we show that Sir2 is up-regulated in response to H2O2 in a dose-dependent manner, with levels increasing up to 50% (supplemental Fig. S1). Sirt1 deacetylates and regulates FoxO1 and 3, which affect the nuclear shift of the transcription factor and the induction of genes involved in stress resistance. Because yeast and mammals share various pathways that are highly conserved throughout evolution, we decided to study a possible connection between Sir2, FKH-TFs, and oxidative stress in yeast.
To that end, we GFP-labeled the FKH-TFs (Fkh1, Fkh2, Hcm1, and Fhl1) (Table 1) and analyzed their cellular localization by fluorescence microscopy (Fig. 1). Fkh1, Fkh2, and Fhl1 were present only in the nucleus, whereas Hcm1 was present both in the nucleus and at the cytoplasm, indicating that it could shift between these compartments. Given this differential trait and that Hcm1 is the FKH-TF with the highest similarity to mammalian FoxOs, we focused our work on the role of Hcm1. First, because HCM1 is periodically transcribed and expressed in the late G1 and early S phases (12), we studied Hcm1 localization at different phases of the cell cycle using α-factor to synchronize the culture (Fig. 2). Hcm1-GFP accumulates to the nucleus during the G1/S transition, when expression is higher, and moves to the cytoplasm at G2/M. Second, we analyzed whether oxidative stress influences Hcm1 localization. In cells exponentially growing in YPD, Hcm1-GFP was present mainly (65%) in the cytoplasm of the cells; 35% had a nuclear label. The addition of 1 mm H2O2 caused an increased nuclear localization, in up to 65% of Hcm1-GFP, after 30 min (Fig. 3, A and B). Upon oxidative stress treatment, 50% of Hcm1-GFP localized to the nucleus after 5–10 min, achieving steady-state levels at ∼15 min, depending upon nuclear activity. Similar results were observed upon addition of 0.25 or 0.5 mm H2O2 (not shown). To study whether Sir2 plays a role in Hcm1 localization, cells submitted to H2O2 were pretreated either with the Sir2 activator isonicotinamide or with Sir2 inhibitors splitomicin and nicotinamide as described (38). The results showed that Sir2 activity contributed to Hcm1 localization (Fig. 3C). Hcm1 was retained in the nucleus when Sir2 was activated with isonicotinamide; before H2O2 addition, 50% of the cells already showed nuclear label. Nicotinamide and splitomicin treatment prevented nuclear accumulation of Hcm1-GFP in response to H2O2. In agreement with these results, HCM1-GFP Δsir2 cultures displayed a lower percentage of cells with nuclear Hcm1-GFP label (supplemental Fig. S2A), and upon H2O2 treatment, the response was similar to that obtained with the use of sirtuins inhibitors (Fig. 3D). To more deeply study the connection of Sir2 with Hcm1, the distribution of Hcm1-GFP along cell cycle was analyzed in synchronized Δsir2 cells. α-Factor could not be used because Δsir2 cells do not show pheromone sensitivity because of a lack of silencing at the mating locus. By centrifugal elutriation, homogenous populations of small G1 phase daughter cells were obtained from HCM1-GFP Δsir2 and HCM1-GFP cultures. Hcm1-GFP localization was analyzed at different phases of the cell cycle. It was clear that lack of Sir2 resulted in lower percentages of cells with nuclear Hcm1-GFP localization throughout cell cycle (supplemental Fig. S2B).
FIGURE 1.
Cellular localization and levels of Fkh1, Fkh2, Hcm1, and Fhl1. A, exponentially growing cells in YPD medium and carrying a chromosomically integrated FKH1-GFP (MJRC01), FKH2-GFP (MJRC02), FHL1-GFP (MJRC04), and HCM1-GFP (MJCR03) fusion were analyzed for GFP fluorescence. Nuclear co-localization was analyzed using DAPI staining. GFP and DAPI staining was analyzed with fluorescence microscopy. B, exponentially growing cells in YPD medium and carrying an integrated HA tag, FKH1–3HA (GRB2409), FKH2–3HA (GRB2411), HCM1–3HA (GRB2405), and FHL1–3HA (GRB2408) were analyzed by Western blot with anti-HA antibodies. Bands corresponding to each FKH-TF were measured with a densitometer, and the relative intensities were calculated. The data are represented as the means ± S.D. from three independent experiments.
FIGURE 2.
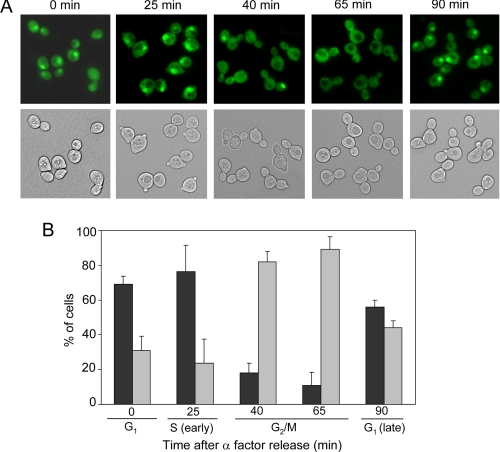
Hcm1 localization shifts from nucleus to the cytoplasm according to its cell cycle regulation. A, HCM1-GFP cells grown exponentially in YPD were treated with α-factor for 2 h. Cells arrested in G1 were released from α-factor, and pictures were taken at different time points by fluorescence microscopy. B, percentage of cells showing nuclear or cytoplasmic localization was quantified. Black bars, nuclear; gray bars, cytoplasmic.
FIGURE 3.
Sir2 is involved in Hcm1 translocation between cytoplasm and nucleus upon oxidative stress. Exponentially growing cells in YPD medium and carrying a chromosomically integrated HCM1-GFP fusion were analyzed for GFP fluorescence. A, images were taken before and after 30 min of 1 mm H2O2 addition. B, time course of nuclear-cytoplasmic translocation of Hcm1 upon 1 mm H2O2 treatment. C, cultures were pretreated for 45 min with 10 mm isonicotinamide (IsoNAM, a Sir2 activator) or with either 1 mm nicotinamide (NAM) or 5 μm splitomicin (Sir2 inhibitors) and submitted to 1 mm H2O2 for 30 min. Hcm1-GFP localization was analyzed every 5 min after oxidative stress. D, HCM1-GFP and HCM1-GFP Δsir2 were submitted to 1 mm H2O2, and Hcm1-GFP localization was analyzed at different time points. In B–D, approximately, 100 cells were counted at each time point by fluorescence microscopy, and Hcm1-GFP was classified as nuclear or cytoplasmic.
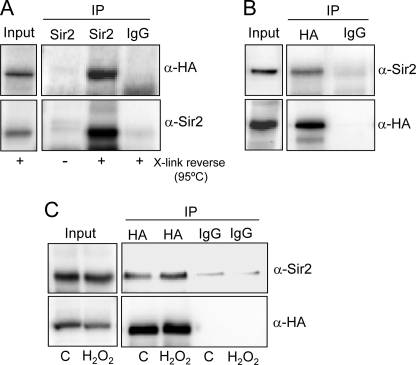
Because the effects described above could be explained by a physical interaction between Sir2 and Hcm1, co-immunoprecipitation experiments were carried out. To make the interaction more stable, PFA was added to the culture. PFA is a reversible cross-linker that efficiently produces protein-protein cross-links in vivo (32). Co-immunoprecipitation experiments indicated that Hcm1 could be clearly detected in Sir2 immunopellets in HCM1-HA cells treated with PFA (Fig. 4A). To better show this interaction, as well as for additional purposes, we constructed a conditional strain in which the HCM1 gene (tagged with HA) was under the doxycycline regulatable tet promoter (tetHCM1-HA) (Table 1). tetHCM1-HA cells displayed levels of Hcm1 five to six times higher than in wild-type cells, and the protein was not detectable after 12 h of treatment with 4 μg/ml doxycycline (supplemental Fig. 3). As shown in Fig. 4B, Sir2 co-immunoprecipitated with Hcm1 when extracts from tetHCM1-HA cells were treated with anti-HA antibodies. The reverse experiment also confirmed these results (not shown). This interaction was difficult to detect in cells with normal levels of Hcm1, probably because of the small amount of Hcm1 involved and/or to transient interactions that could occur between these proteins. However, we were able to detect Sir2 when anti-HA was used to immunoprecipitate Hcm1 in HCM1-HA cells (Fig. 4C).
FIGURE 4.
Hcm1 interacts with Sir2. A, exponentially growing HCM1-HA cells in YPD medium were treated in vivo with PFA for 30 min. Endogenous levels of Hcm1 were immunoprecipitated (IP) with a polyclonal antibody to Sir2 or a control mouse serum (IgG). The immune complexes were checked for the presence of Hcm1-HA by Western blot with an antibody anti-HA. Amounts of the immunoprecipitated Sir2 were assessed by anti-Sir2. Reversal of formaldehyde cross-links was performed by boiling for 20 min at 95 °C prior to SDS-PAGE. B, Hcm1-HA was immunoprecipitated from exponentially growing tetHCM1-HA cells, and the immune complexes were checked for the presence of Sir2 by Western blot. C, Hcm1-HA was immunoprecipitated from exponentially growing HCM1-HA cells (control or treated with 1 mm H2O2 for 30 min), and the immune complexes were checked for the presence of Sir2 by Western blot.
In addition, because oxidative stress may play a role in such a process, the same experiment was performed in cells treated with H2O2 (Fig. 4C). Control experiments, using untagged Hcm1 cells, were also performed (supplemental Fig. S4). Having in mind that mammalian FoxOs can be deacetylated by Sirt1, we analyzed whether Hcm1 might be a substrate for Sir2. However, although several approaches were used, we were unable to detect acetylation of the transcription factor, and as a consequence, deacetylation by Sir2 could not be demonstrated.
Hcm1 Plays a Protecting Role against Oxidative Stress
The observation that Hcm1 is retained in the nucleus upon oxidative stress in a Sir2-dependent process raised the possibility that, as a transcription factor, Hcm1 could have a role in protection against such stress. Cultures of WT, Δhcm1, and tetHCM1 (with and without the doxycycline) strains were exposed either to H2O2 or MD (a superoxide-generating compound), and viabilities were compared (Fig. 5A). Cells overexpressing Hcm1 showed increased resistance, and these differences were especially clear following MD treatment. The addition of doxycycline to these cells completely reversed the acquired resistance. Adding the antibiotic to the WT strain did not affect its resistance to oxidative stress (not shown). Mutant Δhcm1 cells displayed a slight but consistent decreased resistance to oxidative treatment compared with WT cells. supplemental Fig. S5 shows that Hcm1 overexpression increased resistance to high temperature, and this effect was reverted by doxycycline pretreatment, indicating that this transcription factor confers cross-protection against several stresses. Proteins and other macromolecules became oxidized, compromising cell survival as a consequence. Protein carbonyl formation has been extensively used as a marker for protein oxidation and can be easily detected by DNPH derivatization followed by Western blot anti-DNP antibodies (39, 40). After challenging cells with 10 mm MD or 2 mm H2O2, protein carbonylation inversely correlated with Hcm1 levels (Fig. 5B); this indicates that Hcm1 has a protective role against oxidative stress. To analyze whether Sir2 activity might have a role in this increased oxidative stress resistance observed in cells overexpressing Hcm1, WT and tetHCM1 cultures were pretreated with splitomicin and nicotinamide and challenged with H2O2 or MD. Western blot anti-DNP shows that upon Sir2 inactivation, the presence of Hcm1 overexpression did not provide increased resistance to protein carbonylation, supporting the sirtuin-TF connection (Fig. 5C).
FIGURE 5.
Overexpression of Hcm1 results in cells resistant to oxidative stress. Cultures of WT, Δhcm1, and tetHCM1 (minus and plus doxycycline) were grown in YPD medium and treated in each experiment as indicated. A, 2 mm H2O2 or 10 mm MD for 60 min. Viability was measured by plating serial dilutions (1:5) in YPD plates. B, 2 mm H2O2 or 10 mm MD for 60 min. Oxidatively damaged proteins were detected by anti-DNP Western blot. C, pretreated for 45 min with 1 mm nicotinamide (NAM) plus 5 μm splitomicin (Split, Sir2 inhibitors) and submitted to 2 mm H2O2 or 10 mm MD for 60 min.
To understand why HCM1 overdose resulted in such oxidative stress resistance, two of the main antioxidant enzymes (SOD and catalase) were measured. SOD activities with both cytosolic Cu,ZnSOD (Sod1) and mitochondrial MnSOD (Sod2) were analyzed by a zymogram assay and quantified (Fig. 6A). Sod2 activity was 60% higher in tetHCM1 cells. When treated with doxycycline, Sod2 activity decreased to values close to those found in Δhcm1. No differences were detected in Sod1 activity. Catalase activity was measured at the early (A600 = 1) and the late (A600 = 5) exponential phases (Fig. 6B). The differences were statistically significant when cells increased their antioxidant capacity before entering the diauxic phase. At this point, Δhcm1 cells showed a 25% decrease in catalase activity with respect to WT cells. Accordingly, tetHCM1 cells displayed higher catalase activity, which was abrogated by pretreatment with doxycycline. The differences in antioxidant capacity resulted in different rates of superoxide anion production inside the cells. As shown in Fig. 6C, the rate of oxidation of dihydroethidium decreased 1.5-fold in tetHCM1 cells. Again, pretreatment with doxycycline returned superoxide levels to values similar to those in Δhcm1.
FIGURE 6.
Antioxidant activities and intracellular ROS levels are affected by Hcm1 expression. Cultures of WT, Δhcm1, and tetHCM1 (minus and plus doxycycline, doxy) were grown in YPD medium. A, cell lysates were analyzed by native electrophoresis and SOD staining. Total Coomassie Blue protein staining was used as a loading control. Bands corresponding to Mn-SOD activity were measured with a densitometer, and the relative intensity was calculated. B, catalase activity was analyzed in whole extracts from the early exponential (A600 = 1, gray bars) and the late exponential (A600 = 5, black bars) phases. C, the rate of oxidation of dihydroethidium (DHE) by endogenous superoxide was analyzed. The data are represented as the means ± S.D. from three independent experiments. Statistical analysis was performed comparing each mutant with WT cells. *, p < 0.05; **, p < 0.005.
Hcm1 Plays a Role in Mitochondrial Metabolism and Biogenesis
When grown with high glucose concentration, yeast cells obtain their energy from sugar fermentation. The ethanol produced is excreted to the medium. When glucose becomes limiting, the cells enter the diauxic phase, and metabolism shifts from glucose fermentation to ethanol respiration. Finally, after ethanol exhaustion, the cells enter a stationary phase and become extremely resistant to several stresses. Because resistance to oxidative stress plays a preponderant role in increasing cellular fitness during the diauxic shift and survival at stationary phase, growth curves were obtained from WT, Δhcm1, and tetHCM1 cells (Fig. 7A). In rich medium, Δhcm1 stopped sooner and reached lower densities than WT cells. In contrast, cells overproducing Hcm1 were better fitted for diauxic shift and yielded higher culture densities compared with WT. Duplication time values at the exponential phase showed that tetHCM1 cells grew slowly compared with WT cells (t½ = 103 versus 82 min, respectively). The addition of doxycycline reverted cells to generation times similar to WT (85 min) (not shown). No significant differences were observed between Δhcm1 and WT cells.
FIGURE 7.
Increased levels of Hcm1 produce a metabolic shift from glucose fermentation to respiration. Cultures of WT, Δhcm1, and tetHCM1 (minus and plus doxycycline, doxy) were grown in YPD medium. A, growth curves up to 40 h were measured. Duplication times were obtained at the exponential phase. The data are the averages of two independent experiments. B, alcohol dehydrogenase (ADH) activity was measured at the exponential phase. C, oxygen consumption (nmol O2·min−1) was measured by a Clark electrode at exponential phase. B and C, data are represented as the means ± S.D. from three independent experiments. D, amounts of three mitochondrial proteins (α-KGDH, Sod2, and porin) were assessed by immunoblot analysis at exponential phase. Actin was used as a loading control. E, levels of Abf2 were determined by immunoblot analysis at exponential phase. Actin was used as a loading control. The data are represented as the means ± S.D. from three independent experiments. Statistical analysis was performed comparing mutant and WT cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
These differences in duplication time can be explained by the fact that, during exponential growth, tetHCM1 cells display lower alcohol dehydrogenase activity (Fig. 7B) and higher respiration rate, measured as oxygen consumption (Fig. 7C). When doxycycline was added, both parameters reverted to wild-type values, establishing that these differences were due to Hcm1 activity. Taken together, these results indicate that tetHCM1 cells have a metabolic shift from fermentation to increased respiration levels. Because respiratory (mitochondrial) metabolism is slower than fermentative (cytosolic) metabolism, tetHCM1 cells presented slower rates of duplication.
These initial results lead us to study whether Hcm1 had a previously unidentified role in mitochondrial metabolism. Western blot against several mitochondrial proteins, including the E2 subunit of α-ketoglutarate dehydrogenase, Sod2, and porin, showed that these proteins had an increased presence when Hcm1 levels were higher (Fig. 7D). Δhcm1 cells presented a moderately decreased level of all of these proteins. One possible explanation for these results is the difference in the number of mitochondria in these strains. Both fluorescent microscopy, using MitoTracker as a mitochondrial probe (Fig. 8A), and electron microscopy (supplemental Fig. S6) were performed. Citrate synthase has been commonly used as a quantitative marker enzyme for mitochondrial content (41). To that end, citrate synthase activity demonstrated that mitochondrial mass responds to Hcm1 levels (Fig. 8B).
FIGURE 8.
The number of mitochondria responds to Hcm1 levels. Cultures of WT, Δhcm1, and tetHCM1 (with and without doxycycline, doxy) were grown exponentially in YPD medium. A, mitochondria were labeled with the addition of 50 nm MitoTracker and visualized with a fluorescence microscope. B, citrate synthase activity is represented as the means ± S.D. from three independent experiments. Statistical analysis was performed comparing mutant with WT cells. *, p < 0.05; **, p < 0.001.
Microarray Experiments Confirm the Role of Hcm1 in Stress Resistance and Mitochondrial Metabolism
The experiments described above raised the possibility that Hcm1 could have an important new role in mitochondrial biogenesis. To get further insight, we performed microarray experiments to compare the gene expression profile of the whole genome between WT and our mutant strains. The data have been deposited in the NCBI Gene Expression Omnibus (42) and are accessible through series accession number GSE20420.
Although not the aim of our study, it is worth mentioning that genes involved in cell cycle progression were differentially expressed, which agrees with our observations (Fig. 2) and the role of this transcription factor as described previously (7, 12). Compared with WT cells, overexpression of Hcm1 resulted in at least a 1.5-fold increase in the mRNA level of 350 genes involved in a variety of functional categories. Remarkably, 33% of these genes code for proteins localized primarily in mitochondria (Fig. 9A). Fig. 9B shows the up-regulated genes, found in the overexpressed cells, that relate to the main phenotypes described in our mutant strains. Quantitative real time PCR was performed for selected genes to validate the microarray results (Fig. 9C). Because many genes are involved in stress resistance, transcription factors required for stress tolerance were examined in the microarray experiment. Only MSN4 showed increased mRNA levels in the Hcm1-overexpressed cells. No differences were detected in YAP1, SKN7, or HSF1 genes (supplemental Table S1).
FIGURE 9.
Differential gene expression analysis. A, percentage of genes induced >1.5-fold in tetHCM1 compared with the same strain treated with doxycycline and classified by cellular component, according to FatiGO (70). Only those components with a gene percentage higher than 5% are represented. B, genes induced ≥2-fold in tetHCM1 classified by functional categories related to the observed phenotype. The categories were assigned based on information provided by SGD (71) and FatiGO. According to SGD, genes with a documented (double underlined) or potential (underlined) Hcm1-binding site are indicated. Genes in bold type correspond to those validated by RT-PCR and shown in C. C, expression of several genes was determined by quantitative real time PCR analysis. Actin expression was used as an internal control to normalize expression levels. Gray bars, Δhcm1 versus WT; black bars, tetHCM1 versus WT; white bars, tetHCM1 + doxycycline versus WT. For the WT strain, the relative quantity was 1.
One of the metabolic pathways affected by the amount of Hcm1 present was the de novo biosynthesis of NAD+ (Fig. 9, B and C). This important co-factor is produced from tryptophan through the action of BNA genes (BNA1 to BNA6). Four of them: BNA1, BNA2, BNA4, and BNA5, were up-regulated in the tetHCM1 strain, with BNA4, coding for kynurenine monooxigenase, the most induced, by a factor of 5.
Two genes related to activation of respiration (ADR1 and ISF1) seem to be regulated by the levels of Hcm1 (Fig. 9, B and C; supplemental Table S1). ISF1 is a suppressor of Hap2 mutation (43). HAP genes (HAP2/3/4) are important in the expression of genes involved in respiratory metabolism. It is likely that Hcm1-triggered ISF1 expression could participate in the induction of genes involved in respiratory metabolism. ADR1 encodes a transcriptional activator involved in the expression of genes that are regulated by glucose repression (44). In addition, the use of alternative energy sources, such as proline, was clearly regulated by Hcm1 (Fig. 9, B and C) as shown by high induction of PUT1 (coding for proline oxidase), GDH2 (coding for glutamate dehydrogenase), and GAD1 (coding for glutamate decarboxylase) (∼5-, 4.5-, and 2.5-fold, respectively).
Among the genes up-regulated in cells overexpressing Hcm1, it was especially interesting to find ABF2 (autonomous replication sequence-binding factor 2), a nucleoid-associated protein (Fig. 9, B and C). Yeast Abf2 is the ortholog of mammalian mitochondrial Transcription Factor A (TFAM) and plays an important role in the recombination and copy number determination of mtDNA (45). It functions as a histone-like protein in mitochondria, which is particularly important for the maintenance of mtDNA (46, 47). Since the role of Abf2 in mitochondrial biogenesis has been demonstrated (45, 48), we immunodetected Abf2 in WT, Δhcm1, and tetHCM1 (plus and minus doxycycline) cells (Fig. 7E). The results were in accordance with mRNA expression levels and showed that Abf2 protein levels increased when cells presented higher doses of Hcm1. Increased mtDNA copy number via activation of the Mec1/Rad53 pathway can occur independently of Abf2 (49). However, no differences in the mRNA levels of the proteins involved in the Mec1/Rad53 pathway were observed in cells lacking or overexpressing Hcm1.
DISCUSSION
The localization of Hcm1 in both cytoplasm and nucleus was mentioned in a large scale analysis for protein localization in budding yeast (50). Our results show that Hcm1 can shift from cytoplasmic to nuclear localization in the G1/S phase in synchronized cells, in agreement with its known cell cycle-specific role (12). Oxidative stress results in increased nuclear Hcm1 localization. These observations opened the possibility that Hcm1 could interact with sirtuins, as reported for FoxO1 and FoxO3 (3). The results shown in this paper indicate that Sir2 and Hcm1 interact in normal conditions and also under oxidative stress. The physiological relevance of such interaction has been demonstrated in the Δsir2 strain and with the use of activators and inhibitors of situins. Δsir2 cultures presented lower percentage of cells with nuclear Hcm1 and a decreased translocation of Hcm1 in response to oxidative stress and along cell cycle. Moreover, activation or inactivation of sirtuins increases or decreases nuclear trapping of Hcm1, respectively. All of these results strongly support a role for Sir2 activity in Hcm1 localization, increasing its nuclear trapping. The shuttling of mammalian FoxO transcription factors between nucleus and cytoplasm is regulated by a complex series of phosphorylations, acetylations, and ubiquitination (4, 51). In the case of yeast Hcm1, the signal(s) regulating nuclear translocation is still uncertain, although Hcm1 was described as being directly phosphorylated by Cdc28 in whole cell extracts (52). Using the NetNes 1.1 prediction program (53) allows the identification of a putative Leu-rich nuclear export sequence from amino acids 88 to 97 in the Hcm1 sequence. In fact only Leu90 reaches the program threshold, but the sequence L88XL90XXXL94XXL97 is quite similar to the canonical nuclear export sequence signal: LX2–3LX2–3LXL (54).
The fact that Hcm1, as a transcription factor involved in cell cycle regulation, is also involved in the resistance to different stresses uncovers a new function of Hcm1. Previous studies (12) collected microarray data across the cell cycle and identified a late S phase-specific promoter element described as the binding site for Hcm1 required for its cell cycle specific activity. In a recent study (7), analysis of several transcription factors performed by ChIP-on-chip demonstrates that a large proportion of Hcm1 target genes are also involved in bud growth, vesicular trafficking, and cell wall synthesis. Approximately 22% of Hcm1 targets have predicted functions in the synthesis of the cell wall including structural proteins, although 28 of Hcm1 gene targets were involved in carbohydrate metabolism or high affinity glucose transporters. The results presented in this paper indicate that comparing cell viability and protein oxidative damage, increased dosage of Hcm1 results in cells more resistant to thermal stress, H2O2, and especially MD. Accordingly, all of these properties recede in doxycycline-treated tetHCM1 cells. The increased Hcm1 nuclear localization is in agreement with this new protective role against oxidative stress. Sir2 activity plays a role on the increased resistance observed in cells overexpressing Hcm1. Proteins were not protected upon oxidative stress in tetHCM1 cells when pretreated with sirtuin inhibitors.
In Hcm1-overexpressing cells, increased oxygen consumption and decreased alcohol dehydrogenase activity produced a metabolic change, resulting in cells with higher respiration rates. Consequently, this metabolic shift, partially resembling what occurs under calorie restriction, implies that these cells have longer duplication times in glucose-rich medium. Two results, as compared with WT cells, indicate that their mitochondria are fully functional: better fitness entering the diauxic phase, yielding higher cell densities at stationary phase, and lower superoxide levels inside the cells. This may be the sum of two processes: higher Sod2 activity and lower ROS generation. The fact that increased respiratory metabolism results in decreased endogenous stress may be somewhat surprising, because ROS generation mainly occurs in the mitochondrial electron transport chain. However, it has been described that a functionally correct respiratory chain generates less ROS (55, 56).
An important finding of this study describes a new role for Hcm1, participating in both mitochondrial metabolism and biogenesis as shown by an increase in the number of mitochondria, the induction of specific proteins, and mitochondrial metabolic pathways. Induction of mitochondrial metabolism by Hcm1 was demonstrated by the higher respiration rates observed and increased levels of mitochondrial proteins in cells overexpressing Hcm1. Data from microarray analyses also support the induction of a respiratory metabolism. In this context, several genes from the proline utilization pathway are up-regulated. PUT1, coding for the mitochondrial proline oxidase, is highly induced. It catalyzes the conversion of proline into glutamate. PUT1 transcription is induced in the absence of a preferred nitrogen source (57). Glutamate can be sequentially converted into succinate by a mitochondrial pathway involving clearly induced genes such as glutamate decarboxylase GAD1, UGA1, and UGA2 (58). Glutamate can also produce α-ketoglutarate by glutamate dehydrogenase (GDH2) (59), which is also highly induced. These metabolic pathways can be envisaged as a mechanism for replenishing Krebs cycle intermediates necessary for respiratory metabolism displayed by Hcm1-overexpressing cells.
It is also worth mentioning that the ISF1 gene, showing a 4-fold increase, is repressed by fermentative metabolism and was initially described as a possible modulator of mitochondrial biogenesis (15) and later as a suppressor of Hap2 mutants. The transcriptional activator Hap2 is a subunit of the global regulator of respiratory gene expression (43). Moreover, ADR1, a transcription factor that regulates the expression of genes repressed by glucose shows a 3-fold induction. Adr1 also activates the expression of genes required for the mitochondrial metabolism of glycerol, ethanol, and lipids, as well as peroxisome biogenesis (60, 61).
All of these events can be envisaged as a preparation for the diauxic shift, because in this phase cells change from fermentative to respiratory metabolism with the concomitant increase in ROS production. From data in Fig. 7A, it is observed that cells lacking Hcm1 are unable to enter in diauxic shift, in contrast to wild-type cells and those overexpressing Hcm1, which are the best fitted for the transition to diauxic shift and, as a consequence, yield higher culture densities. How can this phenotype be explained? From the microarray analyses, it could be observed that genes important for such transition, such as YGP1 (62), SPI1 (stationary phase induced 1) (63), and GAC1 (64) are up-regulated in the tetHCM1 cells. Molecular chaperones related to response to several stresses are up-regulated in tetHCM1 cells. Examples include HSP26, a cytosolic chaperone induced by heat shock, cell cycle arrest, oxidative stress, or nitrogen or carbon starvation (65), and members of the HSP70 family chaperones (SSA4 and SSB1) and HSP30, which are induced in the diauxic shift and by a variety of stresses.
Another key point of the transcript array analysis is the fact that Hcm1 overexpression induces a set of genes involved in the de novo synthesis of NAD, of which BNA4 showed the highest induction levels. The genes involved in the NAD salvage pathway were unaffected by Hcm1 overexpression. The implications of such activation are crucial for the cell because activating NAD biosynthesis is a key fact for respiratory metabolism as well as for the activity of NAD-dependent histone deacetylases.
Hcm1 not only induces mitochondrial metabolism but also mitochondrial biogenesis. Increased mitochondrial content in tetHCM1 cells was detected with the fluorophor MitoTracker and measured by citrate synthase activity. Consistently, Δhcm1 cells or tetHCM1 cells treated with doxycycline presented citrate synthase activity significantly lower than WT cells. How does Hcm1 regulate mitochondrial content? Among the genes up-regulated in the tetHCM1 cells, ABF2 was especially interesting. Abf2 is a nuclear encoded mitochondrial protein member of the HMG (high mobility group) box, mtDNA-binding protein family and involved in mitochondrial biogenesis. The human homolog of Abf2 is mitochondrial transcription factor A (TFAM) (66). Abf2 has multiple functions in mtDNA metabolism, including stabilization of recombination intermediates (45), segregation, and dynamics of nucleoids (67) and oxidative DNA damage resistance (68). It is also one of the first protein factors proposed to have a direct role in regulating mtDNA copy number and mitochondrial biogenesis (45, 48). By RT-PCR we confirmed that mRNA levels of ABF2 correlate with Hcm1 (Fig. 9C), and more important, we found increased amounts of Abf2 when Hcm1 is overexpressed (Fig. 7E). The increment in Abf2 expression observed is significant because a moderate overexpression of Abf2 has been found to increase mtDNA copy number ∼1.5–2-fold, and deletion of the ABF2 gene results in ∼50% reduction (67). Mec1/Rad53, an alternative pathway to regulate mitochondrial DNA copy number, can increase mtDNA in an abf2 null mutant, suggesting that the two pathways can work independently (69). It could be that both pathways can act in the same direction, with Hcm1 being responsible for the increased amount of Abf2, which, in turn, would trigger the formation of mitochondria.
In conclusion, our results show that Hcm1 is involved in the formation of mitochondria and expressing genes involved in responses to stress, as well as inducing genes that are repressed under high glucose. These metabolic changes allow cells to shift from fermentative to respiratory metabolism conferring higher stress resistance. In yeasts, such metabolic features will be of special importance for entering in the diauxic shift and early stationary phase.
Supplementary Material
Acknowledgment
We thank Dr. Elaine M. Lilly for editing assistance.
This work has been supported by grants BFU2007-66249 and CSD2007-20 Consolider Ingenio 2010 from the Ministerio de Ciencia e Innovación (Spain) and SGR2009-00196 from the Generalitat de Catalunya.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S6.
- FKH-TF
- forkhead transcription factor
- PFA
- p-formaldehyde
- MD
- menadione
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- mtDNA
- mitochondrial DNA.
REFERENCES
- 1.Calnan D. R., Brunet A. (2008) Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
- 2.Arden K. C. (2008) Oncogene 27, 2345–2350 [DOI] [PubMed] [Google Scholar]
- 3.Van der Horst A., Burgering B. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 440–450 [DOI] [PubMed] [Google Scholar]
- 4.Vogt P. K., Jiang H., Aoki M. (2005) Cell Cycle 4, 908–913 [DOI] [PubMed] [Google Scholar]
- 5.Kaestner K. H., Knochel W., Martinez D. E. (2000) Genes Dev. 14, 142–146 [PubMed] [Google Scholar]
- 6.Hollenhorst P. C., Bose M. E., Mielke M. R., Müller U., Fox C. A. (2000) Genetics 154, 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. (2002) Genes Dev. 16, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu G., Spellman P. T., Volpe T., Brown P. O., Botstein D., Davis T. N., Futcher B. (2000) Nature 406, 90–94 [DOI] [PubMed] [Google Scholar]
- 9.Hermann-Le Denmat S., Werner M., Sentenac A., Thuriaux P. (1994) Mol. Cell. Biol. 14, 2905–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinschmidt M., Schulz R., Braus G. H. (2006) Curr. Genet. 49, 218–228 [DOI] [PubMed] [Google Scholar]
- 11.Zhu G., Muller E. G., Amacher S. L., Northrop J. L., Davis T. N. (1993) Mol. Cell. Biol. 13, 1779–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pramila T., Wu W., Miles S., Noble W. S., Breeden L. L. (2006) Genes Dev. 20, 2266–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura Y. I., Kitamura T., Kruse J. P., Raum J. C., Stein R., Gu W., Accili D. (2005) Cell Metab. 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 14.Frescas D., Valenti L., Accili D. (2005) J. Biol. Chem. 280, 20589–20595 [DOI] [PubMed] [Google Scholar]
- 15.Nemoto S., Finkel T. (2002) Science 295, 2450–2452 [DOI] [PubMed] [Google Scholar]
- 16.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. (1997) Nature 389, 994–999 [DOI] [PubMed] [Google Scholar]
- 17.Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A. (2002) J. Biol. Chem. 277, 45099–45107 [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein M., McVey M., Guarente L. (1999) Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 20.Flattery-O'Brien J. A., Dawes I. W. (1998) J. Biol. Chem. 273, 8564–8571 [DOI] [PubMed] [Google Scholar]
- 21.Shapira M., Segal E., Botstein D. (2004) Mol. Biol. Cell 15, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1987) Current Protocols in Molecular Biology, John Wiley and Sons Inc., New York [Google Scholar]
- 23.Gallego C., Garí E., Colomina N., Herrero E., Aldea M. (1997) EMBO J. 16, 7196–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 25.Bellí G., Garí E., Aldea M., Herrero E. (1998) Yeast 14, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 26.Sherman F. (2002) Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 27.Jakubowski W., Biliński T., Bartosz G. (2000) Free Radic Biol. Med. 28, 659–664 [DOI] [PubMed] [Google Scholar]
- 28.Maitra P. K., Lobo Z. (1971) J. Biol. Chem. 246, 475–488 [PubMed] [Google Scholar]
- 29.Shepherd D., Garland P. B. (1969) Biochem. J. 114, 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., Gitlin J. D. (1997) J. Biol. Chem. 272, 23469–23472 [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Manzaneque M. T., Ros J., Cabiscol E., Sorribas A., Herrero E. (1999) Mol. Cell. Biol. 19, 8180–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasilescu J., Guo X., Kast J. (2004) Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 33.Hall D. B., Struhl K. (2002) J. Biol. Chem. 277, 46043–46050 [DOI] [PubMed] [Google Scholar]
- 34.Lieberman H. B. (2003) in Methods in Molecular Biology, pp. 55–76, Humana Press Inc., Totowa, NJ [Google Scholar]
- 35.Viladevall L., Serrano R., Ruiz A., Domenech G., Giraldo J., Barceló A., Ariño J. (2004) J. Biol. Chem. 279, 43614–43624 [DOI] [PubMed] [Google Scholar]
- 36.Irazusta V., Cabiscol E., Reverter-Branchat G., Ros J., Tamarit J. (2006) J. Biol. Chem. 281, 12227–12232 [DOI] [PubMed] [Google Scholar]
- 37.Reverter-Branchat G., Cabiscol E., Tamarit J., Sorolla M. A., Angeles de la Torre M., Ros J. (2007) Microbiology 153, 3667–3676 [DOI] [PubMed] [Google Scholar]
- 38.Sauve A. A., Moir R. D., Schramm V. L., Willis I. M. (2005) Mol. Cell 17, 595–601 [DOI] [PubMed] [Google Scholar]
- 39.Tamarit J., Cabiscol E., Ros J. (1998) J. Biol. Chem. 273, 3027–3032 [DOI] [PubMed] [Google Scholar]
- 40.Cabiscol E., Piulats E., Echave P., Herrero E., Ros J. (2000) J. Biol. Chem. 275, 27393–27398 [DOI] [PubMed] [Google Scholar]
- 41.Williams R. S., Salmons S., Newsholme E. A., Kaufman R. E., Mellor J. (1986) J. Biol. Chem. 261, 376–380 [PubMed] [Google Scholar]
- 42.Edgar R., Domrachev M., Lash A. E. (2002) Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daignan-Fornier B., Nguyen C. C., Reisdorf P., Lemeignan B., Bolotin-Fukuhara M. (1994) Mol. Gen Genet. 243, 575–583 [DOI] [PubMed] [Google Scholar]
- 44.Denis C. L., Young E. T. (1983) Mol. Cell. Biol. 3, 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacAlpine D. M., Perlman P. S., Butow R. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6739–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. (1992) J. Biol. Chem. 267, 3358–3367 [PubMed] [Google Scholar]
- 47.Diffley J. F., Stillman B. (1992) J. Biol. Chem. 267, 3368–3374 [PubMed] [Google Scholar]
- 48.Cho J. H., Lee Y. K., Chae C. B. (2001) Biochim. Biophys. Acta 1522, 175–186 [DOI] [PubMed] [Google Scholar]
- 49.Taylor S. D., Zhang H., Eaton J. S., Rodeheffer M. S., Lebedeva M. A., O'rourke T. W., Siede W., Shadel G. S. (2005) Mol. Biol. Cell 16, 3010–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 51.Qiang L., Banks A. S., Accili D. (2010) J. Biol. Chem. 285, 27396–27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. (2003) Nature 425, 859–864 [DOI] [PubMed] [Google Scholar]
- 53.la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Protein Eng Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 54.Bogerd H. P., Fridell R. A., Benson R. E., Hua J., Cullen B. R. (1996) Mol. Cell. Biol. 16, 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barros M. H., Bandy B., Tahara E. B., Kowaltowski A. J. (2004) J. Biol. Chem. 279, 49883–49888 [DOI] [PubMed] [Google Scholar]
- 56.Barja G. (2007) Rej Res. 10, 215–223 [DOI] [PubMed] [Google Scholar]
- 57.Saxena D., Kannan K. B., Brandriss M. C. (2003) Eukaryot. Cell 2, 552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coleman S. T., Fang T. K., Rovinsky S. A., Turano F. J., Moye-Rowley W. S. (2001) J. Biol. Chem. 276, 244–250 [DOI] [PubMed] [Google Scholar]
- 59.Miller S. M., Magasanik B. (1990) J. Bacteriol. 172, 4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zelenaya-Troitskaya O., Newman S. M., Okamoto K., Perlman P. S., Butow R. A. (1998) Genetics 148, 1763–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Rourke T. W., Doudican N. A., Mackereth M. D., Doetsch P. W., Shadel G. S. (2002) Mol. Cell. Biol. 22, 4086–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin C., Barrientos A., Epstein C. B., Butow R. A., Tzagoloff A. (2007) FEBS Lett. 581, 5658–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., Young E. T. (2005) Mol. Cell. Biol. 25, 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young E. T., Dombek K. M., Tachibana C., Ideker T. (2003) J. Biol. Chem. 278, 26146–26158 [DOI] [PubMed] [Google Scholar]
- 65.Destruelle M., Holzer H., Klionsky D. J. (1994) Mol. Cell. Biol. 14, 2740–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parisi M. A., Clayton D. A. (1991) Science 252, 965–969 [DOI] [PubMed] [Google Scholar]
- 67.Puig S., Perez-Ortin J. E. (2000) Yeast 16, 139–148 [DOI] [PubMed] [Google Scholar]
- 68.Parrou J. L., Enjalbert B., Plourde L., Bauche A., Gonzalez B., François J. (1999) Yeast 15, 191–203 [DOI] [PubMed] [Google Scholar]
- 69.Amorós M., Estruch F. (2001) Mol. Microbiol. 39, 1523–1532 [DOI] [PubMed] [Google Scholar]
- 70.Al-Shahrour F., Minguez P., Tárraga J., Montaner D., Alloza E., Vaquerizas J. M., Conde L., Blaschke C., Vera J., Dopazo J. (2006) Nucleic Acids Res. 34, W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mewes H. W., Frishman D., Gruber C., Geier B., Haase D., Kaps A., Lemcke K., Mannhaupt G., Pfeiffer F., Schüller C., Stocker S., Weil B. (2000) Nucleic Acids Res. 28, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.